Abstract
SNARE protein complexes are key mediators of exocytosis by juxtaposing opposing membranes, leading to membrane fusion. SNAREs generally consist of one or two core domains that can form a four-helix bundle with other SNARE core domains. Some SNAREs, such as syntaxin target-SNAREs and longin vesicular-SNAREs, have independent, folded N-terminal domains that can interact with their respective SNARE core domains and thereby affect the kinetics of SNARE complex formation. This autoinhibition mechanism is believed to regulate the role of the longin VAMP7/TI-VAMP in neuronal morphogenesis. Here we use nuclear magnetic resonance spectroscopy to study the longin-SNARE core domain interaction for VAMP7. Using complete backbone resonance assignments, chemical shift perturbations analysis, and hydrogen/deuterium exchange experiments, we conclusively show that VAMP7 adopts a preferentially closed conformation in solution. Taken together, the closed conformation of longins is conserved, in contrast to the syntaxin family of SNAREs for which mixtures of open and closed states have been observed. This may indicate different regulatory mechanisms for SNARE complexes containing syntaxins and longins, respectively.
Keywords: Membrane Fusion, Neurodevelopment, NMR, Protein Conformation, Vesicles, Longin, Synaptobrevin, Syntaxin
Introduction
The soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)3 complex is the centerpiece of all intracellular membrane fusion reactions. SNARE proteins are localized in opposing membranes, and upon docking, form a trans four-helix bundle. Assembly of this complex promotes membrane juxtaposition and fusion (1, 2). Each SNARE comprises of a characteristic SNARE core domain, a stretch of 60–70 amino acids defining all SNAREs as either Q-SNAREs or R-SNAREs (3–6). SNARE complex formation is a highly controlled process, involving additional regulatory proteins and domains. A major role in modulating SNARE function is provided by N-terminal regulatory protein domains. In particular, the R-SNARE longins and the Q-SNARE syntaxins possess ∼120-amino acid-long N-terminal extensions called longin domain (LD) and Habc, respectively. LD and Habc can fold back on the SNARE core domain and regulate it by inhibiting interactions with other SNARE core domains. The existence of this interaction does not prevent SNARE complex formation, but it affects the kinetics of the assembly (7). For syntaxin, a major role of this interaction is to provide exocytosis with a conformational switch that in combination with SM (Munc18/nSec1) proteins (8, 9) inhibits SNARE complex formation.
The energetics and kinetics of the syntaxin Habc-SNARE core domain interaction are not conserved despite the well conserved domain architecture of these molecules. Although neuronal Syntaxin 1A and yeast Sso1 have been observed in a closed conformation in vitro (i.e. with the N-terminal domain interacting with the SNARE core domain) (10, 11), other syntaxins, such as the endosomal Tlg2p and Pep12p (12) or the yeast, vacuolar Vam3p, are predominantly open (13). Furthermore, varying results have been obtained for the kinetics of neuronal Syntaxin 1A. A single-molecule fluorescence correlation spectroscopy study revealed open-closed conformational transitions with a preference for the open conformation (14), whereas an NMR study suggested a dominant, although unstable, closed conformation (15). In longins, the transmembrane-anchoring region and the SNARE core domain are preceded by the N-terminal LD, a globular fold consisting of five antiparallel β-strands sandwiched by two α-helices on one side and one α-helix on the other (β1β2α1β3β4β5α2α3 topology) (16, 17). The conservation of this domain in all eukaryotes and in several traffic-related protein families other than the SNAREs raises interesting questions on its functional versatility (18–22). Longins comprise three subfamilies of v-SNAREs: Ykt6p, Sec22b, and VAMP7, also known as tetanus insensitive-VAMP (TI-VAMP). The LD targets these SNAREs to their specific traffic networks, where they function in both anterograde and retrograde vesicle fusion events (23–25).
Ykt6 is involved in trafficking to Golgi, the endosome, and the vacuole. It was the first longin for which a NMR study reported an LD-SNARE core intramolecular interaction, decreasing the rate of ternary SNARE complex formation (26). Farnesylation stabilizes this interaction and locks cytosolic Ykt6 in a closed conformation, whereas further lipidation (i.e. palmitoylation) anchors the protein to the vesicle membrane and is suggested to favor the open conformation (27–30). Unlike Ykt6, VAMP7 and Sec22b are inserted in the membrane by a transmembrane domain, and it is unclear whether or not they adopt a closed conformation in their isolated form. A closed conformation has been suggested for Sec22b as a conformational epitope is induced in the LD α2-α3 interface by binding of the SNARE motif to α1-β3. However, in these experiments, Sec22b was in complex with Sec23/24 (24). A closed conformation can be hypothesized for VAMP7 as well because removal of the LD enhances SNARE complex formation both in vitro and in vivo (31). Endocytosis of VAMP7 is mediated by an interaction between its LD and a region of Hrb (25, 32, 33). The crystal structure (Protein Data Bank (PDB) ID 2VX8) of a recombinant chimera of VAMP7 LD fused with Hrb(136–176) shows that Hrb binds the LD and forms a closed conformation that is similar to that of Sec22b and Ykt6. Hrb competes with the VAMP7 SNARE core domain for binding to LD, suggesting a common binding mode also for the VAMP7 LD-SNARE core (32). In contrast to Ykt6, Sec22b, and VAMP7, a predominantly open conformation was found for the yeast-specific longin Nyv1p as no significant LD-SNARE core domain interaction was observed in NMR studies (34).
