Abstract
Nuclear import of the simian virus 40 large tumor antigen (T-ag) is dependent on its nuclear localization signal (NLS) within amino acids 126–132 that is recognized by the importin α/β1 heterodimer, as well as a protein kinase CK2 site at serine 112 upstream of the NLS, which enhances the interaction ∼50-fold. Here we show for the first time that T-ag nuclear import is negatively regulated by N-terminal sequences (amino acids 102–110), which represent the binding site (BS) for the retinoblastoma (Rb) tumor suppressor protein (p110Rb). Quantitative confocal laser scanning microscopic analysis of the transport properties of T-ag constructs with or without Rb binding site mutations in living transfected cells or in a reconstituted nuclear transport system indicates that the presence of the RbBS significantly reduces nuclear accumulation of T-ag. A number of approaches, including the analysis of T-ag nuclear import in an isogenic cell pair with and without functional p110Rb implicate p110Rb binding as being responsible for the reduced nuclear accumulation, with the Ser106 phosphorylation site within the RbBS appearing to enhance the inhibitory effect. Immunoprecipitation experiments confirmed association of T-ag and p110Rb and dependence thereof on negative charge at Ser106. The involvement of p110Rb in modulating T-ag nuclear transport has implications for the regulation of nuclear import of other proteins from viruses of medical significance that interact with p110Rb, and how this may relate to transformation.
Keywords: Nucleus/Nuclear Import, Phosphorylation, Transport/Nuclear, Tumor/Suppressor/Rb, Viral Protein, SV40 Large Tumor Antigen, Negative Regulation
Introduction
Proteins that perform their respective roles in the nucleus generally require a nuclear localization signal (NLS)2 recognized by members of the importin (IMP) family of transporters. In the case of Simian virus 40 (SV40) large tumor antigen (T-ag), the NLS (PKKKRKV132, single letter amino acid code) (1) is recognized by a heterodimer of IMP α and β1, which mediates passage of T-ag through the nuclear envelope-localized nuclear pores and into the nucleus (2–4). Release within the nucleus occurs upon binding of the monomeric guanine nucleotide-binding protein Ran to IMPβ, which dissociates the trimeric IMP-NLS-containing protein complex (2, 4, 5).
Phosphorylation has been shown to be able to regulate nuclear protein import through modulation of NLS-IMP interaction either positively or negatively (2, 6–14). A well characterized example is that of T-ag, which has several phosphorylation residues N-terminal to its NLS, which are phosphorylated during SV40 infection (15, 16), including the CK2 site at Ser111/112, which enhances nuclear uptake by enhancing NLS recognition by IMP α/β (9, 17, 18), and the cyclin-dependent kinase site at Thr124, which effects cytoplasmic retention (6, 19).
The present study reports for the first time the regulatory role of amino acids 102–110, previously shown to represent the “transforming region” of T-ag, corresponding to the binding site (BS) for the tumor suppressor p110Rb protein (retinoblastoma susceptibility factor or Rb) (20–22). Using quantitative confocal laser scanning microscopy (CLSM) in live transfected cells and in an in vitro reconstituted system, we show that the p110Rb binding site (RbBS) inhibits T-ag nuclear import dependent on phosphorylation at Ser106 within the RbBS, a site known to be phosphorylated in infected cells (15, 16). Furthermore, we show that the effect is attributable to binding of p110Rb protein to the RbBS, enhanced by negative charge at the Ser106 phosphorylation site. The results have important implications for the regulation of nuclear transport of the many viral proteins of significance that interact with p110Rb, and how this may relate to transformation.
EXPERIMENTAL PROCEDURES
T-ag Expression Plasmid Construction using Gateway™ Technology
All mammalian and bacterial constructs expressing GFP-T-ag fusion proteins were generated using the Gateway™ system (Invitrogen). Primers including the attB1 and attB2 recombination sites were used to amplify the T-ag sequences of interest using plasmid pPR28 (18), pPR11, and pDAJ, where appropriate, as the templates. PCR fragments were introduced into plasmid vector pDONOR207 (Invitrogen) via the BP recombination reaction, according to the manufacturer's recommendations, to generate the respective entry clones pDONR207-T-ag-(110–135), pDONR207-T-ag-(110-135:NLSmut), pDONR207-T-ag-(102–135), pDONR207- T-ag-(102–135:A106), pDONR207-T-ag-(102–135:D106), pDONR207-T-ag-(102–135:Rb mut1), pDONR207-T-ag-(102–135:NLSmut), pDONR207-T-ag-(87–135), pDONR207-T-ag-(87–135:Rb mut1) pDONR207-T-ag-(87–135:Rb mut2), and pDONR207-T-ag-(87–135:NLSmut).
pDONR207-T-ag constructs were then used to perform LR recombination reactions with the Gateway system compatible expression (“DEST”) vectors pDEST53 (Invitrogen), and pGFPattC (23) according to the manufacturer's recommendations to express GFP-T-ag fusion protein in mammalian and bacterial cell systems, respectively.
Cell Culture and Transfection
The COS-7, CV-1, SAOS-2 (24), and SR-40 (25) cell lines were maintained and seeded into 6- and 12-well plates 1 day prior to transfection for use in CLSM experiments, as previously (26–28). The HTC rat hepatoma cell line was cultured, as previously (29).
In Vitro Nuclear Transport Assay
Nuclear import of the various GFP-T-ag fusion proteins was investigated using an in vitro reconstituted nuclear transports assay as previously (29). Briefly, HTC cells grown on coverslips for 48 h were perforated mechanically, and then inverted onto a microscope slide over a 5-ml chamber of artificial cytoplasm containing reticulocyte lysate (which contains components of the nuclear transport machinery, such as IMPs and Ran), and an ATP-regenerating system, prior to CLSM imaging for up to 30 min. The involvement of the p110Rb protein (present in reticulocyte lysate) in the various GFP-T-ag proteins nuclear import was determined by preincubating the reticulocyte lysate for 15 min at room temperature with a specific mouse monoclonal antibody to p110Rb (OP77-Ab-6; Calbiochem, Gibbstown, NJ) at 47 μg/ml. Anti-GST antibody (Santa Cruz Biotechnology) was also preincubated with reticulocyte lysate and added to the system at the same concentration as a control.
