Abstract
Arabidopsis mutants containing gene disruptions in AHA1 and AHA2, the two most highly expressed isoforms of the Arabidopsis plasma membrane H+-ATPase family, have been isolated and characterized. Plants containing homozygous loss-of-function mutations in either gene grew normally under laboratory conditions. Transcriptome and mass spectrometric measurements demonstrate that lack of lethality in the single gene mutations is not associated with compensation by increases in RNA or protein levels. Selected reaction monitoring using synthetic heavy isotope-labeled C-terminal tryptic peptides as spiked standards with a triple quadrupole mass spectrometer revealed increased levels of phosphorylation of a regulatory threonine residue in both isoforms in the mutants. Using an extracellular pH assay as a measure of in vivo ATPase activity in roots, less proton secreting activity was found in the aha2 mutant. Among 100 different growth conditions, those that decrease the membrane potential (high external potassium) or pH gradient (high external pH) caused a reduction in growth of the aha2 mutant compared with wild type. Despite the normal appearance of single mutants under ideal laboratory growth conditions, embryos containing homozygous double mutations are lethal, demonstrating that, as expected, this protein is absolutely essential for plant cell function. In conclusion, our results demonstrate that the two genes together perform an essential function and that the effects of their single mutations are mostly masked by overlapping patterns of expression and redundant function as well as by compensation at the post-translational level.
Keywords: Arabidopsis, Gene Knockout, H+-ATPase, Mass Spectrometry (MS), Membrane Energetics, Plant, Plasma Membrane Proton Pumps, Protein Phosphorylation, Proton Pumps
Introduction
In animals, the sodium pump is the primary active transport system and creates a membrane potential and sodium gradient that are used by all ion channels and cotransporters (1, 2). In higher plants and fungi, however, the transport of all solutes across the plasma membrane is coupled to a proton gradient rather than a sodium gradient. Thus, in these organisms, a plasma membrane proton pump creates a protonmotive force at the plasma membrane that drives all channels and cotransporters. Given the known importance of transport at the plasma membrane for life functions, it is not surprising that genetic studies of the sodium pump in nematodes, fruit flies, zebrafish, and mice, as well as with the proton pump of yeast, all conclusively demonstrate the lethal effects of loss-of-function mutations for a gene encoding the primary active transporter (Table 1) (3–11). In contrast, although there have been several reports of altered growth of mutant plants containing genetic alterations in the plasma membrane proton pump (12–17), none of these studies have provided evidence indicating that this enzyme performs an essential function for plant life. In this study, we present evidence clearly demonstrating that the plasma membrane proton pump is essential for plant growth. We show that AHA1 and AHA2 (for Arabidopsis H+-ATPase isoforms 1 and 2), the two most highly expressed members of the AHA gene family, perform overlapping functions that mask the lethality in single gene loss-of-function mutants. We also describe phenotypic screening that supports the in planta role of the proton pump in generating a protonmotive force and mass spectrometric methods that allow a more detailed and quantitative analysis of the in vivo regulation of these proteins at the post-translational level.
TABLE 1.
Loss-of-function mutations of plasma membrane P-type ATPase pump maintaining electrochemical gradients cause lethality in many eukaryotic model organisms
| Organisms | Mutagen | Gene | Terminal phenotype | Refs. |
|---|---|---|---|---|
| Yeast | Homologous recombination | PMA1 (H+ pump) | Haploid cell lethality | 3 |
| Thale cress | T-DNA insertion | AHA1 AHA2 double | Embryonic lethality | This study |
| AHA3 (H+ pump) | Male gamete lethality | 4 | ||
| Nematode | Ethyl methane sulfonate | eat-6 (Na+/K+ pump α) | Lethality at a larval stage | 5 |
| Fruit fly | P-element insertion | ATPalpha2206 | Embryonic lethality | 6 |
| Ethyl methane sulfonate | ATPαH64 (Na+/K+ pump α) | 7 | ||
| Zebrafish | N-ethyl N-nitrosourea | atp1a1aαm883 | Embryonic lethality | 8 |
| Spontaneous | had (Na+/K+ pump α1B1) | 9 | ||
| Mouse | Homologous recombination | Na K-ATPase α2, α3(Na+/K+ pump α) | Lethality at embryo and infant | 10 |
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
Mutants (ecotype Columbia) carrying T-DNA insertions in AHA1 (aha1–6, SALK016325; aha1–7, SALK065288; and aha1–8, SALK118350) and AHA2 (aha2–4, SALK082786, and aha2–5, SALK022010) were obtained from the Arabidopsis Biological Resource Center (Ohio State University) (18). Seeds were germinated on plates containing half-strength M&S3 salts, 1% (w/v) sucrose, and 0.7% (w/v) agar. Plants that were transferred to soil/perlite mixture (Jiffy-Mix, Jiffy Products of America, Lorrain, OH; horticultural perlite, The Schundler Co., Metuchen, NJ) were grown at 21 °C under constant light or 22 °C with a regime of 16 h of light/8 h of dark.
T-DNA Mutant Identification and Plant Genotyping
Plant genomic DNA was extracted using the method of Krysan et al. (19), with the elimination of the phenol/chloroform extraction step. The location of the T-DNA insertion in AHA1 or AHA2 was determined by sequencing PCR fragments containing the junction of the T-DNA and plant genomic DNA. The genotypes of plants were determined by PCR with allele-specific primers. The sequences of the PCR primers are provided in the supplemental material.
RNA Extraction, Quantitative RT-PCR, and Microarray Analyses
RNA was extracted from 1-week-old seedlings with the Plant RNeasy kit (Qiagen). For transcriptional profiling with GeneChip Arabidopsis ATH1 Genome Array (Affymetrix), RNA was processed using standard protocols at the Gene Expression Center, University of Wisconsin, Madison. Transcriptome data were normalized and analyzed with the Affymetrix GeneChip operating software and the Robust Multichip Average (RMA) method (20). For quantitative RT-PCR, total RNA was extracted from 10-day-old seedlings, reverse-transcribed (Invitrogen), and subjected to PCR using the iCycler real time PCR system (Bio-Rad) with SYBR Premix (Takara). Sequences of primers used for quantitative RT-PCR are provided in the supplemental material.
Plasma Membrane Preparation
Arabidopsis seedlings were grown in half-strength M&S liquid media as described previously (21). Two-week-old seedlings (20 g) were extracted with 50 ml of buffer (300 mm sucrose, 100 mm Tris-HCl, pH 7.6, 25 mm EDTA-Na2, 25 mm NaF, 1 mm Na2MoO4, 0.5% (w/v) polyvinylpyrrolidone, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin, 1 μg/ml E-64, 1 μm bestatin, 100 μm 1,10-phenanthroline, and 1 mm dithiothreitol), filtered through two layers of Miracloth, and spun 80,000 × g for 50 min at 4 °C. The pellet was resuspended in 9 ml of suspension buffer (300 mm sucrose, 10 mm Tris-HCl, pH 7.5, and 1 mm EDTA-Na2) and subjected to two-phase separation to enrich the plasma membrane fraction (22). The upper layer was spun at 80,000 × g for 50 min at 4 °C, and the pellet was resuspended in 2 ml of suspension buffer. Protein concentration was determined by the BCA assay (Pierce).