The biological role of the LD-SNARE core interaction of VAMP7 has been well established; it negatively regulates membrane fusion during neurite outgrowth in developing neurons and phagocytosis in macrophages, which require plasma membrane growth and remodeling, respectively (25, 35, 36). In contrast, the energetics and dynamics of this interaction are unknown. Here, we use NMR to conclusively show that the longin-SNARE VAMP7 adopts a predominantly closed conformation with remarkable stability. We then present a comparative perspective of the SNARE autoinhibition mechanism for longins and syntaxins.
EXPERIMENTAL PROCEDURES
cDNAs for Bacterial Expression
All cDNAs used for bacterial expression were generated by standard PCR-based techniques from a cDNA encoding full-length human VAMP7 (a kind gift from Maurizio D'Esposito, Institute of Genetics and Biophysics-Consiglio Nazionale delle Ricerche (IGB-CNR), Naples, Italy) and subcloned into pET47b (Novagen) between XmaI and XhoI restriction sites.
Protein Expression and Purification
All recombinant proteins were expressed in BL21(DE3) cells (Invitrogen) for 4 h at 29 °C after induction (A600 ∼ 0.7) with 0.3 mm isopropyl-1-thio-β-d-galactopyranoside at 37 °C. Cells were lysed by sonication in 50 mm Tris-HCl (pH 7.5), 500 mm NaCl, 1 mm dithiothreitol (buffer A), 0.05% Triton X-100, 10 mm imidazole, and protease inhibitors. Crude lysates were centrifuged for 40 min at 48,400 × g, and the supernatant was incubated with Ni2+-nitrilotriacetic acid-agarose beads (Qiagen) for 1 h at 4 °C. The beads were washed with buffer A with 20 mm imidazole and eluted with buffer A and 300 mm imidazole. The His6 tag was removed by PreScission protease (GE healthcare) for 60 h at 4 °C. Further purification was achieved by gel filtration on a Superdex 75 column (GE Healthcare) in 20 mm potassium phosphate (pH 6.5), 100 mm NaCl, 1 mm dithiothreitol. Protein purity and integrity were monitored by SDS-PAGE and mass spectrometry.
Light Scattering
Size exclusion multiangle laser light scattering was performed using a Shodex KW-802.5 column at a flow rate of 0.5 ml/min. Measurements were performed in 20 mm Tris-HCl (pH 7.0), 150 mm NaCl, and 1 mm tris(2-carboxyethyl)phosphine. The elution profile was monitored by UV absorption at 280 nm, light scattering at 690 nm, and differential refractometry using Dawn and OptiLab instruments (Wyatt Technology). Analyses were carried out using the Astra software. For each sample, 100 μl of protein at 3 mg/ml protein were loaded. The differential refractive index increment (dn/dc) value of 0.185 was used in all calculations.
Nuclear Magnetic Resonance Spectroscopy
Uniform 15N and 13C labeling in two combinations (only 15N label or both 15N and 13C) was accomplished by growing Escherichia coli BL21(DE3) in M9 minimal media with 15NH4Cl- and 13C6-labeled glucose as the sole nitrogen and carbon sources, respectively. All samples were prepared in 20 mm potassium phosphate buffer (pH 6.5) containing 100 mm NaCl and 1 mm dithiothreitol, unless otherwise indicated, using a 90:10 (v/v) H2O:D2O mixture. Protein concentrations were assessed by UV spectrophotometry at 280 nm and the Bradford assay (Bio-Rad). For 1H{15N}-HSQC experiments, we used: 18–450 μm VAMP7(1–118), 300–500 μm VAMP7(1–150), 60–320 μm VAMP7(1–160), 11–210 μm VAMP7(1–180)-L43S/Y45S, and 25–300 μm VAMP7(1–180). VAMP7(1–118) was concentrated using Amicon Ultra-5K nominal molecular weight limit devices (Millipore) to 45, 90, and 150 μm when incubated respectively with 100, 280, and 450 μm non-labeled VAMP7(119–180). The hydrogen/deuterium exchange experiments were performed as follows. Samples of VAMP7(1–118), VAMP7(1–150), and VAMP7(1–180) in protonated NMR buffer were lyophilized. The samples, in turn, were dissolved in equivalent volumes of D2O (200 μm final protein concentration), rapidly transferred to the spectrometer, and monitored by a series of fast 1H{15N}-HSQC experiments. The protein concentrations were 450 μm for all the 1H-15N-13C three-dimensional experiments. These experiments were performed using standard sensitivity-enhanced, pulsed-field gradient-based pulse sequences for double and triple resonance spectra (37) and included: three-dimensional 1H-15N NOESY-HSQC, HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, and C(CO)NH, all acquired on the uniformly 15N, 13C-labeled sample. All experiments were acquired on 600- or 800-MHz Varian UNITYINOVA spectrometers at 25 °C, unless otherwise specified. Both instruments were equipped with 5-mm H{CN} triple resonance, gradient probes. All the data were acquired using the VNMR software (Varian, Inc.) and analyzed with SPARKY (38).