CLSM/Image Analysis
Endogenously expressed p110Rb was visualized in CV-1 cells following fixation with 4% (w/v) paraformaldehyde, immunostained using the anti-p110Rb (OP77-Ab-6, Calbiochem; 1:25) mouse monoclonal, followed by Alexafluor 568-labeled goat anti-mouse secondary antibody (Molecular Probes, Eugene, OR; 1:1000). Samples were mounted on coverslips in 4% propylgallate made up in phosphate-buffered saline/glycerol (90% w/v), and imaged using an Olympus Fluorview 1000 CLSM (Olympus, Tokyo, Japan), with a Nikon 60× oil immersion lens (Nikon, Tokyo, Japan). Subcellular localization of GFP-T-ag fusion proteins in living cells was visualized either 8 (COS-7) or 14 h (SAOS-2/SR-40) after transfection by CLSM using a Bio-Rad MRC600 with a ×40 water immersion lens and heated stage. The nuclear to cytoplasmic ratio (Fn/c) was determined as previously (6, 9, 10, 18, 27, 29–32) using the Image J 1.40o public domain software (NIH), from single cell measurements for the nuclear (Fn) and cytoplasmic (Fc) fluorescence, subsequent to the subtraction of fluorescence due to autofluorescence/background.
Fluorescence Recovery after Photobleaching (FRAP)
FRAP was performed essentially as previously (33–35), whereby bleaching of nuclear fluorescence and monitoring of the return of fluorescence to the nucleus enables the nuclear import rate to be determined. Briefly, COS-7 cells transiently expressing GFP-T-ag derivatives were visualized using an Olympus Fluoview 1000 microscope (100× oil immersion lens). Three images were collected using 3% total laser power with excitation at 488 nm (2× zoom, scanned 8 ms/pixel) before photobleaching. To bleach the nucleus, the whole nucleus was selected, and the laser power increased to 100% (10 scans, 12.5 ms/pixel). After bleaching, the cells were immediately scanned and the recovery of fluorescence monitored by acquiring subsequent images at 20-s intervals for 8 min using detector and laser settings identical to those prior to photobleaching. Image analysis was performed as described above. To determine the rate of nuclear import in the nuclear bleaching experiments, the results were expressed in Fn/c over time and analyzed for the initial rate (Fn/c s−1) and fractional recovery of Fn/c expressed as a percent relative to GFP-T-ag-(110–135).
Native PAGE/Fluorimaging
GFP fusion proteins (0.5 mm) were incubated with increasing amounts of IMP α/β for 15 min at room temperature in a final volume of 25 μl prior to loading and native gel electrophoresis, as described (36). Fluorescent bands were visualized using the Wallach Arthur 1422 multiwavelength fluorimager, and quantitative image analysis performed, as previously described (36).
Immunoprecipitation/Western Blot Analysis
GFP fusion proteins were immunoprecipitated using the GFP-Trap® (Chromotek, Germany) bead system, according to the recommendations of the manufacturer. Briefly, cells (10-cm dishes/80% confluent) were harvested 30 h post-transfection and lysed in 300 μl of lysis buffer (150 mm NaCl, 10 mm Tris-HCl (pH 7.5), 0.5 mm EDTA, 0.5% Nonidet P-40, and complete protease inhibitors (Roche, Basel, Switzerland)), and incubated on ice for 30 min. Cell debris was removed by centrifuging at 14,000 rpm for 20 min at 4 °C, and the supernatant adjusted to 750 μl and added to equilibrated GFP-Trap beads for gentle end-over-end mixing for 2 h at 4 °C. The beads were pelleted by centrifugation at 3000 rpm for 2 min, before washing two times for 15 min in 1 ml of dilution buffer (lysis buffer, minus the Nonidet P-40), centrifuging and discarding the supernatant each time. Proteins were eluted from the beads using 100 μl of SDS lysis buffer (0.2% bromphenol blue; 10% glycerol; 200 mm dithiothreitol; 2% SDS; 100 mm Tris; pH 8.3), 25 μl of the eluate was subjected to polyacrylamide gel electrophoresis (8% gel), and the separated proteins then transferred to a nitrocellulose filter (26). The membrane was blocked in buffer A (5% skim milk powder (w/v) and 1× phosphate-buffered saline) overnight at 4 °C and washed three times with buffer B (0.05% Tween 20 and 1× phosphate-buffered saline). Detection of GFP-T-ag fusion proteins was performed by incubating membranes with anti-GFP (Roche; 1:1000) mouse monoclonal primary antibody and horseradish peroxidase (HRP)-coupled goat anti-mouse secondary antibody (CHEMICON, Temecula, CA; 1:10000). Detection of p110Rb was performed analogously, except that anti-p110Rb (4H1, Cell Signaling, Boston, MA; 1:2000 or OP77-Ab-6, Calbiochem; 1:100) specific antibodies were used. αβ-tubulin was detected using anti-αβ-tubulin (Cell Signaling; 1:1000) rabbit primary antibody and HRP-coupled goat anti-rabbit secondary antibody (CHEMICON; 1:10000). The immunoblots were incubated with ECL plus reagent (Amersham Biosciences, Piscataway, NJ), according to the recommendations of the manufacturer and chemiluminescence captured on a photosensitive film.
Densitometric Analysis
Band intensity captured on film was analyzed by performing densitometric scans using the NIH image J (V. 1.41o) public domain software, with the membrane background subtracted. Band intensity was then calculated relative the GFP fusion proteins expression level (loading control) as previously described (19). Results were expressed relative to those for GFP alone.
