Abstract
G-protein-coupled receptor 54 (Gpr54, KISS1 receptor) plays critical roles in puberty regulation, tumor metastasis suppression, and vasoconstriction. Bone morphogenetic protein-7 (Bmp7) is required for kidney organogenesis. However, whether Gpr54 is involved in embryonic kidney development and how Bmp7 expression is regulated in the kidney are largely unknown. Here we report that Gpr54 deletion leads to kidney branching morphogenesis and glomerular development retardation in embryonic kidneys in vivo and in explanted kidneys in vitro. Gpr54 inactivation results in a high risk of low glomerular number in adult kidneys. Gpr54 is expressed in condensed mesenchyme at E12.5 and epithelial cells of proximal and distal tubules and collecting ducts at E17.5 and P0 mouse kidney. Deletion of Gpr54 decreases Bmp7 expression and Smad1 phosphorylation in the developing kidney. Using chromatin immunoprecipitation and luciferase assays, we demonstrate that Gpr54 regulates NFAT2- and Sp1-mediated Bmp7 transcription. Furthermore, we show that NFAT2 cooperates with Sp1 to promote Bmp7 transcription activation. Together, these data suggest that Gpr54 regulates Bmp7 expression through NFAT2 and Sp1 and plays an important role in embryonic kidney branching morphogenesis and glomerular development.
Keywords: Bone Morphogenetic Protein (BMP), Calcineurin, G-protein-coupled Receptors (GPCR), Kidney, NFAT Transcription Factor, Sp1, Gpr54, KISS1 Receptor, KiSS1
Introduction
KISS1 gene encodes a premature 145-amino acid protein that is proteolytically cleaved into polypeptides known as kisspeptins, including Kp-54 (also called metastin), Kp-14, Kp-13, and Kp-10 (1–3). KISS1 was originally identified as a metastasis suppressor in melanoma and breast cancer cells (2, 4–6). Further studies showed that loss of KISS1 gene expression was correlated with increased metastasis and/or tumor progression in a wide variety of tumor types, including malignant pheochromocytoma, esophageal squamous cell carcinoma, bladder tumor, ovarian, gastric, and pancreatic tumors (7–17). Recently, increased interests of Kisspeptins focused on their key roles in the regulation of the hypothalamic-pituitary-gonadal axis during puberty and reproductive development (18–28). KISS1 peptides are natural ligands of a specific G-protein-coupled receptor, called Gpr54, KISS1 receptor (3, 28). G-protein-coupled receptor-54 (Gpr54) is a multifunctional receptor, playing critical roles in puberty development, vasoconstriction, and tumor metastasis suppression (6, 25). KISS1/Gpr54 regulates hypothalamus gonadotropin-releasing hormone (GnRH) expression, controlling the maturation of hypothalamic-pituitary-gonadal axis and puberty (25). Mutation or deletion of Gpr54 causes hypogonadotropic hypogonadism in both human and mice (18, 23, 29, 30). Previous studies demonstrate that activation of Gpr54 by kisspeptin stimulates the phospholipase C-inositol 1,4,5-trisphosphate-calcium cascade signaling pathway, broadly involved in tumor metastasis suppression and GnRH neuron excitation (6, 31). However, the role of KISS1- and Gpr54-mediated signaling in kidney development is still unknown.
The development of the mammalian kidney commences at embryonic day (E) 10.5 in mouse through a series of reciprocal inductive interactions between the Wolffian duct, the ureteric bud, and the surrounding metanephric mesenchyme (32–34). Signals secreted by the metanephric mesenchyme induce the ureteric bud to grow toward and invade the metanephric mesenchyme followed by dichotomous branching morphogenesis at about E11.0 (35, 36). Subsequently, mesenchymal cells are induced to condense around the tip and undergo a mesenchyme-epithelial conversion to form the renal vesicle (37, 38). With renal vesicle elongation and division, the vesicles develop into comma-shaped bodies, S-shaped bodies, and eventually functional nephrons (39, 40). Abnormal kidney branching morphogenesis and glomerular development lead to a broad spectrum of kidney diseases and related syndromes, afflicting millions of people per year worldwide. Severe reduction of branching morphogenesis and nephrogenesis contribute to the major causes of childhood renal failure (33). Low nephron number in adults could lead to essential hypertension, chronic kidney disease, and even chronic renal failure (41).
Bone morphogenetic proteins (Bmps),2 multifunctional growth factors of transforming growth factor β, play important roles in ureteric bud outgrowth, ureteric bud branching, tubule maintenance, and nephrogenesis (42–44). Bmp7 is required for proper kidney formation (45, 46). Deficiency of Bmp7 causes arrest in kidney development after the onset of branching morphogenesis and nephrogenesis (47). Bmp7 activates type I receptor, which phosphorylates a receptor-activated Smad (R-Smad, Smad1, -5, and -8). R-Smads then form heteromeric complexes with the common-mediator Smad (Co-Smad, Smad4) in the cytoplasm and translocate into the nucleus where they interact with other transcription factors or regulate transcription of various target gene themselves. Smad1 is expressed in glomeruli, tubules, and collecting ducts in the kidney (46, 48). Bmp7 is expressed in both ureteric epithelium and mesenchyme of early embryonic kidneys and distal tubules in later stage (46, 49). Bmp7 expression in the human adult normal kidney is predominantly localized to the distal nephron (50), and podocyte-derived BMP7 is essential for nephron development (51). A high level of Bmp7 mRNA expression has been reported in the tubules of the outer medulla, adventitia of renal arteries, and epithelial cell layer of the renal pelvis and the ureter (52). Previous studies reported that histone deacetylase isozyme HDAC5 is involved in the regulation of Bmp7 expression in the proximal tubular cells (53). Retinoic acid and prostaglandin E(2) up-regulate BMP-7 protein expression both in vitro and in vivo (54). An ∼20-kb surrounding exon 1 of Bmp7 is an enhancer element highly conserved between species (55). However, the mechanisms of regulating Bmp7 expression in kidney development are largely unknown.
Here we investigate the roles of Gpr54 in embryonic kidney branching morphogenesis and glomerular development and elucidate how Gpr54 regulates Bmp7 expression in the kidney. Our data indicate that Gpr54 deletion leads to kidney embryonic branching morphogenesis defect, glomerular development retardation, and high risk of low glomerular number in the adult kidney. Furthermore, we demonstrate that Gpr54 regulates Bmp7 expression through the cooperative effect of NFAT2 and Sp1 in the developing kidney.
EXPERIMENTAL PROCEDURES
Antibodies, Constructs, Reagents, and Mice
Anti-Bmp7 antibody (ab 56023), anti-p-Sp1 (phospho-Thr-453, ab 59257), and anti-NFAT2 (ab 2796) antibodies were ordered from Abcam Co. Anti-Sp1 antibody (Upstate®) was kindly provided by Dr. Stephen H. Safe (Center for Environmental and Genetic Medicine, Institute of Biosciences and Technology of Texas A&M University Health science Center). Anti-Smad1 and anti-p-Smad1 antibodies (Cell Signaling) were kind gifts of Dr. Xin-Hua Feng of the Baylor College of Medicine. Anti-KiSS1 and anti-Gpr54 antibodies were ordered from Santa Cruz. The overexpression constructs of GFP-RV-DV-HA-NFAT1 (Addgene-11100), pREP-NFAT2 (Addgene-11788), pREP-NFAT3 (Addgene-11789), and pREP-NFAT4 (Addgene-11790) were kind gifts of Dr. Jinke Cheng (Cardiology Department, The University of Texas M.D. Anderson Cancer Center, Houston, TX). The pcDNA-Sp1 and overexpression constructs of GPR54-HA and NFAT2 mutant were made by our laboratory (56). The pSUPER RNAi SystemTM was ordered from OligoengineTM, and the GPR54 RNAi construct was made by our laboratory. Cyclosporin A (57) was ordered from Calbiochem. KISS-1 (112–121) was ordered from California Peptide Research Inc. Calcineurin autoinhibitory peptide (207001) was ordered from EMD Chemical Inc. Autokit Micro TP and Autokit Glucose were ordered from Wako Diagnostic USA. Histostain® plus (Broad spectrum-DAB) was ordered from Zymed Laboratories Inc.. Gpr54+/− heterozygous male and female mice (strain C57BL/6) were obtained from Dr. Eric L. Gustafson at Schering-Plough Research Institute (58). The genotyping work was done with primers as previously described (58).
Hematoxylin and Eosin Staining
The extracted kidneys were paraffin-embedded, and the 5-μm sections were stained with hematoxylin and eosin as previously described (23). The data were represented as the mean of five or six individual values ±S.D. in the indicated high performance fields.