RESULTS
Resonance Assignments and LD-SNARE Interaction Interface
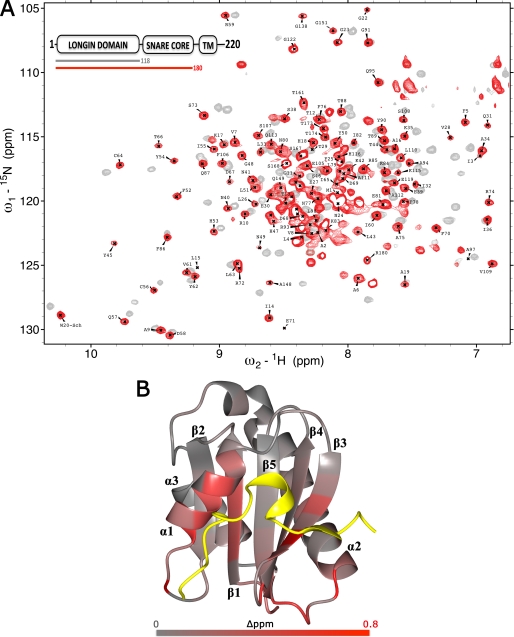
To analyze the structural properties of the VAMP7 LD-SNARE core domain interaction, we acquired, assigned, and compared the 1H{15N}-heteronuclear single-quantum coherence (HSQC) spectra of the isolated LD (VAMP7(1–118)) and of the entire cytoplasmic domain (1–180) of VAMP7. Both spectra showed good dispersion of N-H resonances, which is indicative of the heterogeneity of chemical environments characteristic for folded proteins (Fig. 1A). Many LD peaks show significant chemical shift perturbations in the presence of the SNARE core domain fragment. The change of chemical environment sensed by their nuclei is a direct evidence of the LD-SNARE interaction in VAMP7.
FIGURE 1.
LD-SNARE motif interaction in VAMP7. A, overlay of 1H, 15N-HSQC spectra of VAMP7(1–118) (gray) and VAMP7(1–180) (red). Only the assignments of the LD amide group of VAMP7(1–180) are shown (black labels). The color-coded bar diagram in the top left corner indicates the boundaries of the two constructs for which the spectra are shown with respect to the domain architecture of full-length VAMP7, residues 1–220: LD, SNARE core domain, and transmembrane domain (TM). B, summary of 1H and 15N combined chemical shift changes in the VAMP7 LD induced by the SNARE-motif. The combined 1H and 15N chemical shift changes are mapped on the schematic of the crystal structure of the VAMP7 LD bound to Hrb136–176 (yellow) (PDB ID 2VX8) (32) using a gray/red color scale. They are defined as Δppm = ((ΔδHN)2 + (ΔδN × αN)2)½, where ΔδHN and ΔδN are chemical shift differences of amide proton and nitrogen chemical shifts of the VAMP7 LD with and without the SNARE-motif. The scaling factor (αN) used to normalize the 1H and 15N chemical shifts is 0.17.
To fully interpret the HSQC spectrum we performed full backbone assignments of both constructs. We obtained uniformly 15N, 13C-labeled VAMP7(1–118) and carried out HNCACB, CBCA(CO)NH, C(CO)NH, and three-dimensional 1H-15N NOESY-HSQC triple resonance experiments. We unambiguously connected sequential Cα-Cβ resonances through the comparison of the HNCACB and CBCA(CO)NH spectra in combination with the analysis of side chain carbon resonances in the C(CO)NH (supplemental Fig. S1A). The 1H-15N NOESY-HSQC spectrum was used to review and confirm the assignment by verifying spatial proximity of N-H groups based on the structural information from the crystal structure of VAMP7 in complex with Hrb136–176 (PDB ID 2VX8) (32). All visible backbone resonances in the VAMP7(1–118) 1H{15N}-HSQC spectrum were assigned (supplemental Fig. S2A). Other than the 2 prolines (prolines do not include amides), only 7 residues of the LD were not detected. These residues are distributed in the α2-α3 loop and in the α3-helix. The absence of strong signals for these residues may be due to conformational exchange likely combined with LD-LD self-association. In support of this notion, we observed LD dimerization by size exclusion chromatography multiangle laser light scattering and concentration-dependent perturbations in the LD 1H{15N}-HSQC spectrum, which are consistent with the dimerization interface reported for Sec22b (24, 39) (supplemental Fig. S2). We then transferred all VAMP7(1–118) assignments to the 1H{15N}-HSQC spectrum of a uniformly 15N, 13C-labeled VAMP7(1–180) and acquired HNCA, CA(CO)NH, CBCA(CO)NH, and C(CO)NH triple resonance experiments (supplemental Fig. S1B). VAMP7(1–180) is solely monomeric, as supported by size exclusion chromatography (data not shown) and the lack of change in the 1H{15N}-HSQC spectra acquired in the concentration range from 25 to 750 μm.