RESULTS
Nuclear Accumulation of T-ag Is Reduced in the Presence of the Rb Binding Site
T-ag nuclear accumulation is known to be highly regulated, with phosphorylation adjacent to the NLS playing a key role in enhancing or inhibiting the ability of T-ag to be transported into the nucleus (6, 9, 17, 18). This is in part due to the fact that T-ag is able to bind to many host cell proteins during infection, including members of the Rb family, through the RbBS (amino acids 102–110) N-terminal to the NLS (amino acids 126–132). To test whether the RbBS of T-ag may play a role in modulating T-ag nuclear accumulation, we compared the extent of nuclear localization of various GFP-T-ag fusion protein constructs with and without the RbBS (see Fig. 1) using CLSM on living transfected COS-7 cells at the single cell level (Fig. 2). Determination of the relative levels of specific nuclear and cytoplasmic fluorescence by image analysis of digitized CLSM files revealed that nuclear accumulation of GFP-T-ag-(102–135) and GFP-T-ag-(87–135), both of which contain the RbBS, was significantly lower (p < 0.0058) than that of GFP-T-ag-(110–135) (values for the nuclear to cytoplasmic ratio, Fn/c, of c. 12 and 15 compared with 27, respectively; Fig. 2B), which lacks the RbBS. Significantly, we noted that the same trend was evident for the NLS mutant derivatives where, even though the maximal nuclear accumulation was markedly less than that for the wild-type constructs, the nuclear accumulation of GFP-T-ag-(102–135:NLSmut) and GFP-T-ag-(87–135:NLSmut) was significantly (p < 0.0001) lower than that of GFP-T-ag-(110–135:NLSmut) (Fn/c of c. 1.3 compared with 1.9; Fig. 2, A and B, right). The clear implication was that the presence of the RbBS reduced T-ag nuclear accumulation, and appeared to function independently of the T-ag NLS.
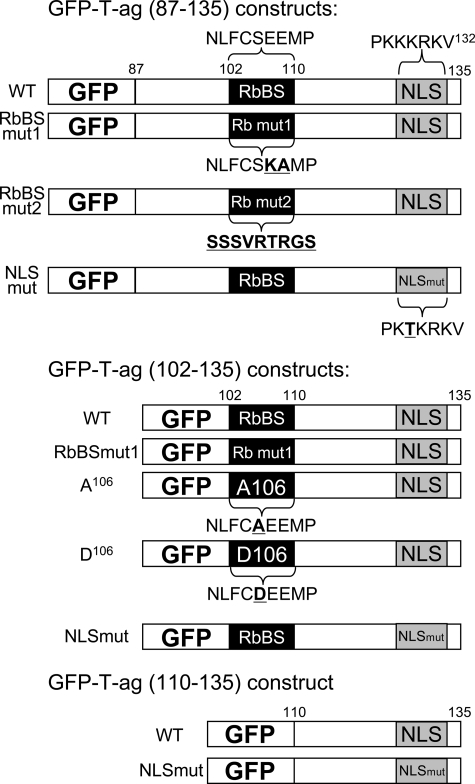
FIGURE 1.
Schematic representation of SV40 T-ag fusion protein constructs used in this study. The position of the NLS (PKKKRKV132, single letter amino acid code) is shown, together with the RbBS and mutants thereof; substituted residues are denoted in bold type and underlined.
FIGURE 2.
The Rb binding site (RbBS-T-ag amino acids 102–110) reduces T-ag maximal nuclear accumulation. A, CLSM images of COS-7 cells transfected to express the indicated GFP-T-ag proteins 8 h post-transfection. B, results of image analysis, performed using the Image J software to determine the nuclear to cytoplasmic ratio (Fn/c), determined from measurements of nuclear (Fn) and cytoplasmic (Fc) fluorescence, subsequent to the subtraction of background fluorescence (autofluorescence). Results represent the mean ± S.E. (n > 52). p values indicate significant differences between Fn/c values in the absence or presence of the RbBS.
The results were extended using the technique of FRAP as previously (33–35), whereby bleaching of nuclear fluorescence and monitoring of the return of fluorescence to the nucleus enables the nuclear import rate to be determined in living cells. Analysis (not shown) for wild-type GFP T-ag-(87-, 102-, and 110–135) proved difficult because of the relatively low amounts of cytoplasmic fluorescence in the transfected cells at steady state, but experimental examination of the GFP-T-ag-(87-, 102- and 110–135) NLS mutant derivatives proved informative (Fig. 3, A and B). Quantification over several experiments showed that the presence of the RbBS in the GFP-T-ag-(87- and 102–135) NLS mutants resulted in significantly (p < 0.012) reduced (>10%) recovery compared with the GFP-T-ag-(110–135) NLS mutant, which lacks the RbBS (Fig. 3C, left); the clear implication was that the presence of the RbBS led to retention in the cytoplasm. The initial rate of nuclear import was not significantly different for these constructs (Fig. 3C, right), indicating that the presence of the RbBS does not alter the rate of nuclear import. This implies that the reduced level of nuclear accumulation of T-ag constructs carrying a functional RbBS is a result of cytoplasmic retention, likely to be via p110Rb binding (see below).
FIGURE 3.
The RbBS causes cytoplasmic retention of T-ag in vivo. A, CLSM images showing the return of nuclear fluorescence after photobleaching (see “Experimental Procedures”) in COS-7 cells expressing the indicated GFP-T-ag constructs. B, quantification of the recovery over time of nuclear fluorescence after photobleaching, shown as Fn/c. C, pooled data (mean, S.E., n > 7) for the fractional recovery (% recovery of Fn/c relative to GFP-T-ag-(110–135); left) and the initial rate (Fn/c s−1; right) into the nucleus for the indicated GFP-T-ag constructs. p values indicate significant differences.