Immunohistochemical Staining
The 5-μm paraffin-embedded kidney sections were performed deparaffinization. The sections were boiled in 10 mm sodium citrate (pH 6.0) for 30∼35 min with an intermitted 10-s cooling in the air followed by cooling at room temperature for 30 min. After washing with 1× PBS (2 × 5 min), the sections were incubated with 1% hydrogen peroxide for 5 min followed by rinsing with 1× PBS (2 × 5 min). After blocking with 1% bovine serum albumin for 30 min, the sections were incubated with fresh anti-Bmp7 antibodies (6 μg/ml) at 4 °C for overnight. After washing with 1× PBS (3 × 5 min), the sections were incubated with fresh second antibodies for 20 min followed by rinsing with 1× PBS (5 × 2 min). Each section was incubated in 2 drops of horseradish peroxidase-streptavidin for 10 min followed by rinsing with 1× PBS (3 × 2 min). Then sections were incubated in DAB chromogen mixture of D1 (DAB substrate buffer), D2 (DAB chromogen solution), and D3 (substrate solution) reagents of Histostain®plus for 3∼10 min. After three washes with double-distilled water, the sections were washed with double-distilled water three times. The sections were then mounted, and images were taken with ZEISS Axioskop 40 photo microscope. For the anti-KiSS1, anti-Gpr54, anti-Smad, anti-p-Smad1 immunohistochemistry assays, most procedures were the same as above-described except for the antibody concentrations (1:100–200 for anti-Gpr54, 1:200–300 for anti-Smad1 and anti-p-Smad1 antibodies).
Glomerular Number Analysis
To estimate the number of glomeruli of the 6-week-old kidney, the average glomerular number of five randomly chosen high performance field (HPF, 20×) in the cortex layer of a section was identified as a glomerular number/HPF/section. The average glomerular number of five continuous third sections of a kidney was identified as a glomerular number/HPF/kidney. The glomerular number/HPF of wild type/Gpr54−/− was identified as the average of five glomerular number/HPF/kidney of wild type or Gpr54−/−, respectively.
Organ Culture
Kidneys were extracted from E12.5 mouse embryos and explanted in the extracellular matrix gel mixture (50% type I collagen, MP Biomedicals) and 50% growth factor-reduced Matrigel (BD Biosciences) followed by incubation at 37 °C with 5% CO2 for 72 h (35). Genotyping was performed at the same time, and wild type and Gpr54−/− kidneys were used. The explanted wild type and Gpr54−/− kidneys (n = 4 for each group) were paraffin-embedded followed by hematoxylin and eosin staining.
Reverse Transcription (RT)-PCR, Quantitative RT-PCR, and Quantitative PCR
RNA was extracted with Trizol reagent (Invitrogen) followed by DNase I (RNase-free, New England BioLabs®) treatment. The cDNA was synthesized with Moloney murine leukemia virus reverse transcriptase (Promega). RT-PCR analyses were performed with 30 cycles using the following primers. Mouse Gpr54 and KiSS1 RT-PCR were performed with primers TACATCGCTAACCTGGCTGCCAC (sense) and ATGCTGAGGCTGACAGCCAGGGC (antisense) and with primers TCCACAGGCCAGCAGTCCGGAC (sense) and GACTGCGGGAGGCACACAG (antisense), respectively. Human GPR54 and KISS1 RT-PCR were performed with primers CGCTGGTCATCTACGTCATC (sense) and GTTGACGAACTTGCACATGAA (antisense) and with primers TGGCAGCTACTGCTTTTCCT (sense) and CAGTAGCAGCTGGCTTCCTC (antisense), respectively. Quantitative real time PCR and quantitative PCR (Q-PCR) were performed in a Stratagene MX 3000PTM system with RT2 Real-TimeTM SYBR Green/Rox PCR master mix (SuperArray Bioscience). The data of the Q-RT-PCR/Q-PCR were represented as the mean of three individual values ± S.D.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assay was performed as previously described with some modification (59). Wild type and Gpr54−/− MEF cells (2 × 107 cells) were cross-linked with formaldehyde, quenched with glycine, resuspended in SDS lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.0, with protease inhibitors and phosphatase inhibitors), sonicated on ice, and centrifuged at 4 °C. Supernatant (400 μl) were diluted to a final volume of 4 ml in a mixture of 9 parts dilution buffer (1% Triton X-100, 150 mm NaCl, 2 mm EDTA, 20 mm Tris-HCl, with protease inhibitors, pH 8.0) and 1 part lysis buffer. Mixtures were incubated with 4 μg of anti-NFAT2 or anti-p-Sp1 or anti-Sp1 antibodies sample with rotating at 4 °C overnight followed with incubation with 100 μl of protein A beads with rotating at 4 °C for 4 h. After gentle centrifugation (2000 rpm), beads were resuspended in 1 ml of wash buffer (1% Triton X-100, 0.1% SDS, 150 mm NaCl, 2 mm EDTA, 20 mm Tris-HCl, with protease inhibitors, pH 8.0) and washed with wash buffer 3 times followed by one wash with a final wash buffer (1% Triton X-100, 0.1% SDS, 500 mm NaCl, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, with protease inhibitors). The immune complexes were eluted with elution buffer (1% SDS, 100 mm NaHCO3) followed by incubation with proteinase K and RNase A (500 μg/ml each) at 37 °C for 30 min. Reverse cross-links were performed by placing the tubes at 65 °C overnight. Immunoprecipitated DNA was extracted and dissolved in sterile water. Q-PCR was performed with RT2 Real-time TM SYBR Green/Rox PCR master mix (SuperArray) in Stratagene MX 3000PTM machine.
Western Blotting
HEK293 cells were cultured, and the transfection was performed as previously described. After the starvation of 10–12 h, the transfected cells were treated with or without 0.1 μm KiSS1 peptide for 10–12 h. For the cyclosporin A (CsA) treatment, the transfected cells were treated with 1 μm CsA or the same concentration of ethanol for 60 min followed by 10–12 starvation and 0.1 μm KiSS1 peptide or PBS treatment for 10–12 h. Cells were washed with cold PBS for twice, and the lysates (in radioimmune precipitation assay buffer with 1 mm EDTA and proteinase inhibitors) were collected by scrapper. The total protein extracts were electrophoresed through a 10 or 12% SDS-polyacrylamide gel and transferred onto polyvinylidene difluoride ImmobilonTM-P membranes. Polyvinylidene difluoride membranes were probed with anti- NFAT2 (1: 500) or anti-Bmp7 (1:500) for 4 h in a cold room, washed in Tris-buffered saline washing buffer, and incubated with peroxidase-conjugated secondary antibodies. ECLTM Western blotting detection reagent was used for visualization.
Transfection and Luciferase Assays
The DNA fragment of −1244 bp ∼ +18 bp of mouse Bmp7 was cloned into XhoI and HindIII sites of pGL3-Basic vector with primers 5′-CTCGAGCTGAGAGACCCAGTATC-3′ (forward) and 5′-AAGCTTGCACGGCAAGGCTAGCAC-3′ (reverse). The DNA fragment of −136 bp ∼ +18 bp of Bmp7 was cloned into XhoI and HindIII sites of pGL3-Basic vector as control with primers 5′-CTCGAGGAGGAGGGAGCTAGGGTTC-3′ (forward) and 5′-AAGCTTGCACGGCAAGGCTAGCAC-3′ (reverse). Indicated constructions were transfected into HEK293 cells with CalphosTM Mammalian transfection kit (Clontech), and luciferase activity was measured using the luciferase assay system (Promega) with Top Count Microplate Scintillation Counter (Canberra).
Image Acquisition
Images were taken with a Nikon digital camera and OLYMPUS-inverted microscope. Representative images were edited on Adobe Photoshop 6.0 software.
RESULTS
Gpr54 Plays a Role in Kidney Branching Morphogenesis
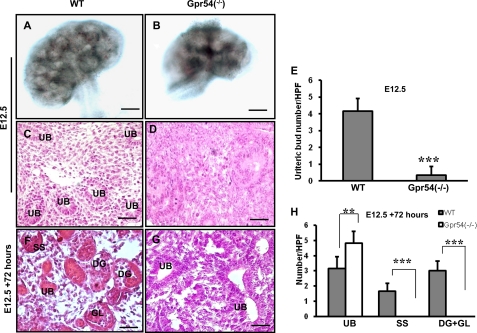
Kidney branching morphogenesis is a key mechanism by which nephrogenesis develops (35). The mouse metanephric kidney commences at approximately E10.5, and branching morphogenesis starts at about E11.5. The rapid increasing period for ureteric bud tips in mouse kidney is from E12.5 to E17.5 with an estimated 52-fold increase (37, 60). To understand whether Gpr54 plays a role in embryonic kidney branching morphogenesis, we examined the size and ureteric bud number of E12.5 wild type and Gpr54−/− kidneys. Gpr54 deletion leads to a smaller kidney in size (Fig. 1, A and B) and a dramatically reduced ureteric bud number at E12.5 (Fig. 1, C–E), indicating that Gpr54 is involved in normal kidney branching morphogenesis.