AHA Peptide Quantitation via Mass Spectrometry
Aliquots of 200 μl, corresponding to ∼120 μg of total protein from each plasma membrane preparation, were precipitated with chloroform/methanol/water. Pellets were washed twice with 80% acetone and resolubilized in 7.2 m urea and 1× PhosStop (Roche Applied Science) in 45 mm ammonium bicarbonate, pH 8. Urea was then diluted to a final concentration of 1 m with 45 mm ammonium bicarbonate, including 1× PhosStop. Solubilized proteins were reduced with 2 mm dithiothreitol and alkylated with 5 mm iodoacetamide. The alkylation reaction was quenched by further addition of dithiothreitol to 2 mm and trypsin (Promega) was added at an E:S ratio of 1:20 (6 μg). Digestion was quenched after 16 h by the addition of neat trifluoroacetic acid to reduce the pH to ∼2. Samples were desalted and concentrated by C18 solid phase extraction using SPEC tips (Varian) with a capacity of 200 μg. Peptides were eluted with 80% (v/v) acetonitrile, 0.5% (v/v) acetic acid in water. Acetonitrile was removed by vacuum centrifugation, and peptides were reconstituted in 60 μl of 0.5% (v/v) acetic acid in water (2 μg/μl). Digests were analyzed by liquid chromatography/MS on an LTQ-Orbitrap hybrid mass spectrometer (ThermoFisher Scientific). HPLC was performed using an Agilent 1100 HPLC system consisting of an isocratic pump for sample loading onto an Agilent Zorbax C18 trapping column (0.3 × 5 mm, 5-μm particles) and a nanopump for elution from the trapping column and onto an in-house built analytical column fabricated from 360 μm outer diameter × 75 μm inner diameter fused silica packed to a length of 15 cm with 3 μm of Magic C18 (Michrom) with an integrated electrospray emitter tip pulled to a diameter of ∼2 μm. Peptides were gradient-eluted using a water/acetonitrile binary solvent system, in which solvent A was 0.1 m acetic acid in water and solvent B was 0.1 m acetic acid, 95% acetonitrile (v/v) in water. Sample loading and desalting were performed over 20 min at a flow rate of 15 μl/min using the isocratic HPLC pump to deliver 0.1 m acetic acid, 1% acetonitrile. Following sample loading, elution was performed at 200 nl/min by increasing solvent B from 1 to 40% over 195 min, 40 to 60% over 20 min, and 60 to 100% over 5 min, after which the stationary phases were re-equilibrated at the initial conditions for 30 min. The LTQ-Orbitrap mass spectrometer was operated in data-dependent mode for automated simultaneous acquisition of MS survey spectra and MS/MS spectra. MS survey spectra were acquired at 100,000 resolving power over the m/z range 300–2000 in the Orbitrap with external mass calibration. MS/MS spectra were performed on the five highest abundance signals present in the survey scan, requiring that they be present in the 2+ or higher charge state and that they pass the dynamic exclusion criteria. Post-acquisition data analysis consisted of data base searching using Mascot 2.2 (Matrix Science) and a data base composed of TAIR release 9 (downloaded on June 19, 2009) to which were added common contaminant proteins such as trypsin and human keratins. The peptides identified by data base searching were imported into Scaffold 2 (Proteome Software), which was used to identify detected peptides with unique sequences to discriminate between the various AHA isoforms. Two such peptides were used to assess abundance of AHA1 and AHA2 among the three genotypes analyzed. These peptides arise from amino acids 407 to 420 (AHA1, +TALTYIDSDGNWHR−; AHA2, +TALTYIDGSGNWHR−) and amino acids 471 to 490 (AHA1, +ESPGGPWEFVGLLPLFDPPR−; AHA2, +ESPGAPWEFVGLLPLFDPPR−). AHA1 and AHA2 protein abundance was determined by calculating the area of extracted ion chromatograms of the isoform-specific peptides from the different plants using the Xcalibur Qual Browser software (ThermoFisher Scientific). A total of four replicate analyses was performed for each genotype.
Selective Reaction Monitoring Mass Spectrometry Analysis
Total proteins from enriched plasma membrane samples (wild type, aha1–6, aha2–4) were isolated from the supernatant using a methanol/chloroform/water extraction as described previously (23). Precipitated proteins were solubilized in 8 m urea containing phosphatase inhibitor mixture (1× PhosStop, Roche Applied Science). Samples were diluted to 1 m urea, and a BCA protein assay was performed to determine protein concentration. Samples were then diluted to contain 1 mg/ml total protein concentration. Disulfide bonds were reduced using dithiothreitol (5 mm, 30 min), alkylated with iodoacetamide (15 mm, 30 min), and protein-digested using trypsin (1:100 trypsin to protein ratio, 37 °C, overnight). Resulting peptide samples were acidified (0.1% trifluoroacetic acid) and concentrated using a solid phase extraction procedure (C18 SepPak, Waters).
Trypsin-digested plasma membrane samples were utilized for creating standard calibration curves for each of the four peptides of interest: AHA1 Thr-948, AHA1 pThr-948, AHA2 Thr-947, and AHA2 pThr-947. Isotope-labeled leucine (13C) peptides were synthesized for each peptide of interest and added to each of the biological samples so that the final concentration of the labeled peptides was 100 amol to 100 fmol per μl. Samples were loaded (2 μg) onto the Agilent Phospho-CHiP. The Agilent Phospho-CHiP consisted of a TiO2 enrichment column sandwiched between two reverse phase C18 columns. This allowed both phosphorylated peptides and nonphosphorylated peptides to be trapped and eluted separately. Trapping, enrichment, and analysis were performed on an Agilent 1200 HPLC system, consisting of a loading capillary pump operating at 4 μl/min flow and a nanoflow pump operating at 300 nl/min. Peptides were trapped at 4 μl/min using 100% solvent A (0.6% acetic acid and 2% formic acid in 2% acetonitrile). The sample was retained on the first reverse phase C18 enrichment column. After switching the enrichment columns in line, all peptides were eluted off the first C18 enrichment column by gradient elution. All peptides were then eluted off the first C18 enrichment column by gradient elution. Phosphorylated peptides bound to the TiO2 section, whereas all other peptides with no TiO2 affinity were transferred to the C18 separation column. Analysis was performed subsequently with a nanoflow analytical pump gradient from 5 to 70% solvent B (100% acetonitrile, 0.6% acetic acid, 0.5% formic acid) over 40 min at 300 nl/min. Elution of phosphorylated peptides from the TiO2 section to the C18 section was then achieved by injection of 15 μl of elution buffer, pH 9.0 (Phospho-CHiP-Kit, Agilent Technologies). Analysis of the eluted (phosphorylated) peptides was performed by switching the precolumn to be in line with the analytical column for a second H2O/acetonitrile gradient. Immediately afterward, an injection of 4 μl of regeneration solution (Phospho-CHiP kit, Agilent Technologies) was performed to wash and re-equilibrate the TiO2 precolumn.
Doubly charged parent ions and at least three to four singly charged fragment y-ions from each of the four peptides of interest were monitored using selective reaction monitoring on the Agilent 6460-QQQ. All data were acquired with a Q1 resolution of 0.7 atomic mass unit and Q3 resolution of 1.2 atomic mass units with each transition monitored with a dwell time of 20 ms. The RF-only q2 collision cell was pressurized with nitrogen gas. All plasma membrane samples were analyzed a minimum of three times with isotope-labeled internal standard added from 100 amol to 100 fmol levels in each analysis.
Peptide responses of both the endogenous peptides and the isotope-labeled internal standards were determined by the area under the curve of the total ion current. Calibration curves were produced by plotting the ion intensity of the isotope-labeled internal standard peptide versus the amount of synthetic standard present, and endogenous peptide levels were determined by comparison with the standard calibration curve. Average concentrations were calculated across technical replicates and standard deviations produced. Coefficients of variance were calculated by taking the ratio of the standard deviation of peptide signal over the technical replicates to the average of the peptide concentration. Coefficients of variance are represented as percentages. Statistically significant differences in peptide levels were determined using a two-tailed Student's t test assuming unequal variance.