Nuclear Accumulation of T-ag Is Increased in the Absence of Functional p110Rb
To test the role of p110Rb binding to the RbBS in inhibiting T-ag nuclear import, various GFP-T-ag fusion proteins with or without the RbBS, and/or with or without mutations inactivating it (see Fig. 1, Refs. 37, 38) were expressed in the isogenic cell pair SAOS-2 and SR-40 (Fig. 4); SAOS-2 cells lack a functional p110Rb possessing a 95 kDa C-terminally truncated non-functional p110Rb protein (see Fig. 5A), whereas SR-40 cells represent SAOS-2 cells that have been transfected to express p110Rb stably (24, 25). Quantitative analysis revealed that GFP-T-ag-(87–135) accumulated to the same extent as GFP-T-ag-(110–135) in the SAOS-2 cell line, but showed significantly (p < 0.0001) reduced accumulation compared with GFP-T-ag-(110–135) in p110Rb-expressing SR-40 cells (see Fig. 4B). No significant difference in nuclear accumulation was evident when comparing the extent of nuclear accumulation of GFP-T-ag-(87–135:Rb mut1) and GFP-T-ag-(87–135:Rb mut2), both of which contain a nonfunctional RbBS, in either SAOS-2 or SR-40 cells (Fig. 4B), in similar fashion to GFP-T-ag-(110–135). The results clearly suggest that the inhibitory effect of the RbBS on T-ag nuclear accumulation requires functional p110Rb binding.
FIGURE 4.
p110Rb expression reduces nuclear import of T-ag constructs containing a functional RbBS. A, CLSM images are shown for the indicated GFP-T-ag proteins 14 h post-transfection in either SAOS-2 and/or SR-40 cell lines. B, results for image analysis, performed as described in the legend to Fig. 2, where n > 38. p values are indicated for significant differences in Fn/c between the 2 cell lines or between constructs. C, CLSM images are shown for the indicated GFP-T-ag proteins 14-h post-transfection in either SAOS-2 and/or SR-40 cell lines. D, results for image analysis, performed as described in the legend to Fig. 2, where n > 17. p values denote significant differences in Fn/c between the two cell lines or between constructs.
FIGURE 5.
Reconstitution of inhibition of T-ag nuclear import by the RbBS in vitro; modulation by Ser106. A, CLSM images for the indicated GFP-T-ag proteins in mechanically perforated HTC cells in the presence of exogenously added cytosol and an ATP-regenerating system in the presence or absence of anti-p110Rb antibody or control (anti-GST antibody) after 20–30 min at room temperature (see “Experimental Procedures”). Western analysis for p110Rb (bottom right) is shown for SAOS-2 and SR-40 cell lysates, with αβ-tubulin as a loading control, as well as for rabbit reticulocyte lysate. B, CLSM files were analyzed using the Image J software (see “Experimental Procedures”) to determine the average Fn/c (± S.E., n > 4), with the kinetics of nuclear import determined by fitting the data to the function Fn/c(t) = Fn/cmax(1 − e−kt). Data are shown for the indicated GFP-T-ag proteins alone (black curves) or with anti-p110Rb antibody (red curves), or control anti-GST antibody (blue curve).
The RbBS contains the Ser106 residue, which is known to be phosphorylated in SV40-infected cells (15, 39); as a first step to assess if this residue was involved in regulating p110Rb effect, quantitative live cell imaging was performed on point mutants designed to prevent (Ala106) or mimic (Asp106) phosphorylation at the site (Fig. 4, C and D). GFP-T-ag-(102–135) and GFP-T-ag-(102–135:D106) accumulated to the same extent as GFP-T-ag-(110–135) in the SAOS-2 cell line but showed a significantly (p < 0.0021) reduced accumulation compared with GFP-T-ag-(110–135) in the p110Rb-expressing SR-40 line (Fig. 4D). There was no significant difference in nuclear accumulation between SAOS-2 and SR-40 cells; however, for GFP-T-ag-(102–135:A106) or when compared with GFP-T-ag-(110–135) (Fig. 4D), suggesting that negative charge (normally supplied by phosphorylation) at Ser106 may facilitate p110Rb binding to the T-ag RbBS, thus resulting in reduced nuclear accumulation.
To confirm these results, we used an established in vitro reconstituted nuclear transport system (40, 41), and bacterially expressed GFP-T-ag-(110–135), GFP-T-ag-(102–135), GFP-T-ag-(102–135:A106), GFP-T-ag-(102–135:D106), and GFP-T-ag-(102–135:Rb mut1). The various proteins were tested for the ability to accumulate in the presence of an ATP-regenerating system, and exogenous cytosol, which supplies IMPs/Ran etc., in the absence and presence of specific antibody to p110Rb (see “Experimental Procedures,” Fig. 5A). Nuclear accumulation was followed over time using CLSM, and the results compared with those for GFP-T-ag-(110–135) (Fig. 5). Quantitative analysis revealed that the GFP-T-ag-(102–135) and GFP-T-ag-(102–135:D106) accumulated to a lower extent than GFP-T-ag-(110–135) in the absence of anti-p110Rb antibody, while GFP-T-ag-(102–135:A106) and GFP-T-ag-(102–135:Rb mut1) accumulated to levels similar to that of GFP-T-ag-(110–135) (Fig. 5B). In the presence of anti-p110Rb antibody, all of the GFP-T-ag-(102–135) fusion proteins showed very similar levels of maximal accumulation, comparable to that of GFP-T-ag-(110–135) in the absence of antibody. In the case of GFP-T-ag-(102–135) and GFP-T-ag-(102–135:D106) with a functional RbBS, this represented a significant (p = 0.0025 and 0.0192, respectively) increase in maximal nuclear accumulation (Fig. 5B; see Table 1 for pooled data); that the results were specific to p110Rb, was indicated by the fact that a control antibody (anti-GST) had no effect on the nuclear accumulation of these proteins (Fig. 5B). This implies that the lower nuclear accumulation of T-ag constructs carrying a functional RbBS is a result of p110Rb binding hindering the transport process (see above).
TABLE 1.
Pooled data for nuclear import kinetics for GFP-T-ag proteins in vitro
Results are for the mean ± S.E. (number in parentheses) for in vitro nuclear transport assays as per Fig. 5. p values are shown for significant differences.
| Protein | Maximum Fn/c |
p values | |
|---|---|---|---|
| No addition | Anti-p110RbAb | ||
| GFP-T-ag-(110–135) | 2.7 ± 0.05 (4) | 2.6 ± 0.14 (4) | NSa |
| GFP-T-ag-(102–135) | 1.9 ± 0.06 (4) | 2.4 ± 0.08 (4) | 0.0041 |
| GFP-T-ag-(102–135:Ala106) | 2.5 ± 0.23 (3) | 2.5 ± 0.08 (3) | NS |
| GFP-T-ag-(102–135:Asp106) | 1.8 ± 0.09 (3) | 2.4 ± 0.13 (3) | 0.0321 |
| GFP-T-ag-(102–135:RbBSmut) | 2.8 ± 0.22 (4) | 2.9 ± 0.14 (4) | NS |
a NS, not significant.