FIGURE 1.
Gpr54 regulates kidney branching morphogenesis. A and B, Gpr54 deletion leads to smaller kidney size at E12.5. A representative image is shown. C–E, Gpr54 deletion leads to retarded branching morphogenesis in vivo. E12.5 wild type kidney has many ureteric buds (UB) (C), whereas E12.5 Gpr54−/− kidneys have very few ureteric buds (D). The average ureteric bud number of wild type versus Gpr54−/− was 4.2 ± 0.75 ureteric buds versus 0.3 ± 0.5 ureteric buds/HPF (20×) (E). F–H, Gpr54 deletion results in retarded branching morphogenesis and glomerular development in vitro. E12.5 wild type (WT) and Gpr54−/− kidneys were extracted and explanted in extracellular matrix gel mixture for 72 h followed by hematoxylin and eosin staining. F, s-shaped bodies (SS), developing glomeruli (DG), and glomeruli (GL) are presented in the explanted wild type kidney. G, ureteric buds were found in the explanted Gpr54−/− kidneys, but not S-shaped body, developing glomeruli or glomeruli. H, shown is a significant decrease in the number of developing glomeruli and glomeruli with an increase in the number of ureteric buds in Gpr54−/− kidney compared with wild type. Ureteric bud number/HPF in WT versus Gpr54−/− kidney was 3.2 ± 0.7 versus 4.8 ± 0.7; S-shaped body number/HPF in WT versus Gpr54−/− kidney was 1.6 ± 0.5 versus 0; developing glomeruli and glomerular number (developing glomeruli + glomeruli)/HPF in WT versus Gpr54−/− kidney was 3 ± 0.6 versus 0. The data are represented as the mean of four or five individual values ± S.D. **, p < 0.01; ***, p < 0.001. Scale bar = 100 μm in Chen et al. (77) and 10 μm in C–G.
Ureteric bud branching morphogenesis is controlled by both intrinsic (cell autonomous) in the kidney and secreted extrinsic (non-cell autonomous) factors outside the kidney (36). To test whether the kidney branching morphogenesis defects found in Gpr54−/− mice are regulated by intrinsic factors in the kidney, we extracted E12.5 wild type and Gpr54−/− null kidneys and explanted them in extracellular matrix culture mixtures. After culturing at 37 °C for 72 h, the explanted kidneys were embedded with paraffin, and a hematoxylin and eosin staining analysis was performed. In the explanted wild type kidneys, we found the existence of the ureteric buds, S-shaped bodies, developing glomeruli, and glomeruli, indicating that continuous development occurs in the Gpr54 wild type kidney in vitro (Fig. 1F). However, we can only find the ureteric buds, not the S-shaped bodies or the glomeruli in the explanted Gpr54−/− kidneys (Fig. 1, G and H), suggesting a key for Gpr54 in embryonic kidney-branching morphogenesis.
Gpr54 Is Involved in Proper Embryonic Glomerular Development
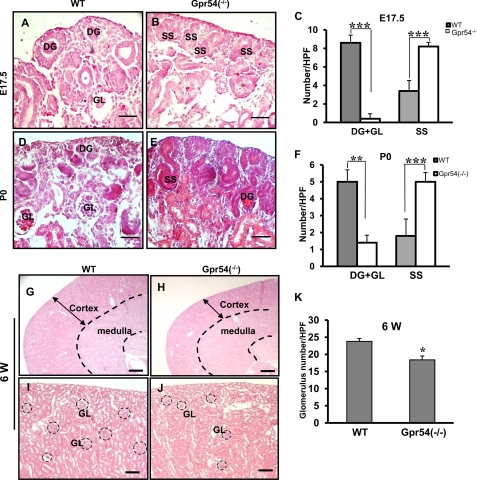
Early branching morphogenesis maturation has important effects on the development of S-shaped bodies and glomeruli. Glomeruli form approximately at E13.5 with a subsequent increase in number up to 54-fold from E13.5 to E17.5. Glomerular number increases about 2.5-fold per day from E17.5 to P0 in mouse kidney (60). To investigate whether Gpr54 is involved in glomerular development, we analyzed the glomerular number in wild type and Gpr54−/− kidneys at E17.5 and P0. As shown in Fig. 2, A–C, the developing glomeruli is the majority in the nephrogenic zone, and glomeruli exist in the cortex layer of E17.5 wild type kidneys. However, the S-shaped bodies were prominent in the E17.5 Gpr54−/− nephrogenic zone, indicating that Gpr54 deficiency delays glomerular development at E17.5 stage. At P0, developing glomeruli and mature glomeruli are prominent in wild type cortex layers, whereas S-shaped bodies and developing glomeruli are prominent in Gpr54−/− cortex layers (Fig. 2, D–F), suggesting that Gpr54 is involved in glomerular development at P0 stage. Together, the above data indicate that Gpr54 plays a role in embryonic glomerular development. To trace the role of Gpr54 in glomerular development in adult kidney, we further examined the glomerular number in 6-week-old wild type and Gpr54−/− kidneys. We observed that 62% of Gpr54−/− mice (13 of 21 mice) had smaller kidneys in size and thinner cortical layers compared to the same structure in wild type kidneys (Fig. 2, G and H). The average glomerular number in the smaller Gpr54−/− kidneys was lower than that of wild type kidneys (Fig. 2, I–K), suggesting that Gpr54 deletion leads to a high risk of low nephron number in adult kidneys.
FIGURE 2.
Gpr54 is involved in embryonic glomerular development. Gpr54−/− kidneys exhibited retarded glomerular development at E17.5 (A–C). A and B, developing glomeruli (DG) in the nephrogenic zone and glomeruli (GL) in the cortex layer were found in E17.5 wild type kidneys, whereas S-shaped body (SS) was found in the nephrogenic zone of E17.5 Gpr54−/− kidneys. C, developing glomeruli and glomeruli number (DG+GL)/HPF in wild type (WT) versus Gpr54−/− kidneys was 7.8 ± 0.8 versus 0.4 ± 0.5. S-shaped bodies/HPF in WT versus Gpr54−/− kidney was 3.4 ± 1.1 versus 8.2 ± 0.8. D–F, Gpr54 deletion leads to retarded glomerular development at P0. A representative image shows that the developing glomeruli and mature glomeruli are the prominent in nephrogenic zone of P0 wild type kidneys (D), whereas S-shaped bodies (SS) and developing glomeruli (DG) are the majority in P0 Gpr54−/− nephrogenic zones (E). F, developing glomeruli and glomerular (DG+GL)/HPF in WT versus Gpr54−/− kidney was 5 ± 0.7 versus 1.4 ± 0.5. S-shaped body number/HPF in WT versus Gpr54−/− kidney was 1.8 ± 0.8 versus 5 ± 0.7. Gpr54 deletion increases low glomerular number risk in adult mouse kidneys. G and H, ∼62% Gpr54−/− kidneys (13 of 21 mice) displayed a thinner cortex layer (arrow) compared with that of wild type at 6 weeks (6 W) postnatal. I–K, shown is the average glomerular number/HPF in wild type (WT) versus Gpr54−/− kidney with a thinner cortex layer was 23.8 ± 0.8 versus 18.4 ± 1.1. The glomerular number was counted as described under “Experimental Procedures.” Black circles show the glomeruli. The data are represented as the mean of five individual values ± S.D. *, p < 0.1; **, p < 0.01; ***, p < 0.001. Scale bar = 10 μm in (A–E), 150 μm in (G and H), and 40 μm in (I and J).