Microscopy and Staining
To visualize the frequency of aborted seed development, fully elongated green siliques of wild type or mutant plants were harvested, manually opened, and viewed using a Nikon SMZ1500 stereomicroscope. Images were captured using a Spot Insight color digital camera (Diagnostic Instruments, Inc.). Developing seeds from aha1–6/aha1–6;AHA2/aha2–4 and AHA1/aha1–6;aha2–4/aha2–4 plants were fixed in 2% glutaraldehyde, dehydrated in ethanol series, and embedded in LR White. Sections were stained with 1% toluidine Blue.
β-Glucuronidase (GUS) Reporter Assay
A 3.5-kb DNA fragment containing the AHA1 promoter region and the first exon (14 amino acids) was PCR-amplified and cloned into pCAMBIA1301 at the EcoRI and NcoI sites to generate an AHA1 promoter-GUS fusion construct. Plants carrying this transcriptional reporter were generated via Agrobacterium-mediated transformation (strain GV3101) described in Ref. 24. Multiple transgenic plants were subjected to GUS immunoblotting to verify the production of GUS protein. T2 plants homozygous for a single insertion of the transgene were selected and used for GUS assay. Siliques of various developmental stages were harvested and incubated at 37 °C for 16 h in the following reaction mixture: 50 mm sodium phosphate buffer, pH 7.2, 0.2% Triton X-100, 2 mm potassium ferrocyanide, 2 mm potassium ferricyanide, 15% methanol, and 2 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid. The stained tissues were subjected to washing with a series of ethanol concentrations, fixation, embedding, and sectioning.
Molecular Complementation
A 9.9-kb genomic DNA fragment containing the AHA1 promoter region (3377 bp), AHA1, and 1100 bp downstream from the stop codon was amplified by PCR from BAC clone F19F24 (the Arabidopsis Biological Resource Center), using the following primers: 5′-GCCCTCGCTATATTGATGTCAG-3′ and 5′-CTTTGGACGTGACTCGCAGA-3′. The PCR fragment was first cloned into the pCR2.1 vector with TA cloning (Invitrogen) and then subcloned into the pCAMBIA1200 binary vector, which confers hygromycin resistance as the plant selection marker, at the SalI and SacI restriction sites. The resulting plasmid was used to introduce into aha1–6/aha1–6;AHA2/aha2–4 plants as describe above. Transgenic seedlings were selected on M&S plates containing 25 μg/ml hygromycin. HygR aha1–6; aha2–4 homozygous double mutant plants whose embryonic lethality was complemented by the presence of an AHA1 transgene were identified using PCR with locus-specific primers. Primers used to identify the endogenous AHA1 allele were 5′-CGACGAATGATCTAATCACAAGC-3′ and 5′-CAAAGTCCGGTTAAGGGTATTTC-3′, and those used to identify the AHA1 transgene were 5′-TGTAAAACGACGGCCAGTG-3′ and 5′-TCATGTGATTTCTTGCCATC-3′. For complementation with the AHA2 transgene, a 10-kb genomic DNA fragment containing the AHA2 promoter region (4337 bp), AHA2, and 586-bp downstream from the stop codon was obtained by PmlI-digesting BAC clone F9N11. The resulting DNA fragment was cloned into pCR2.1 at the EcoRV site and again subcloned into the pCAMBIA1200 at the SalI and SacI sites. The resulting plasmid was introduced into AHA1/aha1–6;aha2–4/aha2–4 plants as described previously. Complemented plants were identified as described above. Primers used to identify the AHA2 transgene were 5′-CATGTTGGCAAGCTGCTC-3′ and 5′-AAATTAGCAGGCGCATGT-3′.
Site-directed Mutagenesis
A restriction enzyme site XbaI was introduced to AHA2 via the QuikChange II XL system (Stratagene) using the following pair of primers: 5′-CGGTTTACGCCAATCTAGAGGAATTTGCAAAG-3′ and 5′-CTTTGCAAATTCCTCTAGATTGGCGTAAACCG-3′. This insertion resulted in a mutation at the Trp-807 to Leu-Glu. Into the XbaI site, green fluorescent protein gene was inserted to create putative an AHA2-GFP translational fusion protein with a molecular mass, ∼125 kDa. The AHA2-GFP fusion construct was transformed to the mutant plants via Agrobacterium as described above.
Root Growth Phenotyping
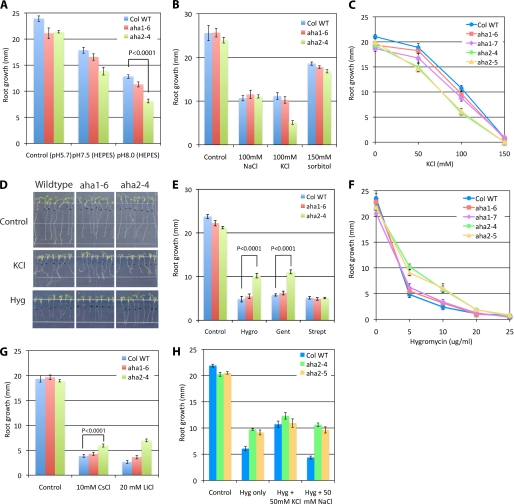
Seeds were germinated on vertically positioned half-strength M&S plates for 3 days. Seedlings were transferred to the media supplemented with additives and grown for 4 days. During the entire course of experiments, seedlings were grown under continuous light (25 μmol m−2 s−1). Root growth was quantified with the ImageJ software (www.nih.gov). Root growth results shown in Fig. 4 are one representative of triplicate analyses with each containing 14 seedlings. Growth conditions used for screening root phenotypes are provided in the supplemental material.
FIGURE 4.
Root growth phenotypes of aha1 and aha2 mutants in response to treatments that perturb the protonmotive force. A, high pH sensitivity of root growth in aha2 mutant. B, high potassium sensitivity of root growth in aha2 mutant. C, dose-response effects of potassium on aha1 and aha2 root growth. D, images of root growth on control, KCl, and hygromycin media. E, effects of aminoglucoside antibiotics on aha2 root growth. Hygro, 5 μg/ml hygromycin; Gent, 10 μg/ml gentamicin; Strept, 100 μg/ml streptomycin. F, dose-response effects of hygromycin B on aha1 and aha2 root growth. G, cesium and lithium tolerance in aha2 mutant. H, suppression of hygromycin inhibitory effect by potassium. Col WT, wild type plant of A. thaliana, ecotype Columbia.
Extracellular Acidification Assay
A five-day-old single seedling were incubated in 250 μl of ¼ strength M&S media (pH 6.2, unbuffered), including 1% sucrose and 30 μg/ml fluorescein isothiocyanate/dextran (Mr 10,000, Sigma) for 16 h. Fluorescent emission was detected at 535 nm with excitation at 485 nm, using a microplate reader (Tecan SpectraFluor Plus). Media pH were calibrated by a standard curve with a range from 5.0 to 6.6.