Negative Charge at the Ser106 Phosphorylation Site Enhances p110Rb Binding to T-ag
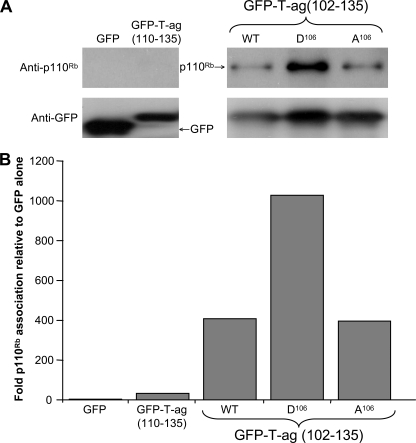
To confirm that the effects observed were due to p110Rb binding, COS-7 cells were transfected to express GFP-T-ag-(102–135; WT, Ala106, or Asp106), with GFP-T-ag-(110–135), and GFP alone as negative control, and the GFP-Trap® system used to immunoprecipitate the proteins (see “Experimental Procedures”), followed by Western analysis to assess the extent of association of endogenous p110Rb to the GFP-T-ag-(102–135) derivatives (Fig. 6, A and B). As expected, p110Rb was detected in pull-downs with the T-ag-(102–135) proteins; significantly, T-ag-(102–135:D106) pulled down markedly higher amounts of endogenous p110Rb protein than the GFP-T-ag-(102–135:A106) or GFP-T-ag-(102–135) WT. Densitometric analysis correcting for the extent of GFP/GFP-T-ag expression indicated that p110Rb bound GFP-T-ag-(102–135) WT and Ala106 to an extent almost 400-fold that of GFP alone. In contrast, GFP-T-ag (102–135:Asp106) showed p110Rb binding >1000-fold that of GFP alone, 2.5-fold higher than that of GFP-T-ag-(102–135) Ala106 (Fig. 6B). Clearly, association of p110Rb with T-ag was strongly dependent on a negative charge at Ser106, consistent with the in vivo (Fig. 4, C and D) and in vitro transport data (Fig. 5).
FIGURE 6.
Binding of T-ag to p110Rb is enhanced by negative charge at the Ser106 phosphorylation site. A, lysates of COS-7 cells transfected to express either GFP, GFP-T-ag-(110–135), or the indicated GFP-T-ag-(102–135) derivatives were immunoprecipitated using GFP-Trap® as described under “Experimental Procedures” prior to Western detection of p110Rb (top) or the GFP fusion proteins themselves (bottom) using specific antibodies. B, densitometric analysis to estimate levels of protein in A, where the value for p110Rb above background was calculated relative to that of GFP/GFP fusion protein, with values then expressed as a percentage of the value for GFP alone.
The RbBS Does Not Impair Imp α/β Binding to the T-ag NLS
To eliminate the possibility that the results for reduced nuclear import through the RbBS were not the result, through steric effects, of reduced binding of IMP α/β to the T-ag NLS, we assessed the various GFP-T-ag fusion proteins for IMP α/β binding using native gel electrophoresis/fluorography (36). Briefly, the proteins with or without the RbBS were incubated with increasing concentrations of predimerized Imp α/β complex, and then electrophoresed on a native gel followed by fluoroimaging (Fig. 7); Imp α/β binding results in reduced electrophoretic mobility, enabling quantitation of the extent of binding by image analysis, and estimation of the apparent dissociation constant (Kd). High affinity binding was evident for all of the GFP fusion products used (Kd of 210–340 nm; see Fig. 7; Table 2), with binding affinities in keeping with those determined previously using this assay (36). This clearly implies that the GFP-T-ag constructs carrying the RbBS are still able to bind IMP α/β with high affinity, meaning that impaired IMP binding is not the basis of the ability of the RbBS to impair T-ag nuclear import; rather, binding of p110Rb is the mechanism by which nuclear import is reduced (see above).
FIGURE 7.
Imp α/β binding to T-ag NLS-containing proteins is not influenced by the RbBS as determined by native gel electrophoresis/fluorography. A, increasing concentrations of predimerized IMP α/β were added to 0.5 μm of GFP-T-ag-(110–135) (top panel) or GFP-T-ag-(102–135) (bottom panel), incubated for 15 min at 30 °C, and then subjected to native gel electrophoresis and fluorography as described under “Experimental Procedures”. B, quantification of IMP α/β binding affinity. The Image J software was used to determine the percentage of protein with altered mobility for both GFP-T-ag-(110–135) (left panel) and GFP-T-ag-(102–135) (right panel). Apparent dissociation constants, determined by sigmoidal curve fitting, are indicated (see Table 2 for pooled data).
TABLE 2.