Gpr54 Is Spatially Expressed in the Developing Kidney
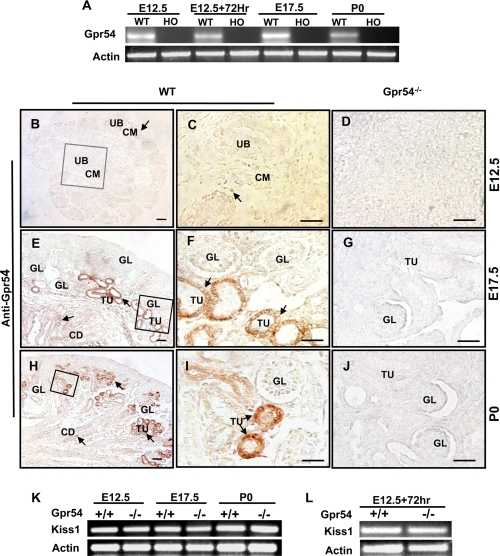
In the RT-PCR analysis, we found that Gpr54 mRNA is consistently expressed in E12.5, E17.5, and P0 wild type kidneys as well as in explanted E12.5 wild type kidneys (Fig. 3A). To examine the distribution of Gpr54 protein in the developing kidney, we performed immunohistochemistry assays with Gpr54-specific antibody and found Gpr54 immuno-staining signals present in the cell membrane of condensed mesenchyme at E12.5 (Fig. 3, B–D), the epithelial cells of proximal and distal tubules, and the collecting ducts at E17.5 (Fig. 3, E–G) and P0 (Fig. 3, H–J) kidney but not in the Gpr54−/− kidneys at the same periods. These data indicated that Gpr54 is spatially expressed in the developing kidneys. Previous studies reported that KISS1 (the ligand for Gpr54) is expressed in diverse organs such as brain (61), placenta (62), and ovary (63). Here we found that KISS1 mRNA is consistently expressed in E12.5, E17.5, and P0 wild type and Gpr54−/− kidneys (Fig. 3K) and in explanted wild type and Gpr54−/− kidneys (Fig. 3L), suggesting that KISS1 mRNA is not affected by the deletion of Gpr54 in the mouse.
FIGURE 3.
Gpr54 is expressed in developing kidney. A, Gpr54 mRNA is expressed in wild type (WT) developing and explanted kidneys but not in the Gpr54−/− kidneys (homology, HO). B–J, Gpr54 protein immunostaining signals are present in cell membranes of condensed mesenchyme (CM) in E12.5 WT kidney (B–D), epithelial cells of proximal and distal tubules (TU) and collecting ducts (CD) in E17.5 (E–G), and P0 (H–J) wild type kidneys but not in Gpr54−/− kidney at the same periods. UB, ureteric buds; GL, glomeruli. K, KISS1 mRNA is expressed in E12.5, E17.5, and P0 wild type and Gpr54−/− kidneys. Deletion of Gpr54 had no effect on KISS1 mRNA in the kidneys.
Gpr54 Regulates Bmp7 Expression in the Developing Kidney
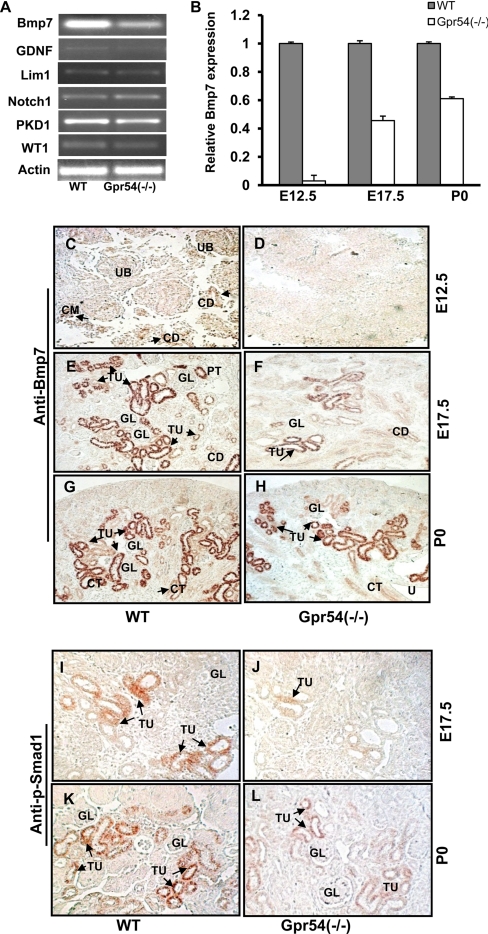
Kidney branching morphogenesis and glomerular development are regulated by sophisticated regulatory networks and genes, such as glial cell-derived neurotrophic factor (GDNF), Lim1, Notch 1, PKD1, WT1, and Bmp7 (64, 65). Endogenous bone morphogenetic protein-7 (Bmp7) plays a crucial role in early metanephric kidney development in vivo. Bmp7 deficiency causes an arrest in branching morphogenesis and nephrogenesis (37). Bmp7 null mice are born alive but most die within 24 h due to renal failure (37). To understand the underlying molecular mechanism of the retarded branching morphogenesis and glomerular development defects in Gpr54−/− mouse kidneys, we isolated the total RNAs from E12.5 wild type and Gpr54−/− mutant kidneys and screened the gene expression profiles of the above listed genes by RT-PCR. Interestingly, the expression of Bmp7 mRNA is significantly decreased in E12.5 Gpr54−/− mouse kidneys compared with wild type (Fig. 4, A and B). To determine whether Gpr54 regulates Bmp7 mRNA expression in developing kidneys, we examined the expression levels of Bmp7 mRNA at E12.5, E17.5, and P0 wild type and Gpr54−/− mouse kidneys with real-time PCR. Our data indicate that Bmp7 mRNA expression decreased in Gpr54−/− kidneys compared with that in wild type kidney at all developmental stages (Fig. 4B). To determine the regulation of Bmp7 at the protein level, we further examined Bmp7 protein expression using immunohistochemistry analysis with specific anti-Bmp7 antibody. As shown in Fig. 4, C–H, deletion of Gpr54 significantly decreased the protein expression levels of Bmp7 at different developmental stages of Gpr54−/− embryonic kidneys.
FIGURE 4.
Deletion of Gpr54 down-regulates Bmp7 expression and Smad1 phosphorylation in developing kidney. A, Bmp7 mRNA expression was decreased in E12.5 Gpr54−/− kidneys. Total RNA was isolated from E12.5 wild type (WT) and Gpr54−/− kidneys, and key genes involved in kidney development were examined by RT-PCR. GDNF, glial cell-derived neurotrophic factor. B, real-time PCR analysis of Bmp7 mRNA expression in E12.5, E17.5, and P0 wild type and Gpr54−/− kidneys is shown. Decreased Bmp7 mRNA expressions were observed in E12.5, E17.5, and P0 Gpr54−/− kidneys compared with wild type kidneys. C-H, Gpr54 deletion leads to decreased Bmp7 protein expression in E12.5, E17.5, and P0 kidneys. Immunohistochemistry staining was performed in E12.5, E17.5, and P0 wild type and Gpr54−/− kidneys with anti-Bmp7 antibody. C and D, detectable Bmp7 immunostaining was found in ureteric buds (UB), metanephric mesenchymal cells (CM), and the epithelial cells of collecting buds (CD) in E12.5 wild type kidneys but not in E12.5 Gpr54−/− kidneys. E and F, Bmp7 is expressed in the epithelial cells of proximal and distal tubules (PT and TU) and collecting ducts (CD) in E17.5 wild type kidneys. GL, glomeruli. Bmp7 immunostaining was significantly decreased in Gpr54−/− kidneys. G and H, Bmp7 is expressed in the epithelial cells of proximal and distal tubules (TU) and collecting ducts (CD) in P0 wild type and Gpr54−/− kidneys. I–L, immunostaining of phosphorylated Smad1 (p-Smad1) in E17.5 and P0 kidney is shown. p-Smad1 was decreased in the epithelial cells of proximal and distal tubules (TU) and the collecting ducts (CD) in E17.5 Gpr54−/− kidneys (J and L) compared with wild type kidneys (I and K). Scare bar = 10 μm. Arrows show detectable Bmp7 or p-Smad1 immunostaining.
Because Bmp7-activated receptors will induce the phosphorylation and activation of Smad1, we further investigated whether Gpr54 deletion regulated Smad1 phosphorylation using immunohistochemistry analyses with anti-Smad1 and anti-p-Smad1 antibodies. We demonstrate that Smad1 phosphorylation is decreased in E17.5 and P0 Gpr54−/− kidneys compared with wild type (Fig. 4, I–L). There was no difference in the total Smad1 protein expression between wild type and Gpr54−/− kidneys (data not shown). Taken together, these data indicate that Gpr54 regulates Bmp7 expression in the developing kidney.