RESULTS
Isolation and Molecular Characterization of A. thaliana Mutant Plants Containing AHA1 and AHA2 Gene Disruptions
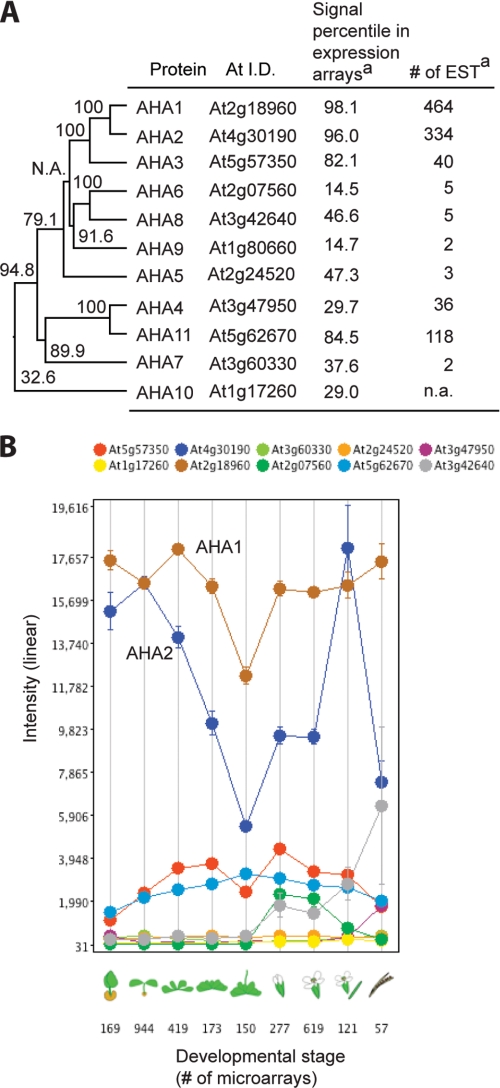
The Arabidopsis genome carries 11 similar but discrete members of the plasma membrane proton pump family (AHA1–11, for Arabidopsis H+-ATPase isoforms 1–11). The AHAs are a subfamily of the P-type ATPase superfamily, which has 46 members in Arabidopsis, and the other members transport specific solutes other than protons (25). Thus, only the 11 AHA isoforms appear to catalyze the ATP-dependent active efflux of protons across the plasma membrane, resulting in a protonmotive force composed of both a membrane potential with inside negative values of −140 to −210 mV (26, 27), and a pH gradient generated by apoplastic pH 5–6 and cytoplasmic pH 7.5. There are no genes in Arabidopsis encoding a sodium pump, or any other known primary active transport system, suggesting that members of the AHA family satisfy all of the primary active transport requirements of this organism. Transcriptome analysis of the 11 AHA members based on hybridization to DNA chips or the number of expressed sequence tags reflecting mRNA abundances indicates that AHA1 and AHA2 are by far the most highly expressed members of this family throughout plant life (Fig. 1, A and B). In addition, when the 11 protein sequences are compared for sequence similarities via a phylogenetic tree, it is clear that AHA1 and AHA2 are the most closely related of all the members, suggesting that they arose from the recent duplication of a common ancestor (Fig. 1A). Expression studies with AHA3 suggest that this enzyme plays a role in phloem transport, but because homozygous AHA3 knock-out mutant plants are male-sterile, there is no clear genetic evidence of a critical role for AHA3 in vegetative growth (4). Genetic and expression studies with AHA4 and -10 suggest that they play roles in salt stress tolerance and seed coat pigmentation, but again, genetic evidence for an essential function is lacking (15, 16).
FIGURE 1.
Phylogenetic and expression analyses of 11 AHA members. A, phylogenetic tree and expression level of AHA members based on microarray and expression sequence tag analyses. Protein sequences were aligned by an algorithm, ClustalV (PAM250), provided by Megalign (Lasergene, DNASTAR, Inc., Madison, WI). Bootstrap values (%) obtained by resampling 1000 times were indicated at the branches of the tree. n.a., not available. a, data were obtained on line. B, developmental expression study of AHA members with Genevestigator. Data were obtained on line.
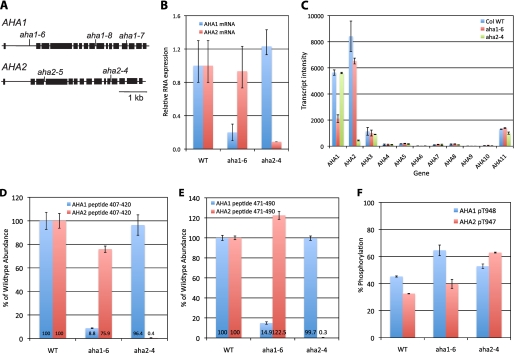
To critically test whether AHA1 or AHA2 proteins are essential, we examined the growth of various insertional mutant alleles of each gene using available T-DNA collections. All of the mutants used in this study were obtained from the Salk Collection in the Columbia ecotype, and sequence analysis of the T-DNA/plant genomic DNA junction demonstrated that the insertion occurred either within an intron (aha1–6, aha1–8, aha2–4, and aha2–5) or within an exon (aha1–7) (Fig. 2A).
FIGURE 2.
Molecular characterization of loss-of-function mutants of AHA1 and AHA2. A, description of the T-DNA insertional alleles of AHA1 and AHA2. Black boxes represent exons. Vertical lines indicate the location of T-DNAs. aha1–6, SALK016325; aha1–7, SALK065288; aha1–8, SALK118350; aha2–4, SALK082786; and aha2–5, SALK022010. B, quantitative RT-PCR of AHA1 and AHA2 in wild type (WT), aha1–6, and aha2–4 mutants. C, transcript signal intensities of the 11 AHA members in WT, aha1–6, and aha2–4 mutants determined by microarray analyses. Graph shows mean values with S.D. D and E, AHA1 and AHA2 protein abundances in WT, aha1–6, and aha2–4 plants. Isoform-specific polypeptides (the amino acid position 407–420 and 471–490) were determined by quantifying the peak areas of the peptide ions found in the total ion chromatogram of mass spectrometric analyses. Data are shown as means ± S.E. of three separate measurements. F, selective reaction monitoring mass spectrometry analysis for AHA1 and -2 C-terminal peptides of nonphosphorylated and phosphorylated forms. Proportion of AHA1 phosphopeptide Thr(P)-948 and AHA2 phosphopeptide Thr(P)-947 was quantified and shown ± S.D. Col WT, wild type plant of A. thaliana, ecotype Columbia.
To determine whether the insertional mutations resulted in growth alterations, mutants were selfed, and homozygous plants lacking wild type copies of either AHA1 or AHA2 genes were obtained. Growth measurements under ideal laboratory conditions revealed no growth defect in aha1 or aha2 mutant plants. To determine how severe the insertions were at the level of gene expression, AHA1 and AHA2 RNA abundances were quantified via various methods, including microarray and quantitative RT-PCR (Fig. 2). The results of these experiments demonstrated that aha1–6 and aha1–8 mutants carry knockdown AHA1 alleles, and the aha1–7 mutant appears to express a truncated AHA1 transcript whose abundance at the post-insertion region was below the detection threshold. Although aha1–7 lacks the AHA1 full-length transcript, we were unable to determine whether its predicted protein that is truncated adjacent to amino acid 739 within the M7–M8 extracellular loop has catalytic activity. AHA2 expression in the aha2–4 and aha2–5 mutants was reduced by 90–95% in both cases, compared with wild type plants. In addition, these studies demonstrated that in all of the insertional mutant plants showing reduced AHA1 or -2 levels, there was no compensation observed at the transcript level. In other words, reduced levels of AHA1 or AHA2 did not result in increased levels of any other AHA isoform (Fig. 2, B and C).
To determine whether compensation was occurring at the translational level, protein abundance was analyzed via mass spectrometric approaches. Plants were grown and plasma membranes were isolated from wild type, aha1–6 and aha2–4 mutants. After trypsinization, peptides were subjected to analysis on an ESI-LTQ-Orbitrap tandem mass spectrometer. Because AHA1 and AHA2 are very conserved at the amino acid sequence level (94.4% identity), it was essential to identify and quantify the tryptic peptides with isoform-specific peptide sequences. Two tryptic peptides, from amino acid 407 to 420 and 471 to 490, in AHA1 and AHA2 were particularly informative in this regard. As shown in Fig. 2, D and E, by quantifying the relative intensity of these peptide ions in the extracted chromatogram, we were able to determine that, consistent with the mRNA studies, the mutants have reduced levels of AHA1 or AHA2 proteins. In addition, the mass spectrometric measurements demonstrated that there were no increases in the AHA2 protein nor any other AHA isoforms in the aha1–6 mutants and vice versa, i.e. there was no compensation occurring at the translational level in the aha1–6 or aha2–4 mutants.