Pooled data for T-ag NLS-containing protein-Imp α/β interaction as determined using gel mobility shift assays
Results are for the mean ± S.E. (number in parentheses) for gel mobility shift assays as per Fig. 7. No significant difference was observed seen between any of the protein Kd values.
| Protein | Kd |
|---|---|
| μm | |
| GFP-T-ag-(111–135) | 0.21 ± 0.06 (3) |
| GFP-T-ag-(102–135) | 0.24 ± 0.04 (4) |
| GFP-T-ag-(102–135:Ala106) | 0.33 ± 0.11 (4) |
| GFP-T-ag-(102–135:Asp106) | 0.34 ± 0.07 (3) |
| GFP-T-ag-(87–135) | 0.28 ± 0.16 (3) |
| GFP-T-ag-(87–135:Rb mut1) | 0.34 ± 0.10 (3) |
Nuclear Localization of Endogenous p110Rb Is Reduced in the Presence of T-ag
The results above indicate that complexation of p110Rb with T-ag results in cytoplasmic retention and consequently reduced nuclear localization on the part of T-ag. To assess the potential effect T-ag, in turn, may have on the localization of p110Rb, we overexpressed GFP-T-ag-(102–135: WT, Ala106, or Asp106) in CV-1 cells and subsequently fixed and immunostained the cells for p110Rb followed by CLSM analysis (Fig. 8A). Decreased nuclear localization of p110Rb was evident in the case of CV-1 cells expressing GFP-T-ag-(102–135) WT or Asp106, compared with those either not expressing T-ag, or GFP-T-ag-(102–135)Ala106; this was mirrored by the presence of more cytoplasmic fluorescence due to GFP-T-ag-(102–135) in the case of the WT or Asp106 derivatives, but not the Ala106 derivative. Image analysis confirmed the results, whereby the average nuclear fluorescence (Fn) of p110Rb in the presence of GFP-T-ag-(102–135) WT and Asp106, but not Ala106, was significantly (p < 0.0001) reduced (>30%), accompanied by a significant (p < 0.0001) increase (>25%;) in the average cytoplasmic fluorescence (Fc) compared with nontransfected cells (Fig. 8B, left and center panels). Determination of the nuclear to cytoplasmic ratio (Fn/c) indicated the significantly (p < 0.0001) lower (>50%) levels of nuclear accumulation of p110Rb in CV-1 cells expressing GFP-T-ag-(102–135) WT and Asp106, but not Ala106, when compared with nontransfected cells (Fig. 8B, right panel); this closely followed the results for GFP-T-ag, where GFP-T-ag-(102–135:A106) showed significantly higher nuclear accumulation compared with the WT and Asp106 derivative (Fig. 8C, compare with results for SR40 cells, Fig. 4D). These results indicate that negative charge at Ser106, either through phosphorylation or substitution with the phosphomimetic Asp106, results in reduced nuclear accumulation of p110Rb, in parallel to that of GFP-T-ag-(102–135); complexation between T-ag and p110Rb thus appears to be critical to cytoplasmic retention of both proteins, consistent with the results in Figs. 2–4.
FIGURE 8.
Nuclear accumulation of p110Rb is reduced in the presence of overexpressed T-ag dependent on negative charge at the Ser106 phosphorylation site. A, CLSM images are shown for CV-1 cells transfected to express GFP-T-ag-(102–135) constructs as indicated, followed by fixation 16-h post-transfection, and immunostaining using a specific anti-p110Rb antibody followed by an Alexa568-labeled secondary antibody. B, results for image analysis of CLSM files such as those in A, performed as per Fig. 2, showing the average Fn (left), Fc (middle), and Fn/c ratio (right) (n > 20) of p110Rb in the presence of the indicated GFP-T-ag derivatives; Fn and Fc values are expressed in percent relative to those for p110Rb in the absence of exogenous T-ag expression. Significant differences in the absence or presence of the T-ag constructs are indicated by the p values. C, results for image analysis of CLSM files such as those in A, performed as per Fig. 2, showing the average Fn/c ratio (n > 20) of the GFP-T-ag derivatives. Significant differences (p < 0.0001) are evident between the values for both the WT and Asp106 derivative, compared with that for GFP-T-ag-(102–135:A106).
DISCUSSION
Previous studies have highlighted the regulatory impact on T-ag nuclear accumulation of the various phosphorylation sites flanking the NLS (amino acids 126–132) (9, 10, 17, 18), but this is first study to document the inhibitory effect of the RbBS (amino acids 102–110, including Ser106). We show here for the first time that the presence of the RbBS reduces nuclear accumulation over 50%, both in living transfected cells, and in an in vitro reconstituted system; reduced nuclear accumulation was not observed in cells not expressing functional p110Rb, for T-ag proteins lacking a functional RbBS, or for RbBS-containing constructs in vitro in the presence of an anti-p110Rb antibody. The presence of the RbBS does not alter in any way recognition of the T-ag NLS by IMP α/β (Fig. 7), with Rb-T-ag interactions not influenced by mutations of the T-ag NLS (Fig. 2), implying that conformational effects are not involved. The clear implication is that binding of p110Rb to the RbBS reduces T-ag nuclear import, almost certainly by retaining T-ag in the cytoplasm, as supported by a number of observations. These include the FRAP analysis (Fig. 3), which indicates a reduction in fractional recovery of T-ag in the presence of the RbBS compared with in its absence, but no effect on the transport rate, as an indication of a lack of cotransport of T-ag and p110Rb to the nucleus, as well as a lack of masking effects of p110Rb on the T-ag NLS. The results for reduced nuclear localization of p110Rb upon overexpression of T-ag in CV-1 cells (Fig. 8) similarly imply that complexation of p110Rb and T-ag in the cytoplasm leads to cytoplasmic retention of the p110Rb-T-ag complex; the clear implication is that the p110Rb-T-ag complex is not functional for nuclear import, presumably as a result of impaired recognition by IMP α/β. Intriguingly, Ser106 within the RbBS, known to be phosphorylated in SV40-infected cells, appears to modulate binding of p110Rb to the RbBS (see Fig. 6), whereby negative charge at the site enhances binding and thereby inhibition of nuclear transport, while a nonphosphorylatable alanine at the site proves to be as effective as mutations perturbing p110Rb binding (e.g. KA108), eliminating the inhibition seen in the presence of p110Rb.