Gpr54 Regulates Bmp7 Expression through NFAT2
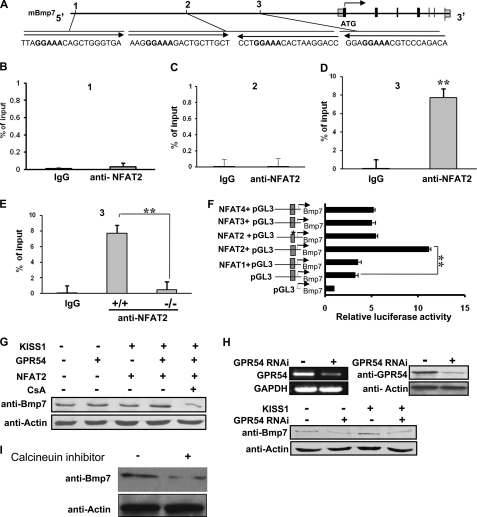
Activation of Gpr54 by KISS1 peptide stimulates an intracellular phospholipase C- inositol 1,4,5-trisphosphate-calcium cascade via activation of Gαq (6, 31), suggesting that Gpr54 may induce a calcium-dependent downstream signaling pathway. NFAT transcription factors are sequestered in the cytosol in an inactive hyperphosphorylated form. Intracellular Ca2+ release leads to calcineurin activation, NFAT dephosphorylation, and the translocation of dephosphorylated NFATs to the nucleus where they regulate target genes (66). NFATc1 (NFAT2), not NFATc2 (NFAT1) or NFTAc3 (NFAT4), plays a key role in G-protein-coupled receptor agonist-induced NFAT-DNA binding and cell proliferation (67). Previous data showed that NFAT2 is expressed in the nephrogenic zone in vesicles, ureteric buds, S-shaped bodies, and glomeruli of embryonic kidneys (68). To understand the molecular mechanism of Bmp7 regulation by the Gpr54-Ca2+-signaling pathway, we examined whether Bmp7 can be regulated by the transcription factor NFAT, the downstream target of Gpr54-Ca2+ signaling. First, we searched the upstream 3-kb region in mouse Bmp7 promoter region (see the Ensembl genome browser and Genomatix databases) and found four conserved potential NFAT binding sites (Fig. 5A). Then we investigated the binding affinity between NFAT2 and the four potential NFAT2 binding regions of the Bmp7 promoter by CHIP assays with anti-NFAT2 antibody and Q-PCR. Our data indicate that region 3 (containing the fragment from −1159 bp to −1265 bp) showed a high binding affinity with NFAT2, whereas region 1 and region 2 showed almost no binding affinity, suggesting that NFAT2 binds to the region 3 of Bmp7 promoter (Fig. 5, B–D). Because NFAT2 is a calcium/calcineurin-dependent transcription factor and Gpr54 signaling regulates intracellular calcium release, it is possible that Gpr54 deletion will suppress the binding affinity between calcium-dependent NFAT2 and the Bmp7 promoter. To test this hypothesis, we compared the binding affinity between the NFAT2 and region 3 of the Bmp7 promoter in wild type and Gpr54−/− MEF cells and found that deletion of Gpr54 significantly decreased the binding affinity between NFAT2 and the region 3 in Bmp7 promoter in Gpr54−/− null MEF cells (Fig. 5E), suggesting that Gpr54 regulates the binding of NFAT2 to the promoter region of Bmp7. To further examine whether NFAT2 regulates Bmp7 transcription in kidney cells, we performed luciferase assays in HEK 293 cells with the Bmp7 promoter region (−234 ∼−1506 bp) containing region 3 (−1159 ∼ −1265 bp). Our data indicate that NFAT2, not NFAT1, NFAT2, NFAT3, or NFAT4, enhanced Bmp7 promoter-induced luciferase activity. To confirm that region 3 contains the NFAT2 binding site, we performed site-directed mutagenesis and mutated the specific Bmp7 promoter from GGAAA to GGAGA in the potential NFAT2 site. Our data indicate that mutation at region 3 of the NFAT2 site significantly decreased the luciferase activity compared to the wild type Bmp7 promoter, suggesting that region 3 is an NFAT2 specific affinity binding site (Fig. 5F, stars). To investigate the effects of KISS1/GPR54/NFAT2 on Bmp7 expression, we overexpressed GPR54 and NFAT2 in HEK293 cells with or without KiSS1 treatment together with calcineurin-specific inhibitor CsA. We found that CsA inhibited the up-regulation effects of KiSS1/Gpr54/NFAT2 (Fig. 5G, last column). Furthermore, we found GPR54 and KISS1 are expressed in HEK293 cells (data not shown), and down-regulation of GPR54 by GPR54 RNAi decreased Bmp7 expression in HEK293 cells (Fig. 5H). Together, these data indicate that Gpr54 regulates Bmp7 expression through the transcription factor NFAT2 in the kidney.
FIGURE 5.
Gpr54 regulates NFAT2-mediated Bmp7 transcription. A, shown is the gene structure of mouse Bmp7 mBmp7 (NP_031583). Black boxes represent coding exons, and gray boxes represent non-coding exons. Arrows show the 5′ to 3′ direction. Sequences are potential NFAT transcription factor binding sites in the 3-kb Bmp7 promoter region. Boldface sequences of GGAAA are the minimal binding sites for NFAT2. Regions 1–3 show three investigated regions containing the presented sequences in the CHIP and Q-PCR assays. B–D, region 3 of Bmp7 promoter shows relative high binding affinity to NFAT2 by CHIP assays with anti-NFAT2 antibody in MEF cells followed by Q-PCR. IgG was used as a control, and the relative binding affinity was expressed as percent of the input. E, Gpr54 deletion decreased the binding affinity between NFAT2 and the region 3 of Bmp7 promoter in Gpr54−/−MEF cells. F, NFAT2 enhanced Bmp7 transcriptional activity in luciferase assays is shown. Gray boxes in the promoter represent NFAT binding region 3. The indicated constructs were co-transfected into HEK293 cells followed by luciferase activity analyses. NFAT2, but not NFAT1, NFAT2, NFAT3, or NFAT4, increased Bmp7 promoter-induced luciferase activity. Bmp7 promoter region 3 mutation (*, GGAAA was mutated to GGAGA) decreased luciferase activity. G, overexpression of GPR54 and NFAT2 together with KISS1 (Kisspeptin-10 (Kp-10)) treatment increased Bmp7 expression in HEK293 cells, whereas calcineurin/NFAT signaling-specific inhibitor CsA inhibited these enhanced effects. H, GPR54 was knocked-down by shRNA in HEK293 cells at mRNA (top panel, left) and protein levels (top panel, right). Down-regulation of Gpr54 decreased Bmp7 expression in HEK293 cells (bottom panel). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. I, calcineurin inhibitor decreased the expression of Bmp7 in HEK293 cells. Data represent three individual repeats ± S.D. **, p < 0.01.
Gpr54 Regulates Bmp7 Transcription via Sp1
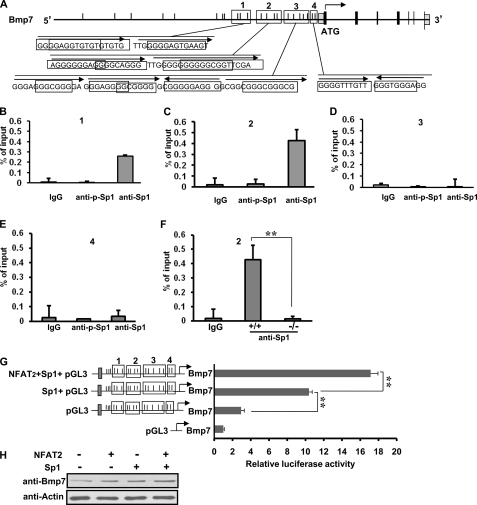
Elevation of intracellular Ca2+ level triggers diverse signal transduction networks by activating calcium-dependent pathways, such as Ca2+-calmodulin-dependent kinases and phosphatases (69). Previous studies reported that calcium/calcineurin enhance the activity of transcription factor Sp1 on p21WAF1/CIP1 expression in keratinocyte cells (70). Sp1 regulates the expression of many target genes by binding to the G-rich elements, such as GC-box (GGGGCGGGG) and the related GT/CACCC-box (GGTGTGGGG) (71). Phosphorylation or dephosphorylation of Sp1 can regulate target genes in either a positive or negative manner (72). To examine whether Gpr54 regulates Bmp7 transcription by Sp1 protein in developing kidneys, we analyzed the upstream 3-kb area of mouse Bmp7 promoter and found 22 potential Sp1 binding sites (Fig. 6A). To test the potential functional Sp1 binding regions in the Bmp7 promoter and examine whether phosphorylation of Sp1 is necessary for the binding affinity, we examined the binding of Sp1 to the 1-kb upstream region of Bmp7 promoter, which contains 14 potential Sp1 binding sites, by CHIP assay with anti-Sp1 (for both phosphorylated and non-phosphorylated Sp1), anti-p-Sp1 antibodies, and by Q-PCR in wild type MEF cells. Our data indicate that Sp1 binds to region 1 (−763 ∼ −916 bp) and region 2 (−588 bp ∼ −784 bp) with relatively high affinity but with little binding affinity to region 3 (−147 ∼ −345 bp) and region 4 (−1 ∼ −122 bp), indicating that regions 1 and 2 are the potential Sp1 binding regions in Bmp7 promoter. Region 2 showed relative stronger binding affinities than region 1 with Sp1 (Fig. 6, B and C). Interestingly, we found that the phosphorylated Sp1 showed no binding affinity to all four regions (Fig. 6, B–E), suggesting that the non-phosphorylated Sp1 is the active Sp1 to bind to Bmp7 promoter. To further investigate if Gpr54 plays a role in Sp1-regulated Bmp7 transcription, we performed CHIP and Q-PCR assays with region 2 in wild type and Gpr54−/− MEF cells. Our data demonstrated that deletion of Gpr54 significantly decreased the binding affinity between Sp1 and Bmp7 promoter in Gpr54−/− MEF cells (Fig. 6F). To confirm that Sp1 regulates Bmp7 transcription in kidney cells, we performed luciferase assays in HEK 293 cells and found that Sp1 significantly increased Bmp7 promoter induced luciferase activity (Fig. 6G).