We mapped the LD chemical shift perturbations observed between VAMP7(1–118) and VAMP7(1–180) onto the VAMP7 LD-Hrb136–176 crystal structure (PDB ID 2VX8) (Fig. 1B) (32). The largest perturbations for LD residues are found in the Hrb136–176-binding region. The most significant perturbations correspond to residues of the α1-β3 pocket, supporting a direct contact with the SNARE core domain with this pocket. Other chemical shift perturbations are observed in the β5-α2 loop and in the N-terminal half of α2, likely as a combined result of spatial proximity to the N terminus of the SNARE core domain and to β3-β4 backbone adjustment due to the SNARE core domain interaction with the α1-β3 pocket. Similarly, a backbone adjustment induced on α1 propagates through the central strand β1 (residues Phe-5, Ala-6, Val-7) to the α2-α3 pocket. The interaction between LD and the SNARE core domain is also consistent with the reported LD-SNARE motif interactions for Ykt6 and Sec22b (24, 26, 27).
VAMP7 Adopts a Closed Conformation
The sharp lines obtained in the HSQC spectra of both the LD (VAMP7(1–118)) and the entire cytoplasmic domain (1–180) of VAMP7 indicate homogeneous chemical environments sensed by the amide protons of these proteins and thus unique conformations. Together with the observed chemical shift perturbations in the presence of SNARE core domain, this observation suggests a predominantly closed state for VAMP7(1–180). To validate this hypothesis, we performed hydrogen/deuterium exchange experiments to compare the protection from exchange with solvent deuterons in VAMP7(1–118), VAMP7(1–150), and VAMP7(1–180). When compared with VAMP7(1–118), VAMP7(1–150) showed modest extra protection of Lys-35, Ile-36, Cys-56, Gln-57, and Arg-59 in the α1-β3 pocket and of Phe-76 and Phe-78 in the stretch of α2 likely facing the N-terminal portion of the SNARE core domain (data not shown). Remarkably, the VAMP7(1–180) construct exhibited a strong increase of protection for these residues as well as new protection for the α1-β3 residues Val-28, Gln-31, Ala-34, Ile-36, Leu-43, Thr-44, and Tyr-45 (Fig. 2) with the latter being protected over 150 min at 25 °C (data not shown). No other surfaces that were solvent-exposed in the isolated LD show any protection in VAMP7(1–180), supporting the notion that the C-terminal 20 residues of the SNARE core domain do not interact with the LD. To further validate that the LD-SNARE core domain interaction results in a stable, closed conformation, we mutated 2 key residues on the LD β3 strand, i.e. VAMP7(1–180)-L43S/Y45S. Residues Leu-43 and Tyr-45 (corresponding to Leu-86 and Tyr-88 in chain A of the VAMP7 LD-Hrb136–176 crystal structure) contribute the hydrophobic pocket on the surface of the LD that binds Hrb, and mutation of these residues into serines results in inhibition of the VAMP7-Hrb interaction (32). In agreement with the hypothesis that the SNARE core domain binds to the same surface region of the LD, residues Leu-43 and Tyr-45 have strong chemical shift changes (in Fig. 1B, they correspond to the red-labeled residues in the β3), and they are protected against solvent in the presence of the SNARE core domain (Fig. 2B). The L43S/Y45S mutation results in a reduction of the LD resonance perturbations observed for wild-type VAMP7 (Fig. 3A), confirming the key role of these residues in the LD-SNARE core domain interaction.