This study shows that, in the context of functional p110Rb, T-ag has lower maximal nuclear accumulation, implying that, additional to growth/cell cycle regulation and various other host cell functions, p110Rb may have a role in combating SV40 virus infection by reducing T-ag nuclear accumulation by binding T-ag in the cytoplasm; that p110Rb is present at detectable levels in the cytoplasm throughout the cell cycle has been reported by Ferecatu et al. (42). Importantly, studies of SV40 itself suggest that various mutations (e.g. E107K, E108K) in the RbBS that impair transformation (20, 37, 38) (and hence p110Rb binding) do not inhibit viral replication (37), precisely as would be predicted from the results here; whether virus production is enhanced remains to be determined. T-ag is believed to bind exclusively to hypophosphorylated p110Rb, which is normally predominantly found in the G1 phase of the cell cycle (43), so that it is significant that SV40 infection is known to perturb the cell cycle, resulting in progression to G2-M phase (44, 45), where p110Rb binding to T-ag would be less likely. Intriguingly in this context, however, the cellular response to SV40 infection/stress is to increase the level of hypophosphorylated p110Rb but not p107 as the cells move through G2 (46–48). That p110Rb appears to be the key Rb family member in this context is consistent with our preliminary data (not shown) that antibodies to p130 do not alter the nuclear import kinetics of RbBS-containing T-ag to a marked extent in our in vitro transport system, as well as with our observations for the SAOS-2/SR-40 isogenic cells pair (Fig. 4), where SAOS-2 cells, which lack functional p110Rb, fail to show an effect of the RbBS on T-ag nuclear accumulation. Because various other DNA tumor virus gene products of significance such as adenovirus E1A (49–51), human papilloma virus E7 (41, 52–54), the large tumor antigens of the JC and Bk viruses (55–57), pUL97 from human cytomegalovirus virus (58), and the MC007L gene product of the molluscum contagiosum virus (59) are all known to interact with p110Rb, it seems likely that p110Rb may act generally in infection by different transforming viruses to modulate the nuclear import of diverse viral proteins. With this in mind, the future focus of this laboratory to establish the precise mechanism of p110Rb-mediated inhibition of nuclear import of T-ag and potentially other viral proteins, looms as having important wider significance in the general area of DNA virus-induced transformation.
This work was supported, in whole or in part, by the NHMRC (National Health and Medical Research Council) (ID 143710 and 384109).
- NLS
- nuclear localization signal
- GFP
- green fluorescent protein
- FRAP
- fluorescence recovery after photobleaching
- IMP
- importin
- Rb
- retinoblastoma
- SV40
- simian virus 40
- T-ag
- large tumor antigen
- BS
- binding site
- CLSM
- confocal laser scanning microscopy
- GST
- glutathione S-transferase
- WT
- wild type.
REFERENCES
- 1.Kalderon D., Richardson W. D., Markham A. F., Smith A. E. (1984) Nature 311, 33–38 [DOI] [PubMed] [Google Scholar]
- 2.Jans D. A., Xiao C. Y., Lam M. H. (2000) Bioessays 22, 532–544 [DOI] [PubMed] [Google Scholar]
- 3.Terry L. J., Shows E. B., Wente S. R. (2007) Science 318, 1412–1416 [DOI] [PubMed] [Google Scholar]
- 4.Görlich D., Kutay U. (1999) Annu. Rev. Cell Dev. Biol. 15, 607–660 [DOI] [PubMed] [Google Scholar]
- 5.Fried H., Kutay U. (2003) Cell Mol. Life Sci. 60, 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jans D. A., Ackermann M. J., Bischoff J. R., Beach D. H., Peters R. (1991) J. Cell Biol. 115, 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jans D. A., Moll T., Nasmyth K., Jans P. (1995) J. Biol. Chem. 270, 17064–17067 [DOI] [PubMed] [Google Scholar]
- 8.Jans D. A., Hübner S. (1996) Physiol. Rev. 76, 651–685 [DOI] [PubMed] [Google Scholar]
- 9.Hübner S., Xiao C. Y., Jans D. A. (1997) J. Biol. Chem. 272, 17191–17195 [DOI] [PubMed] [Google Scholar]
- 10.Xiao C. Y., Hübner S., Jans D. A. (1997) J. Biol. Chem. 272, 22191–22198 [DOI] [PubMed] [Google Scholar]
- 11.Briggs L. J., Stein D., Goltz J., Corrigan V. C., Efthymiadis A., Hübner S., Jans D. A. (1998) J. Biol. Chem. 273, 22745–22752 [DOI] [PubMed] [Google Scholar]
- 12.Harreman M. T., Kline T. M., Milford H. G., Harben M. B., Hodel A. E., Corbett A. H. (2004) J. Biol. Chem. 279, 20613–20621 [DOI] [PubMed] [Google Scholar]
- 13.Poon I. K., Jans D. A. (2005) Traffic 6, 173–186 [DOI] [PubMed] [Google Scholar]
- 14.Poon I. K., Oro C., Dias M. M., Zhang J., Jans D. A. (2005) Cancer Res. 65, 7059–7064 [DOI] [PubMed] [Google Scholar]
- 15.Scheidtmann K. H., Echle B., Walter G. (1982) J. Virol. 44, 116–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheidtmann K. H., Hardung M., Echle B., Walter G. (1984) J. Virol. 50, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jans D. A., Jans P. (1994) Oncogene 9, 2961–2968 [PubMed] [Google Scholar]
- 18.Rihs H. P., Jans D. A., Fan H., Peters R. (1991) EMBO J. 10, 633–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fulcher A. J., Roth D. M., Fatima S., Alvisi G., Jans D. A. (2009) FASEB J., in press [DOI] [PubMed] [Google Scholar]