It is well established that protein kinase-mediated phosphorylation of the penultimate threonine in the plasma membrane proton pump can result in a hyperactive enzyme, with increased catalytic activity (28). We thus sought a method to measure the stoichiometry of phosphorylation for AHA1 and AHA2 at this residue, to determine whether the homozygous mutant plants were compensating for the loss of one of the isoforms by an increase in the level of phosphorylation of the other, and/or of the small amount of residual ATPase remaining in the insertional mutants. For this purpose, we chose a highly sensitive and quantitative method, selected reaction monitoring, with heavy isotope synthetic peptides directed against a C-terminal tryptic fragment as follows: AHA1, +GLDIDTAGHHYTV−; AHA2, +GLDIDTAGHYTV−. Heavy isotope-labeled peptides with and without phosphorylation at AHA1 Thr-948 or AHA2 Thr-947 were synthesized and used as internal standards for absolute quantitation of the C-terminal tryptic peptide from both AHA1 and AHA2 with a triple quadrupole mass spectrometer. By inferring the abundance of AHA1 and AHA2 proteins as the sum of phosphorylated and unphosphorylated forms of the C-terminal peptides, we confirmed our earlier result of untargeted analysis with the LTQ-Orbitrap mass spectrometer that there was no compensation at the translational level observed in the aha1–6 or aha2–4 mutants. With the targeted analysis, however, we observed significant increases in AHA1 phosphorylation level of the residual AHA1 protein in aha1–6 mutants (Fig. 2F). Similarly, AHA2 phosphorylation level of the residual AHA2 protein was increased in aha2–4 mutant. There were also small increases in the phosphorylation of AHA1 protein in aha2–4 mutant as well as in that of AHA2 protein in aha1–6 mutant. Our results are consistent with an increase in kinase-mediated phosphorylation of the penultimate Thr residue behaving as a post-translational means of compensating for the loss of AHA proteins. Although the trends were clear, the errors associated with the measurements do not allow a clear estimate of the degree of compensation attained.
Simultaneous Disruption of both AHA1 and AHA2 Genes in a Single Plant Is Lethal
To obtain a single plant containing both AHA1 and AHA2 gene disruptions, various aha1 and aha2 mutants were crossed, and seeds resulting from the dihybrid plants were selfed. Progeny from this population was then examined to determine whether any plants lacking both AHA1 and 2 wild type alleles could be obtained. As shown in Table 2, with three different pairs of mutants crossed (i.e. aha1–6 and aha2–4; aha1–7 and aha2–5; and aha1–8 and aha2–5), a homozygous double mutant was never recovered in the progeny, suggesting that the double mutation was lethal at some point during gametogenesis or embryogenesis.
TABLE 2.
Plant lines segregating aha1 and aha2 mutant alleles failed to produce homozygous double mutants
The genotypes of progeny from AHA1+/−AHA2+/− mutants were determined using a PCR-based method.
| Parent genotype | No. of progeny genotyped | No. of homozygous double mutants observed | No. of homozygous double mutants expected |
|---|---|---|---|
| AHA1/aha1–6 AHA2/aha2–4 | 243 | 0 | 15 |
| AHA1/aha1–7 AHA2/aha2–5 | 52 | 0 | 3 |
| AHA1/aha1–8 AHA2/aha2–5 | 55 | 0 | 3 |
To identify the developmental stage first affected by the double mutant condition, we analyzed the segregation pattern of the aha1–6 allele from plants homozygous for the aha2–4 mutations and heterozygous for the aha1–6 mutation. The observed ratios of progeny allele frequencies were subjected to the χ2 test to examine whether our hypothesis for embryonic lethality in the homozygous double mutants is statistically supported. The results indicate that lethality is likely to occur during embryo development rather than during gamete development (Table 3). A similar result was obtained in reciprocal experiments testing the aha2–4 allele segregation pattern from double mutants homozygous for the aha1–6 mutation and heterozygous for the aha2–4 mutation. We also analyzed the transmission efficiencies of the insertional mutant alleles through the male and female gametes. We performed two sets of reciprocal crosses between wild type plants and plants heterozygous for one AHA insertion allele and homozygous for the other. In the subsequent progeny, we observed normal transmission efficiencies of the aha1–6 and aha2–4 mutations through both pollen and ovule, indicating that the double mutations do not affect female or male gametogenesis (Table 4).
TABLE 3.
Progeny of AHA1/aha1–6 aha2–4/aha2–4 and aha1–6/aha1–6 AHA2/aha2–4 parents
The genotypes of F3 progeny were determined by PCR. Ob. indicates the number of individuals observed; Ex. 1 indicates the expected number based on Mendelian inheritance. Ex. 2 indicates the expected number when the homozygous double mutant plants are not viable due to lethality during embryogenesis. Ex. 3 indicates the expected number when the double mutant plants are not viable due to lethality during gametogenesis. χ 2 test was used to determine the probability of which the deviation of the observed value from the expected value was due to chance.
| Parent genotype | Progeny genotype | Ob. | Ex. 1 | Ex. 2 | Ex. 3 |
|---|---|---|---|---|---|
| AHA1/aha1–6 aha2–4/aha2–4 | AHA1/AHA1 aha2–4/aha2–4 | 28 | 20.5 | 27.3 | 41 |
| AHA1/aha1–6 aha2–4/aha2–4 | 54 | 41 | 54.7 | 41 | |
| aha1–6/aha1–6 aha2–4/aha2–4 | 0 | 20.5 | 0 | 0 | |
| p < 0.001 | p > 0.95 | p < 0.01 | |||
| aha1–6/aha1–6 AHA2/aha2–4 | aha1–6/aha1–6 AHA2/AHA2 | 13 | 10.5 | 14 | 21 |
| aha1–6/aha1–6 AHA2/aha2–4 | 29 | 21 | 28 | 21 | |
| aha1–6/aha1–6 aha2–4/aha2–4 | 0 | 10.5 | 0 | 0 | |
| p < 0.001 | p > 0.95 | p < 0.05 |
TABLE 4.