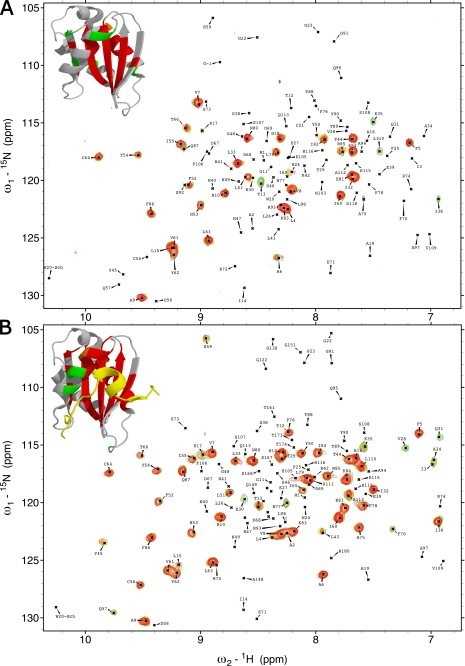
FIGURE 2.
Deuterium solvent-exchange protection in VAMP7(1–118) and VAMP7(1–180). A, overlay of 1H, 15N-HSQC spectra of VAMP7(1–118) after 1 min and 25 s (green), 6 min and 15 s (orange), and 11 and 5 min and 5 s (red) from resuspension in deuterated solvent. Top left corner, the detectable resonance peaks of these three spectra are mapped on the schematic of the LD-Hrb136–176 crystal structure (PDB ID 2VX8) (32) using the colors corresponding to the three spectra. The LD peaks that were not detected due to rapid deuterium exchange are colored gray. B, overlay of 1H, 15N-HSQC spectra of VAMP7(1–180) after 1 min and 20 s (green), 6 min and 5 s (orange), and 11 min (red) from resuspension in deuterated solvent. Top left corner, the detectable resonance peaks of these three spectra are colored in the LD-Hrb136–176 as described for panel A.
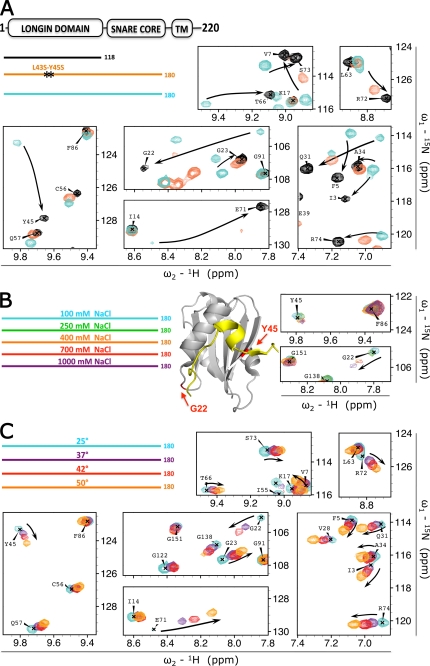
FIGURE 3.
Stability of the VAMP7 closed conformation. A, effect of different 15N-labeled recombinant constructs: VAMP7(1–118) (black), VAMP7(1–180)-L43S/Y45S (orange), and VAMP7(1–180) (cyan). The color-coded bar diagram in the top left corner indicates the boundaries of these three constructs for which the spectra are shown with respect to the domain architecture of full-length VAMP7, residues 1–220: LD, SNARE core domain, and transmembrane domain (TM). Black stars on the orange bar represent the L43S/Y45S point mutations. An overlay of the 1H{15N}-HSQC spectra of these proteins is shown with matching colors for seven representative regions of the spectra. Black arrows indicate resonance perturbation trends from closed to open conformation. B, effect of ionic strength on 15N-labeled VAMP7(1–180) in 100 mm NaCl (cyan), 250 mm NaCl (green), 400 mm NaCl (orange), 700 mm NaCl (red), and 1000 mm NaCl (purple) (legend in top left corner). Two red arrows on the ribbon diagram point to the position of residues Gly-22 and Tyr-45 in the LD (gray) as found in the crystal structure of VAMP7 LD in complex with the Hrb136–176 fragment of the protein (yellow) (PDB ID 2VX8) (32). The 1H{15N}-HSQC resonances for these 2 residues at the five saline conditions are superimposed and shown on the right with colors matching the legend. C, effect of temperature on 15N-labeled VAMP7(1–180). Spectra were acquired at 25 °C (cyan), 37 °C (purple), 42 °C (red), and 50 °C (orange) (legend in top left corner). An overlay of the 1H{15N}-HSQC spectra of VAMP7(1–180) at these temperatures is shown with the same spectral regions of panel A to illustrate the opposite shift perturbation trend (from closed toward open state) indicated by the black arrows.