FIGURE 6.
Gpr54 regulates Bmp7 transcription through Sp1 and NFAT2. A, Sp1 binding sites of mouse Bmp7 promoter (NP_031583) are shown. Black boxes represent coding exons. There are 22 potential Sp1 binding sites (black line) in the 3-kb area of Bmp7 promoter. Black frames (1–4) represent the investigated regions in CHIP assays. Sequences are shown in the GC/GT boxes. Arrows show the 5′ to 3′ directions. B–E, Sp1, not phosphorylated Sp1, binds to the Sp1 regions of Bmp7 gene in our CHIP assays with anti-Sp1 and anti-p-Sp1 antibodies in wild type MEF cells followed by Q-PCR. IgG was used as a control, and the relative binding affinity was expressed as percent of the input. Phosphorylated Sp1 showed almost no binding affinity to the indicated regions. Regions 1 and 2 showed the relative high binding affinity between Sp1 and Bmp7 promoter. F, deletion of Gpr54 decreased the binding affinity between Sp1 and the region 2 of Bmp7 promoter. G, Sp1 increased Bmp7 promoter induced luciferase activity and addition of NFAT2 enhanced the Sp1 activity. Gray boxes represent NFAT2 binding region 3 (in Fig. 5). H, Sp1 and NFAT2 overexpression enhanced Bmp7 protein expression (lanes 2 and 3). Cotransfection of NFAT2 and Sp1 has additive effect on Bmp7 expression (lane 4). Data represent three individual repeats ± S.D. **, p < 0.01.
NFAT2 Cooperates with Sp1 in Regulating Bmp7 Expression
Previous studies reported that NFAT2 associates with Sp1 in a calcineurin-dependent manner in HEK293 cells (70). NFAT2 cooperates with Sp1 in mediating MT1-MMP (membrane type 1 matrix metalloproteinase) transcription in glomerular mesangial cells (73). To examine whether NFAT2 cooperates with Sp1 in Bmp7 transcription regulation, we performed luciferase assays with NFAT2, Sp1, and Bmp7 reporter in HEK293 cells. We found that NFAT2 increased the Sp1-mediated Bmp7 promoter luciferase activity, suggesting that NFAT2 cooperates with Sp1 to regulate Bmp7 transcriptional activation (Fig. 6G). Furthermore, we found that coexpression of NFAT2 and Sp1 increased Bmp7 expression using Western blotting assays (Fig. 6H). Together, all the above data suggest that Gpr54 regulates Bmp7 transcription through NFAT2 and Sp1 transcription factors in the kidney.
DISCUSSION
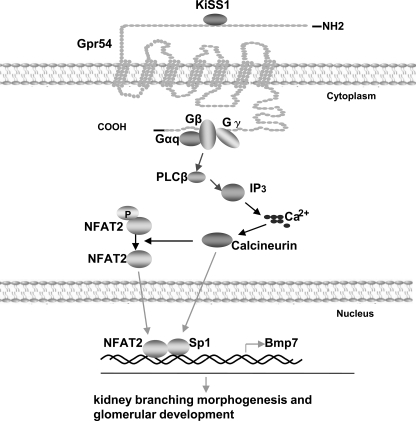
We demonstrate that Gpr54 (KISS1 receptor) plays a role in kidney-branching morphogenesis and embryonic glomerular development, expanding the understanding of the biological functions of Gpr54 to kidney development. For the first time, our data provide a molecular mechanism to show that a G-protein-coupled receptor regulates the expression of Bmp7, a key regulator for proper kidney formation (Fig. 7). These findings shield light to the previously unknown Gpr54 roles in kidney development and in the regulation of Bmp7 expression.
FIGURE 7.
The proposed model for KISS1 and Gpr54 function in kidney branching morphogenesis and glomerular development. Activation of Gpr54 by KISS1 leads to the activation of Gαq and phospholipase Cβ, which increases intracellular Ca2+ concentration and activation of calcium- calmodulin-dependent calcineurin. Calcineurin-dephosphorylated NFAT2 and Sp1 bind to Bmp7 promoter and regulate Bmp7 expression, which plays a critical role in kidney branching morphogenesis and glomerular development. Blue arrows come from previous study of our laboratory, black arrows come from the previous reports of other groups, and red arrows come from this study. PLCβ, phospholipase Cβ; IP3, inositol 1,4,5-trisphosphate.
More than 10 years ago Jena et al. (45) found that Bmp7 was critical for kidney development. Numerous studies have demonstrated the expression patterns of Bmp7 and its downstream signaling pathways in the developing kidney. Although Wnt is one of the upstream candidates that regulate Bmp7 transcription (55), the signaling pathways that regulate Bmp7 expression are largely not understood (37, 46). Our studies revealed that NFAT2 and Sp1 are two transcription factors involved in Bmp7 transcription regulation. Activation of Gpr54 by KISS1 leads to intracellular Ca2+ increase and the activation of the Ca2+-dependent phosphatase, calcineurin, which dephosphorylates NFAT2 and Sp1 proteins in order to promote their binding activities. Dephosphorylated NFAT2 and Sp1 proteins can bind to the promoter regions of Bmp7 and regulate Bmp7 transcription (Fig. 7). These findings provide new insights into the upstream signals that regulate the expression of Bmp7, which is critical for kidney development.
Beside the crucial function of the calcium-dependent NFAT signaling in immune system, NFAT is also essential for the development of many diverse organs, such as kidney (66, 68). NFAT transcription factors are sequestered in the cytosol in an inactive hyperphosphorylated form. Ca2+ release leads to activation of Ca2+-dependent calcineurin, which can dephosphorylate NFATs. Dephosphorylated NFATs can translocate to the nucleus where they regulate target genes (66). NFATc1 (NFAT2), but not NFATc2 (NFAT1) or NFTAc3 (NFAT4), is a calcium-signaling-dependent transcript factor and plays a role in G-protein-coupled receptor agonist-induced NFAT-DNA binding and cell proliferation (67). NFAT2 has been reported to regulate the development of glomeruli through directly binding to the minimal consensus promoter sequence of GGAAA (68, 74). In our studies we demonstrate that Gpr54 plays a role in embryonic kidney branching morphogenesis and glomeruli development through NFAT2. Our previous studies have shown that activation of Gpr54 by KiSS1 leads to the activation of Gαq- phospholipase C β and an increase of intracellular Ca2+ (6), which can activate Ca2+-dependent calcineurin. Therefore, Gpr54-regulated calcium/calcineurin can dephosphorylate and activate NFAT2 to regulate the expression of Bmp7, a key signaling molecule in embryonic kidney development (Fig. 7).
Previous evidences show that non-phosphorylated Sp1 always associates with rapid cell growth (75). Calcineurin enhances Sp1-induced gene transcription in keratinocytes (67, 76). Many studies show that phosphatases can promote Sp1 binding to DNA by dephosphorylating Sp1 protein (72). Here we demonstrate that non-phosphorylated Sp1 effectively binds to the Bmp7 promoter in CHIP assays and deletion of Gpr54 leads to decreased binding affinity of Sp1 and decreased Bmp7 expression. In addition, we elucidate that NFAT2 cooperates with Sp1 in mediating Bmp7 transcription in kidney cells. These data indicate that activation of Gpr54 increases intracellular Ca2+ and leads to activation of Ca2+-dependent phosphatase calcineurin and de-phosphorylation of Sp1 in regulating Bmp7 transcription in developing kidney.
Kidney branching morphogenesis, S-shaped body formation, and glomerular development are all important phases of normal glomeruli number formation at birth (78, 79). Disruption of these processes leads to a series of diseases, such as rare hereditary syndromes, primary hypertension, and chronic kidney failure (41). Here we demonstrated that the KISS1-activated G-protein-coupled receptor Gpr54 (KISS1R) regulates embryonic kidney development and morphogenesis. Deletion of Gpr54 leads to retarded kidney branching morphogenesis and embryonic glomerular development and a high risk of low nephron number in adult kidneys, suggesting that Gpr54 inactivation could be a previously unreported mechanism of kidney pathogenesis.