Gametophytic transmission of aha1–6 and aha2–4 alleles determined by reciprocal crosses
Two sets of reciprocal crosses were performed to examine the transmission of aha1–6 aha2–4 double mutations through the male or female gametes. The mutant parents are AHA1/aha1–6 aha2–4/aha2–4 or aha1–6/aha1–6 AHA2/aha2–4, and the other parent is wild type (AHA1/AHA1 AHA2/AHA2).
| Female parent genotype | Male parent genotype | Progeny genotype | |
|---|---|---|---|
| AHA1/AHA1 AHA2/aha2–4 | AHA1/aha1–6 AHA2/aha2–4 | ||
| AHA1/aha1–6 aha2–4/aha2–4 | AHA1/AHA1 AHA2/AHA2 | 24 | 29 |
| AHA1/AHA1 AHA2/AHA2 | AHA1/aha1–6 aha2–4/aha2–4 | 18 | 14 |
| AHA1/aha1–6 AHA2/AHA2 | AHA1/aha1–6 AHA2/aha2–4 | ||
| aha1–6/aha1–6 AHA2/aha2–4 | AHA1/AHA1 AHA2/AHA2 | 14 | 14 |
| AHA1/AHA1 AHA2/AHA2 | aha1–6/aha1–6 AHA2/aha2–4 | 13 | 15 |
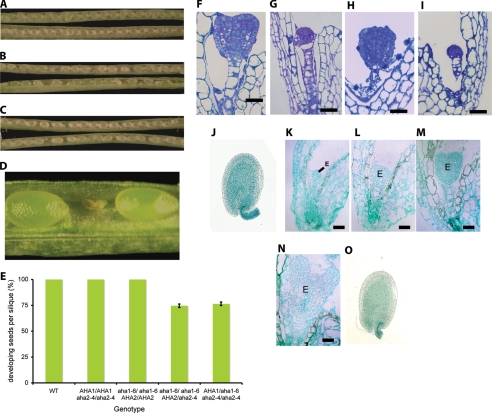
In the course of our experiments, we observed aborted developing seeds within the siliques of self-pollinated flowers from the double mutant plants that carry only one wild type copy of either AHA1 or AHA2. The aborted seeds were randomly distributed and accounted for approximately one-quarter of the total number of seeds within the silique (Fig. 3, A–D). Wild type plants or either aha1–6 or aha2–4 single mutants grown in the same environment rarely showed aborted seeds within their siliques. To investigate early seed abortion in putative homozygous double mutants, we microscopically examined thin tissue sections of developing seeds from self-pollinated plants that are homozygous mutants at the AHA1 locus and heterozygous mutants at the AHA2 locus. Whereas ∼75% of the seeds within the analyzed siliques contained embryos at the globular heart transition stage, the remaining 25% contained embryos arrested at the pre-globular stage (Fig. 3, E–I). We hypothesized that these arrested embryos are the nonviable homozygous double mutants. The same results were observed with reciprocal sets of double mutants.
FIGURE 3.
Seed developments in the double mutants and AHA1 and AHA2 promoter activity. A, phenotype of seed development in WT viewed under a stereomicroscope. B, immature siliques of aha1–6/aha1–6 AHA2/aha2–4 plants containing empty spaces. C, immature siliques of AHA1/aha1–6 aha2–4/aha2–4 plants containing empty spaces. D, magnified view of an aborted seed in a silique of aha1–6/aha1–6 AHA2/aha2–4 plant. E, percentage of developing seeds from the double mutants compared with WT or the AHA single mutant plants. Bars indicate S.E. F, normal growing embryo from self-pollinated aha1–6/aha1–6 AHA2/aha2–4 plants. G, arrested embryos from the same silique as in F. H, normal growing embryo from self-pollinated AHA1/aha1–6 aha2–4/aha2–4 plants. I, arrested embryos from the same silique as in H. Scale bar, 20 μm. J–N, GUS staining in embryos for AHA1 promoter activity. J, developing seed; K, 8-cell stage; L, early globular stage; M, late globular stage; N, heart stage. Scale bar, 20 μm. O, GUS staining in developing seed for AHA2 promoter activity. Scale bar, 10 μm. E; embryo.
To test whether there is a causal link between the homozygous double mutant genotype and the lethal phenotype, we carried out molecular complementation. A 10-kb genomic DNA fragment, including the AHA1 gene and the entire regulatory region, was introduced into double mutant plants that lack wild type copies of the AHA1 gene. A total of 184 independently transformed plants (T1) were recovered and further screened. We obtained 11 homozygous double mutant plants carrying wild type AHA1 transgenes among the T1 population (Table 5). Genotyping of the T2 progeny resulting from self-fertilization of the rescued lines confirmed the existence of plants homozygous for the aha1–6 and aha2–4 alleles that are viable due to the presence of the AHA1 transgene. Similar results were obtained when we transformed double mutant plants that lack wild type copies of the AHA2 gene with a genomic DNA fragment, including the AHA2 gene and regulatory region (Table 5).
TABLE 5.
Molecular complementation of aha1–6/aha1–6;aha2–4/aha2–4 plants with wild type or modified transgenes of AHA1 or AHA2
| Transgene | % of rescued plants in T1 populationa | % of rescued plantsb in T2 population |
|---|---|---|
| Wild type AHA1 genomic clonec | 5.98% (n = 184) | 21.4% (n = 42) |
| 16.7% (n = 42) | ||
| Wild type AHA2 genomic cloned | 5.56% (n = 90) | 15.4% (n = 39) |
| 23.3% (n = 30) | ||
| AHA2(W807LE)e | 4.3% (n = 23) | 20.0% (n = 30) |
| 15.3% (n = 32) | ||
| AHA2(W807GFP)f | 0% (n = 82) | 0% (n = 30) |
| 0% (n = 30) |
a Numbers in parentheses indicate total numbers of transformed plants that were PCR-genotyped.
b T2 progeny of two independently transformed lines from AHA1/aha1–6;aha2–4/aha2–4 or aha1–6/aha1–6;AHA2/aha2–4 plants carrying transgenes were genotyped to identify rescued plants.
c aha1–6/aha1–6;AHA2/aha2–4 plants were transformed with wild type AHA1 gene.
d AHA1/aha1–6;aha2–4/aha2–4 plants were transformed with wild type AHA2 gene.
e AHA2 genomic clone with a mutation of Trp-807 to Leu-Glu to create an XbaI restriction enzyme site.
f AHA2 genomic clone with a translational fusion to green fluorescent protein that is inserted at the Trp-807.
Our successful complementation of the double mutants with either AHA1 or AHA2 transgenes provides us a system for further investigating the structure-function relationship of AHA proteins. The double mutant plants whose endogenous AHA1 and AHA2 genes are replaced with tagged transgenes would be useful for analyzing AHA protein localization and post-translational modifications. To explore a potential site for inserting a tag within AHA2 protein, we created a point mutation in this transgene. To avoid generating mutations within the activity regulatory domains at the N- or C-terminal ends (29), the M9–M10 extracellular loop was chosen as a target site. A mutation was created to introduce a restriction enzyme (XbaI) site for cloning purposes at the amino acid Trp-807 position, which converts this residue into a dimer, Leu-Glu (W807L/W807E mutation). This AHA2 transgene carrying the W807L/W807E mutation successfully rescued the embryo lethality of the homozygous double mutants (Table 5), and the rescued plants appeared to be normal, suggesting this single amino acid insertion at the loop does not influence the AHA2 catalytic activity. Next, we transformed the double mutants with the AHA2 transgene that carries green fluorescent protein as an in-frame insertion at this Trp-807 position to produce an ∼125-kDa AHA2-GFP fusion pump protein; however, no rescued homozygous double mutants were recovered from the transgenic population, suggesting that this modified AHA2(W807GFP) transgene did not complement the embryo lethality and that the AHA2 protein conformation was unlikely to be able to tolerate a large polypeptide insertion at this extracellular location.
The fact that the aha1 aha2 homozygous double mutant embryos failed to develop properly indicates that AHA1 and AHA2 are transcriptionally active during embryogenesis. To test this prediction, we studied AHA1 and AHA2 promoter activities using a GUS reporter gene assay. Plants carrying a 3.4-kb AHA1 promoter or a 3.2-kb AHA2 promoter region and the first exon fused with GUS were generated and assayed for GUS activity. Histochemical staining for GUS activity indicates that the AHA1 promoter is active on the surface of developing seeds and from 8- to 16-cell stages to the heart stage of embryo development (Fig. 3, J–N). In contrast, GUS staining for the AHA2 promoter activity was observed much lower than the AHA1 promoter on the surface of developing seeds (Fig. 3O) and was detected up to the early globular stage of embryo development.