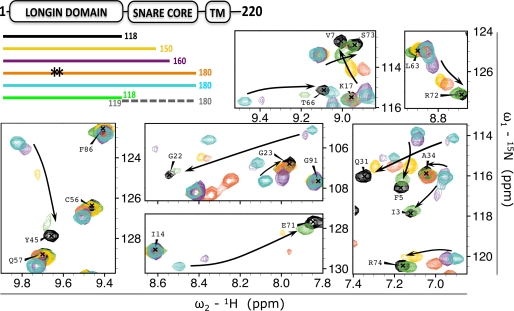
To determine the minimal LD-binding site, we designed two truncations, VAMP7(1–160) and VAMP7(1–150), corresponding to the same length of the LD-binding sites found in the crystal structures of VAMP7-Hrb and Sec22b (24, 32). We also tested the contribution of the covalent linker between LD and SNARE core domain by acquiring the 1H{15N}-HSQC spectrum of 15N-labeled LD with an excess of non-labeled SNARE core domain. With the exception of the VAMP7(1–160) construct, whose HSQC spectrum is essentially that of VAMP7(1–180), these constructs result in smaller chemical shift perturbations and peak broadening when compared with VAMP7(1–180) (Fig. 4), supporting fast exchange between open and closed states. Our data show that the native, full-length cytoplasmic domain of VAMP7 adopts a predominantly closed conformation that requires at least the N-terminal 160 residues of VAMP7 to be detectable and that VAMP7 is fully stabilized in the closed state by the C-terminal 20 residues preceding the transmembrane domain.
FIGURE 4.
Effect of sequence specificity, construct length, and covalent linkage on the LD-SNARE core interaction. The effect of truncation and mutation of 15N-labeled VAMP7 constructs on LD resonances is shown as follows: VAMP7(1–118) (black), VAMP7(1–150) (yellow), VAMP7(1–160) (purple), VAMP7(1–180)-L43S/Y45S (orange), VAMP7(1–180) (cyan), and VAMP7(1–118) (green) incubated with non-labeled VAMP7(119–180) (dashed gray line). The color-coded bar diagram in the top left corner indicates the boundaries of these six constructs for which the spectra are shown with respect to the domain architecture of full-length VAMP7, residues 1–220: LD, SNARE core domain, and transmembrane domain (TM). Black stars on the orange bar represent the L43S/Y45S mutation from the canonical amino acid sequence of VAMP7. An overlay of the 1H{15N}-HSQC spectra of these proteins is shown with matching colors in seven representative regions of the spectra (same regions shown in Fig. 3, A and C). The three panels on the left are shown with lower contour levels to display broadened resonances. Black arrows indicate the chemical shift perturbation trends from the closed to the open conformation.
VAMP7 adopts a stable, closed conformation without any known auxiliary proteins required to stabilize it. By contrast, a modest increase in ionic strength (150 mm NaCl) is sufficient to destabilize the closed conformation of Syntaxin 1A in the absence of Munc18 (15). Hence, we investigated the stability of the closed conformation of VAMP7 with respect to the ionic strength to compare these two systems. We titrated VAMP7(1–180) in 100, 250, 400, 700, and 1000 mm NaCl and recorded a 1H{15N}-HSQC spectrum for each condition. Intriguingly, none of these conditions achieved an opening of the closed conformation (Fig. 3B). Such resistance to ionic strength is likely due to the hydrophobic character of the α1-β3 pocket, in particular, residues Phe-25, Val-28, Ile-32, Leu-43, Tyr-45, and Phe-52.
We further investigated the stability of the closed conformation of VAMP7 by monitoring the effect of increasing temperature (25, 37, 42, and 50 °C) on the 1H{15N}-HSQC spectrum of VAMP7(1–180) (Fig. 3C). The increase of temperature causes a progressive although incomplete shift of the LD resonances toward their respective “open state” values (i.e. isolated LD), with the 37, 42, and 50 °C spectra representing a predominantly closed, an intermediate, and a predominantly open conformation, respectively. In analogy with the increase of ionic strength, the temperature effect is larger for Gly-22 than for other residues such as Tyr-45, which still shows interaction with the SNARE core domain at 50 °C. Taken together, VAMP7 adopts a stable, closed conformation in solution even in the absence of any known accessory proteins.
We observed spontaneous SNARE complex assembly during incubation of either VAMP7(1–160) or VAMP7(1–180) with SNAP25 and Syntaxin 1A in vitro (data not shown), consistent with a previous report (31). Our hydrogen/deuterium exchange experiments show poor protection for LD-α1 residues Val-28 and Gln-31 (Fig. 2B, green residues in the ribbon diagram) and no protection for Phe-25 and Gly-22, located in the N-terminal portion of α1 and the β2-α1 loop, respectively. This is consistent with higher sensitivity of Gly-22 to salt and temperature increases (Fig. 3, B and C). We speculate that the C-terminal LD-binding site would therefore be available for initiating SNARE complex assembly. Once interactions with the other SNAREs have been initiated, the remainder of the VAMP7 SNARE core domain will then participate in the complex assembly as a result of local conformational fluctuations.
DISCUSSION
The longin v-SNAREs Ykt6, VAMP7, and Sec22b are characterized by an autoinhibitory intramolecular interaction between the LD and the SNARE core domain. In Ykt6, lipidation contributes to regulate this interaction by stabilizing a cytosolic closed conformation and likely allowing opening transitions upon anchoring to the membrane (27, 29, 30). Instead, Sec22b and VAMP7 are permanently inserted in the membrane through a transmembrane region (24, 30, 32, 39).