Acknowledgments
We greatly appreciate Dr. Eric L. Gustafson and Galya Vassileva of the Functional Genomics Department of Schering-Plough Research Institute (Kenilworth, NJ 07033) for providing the Gpr54+/− mouse (male and female) line. We thank Dr. Stephen H. Safe at Texas A&M Health Science Center and Dr. Xin-Hua Feng at the Baylor College of Medicine for antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01CA106479 (NCI).
- Bmp
- bone morphogenetic protein
- HPF
- high performance field
- RT
- reverse transcription
- Q-PCR
- quantitative PCR
- CsA
- cyclosporin A
- CHIP
- chromatin immunoprecipitation
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Li D., Yu W., Liu M. ( 2009) Peptides 30, 130– 138 [DOI] [PubMed] [Google Scholar]
- 2.Kotani M., Detheux M., Vandenbogaerde A., Communi D., Vanderwinden J. M., Le Poul E., Brézillon S., Tyldesley R., Suarez-Huerta N., Vandeput F., Blanpain C., Schiffmann S. N., Vassart G., Parmentier M. ( 2001) J. Biol. Chem. 276, 34631– 34636 [DOI] [PubMed] [Google Scholar]
- 3.Ohtaki T., Shintani Y., Honda S., Matsumoto H., Hori A., Kanehashi K., Terao Y., Kumano S., Takatsu Y., Masuda Y., Ishibashi Y., Watanabe T., Asada M., Yamada T., Suenaga M., Kitada C., Usuki S., Kurokawa T., Onda H., Nishimura O., Fujino M. ( 2001) Nature 411, 613– 617 [DOI] [PubMed] [Google Scholar]
- 4.Lee J. H., Miele M. E., Hicks D. J., Phillips K. K., Trent J. M., Weissman B. E., Welch D. R. ( 1996) J. Natl. Cancer Inst. 88, 1731– 1737 [DOI] [PubMed] [Google Scholar]
- 5.Lee J. H., Welch D. R. ( 1997) Cancer Res. 57, 2384– 2387 [PubMed] [Google Scholar]
- 6.Stafford L. J., Xia C., Ma W., Cai Y., Liu M. ( 2002) Cancer Res. 62, 5399– 5404 [PubMed] [Google Scholar]
- 7.Sanchez-Carbayo M., Capodieci P., Cordon-Cardo C. ( 2003) Am. J. Pathol. 162, 609– 617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dittmer A., Vetter M., Schunke D., Span P. N., Sweep F., Thomssen C., Dittmer J. ( 2006) J. Biol. Chem. 281, 14563– 14572 [DOI] [PubMed] [Google Scholar]
- 9.Jiang T., Zhang S. L., Lin B., Meng L. R., Gao H. ( 2005) Zhonghua Zhong Liu Za Zhi 27, 229– 231 [PubMed] [Google Scholar]
- 10.Nicolle G., Comperat E., Nicolaïew N., Cancel-Tassin G., Cussenot O. ( 2007) Ann. Oncol. 18, 605– 607 [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Carbayo M., Belbin T. J., Scotlandi K., Prystowsky M., Baldini N., Childs G., Cordon-Cardo C. ( 2003) Lab. Invest. 83, 507– 517 [DOI] [PubMed] [Google Scholar]
- 12.Nash K. T., Phadke P. A., Navenot J. M., Hurst D. R., Accavitti-Loper M. A., Sztul E., Vaidya K. S., Frost A. R., Kappes J. C., Peiper S. C., Welch D. R. ( 2007) J. Natl. Cancer Inst. 99, 309– 321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta S., Lai E. W., Pang A. L., Brouwers F. M., Chan W. Y., Eisenhofer G., de Krijger R., Ksinantova L., Breza J., Blazicek P., Kvetnansky R., Wesley R. A., Pacak K. ( 2005) Int. J. Cancer 114, 139– 143 [DOI] [PubMed] [Google Scholar]
- 14.Ikeguchi M., Yamaguchi K., Kaibara N. ( 2004) Clin. Cancer Res. 10, 1379– 1383 [DOI] [PubMed] [Google Scholar]
- 15.Jiang Y., Berk M., Singh L. S., Tan H., Yin L., Powell C. T., Xu Y. ( 2005) Clin. Exp. Metastasis 22, 369– 376 [DOI] [PubMed] [Google Scholar]
- 16.Dhar D. K., Naora H., Kubota H., Maruyama R., Yoshimura H., Tonomoto Y., Tachibana M., Ono T., Otani H., Nagasue N. ( 2004) Int. J. Cancer 111, 868– 872 [DOI] [PubMed] [Google Scholar]
- 17.Masui T., Doi R., Mori T., Toyoda E., Koizumi M., Kami K., Ito D., Peiper S. C., Broach J. R., Oishi S., Niida A., Fujii N., Imamura M. ( 2004) Biochem. Biophys. Res. Commun. 315, 85– 92 [DOI] [PubMed] [Google Scholar]
- 18.Seminara S. B., Messager S., Chatzidaki E. E., Thresher R. R., Acierno J. S., Jr., Shagoury J. K., Bo-Abbas Y., Kuohung W., Schwinof K. M., Hendrick A. G., Zahn D., Dixon J., Kaiser U. B., Slaugenhaupt S. A., Gusella J. F., O'Rahilly S., Carlton M. B., Crowley W. F., Jr., Aparicio S. A., Colledge W. H. ( 2003) N. Engl. J. Med. 349, 1614– 1627 [DOI] [PubMed] [Google Scholar]
- 19.Zhang C., Roepke T. A., Kelly M. J., Rønnekleiv O. K. ( 2008) J. Neurosci. 28, 4423– 4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J., Jia Z., Li Q., Wang L., Rashid A., Zhu Z., Evans D. B., Vauthey J. N., Xie K., Yao J. C. ( 2007) Cancer 109, 1478– 1486 [DOI] [PubMed] [Google Scholar]
- 21.Colledge W. H. ( 2004) Trends Endocrinol. Metab. 15, 448– 453 [DOI] [PubMed] [Google Scholar]
- 22.Seminara S. B., Kaiser U. B. ( 2005) Endocrinology 146, 1686– 1688 [DOI] [PubMed] [Google Scholar]
- 23.d'Anglemont de Tassigny X., Fagg L. A., Dixon J. P., Day K., Leitch H. G., Hendrick A. G., Zahn D., Franceschini I., Caraty A., Carlton M. B., Aparicio S. A., Colledge W. H. ( 2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10714– 10719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.d'Anglemont de Tassigny X., Fagg L. A., Carlton M. B., Colledge W. H. ( 2008) Endocrinology 149, 3926– 3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popa S. M., Clifton D. K., Steiner R. A. ( 2008) Annu. Rev. Physiol. 70, 213– 238 [DOI] [PubMed] [Google Scholar]
- 26.Gottsch M. L., Cunningham M. J., Smith J. T., Popa S. M., Acohido B. V., Crowley W. F., Seminara S., Clifton D. K., Steiner R. A. ( 2004) Endocrinology 145, 4073– 4077 [DOI] [PubMed] [Google Scholar]
- 27.Navarro V. M., Castellano J. M., Fernández-Fernández R., Barreiro M. L., Roa J., Sanchez-Criado J. E., Aguilar E., Dieguez C., Pinilla L., Tena-Sempere M. ( 2004) Endocrinology 145, 4565– 4574 [DOI] [PubMed] [Google Scholar]
- 28.Navarro V. M., Castellano J. M., Fernández-Fernández R., Tovar S., Roa J., Mayen A., Nogueiras R., Vazquez M. J., Barreiro M. L., Magni P., Aguilar E., Dieguez C., Pinilla L., Tena-Sempere M. ( 2005) Endocrinology 146, 156– 163 [DOI] [PubMed] [Google Scholar]
- 29.Teles M. G., Bianco S. D., Brito V. N., Trarbach E. B., Kuohung W., Xu S., Seminara S. B., Mendonca B. B., Kaiser U. B., Latronico A. C. ( 2008) N. Engl. J. Med. 358, 709– 715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Roux N., Genin E., Carel J. C., Matsuda F., Chaussain J. L., Milgrom E. ( 2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10972– 10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X., Lee K., Herbison A. E. ( 2008) Endocrinology 149, 4605– 4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Self M., Lagutin O. V., Bowling B., Hendrix J., Cai Y., Dressler G. R., Oliver G. ( 2006) EMBO J. 25, 5214– 5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah M. M., Sampogna R. V., Sakurai H., Bush K. T., Nigam S. K. ( 2004) Development 131, 1449– 1462 [DOI] [PubMed] [Google Scholar]