Growth of Wild Type and AHA Mutants under a Large Number of Environmental Challenges
The fact that the homozygous double mutants are embryo-lethal supports the hypothesis that, collectively, the two genes perform an essential function. Their roles in vegetative growth, however, remain to be determined. To investigate the functions of AHA1 and -2 during vegetative growth, we grew aha1 and aha2 homozygous single mutants under >100 different environmental conditions (supplemental material). Although there are many different growth/developmental analyses one could make, we chose to measure root length because this can be done quantitatively under a large number of conditions. The list of environmental challenges was chosen based on known or suspected functions of the pump. For example, a role of the pump in auxin-mediated cell elongation has long been considered important for mechanistic models of plant cell elongation. Thus, studies were performed with aha1 and aha2 mutant plants grown in the presence of various concentrations of added auxin and other hormones. Surprisingly, in root elongation assays, only a few out of over 100 conditions elicited a difference between wild type and mutant plants. Notably, none of the known phytohormones (gibberellic acid, abscisic acid, kinetin, ethylene (1-aminocyclopropane-1-carboxylic acid), and iodole-3-acetic acid) nor plant growth regulators (brassinolide and jasmonate) produced a difference in root growth among the three genotypes. Of all the many changes to the medium, the only two in which the mutants fared worse than wild type were those expected to alter the protonmotive force across the plasma membrane (a high external potassium concentration and high external pH) (Fig. 4A). As shown in Fig. 4, B and C, the effect of potassium was only observed at concentrations ≥50 mm and was specific for potassium because neither sodium chloride nor sorbitol produced a differential effect on root growth of the two mutants. It is well known that high concentrations of potassium cause a decrease in membrane potential (26), which is not elicited by high concentrations of less permeable cations like sodium or with neutral solutes like sorbitol, which also eliminates an effect of osmotic response because all three compounds (potassium, sodium and sorbitol) are capable of causing osmotic stresses. Similarly, it is likely that at high external pH, the protonmotive force is compromised because there is a 100-fold decrease in proton concentration going from pH 6.0 to 8.0 in the external medium. It is likely that both the potassium-induced decrease in membrane potential and the alkaline external pH exerted stresses on the ability of the proton pump to maintain a sufficient protonmotive force to support growth, which could not be overcome in the AHA-impaired mutants. In all cases, the phenotypic differences are more pronounced in the aha2 mutants, which is consistent with the greater impairment in pump mRNA and protein levels found in these mutants, and in the greater importance that this isoform is likely to have in roots, given its pattern of expression (30).
It is interesting to note that the only other environmental conditions that elicited a difference between genotypes were the additions of a variety of toxic cations. In all of these cases, there was a marked advantage in growth in the mutants, compared with the wild type. This phenomenon was observed with aminoglucoside antibiotics, including hygromycin and gentamicin, and with alkali metal ions, including lithium and cesium, and is consistent with the reduced uptake of a toxic compound in roots of plants containing reduced protonmotive force (Fig. 4, D–G). Consistent with this interpretation, we demonstrated that the hygromycin effect could be reduced by the presence of 50 mm potassium chloride but not 50 mm sodium chloride (Fig. 4H). In other words, when the overall membrane potential was reduced by potassium, so that little hygromycin was entering root cells, there was no difference among the three genotypes. In the absence of high potassium, however, when the membrane potential was driving hygromycin transport into root cells, the aha2 mutants showed less sensitivity to the toxic cation. A similar observation has been made in yeast mutants with impaired PMA1 activity. In fact, hygromycin-insensitive mutants were the first plasma membrane proton pump mutants to be found in early work with Saccharomyces cerevisiae (31).
Measurements of AHA1 and AHA2 Pump Activity
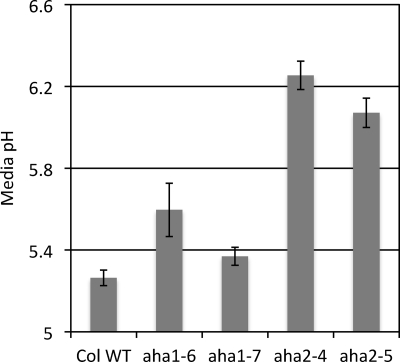
The above experiments demonstrate the growth consequences caused by reduced proton pump activity in the aha2 and aha1 mutants, although there were no such apparent consequences when the plants were grown under ideal laboratory conditions without environmental challenges such as high potassium or high external pH. In addition, although we were unable to ascertain to what extent the increased phosphorylation status of the penultimate threonine could compensate for loss of an isoform, it is possible that this kinase-mediated mechanism allows plant tissues to maintain a wild type level of pump activity in each of the single mutants. Thus, to provide a more direct answer to the question of whether the mutant plants contain reduced catalytic activity, we measured the bathing medium pH during root growth. We used an indicator dye, fluorescein, to measure and compare pH changes in root rhizosphere from the wild type versus mutant plants. This assay has been used previously in higher plants as a measure of root surface pH (32). As shown in Fig. 5, we observed that the aha2 mutant plant was clearly impaired in proton-secreting activities, compared with wild type, and that the aha1 mutants appeared normal compared with wild type. These results reinforce our previous demonstrations that the aha2 mutants are more severe in their root growth defects compared with aha1 and, more importantly, are a direct demonstration that pump activity is impaired in aha2 mutants even under ideal laboratory conditions.
FIGURE 5.
Proton-secreting activity in roots of wild type and aha1 and aha2 mutants. Representative extracellular pH assay reflecting plasma membrane proton pump activity. pH of bathing media was quantified with fluorescein dye. n = 7; error bars indicate S.E. Col WT, wild type plant of A. thaliana, ecotype Columbia.
DISCUSSION
In this study, we characterize the function of the two most highly expressed members, AHA1 and AHA2, from the plasma membrane proton pump gene family, using Arabidopsis loss-of-function mutant plants. Our experiments demonstrate that although there is little consequence of impairing the function of either AHA1 or AHA2 in Arabidopsis plants grown under ideal conditions, the loss of both genes is absolutely lethal, blocking embryogenesis at an early stage of development. The successful complementation of the embryo lethality with either AHA1 or AHA2 transgenes proves that a single copy of either gene is equally sufficient to maintain the protonmotive force in these plants. Unlike work in animals with the Na+/K+-ATPase where there is a specific inhibitor available (33), there are no selective inhibitors for the plasma membrane proton pump. Thus, the loss-of-function mutants characterized here represent useful tools to investigate in planta roles of the proton pump. Consistent with the genetic experiments performed in other model organisms, our results demonstrate the prerequisite of the plasma membrane electrochemical gradient for life. Whereas a single gene deletion is sufficient to reveal the essential role of the primary active transport pumps in other organisms (Table 1), the necessity of deleting the two pump genes in Arabidopsis may indicate that this redundancy is a useful part of evolutionary adaptation of the higher plants as sessile multicellular organisms.
We further show that although plants harboring aha1 or aha2 single gene mutations do not compensate for the loss by increasing the expression of the other AHA members at the RNA or protein levels, there is a post-translational compensatory measure. Phosphorylation of the penultimate threonine in the remaining AHA1 protein in the aha1 mutant or that in remaining AHA2 proteins in aha2 mutant is significantly up-regulated. Phosphorylative compensation for the loss of one isoform in either aha1 or aha2 mutants, however, was observed only to a small extent. Despite this increase in the level of regulatory phosphorylation, aha2 mutants contain reduced levels of overall catalytic activity in their roots, as demonstrated directly by measurements of proton secretion and indirectly by impaired reaction to conditions that reduce the protonmotive force and place strain on the ability of the cell to maintain a sufficient driving force for transport-related growth requirements. The growth advantage that the aha2 mutants show over wild type in the presence of toxic cations like hygromycin, cesium, and lithium is also a clear indicator that the protonmotive force is impaired. The differential phenotypic responses in the aha1 and aha2 mutants to a variety of stresses are likely due to isoform-specific expression in root cells, because AHA2 is more highly expressed in the root epidermal cells compared with AHA1.