In this study, we conclusively show that VAMP7 adopts a stable, closed conformation in solution. We also show how its stability depends on the full extent of the SNARE core domain, although not all residues of the SNARE core domain contribute equally to the interaction. Although lipidation provides cytosolic Ykt6 with a larger interaction surface between the LD and the SNARE core, we show here that a stable, dominantly closed conformation is also possible for transmembrane longin v-SNAREs in the absence of lipidation; unlike what was shown for Ykt6 (30), we do not observe dodecylphosphocholine binding to VAMP7 (data not shown). This observation is consistent with the hypothesis that cytosolic longin v-SNAREs require a stricter autoinhibition than membrane-bound ones to prevent unspecific SNARE core pairing (30). Available data are consistent with the conservation of a closed conformation of longins (with the exception of the Saccharomyces-specific Nyv1p) (34). Together with previous studies of syntaxins, we can compare the two different autoinhibitory SNAREs. Despite the conceptual analogy of open and closed states, these two SNAREs differ in terms of conservation and stability of the closed conformation. In contrast with longins, the existence of a closed conformation in solution is not conserved in syntaxins (10–13). Additionally, in a subset of syntaxins that adopt a closed conformation, the closed conformation is only marginally stable and can be easily disrupted, e.g. by favoring intermolecular SNARE core pairing at higher ionic strength (15). The sensitivity to solution conditions and the intrinsic flexibility of the syntaxin C terminus likely explain the differences in energetics and kinetics between open and closed states obtained in previous fluorescence correlation spectroscopy and NMR studies (14, 15). Syntaxins require accessory SM proteins to be held in an “arrested” closed state that inhibits SNARE complex formation.
In vitro, VAMP7 can form SNARE complex with the specific target-SNAREs. The higher stability of the closed state for VAMP7 when compared with syntaxins poses the question of whether longins require accessory proteins to further stabilize an arrested state to inhibit SNARE complex formation in vivo or whether the LD is sufficient to provide a similar inhibiting role to syntaxins, even without additional SM proteins. Although the interaction between LD and SNARE core might be sufficient to modulate SNARE complex assembly, we cannot exclude that an unknown protein factor destabilizes this interaction to achieve the timely SNARE complex assembly in vivo as suggested (35). It is also possible that SNARE assembly regulation is entirely different for SNARE complexes involving VAMP7. Our results have provided the molecular foundation to address these questions in future experiments.
Supplementary Material
Acknowledgment
We thank Thierry Galli for critical reading.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1-MH63105 (to A. T. B.). This work was also supported by grants from the MIUR-PRIN2005 and the Padua University (PRAT2007 Project CPDA077345/07) (to F. F.).
The HSQC spectra of VAMP7(1–118) and VAMP7(1–180) have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) with accession numbers 16866 and 16867, respectively.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- SNARE
- soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- v-SNARE
- vesicular-SNARE
- VAMP
- vesicle-associated membrane protein
- LD
- longin domain
- HSQC
- heteronuclear single quantum correlation
- NOESY
- nuclear Overhauser effect spectroscopy.
REFERENCES
- 1.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. (1998) Nature 395, 347–353 [DOI] [PubMed] [Google Scholar]
- 2.Liu W., Parpura V. (2009) Front. Neuroenergetics 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fasshauer D., Sutton R. B., Brunger A. T., Jahn R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15781–15786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunger A. T. (2005) Q. Rev. Biophys. 38, 1–47 [DOI] [PubMed] [Google Scholar]
- 5.Jahn R., Scheller R. H. (2006) Nat. Rev. Mol. Cell Biol. 7, 631–643 [DOI] [PubMed] [Google Scholar]
- 6.Wickner W., Schekman R. (2008) Nat. Struct. Mol. Biol. 15, 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fasshauer D., Antonin W., Subramaniam V., Jahn R. (2002) Nat. Struct. Biol. 9, 144–151 [DOI] [PubMed] [Google Scholar]