- 34.Grobstein C. ( 1955) Ann. N.Y. Acad. Sci. 60, 1095– 1107 [DOI] [PubMed] [Google Scholar]
- 35.Qiao J., Sakurai H., Nigam S. K. ( 1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7330– 7335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dressler G. ( 2002) Trends Cell Biol. 12, 390– 395 [DOI] [PubMed] [Google Scholar]
- 37.Cain J. E., Hartwig S., Bertram J. F., Rosenblum N. D. ( 2008) Differentiation 76, 831– 842 [DOI] [PubMed] [Google Scholar]
- 38.Dressler G. R. ( 2006) Annu. Rev. Cell Dev. Biol. 22, 509– 529 [DOI] [PubMed] [Google Scholar]
- 39.Saxén L., Sariola H. ( 1987) Pediatr. Nephrol. 1, 385– 392 [DOI] [PubMed] [Google Scholar]
- 40.Vainio S., Lin Y. ( 2002) Nat. Rev. Genet. 3, 533– 543 [DOI] [PubMed] [Google Scholar]
- 41.Elliott W. J., Black H. R. ( 2007) Nat. Clin. Pract. Cardiovasc. Med. 4, 538– 548 [DOI] [PubMed] [Google Scholar]
- 42.Martinez G., Mishina Y., Bertram J. F. ( 2002) Int. J. Dev. Biol. 46, 525– 533 [PubMed] [Google Scholar]
- 43.Piscione T. D., Phan T., Rosenblum N. D. ( 2001) Am. J. Physiol. Renal Physiol. 280, F19– F33 [DOI] [PubMed] [Google Scholar]
- 44.Godin R. E., Robertson E. J., Dudley A. T. ( 1999) Int. J. Dev. Biol. 43, 405– 411 [PubMed] [Google Scholar]
- 45.Jena N., Martín-Seisdedos C., McCue P., Croce C. M. ( 1997) Exp. Cell Res. 230, 28– 37 [DOI] [PubMed] [Google Scholar]
- 46.Godin R. E., Takaesu N. T., Robertson E. J., Dudley A. T. ( 1998) Development 125, 3473– 3482 [DOI] [PubMed] [Google Scholar]
- 47.Wu X., Ferrara C., Shapiro E., Grishina I. ( 2009) Gene Expr. Patterns 9, 224– 230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitu G. M., Wang S., Hirschberg R. ( 2007) Am. J. Physiol. Renal Physiol. 293, F1641– F1648 [DOI] [PubMed] [Google Scholar]
- 49.Patel S. R., Dressler G. R. ( 2005) Trends Mol. Med. 11, 512– 518 [DOI] [PubMed] [Google Scholar]
- 50.Wetzel P., Haag J., Câmpean V., Goldschmeding R., Atalla A., Amann K., Aigner T. ( 2006) Kidney Int. 70, 717– 723 [DOI] [PubMed] [Google Scholar]
- 51.Kazama I., Mahoney Z., Miner J. H., Graf D., Economides A. N., Kreidberg J. A. ( 2008) J. Am. Soc. Nephrol. 19, 2181– 2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simon M., Maresh J. G., Harris S. E., Hernandez J. D., Arar M., Olson M. S., Abboud H. E. ( 1999) Am. J. Physiol. 276, F382– F389 [DOI] [PubMed] [Google Scholar]
- 53.Marumo T., Hishikawa K., Yoshikawa M., Fujita T. ( 2008) J. Am. Soc. Nephrol. 19, 1311– 1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paralkar V. M., Grasser W. A., Mansolf A. L., Baumann A. P., Owen T. A., Smock S. L., Martinovic S., Borovecki F., Vukicevic S., Ke H. Z., Thompson D. D. ( 2002) J. Cell Physiol. 190, 207– 217 [DOI] [PubMed] [Google Scholar]
- 55.Adams D., Karolak M., Robertson E., Oxburgh L. ( 2007) Dev. Biol. 311, 679– 690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li D., Mitchell D., Luo J., Yi Z., Cho S. G., Guo J., Li X., Ning G., Wu X., Liu M. ( 2007) Endocrinology 148, 4821– 4828 [DOI] [PubMed] [Google Scholar]
- 57.Fulop T., Jr., Barabas G., Varga Z., József C., Csabina S., Szucs S., Seres I., Szikszay E., Jeney Z., Penyige A. ( 1993) Cell. Signal. 5, 593– 603 [DOI] [PubMed] [Google Scholar]
- 58.Funes S., Hedrick J. A., Vassileva G., Markowitz L., Abbondanzo S., Golovko A., Yang S., Monsma F. J., Gustafson E. L. ( 2003) Biochem. Biophys. Res. Commun. 312, 1357– 1363 [DOI] [PubMed] [Google Scholar]
- 59.Peters A. H., Kubicek S., Mechtler K., O'Sullivan R. J., Derijck A. A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., Martens J. H., Jenuwein T. ( 2003) Mol. Cell 12, 1577– 1589 [DOI] [PubMed] [Google Scholar]
- 60.Cebrián C., Borodo K., Charles N., Herzlinger D. A. ( 2004) Dev. Dyn. 231, 601– 608 [DOI] [PubMed] [Google Scholar]
- 61.Brailoiu G. C., Dun S. L., Ohsawa M., Yin D., Yang J., Chang J. K., Brailoiu E., Dun N. J. ( 2005) J. Comp. Neurol. 481, 314– 329 [DOI] [PubMed] [Google Scholar]
- 62.Terao Y., Kumano S., Takatsu Y., Hattori M., Nishimura A., Ohtaki T., Shintani Y. ( 2004) Biochim. Biophys. Acta 1678, 102– 110 [DOI] [PubMed] [Google Scholar]
- 63.Castellano J. M., Gaytan M., Roa J., Vigo E., Navarro V. M., Bellido C., Dieguez C., Aguilar E., Sánchez-Criado J. E., Pellicer A., Pinilla L., Gaytan F., Tena-Sempere M. ( 2006) Endocrinology 147, 4852– 4862 [DOI] [PubMed] [Google Scholar]
- 64.van Adelsberg J. ( 1999) Dev. Genet. 24, 299– 308 [DOI] [PubMed] [Google Scholar]
- 65.Bates C. M. ( 2000) Mol. Genet. Metab. 71, 391– 396 [DOI] [PubMed] [Google Scholar]
- 66.Wu H., Peisley A., Graef I. A., Crabtree G. R. ( 2007) Trends Cell Biol. 17, 251– 260 [DOI] [PubMed] [Google Scholar]
- 67.Yellaturu C. R., Ghosh S. K., Rao R. K., Jennings L. K., Hassid A., Rao G. N. ( 2002) Biochem. J. 368, 183– 190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puri S., Magenheimer B. S., Maser R. L., Ryan E. M., Zien C. A., Walker D. D., Wallace D. P., Hempson S. J., Calvet J. P. ( 2004) J. Biol. Chem. 279, 55455– 55464 [DOI] [PubMed] [Google Scholar]
- 69.Clapham D. E. ( 2007) Cell 131, 1047– 1058 [DOI] [PubMed] [Google Scholar]
- 70.Santini M. P., Talora C., Seki T., Bolgan L., Dotto G. P. ( 2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9575– 9580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao C., Meng A. ( 2005) Dev. Growth Differ. 47, 201– 211 [DOI] [PubMed] [Google Scholar]
- 72.Chu S., Ferro T. J. ( 2005) Gene 348, 1– 11 [DOI] [PubMed] [Google Scholar]
- 73.Alfonso-Jaume M. A., Mahimkar R., Lovett D. H. ( 2004) Biochem. J. 380, 735– 747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sugimoto T., Haneda M., Sawano H., Isshiki K., Maeda S., Koya D., Inoki K., Yasuda H., Kashiwagi A., Kikkawa R. ( 2001) J. Am. Soc. Nephrol. 12, 1359– 1368 [DOI] [PubMed] [Google Scholar]
- 75.Lacroix I., Lipcey C., Imbert J., Kahn-Perlès B. ( 2002) J. Biol. Chem. 277, 9598– 9605 [DOI] [PubMed] [Google Scholar]
- 76.Sakaguchi M., Miyazaki M., Sonegawa H., Kashiwagi M., Ohba M., Kuroki T., Namba M., Huh N. H. ( 2004) J. Cell Biol. 164, 979– 984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen J., Tucker C. L., Woodford B., Szél A., Lem J., Gianella-Borradori A., Simon M. I., Bogenmann E. ( 1994) Proc. Natl. Acad. Sci. U.S.A. 91, 2611– 2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dudley A. T., Lyons K. M., Robertson E. J. ( 1995) Genes Dev. 9, 2795– 2807 [DOI] [PubMed] [Google Scholar]
- 79.Luo G., Hofmann C., Bronckers A. L., Sohocki M., Bradley A., Karsenty G. ( 1995) Genes Dev. 9, 2808– 2820 [DOI] [PubMed] [Google Scholar]