Finally, we report on a method using heavy isotope-labeled synthetic peptides corresponding to the phosphorylated and nonphosphorylated versions of the C-terminal tryptic fragment of the AHAs, in which selected reaction monitoring with a triple quadrupole mass spectrometer is used to measure the stoichiometry of phosphorylation of the pump. This method is amenable to high throughput use under a range of conditions and, together with transgene-mediated genetic complementation studies using the double mutant lines, will provide a new approach for understanding the in planta role played by the plasma membrane proton pump in higher plants.
Supplementary Material
Acknowledgments
We gratefully acknowledge Ed Huttlin and Gary Case for peptide designing and synthesis; Patrick Krysan for assistance with microscopy; David C. Nelson, Melissa LeBlanc, and Jeffrey Harper for constructive discussions; Fernando I. Rodríguez and Angie Bewell for technical assistance; and Wayne Davis and Sandra Splinter BonDurant for assistance with the microarray experiment.
This work was supported by grants from the Department of Energy and the National Science Foundation (to M. R. S.) and by National Science Foundation Grant MCB-0619736 (to M. S. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental sequences and additional references.
- M&S
- Murashige and Skoog
- MS
- mass spectrometry
- HPLC
- high pressure liquid chromatography
- RT
- reverse transcription
- WT
- wild type
- GUS
- β-glucuronidase.
REFERENCES
- 1.Lingrel J. B., Williams M. T., Vorhees C. V., Moseley A. E. (2007) J. Bioenerg. Biomembr. 39, 385–389 [DOI] [PubMed] [Google Scholar]
- 2.de Carvalho Aguiar P., Sweadner K. J., Penniston J. T., Zaremba J., Liu L., Caton M., Linazasoro G., Borg M., Tijssen M. A., Bressman S. B., Dobyns W. B., Brashear A., Ozelius L. J. (2004) Neuron 43, 169–175 [DOI] [PubMed] [Google Scholar]
- 3.Serrano R., Kielland-Brandt M. C., Fink G. R. (1986) Nature 319, 689–693 [DOI] [PubMed] [Google Scholar]
- 4.Robertson W. R., Clark K., Young J. C., Sussman M. R. (2004) Genetics 168, 1677–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis M. W., Somerville D., Lee R. Y., Lockery S., Avery L., Fambrough D. M. (1995) J. Neurosci. 15, 8408–8418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Y., Huynh L., Takeyasu K., Fambrough D. M. (1997) Genes Funct. 1, 99–117 [DOI] [PubMed] [Google Scholar]
- 7.Palladino M. J., Bower J. E., Kreber R., Ganetzky B. (2003) J. Neurosci. 23, 1276–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellertsdottir E., Ganz J., Dürr K., Loges N., Biemar F., Seifert F., Ettl A. K., Kramer-Zucker A. K., Nitschke R., Driever W. (2006) Dev. Dyn. 235, 1794–1808 [DOI] [PubMed] [Google Scholar]
- 9.Shu X., Cheng K., Patel N., Chen F., Joseph E., Tsai H. J., Chen J. N. (2003) Development. 130, 6165–6173 [DOI] [PubMed] [Google Scholar]
- 10.James P. F., Grupp I. L., Grupp G., Woo A. L., Askew G. R., Croyle M. L., Walsh R. A., Lingrel J. B. (1999) Mol. Cell 3, 555–563 [DOI] [PubMed] [Google Scholar]
- 11.Moseley A. E., Williams M. T., Schaefer T. L., Bohanan C. S., Neumann J. C., Behbehani M. M., Vorhees C. V., Lingrel J. B. (2007) J. Neurosci. 27, 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merlot S., Leonhardt N., Fenzi F., Valon C., Costa M., Piette L., Vavasseur A., Genty B., Boivin K., Müller A., Giraudat J., Leung J. (2007) EMBO J. 26, 3216–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao R., Dielen V., Kinet J. M., Boutry M. (2000) Plant Cell 12, 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gévaudant F., Duby G., von Stedingk E., Zhao R., Morsomme P., Boutry M. (2007) Plant Physiol. 144, 1763–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vitart V., Baxter I., Doerner P., Harper J. F. (2001) Plant J. 27, 191–201 [DOI] [PubMed] [Google Scholar]
- 16.Baxter I. R., Young J. C., Armstrong G., Foster N., Bogenschutz N., Cordova T., Peer W. A., Hazen S. P., Murphy A. S., Harper J. F. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2649–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santi S., Schmidt W. (2009) New Phytol. 183, 1072–1084 [DOI] [PubMed] [Google Scholar]
- 18.Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., Parker H., Prednis L., Ansari Y., Choy N., Deen H., Geralt M., Hazari N., Hom E., Karnes M., Mulholland C., Ndubaku R., Schmidt I., Guzman P., Aguilar-Henonin L., Schmid M., Weigel D., Carter D. E., Marchand T., Risseeuw E., Brogden D., Zeko A., Crosby W. L., Berry C. C., Ecker J. R. (2003) Science 301, 653–657 [DOI] [PubMed] [Google Scholar]
- 19.Krysan P. J., Young J. C., Tax F., Sussman M. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8145–8150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 21.Nelson C. J., Hegeman A. D., Harms A. C., Sussman M. R. (2006) Mol. Cell. Proteomics 5, 1382–1395 [DOI] [PubMed] [Google Scholar]
- 22.Larsson C., Sommarin M., Widell S. (1994) Methods Enzymol. 228, 451–469 [Google Scholar]
- 23.Wessel D., Flügge U. I. (1984) Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 24.Clough S. J., Bent A. F. (1998) Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 25.Baxter I., Tchieu J., Sussman M. R., Boutry M., Palmgren M. G., Gribskov M., Harper J. F., Axelsen K. B. (2003) Plant Physiol. 132, 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirsch R. E., Lewis B. D., Spalding E. P., Sussman M. R. (1998) Science 280, 918–921 [DOI] [PubMed] [Google Scholar]
- 27.Fuglsang A. T., Guo Y., Cuin T. A., Qiu Q., Song C., Kristiansen K. A., Bych K., Schulz A., Shabala S., Schumaker K. S., Palmgren M. G., Zhu J. K. (2007) Plant Cell 19, 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lecchi S., Nelson C. J., Allen K. E., Swaney D. L., Thompson K. L., Coon J. J., Sussman M. R., Slayman C. W. (2007) J. Biol. Chem. 282, 35471–35481 [DOI] [PubMed] [Google Scholar]
- 29.Ekberg K., Palmgren M. G., Veierskov B., Buch-Pedersen M. J. (2010) J. Biol. Chem. 285, 7344–7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harper J. F., Manney L., DeWitt N. D., Yoo M. H., Sussman M. R. (1990) J. Biol. Chem. 265, 13601–13608 [PubMed] [Google Scholar]
- 31.McCusker J. H., Perlin D. S., Haber J. E. (1987) Mol. Cell. Biol. 7, 4082–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monshausen G. B., Bibikova T. N., Messerli M. A., Shi C., Gilroy S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20996–21001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yatime L., Buch-Pedersen M. J., Musgaard M., Morth J. P., Lund Winther A. M., Pedersen B. P., Olesen C., Andersen J. P., Vilsen B., Schiøtt B., Palmgren M. G., Møller J. V., Nissen P., Fedosova N. (2009) Biochim. Biophys. Acta 1787, 207–220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.