- 20.DeCaprio J. A., Ludlow J. W., Figge J., Shew J. Y., Huang C. M., Lee W. H., Marsilio E., Paucha E., Livingston D. M. (1988) Cell 54, 275–283 [DOI] [PubMed] [Google Scholar]
- 21.DeCaprio J. A., Ludlow J. W., Lynch D., Furukawa Y., Griffin J., Piwnica-Worms H., Huang C. M., Livingston D. M. (1989) Cell 58, 1085–1095 [DOI] [PubMed] [Google Scholar]
- 22.Ewen M. E., Ludlow J. W., Marsilio E., DeCaprio J. A., Millikan R. C., Cheng S. H., Paucha E., Livingston D. M. (1989) Cell 58, 257–267 [DOI] [PubMed] [Google Scholar]
- 23.Baliga B. C., Colussi P. A., Read S. H., Dias M. M., Jans D. A., Kumar S. (2003) J. Biol. Chem. 278, 4899–4905 [DOI] [PubMed] [Google Scholar]
- 24.Fogh J., Fogh J. M., Orfeo T. (1977) J. Natl. Cancer Inst. 59, 221–226 [DOI] [PubMed] [Google Scholar]
- 25.Huang H. J., Yee J. K., Shew J. Y., Chen P. L., Bookstein R., Friedmann T., Lee E. Y., Lee W. H. (1988) Science 242, 1563–1566 [DOI] [PubMed] [Google Scholar]
- 26.Alvisi G., Jans D. A., Ripalti A. (2006) Biochemistry 45, 6866–6872 [DOI] [PubMed] [Google Scholar]
- 27.Poon I. K., Oro C., Dias M. M., Zhang J. P., Jans D. A. (2005) J. Virol. 79, 1339–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuusisto H. V., Wagstaff K. M., Alvisi G., Jans D. A. (2008) Int. J. Cancer 123, 2965–2969 [DOI] [PubMed] [Google Scholar]
- 29.Hearps A. C., Jans D. A. (2006) Biochem. J. 398, 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghildyal R., Ho A., Wagstaff K. M., Dias M. M., Barton C. L., Jans P., Bardin P., Jans D. A. (2005) Biochemistry 44, 12887–12895 [DOI] [PubMed] [Google Scholar]
- 31.Hübner S., Eam J. E., Wagstaff K. M., Jans D. A. (2006) J. Cell. Biochem. 98, 810–826 [DOI] [PubMed] [Google Scholar]
- 32.Hearps A. C., Wagstaff K. M., Piller S. C., Jans D. A. (2008) Biochemistry 47, 2199–2210 [DOI] [PubMed] [Google Scholar]
- 33.Lam M. H., Henderson B., Gillespie M. T., Jans D. A. (2001) Traffic 2, 812–819 [DOI] [PubMed] [Google Scholar]
- 34.Lam M. H., Thomas R. J., Loveland K. L., Schilders S., Gu M., Martin T. J., Gillespie M. T., Jans D. A. (2002) Mol. Endocrinol. 16, 390–401 [DOI] [PubMed] [Google Scholar]
- 35.Roth D. M., Moseley G. W., Glover D., Pouton C. W., Jans D. A. (2007) Traffic 8, 673–686 [DOI] [PubMed] [Google Scholar]
- 36.Wagstaff K. M., Dias M. M., Alvisi G., Jans D. A. (2005) J. Fluoresc. 15, 469–473 [DOI] [PubMed] [Google Scholar]
- 37.Kalderon D., Smith A. E. (1984) Virology 139, 109–137 [DOI] [PubMed] [Google Scholar]
- 38.Cherington V., Brown M., Paucha E., St Louis J., Spiegelman B. M., Roberts T. M. (1988) Mol. Cell. Biol. 8, 1380–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grässer F. A., Scheidtmann K. H., Tuazon P. T., Traugh J. A., Walter G. (1988) Virology 165, 13–22 [DOI] [PubMed] [Google Scholar]
- 40.Hu W., Jans D. A. (1999) J. Biol. Chem. 274, 15820–15827 [DOI] [PubMed] [Google Scholar]
- 41.Hu W., Kemp B. E., Jans D. A. (2005) J. Cell. Biochem. 95, 782–793 [DOI] [PubMed] [Google Scholar]
- 42.Ferecatu I., Le Floch N., Bergeaud M., Rodríguez-Enfedaque A., Rincheval V., Oliver L., Vallette F. M., Mignotte B., Vayssière J. L. (2009) BMC Cell Biol. 10, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchkovich K., Duffy L. A., Harlow E. (1989) Cell 58, 1097–1105 [DOI] [PubMed] [Google Scholar]
- 44.Sladek T. L., Jacobberger J. W. (1992) J. Virol. 66, 1059–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ordenes G. E., Beck I., Carmona M. T., Santos M. (1989) J. Cell Sci. 93, 525–532 [DOI] [PubMed] [Google Scholar]
- 46.Yen A., Sturgill R. (1998) Exp. Cell Res. 241, 324–331 [DOI] [PubMed] [Google Scholar]
- 47.Friedrich T. D., Laffin J., Lehman J. M. (1993) Oncogene 8, 1673–1677 [PubMed] [Google Scholar]
- 48.Cobrinik D. (2005) Oncogene 24, 2796–2809 [DOI] [PubMed] [Google Scholar]
- 49.Whyte P., Buchkovich K. J., Horowitz J. M., Friend S. H., Raybuck M., Weinberg R. A., Harlow E. (1988) Nature 334, 124–129 [DOI] [PubMed] [Google Scholar]
- 50.Whyte P., Williamson N. M., Harlow E. (1989) Cell 56, 67–75 [DOI] [PubMed] [Google Scholar]
- 51.Herrmann C. H., Su L. K., Harlow E. (1991) J. Virol. 65, 5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dyson N., Buchkovich K., Whyte P., Harlow E. (1989) Cell 58, 249–255 [DOI] [PubMed] [Google Scholar]
- 53.Gage J. R., Meyers C., Wettstein F. O. (1990) J. Virol. 64, 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davies R., Hicks R., Crook T., Morris J., Vousden K. (1993) J. Virol. 67, 2521–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bollag B., Chuke W. F., Frisque R. J. (1989) J. Virol. 63, 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haggerty S., Walker D. L., Frisque R. J. (1989) J. Virol. 63, 2180–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harris K. F., Christensen J. B., Imperiale M. J. (1996) J. Virol. 70, 2378–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prichard M. N., Sztul E., Daily S. L., Perry A. L., Frederick S. L., Gill R. B., Hartline C. B., Streblow D. N., Varnum S. M., Smith R. D., Kern E. R. (2008) J. Virol. 82, 5054–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mohr S., Grandemange S., Massimi P., Darai G., Banks L., Martinou J. C., Zeier M., Muranyi W. (2008) J. Virol. 82, 10625–10633 [DOI] [PMC free article] [PubMed] [Google Scholar]