- 8.Burkhardt P., Hattendorf D. A., Weis W. I., Fasshauer D. (2008) EMBO J. 27, 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerber S. H., Rah J. C., Min S. W., Liu X., de Wit H., Dulubova I., Meyer A. C., Rizo J., Arancillo M., Hammer R. E., Verhage M., Rosenmund C., Südhof T. C. (2008) Science 321, 1507–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholson K. L., Munson M., Miller R. B., Filip T. J., Fairman R., Hughson F. M. (1998) Nat. Struct. Biol. 5, 793–802 [DOI] [PubMed] [Google Scholar]
- 11.Fiebig K. M., Rice L. M., Pollock E., Brunger A. T. (1999) Nat. Struct. Biol. 6, 117–123 [DOI] [PubMed] [Google Scholar]
- 12.Dulubova I., Yamaguchi T., Gao Y., Min S. W., Huryeva I., Südhof T. C., Rizo J. (2002) EMBO J. 21, 3620–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dulubova I., Yamaguchi T., Wang Y., Südhof T. C., Rizo J. (2001) Nat. Struct. Biol. 8, 258–264 [DOI] [PubMed] [Google Scholar]
- 14.Margittai M., Widengren J., Schweinberger E., Schröder G. F., Felekyan S., Haustein E., König M., Fasshauer D., Grubmüller H., Jahn R., Seidel C. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15516–15521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X., Lu J., Dulubova I., Rizo J. (2008) J. Biomol. NMR 41, 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filippini F., Rossi V., Galli T., Budillon A., D'Urso M., D'Esposito M. (2001) Trends Biochem. Sci 26, 407–409 [DOI] [PubMed] [Google Scholar]
- 17.Rossi V., Banfield D. K., Vacca M., Dietrich L. E., Ungermann C., D'Esposito M., Galli T., Filippini F. (2004) Trends Biochem. Sci 29, 682–688 [DOI] [PubMed] [Google Scholar]
- 18.Collins B. M., McCoy A. J., Kent H. M., Evans P. R., Owen D. J. (2002) Cell 109, 523–535 [DOI] [PubMed] [Google Scholar]
- 19.Kinch L. N., Grishin N. V. (2006) Protein Sci. 15, 2669–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlenker O., Hendricks A., Sinning I., Wild K. (2006) J. Biol. Chem. 281, 8898–8906 [DOI] [PubMed] [Google Scholar]
- 21.Kim Y. G., Raunser S., Munger C., Wagner J., Song Y. L., Cygler M., Walz T., Oh B. H., Sacher M. (2006) Cell 127, 817–830 [DOI] [PubMed] [Google Scholar]
- 22.Vedovato M., Rossi V., Dacks J. B., Filippini F. (2009) BMC Genomics 10, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meiringer C. T., Auffarth K., Hou H., Ungermann C. (2008) Traffic 9, 1510–1521 [DOI] [PubMed] [Google Scholar]
- 24.Mancias J. D., Goldberg J. (2007) Mol. Cell 26, 403–414 [DOI] [PubMed] [Google Scholar]
- 25.Chaineau M., Danglot L., Galli T. (2009) FEBS Lett. 583, 3817–3826 [DOI] [PubMed] [Google Scholar]
- 26.Tochio H., Tsui M. M., Banfield D. K., Zhang M. (2001) Science 293, 698–702 [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa H., Yang Z., Oltedal L., Davanger S., Hay J. C. (2004) J. Cell Sci. 117, 4495–4508 [DOI] [PubMed] [Google Scholar]
- 28.Hasegawa H., Zinsser S., Rhee Y., Vik-Mo E. O., Davanger S., Hay J. C. (2003) Mol. Biol. Cell 14, 698–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pylypenko O., Schönichen A., Ludwig D., Ungermann C., Goody R. S., Rak A., Geyer M. (2008) J. Mol. Biol. 377, 1334–1345 [DOI] [PubMed] [Google Scholar]
- 30.Wen W., Yu J., Pan L., Wei Z., Weng J., Wang W., Ong Y. S., Tran T. H., Hong W., Zhang M. (2010) Mol. Cell 37, 383–395 [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Arca S., Rudge R., Vacca M., Raposo G., Camonis J., Proux-Gillardeaux V., Daviet L., Formstecher E., Hamburger A., Filippini F., D'Esposito M., Galli T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9011–9016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pryor P. R., Jackson L., Gray S. R., Edeling M. A., Thompson A., Sanderson C. M., Evans P. R., Owen D. J., Luzio J. P. (2008) Cell 134, 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaineau M., Danglot L., Proux-Gillardeaux V., Galli T. (2008) J. Biol. Chem. 283, 34365–34373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen W., Chen L., Wu H., Sun X., Zhang M., Banfield D. K. (2006) Mol. Biol. Cell 17, 4282–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez-Arca S., Alberts P., Zahraoui A., Louvard D., Galli T. (2000) J. Cell Biol. 149, 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braun V., Fraisier V., Raposo G., Hurbain I., Sibarita J. B., Chavrier P., Galli T., Niedergang F. (2004) EMBO J. 23, 4166–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang O., Kay L. E., Olivier J. P., Forman-Kay J. D. (1994) J. Biomol. NMR 4, 845–858 [DOI] [PubMed] [Google Scholar]
- 38.Goddard T. D., Kneller D. G.SPARKY 3 (2007) University of California, San Francisco, CA [Google Scholar]
- 39.Gonzalez L. C., Jr., Weis W. I., Scheller R. H. (2001) J. Biol. Chem. 276, 24203–24211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.