Abstract
Purpose of Review
A systematic approach to studying gene-environment interaction can have immediate impact on our understanding of how environmental factors induce developmental disease and toxicity and provide biological insight for potential treatment and prevention measures.
Recent Findings
Because DNA sequence is static, genetic studies typically are not conducted prospectively. This limits the ability to incorporate environmental data into an analysis, as such data is usually collected cross-sectionally. Prospective environmental data collection could account for the role of critical windows of susceptibility that likely corresponds to the expression of specific genes and gene pathways. The use of large scale genomic platforms to discover genetic variants that modify environmental exposure in conjunction with a priori planned replication studies would reduce the number of false positive results.
Summary
Using a genome-wide approach, combined with a prospective longitudinal of environmental exposure at critical developmental windows is the optimal design for gene-environment interaction research. This approach would discover susceptibility variants, then validate the findings in an independent sample of children. Designs which combine the strengths and methodologies of each field will yield data which can account for both genetic variability and the role of critical developmental windows in the etiology of childhood disease and development.
Keywords: genetics, pediatrics, prenatal environment
Introduction and Purpose of Review
Rapid advances in genetic technology allow the measurement of genomic variation. However, with only a few exceptions, our genetic makeup does solely not determine disease or even health. It only represents our potential for each. The promise of the Human Genome Project to prevent or treat disease will not be fulfilled until the interactions between our environment and our genetic makeup are understood. This review provides a framework to study and understand the biology underlying gene-environment interactions. Environment, in this context, can be defined broadly and include nutritions, chemical and physical exposures, as well as social factors. The principles described, could apply to any childhood disease or developmental issue. The main thrust of this paper is to highlight the inherent differences in approach used by geneticists and environmental scientists in the study of childhood diseases and to propose a framework in which the 2 fields can be combined to optimize the strengths of each.
Recent Findings
Disease and development are the sum of both genetic and environmental factors.[1] Rothman and Greenland, in fact, have argued that all diseases are 100% genetic and 100% environmental.[1] While this may seem incongruous at first, the principle can be understood if we accept that the effects of each are not independent but are instead interactive. If one accepts this premise, a case can be made that even extreme examples have root causes that are both genetic and environmental. For example, death from a gunshot wound to the chest may seem to be 100% environmental, but genetic factors may have predisposed to behaviors that led to the shooting, or may have determined the extent of bleeding from the wound, or the ability of the cardiovascular system to maintain blood pressure long enough to survive transport to a hospital. If we count these genetic factors as contributing to death, then what percent of death is due to environment and what percent due to genetics? Likewise, diseases we believe are purely genetic have environmental components. Hemochromatosis is a genetic disease of excess iron absorption leading to heart disease, liver disease and diabetes. It is not difficult to imagine, however, that a vegan lifestyle would likely mitigate the role of any genetic predisposition to absorb iron, thereby preventing the disease. If one accepts that disease and development are multi-factorial, and that these factors are not independent, one can view development or disease causation, as a set ofinteracting causal components changing over time.1 If we understand how genetics and environment interact, we can understand the causative and biological mechanisms behind our observations.
Development, the environment and genetic susceptibility: the role of timing
Fetal life and early childhood appear to be life stages that are critical for programming health effects which manifest years later. There are likely multiple developmental life stages that are sensitive to certain environmental factors because many genes are only expressed during specific developmental stages and are subsequently turned off. Growth factors, in particular, are activated in childhood and limited in expression thereafter. Environmental exposures occurring during different life stages will have different sets of expressed gene products with which to interact. Therefore, interactions likely depend on the life stage at which exposure occurs. For example, growing evidence from animal research indicates that the CNS is highly vulnerable to chemical injury during development.[2],[3] This is due to interference with processes critical to neurodevelopment such as neuronal growth, synaptic network formation, neuronal migration,and development of receptor numbers. These processes are most active in childhood and subside during adult life. If toxic exposures impact these processes, they can alter the trajectory of brain development, but would be less toxic once development is complete. Differential toxicity in children vs adults has been shown for many chemicals including methyl mercury,[4, 5] PCBs,[6] organophosphate pesticides,[7] and lead.[8, 9] Genetic variants that produce gene-environment interactions may only do so when the exposure timing corresponds to a critical developmental window during which the gene is highly expressed. A design that cannot address the timing of environmental exposures cannot properly assess gene-environment interactions. This issue is important because understanding how certain individuals are genetically predisposed to adverse outcomes following toxic exposures is the key to understanding the mechanisms of action for chemicals and toxicants in humans and to creating effective interventions for prevention and treatment of toxic exposures.[10]
Genetics as a predictor of disease and development
With the advent of fixed content and customizable high density genotyping techniques, investigators can now screen thousands of genetic variants in population based studies. Despite these advances the predictive value of common genetic variants for human health and disease has been modest. Although the complexity of multi-gene diseases may partly explain this, the primary reason the predictive power of genetic factors is low is because few studies have addressed gene-environment interactions. Incorporating environmental factors into genetic studies of complex disease will not be simple and could even cause conflict with respect to how to design and conduct these studies. For example, taking advantage of the static nature of DNA sequence, genetic main effects are typically studied by first identifying subjects with the disease. Genetic studies will typically employ case-control designs, comparing DNA sequence between groups of individuals with a disease to a control group. Because DNA sequence is unchanged from birth till death, geneticists can be confident that genetic variants existed prior to the disease expression. There is no incentive to conduct a genetic association study prospectively. Even family based designs identify subjects based on disease status. However, environmental exposures vary over time. This makes case-control or family based designs impractical to study environmental toxicants, as to do so would mean the environmental exposure is only measured cross-sectionally. For this reason, case control designs are limited in their ability to detect gene environment interactions. Yet, the vast majority of genetic association studies (including those that test for gene-environment interaction) are case-control designs.
The prospective cohort design can address many of the limitations of case control studies. Recently, a series of articles have highlighted both the paucity of genetic data collected thus far in prospective cohorts and the advantages of a prospective cohort design for gene-environment interaction studies. Advocates of this approach include the former director of the National Human Genome Research Institute and current NIH director, Dr. Francis Collins.[11-15] Although cohort studies require long follow-up and are costly, they have important strengths in characterizing exposures and risk factors before phenotype onset, which reduces important biases common in case control studies.
Also, as previously explained, substantial evidence shows that particular life stages, specifically the in utero and early life windows, may be more sensitive to chemical exposures than other life stages. Because environmental exposures cannot be accurately reconstructed retrospectively, prospective environmental data collection is critical to understanding the relationship between environment and phenotype. What has not been previously studied is whether gene-environment interactions occur only when exposure is experienced during these developmental windows. Therefore, the best method for studying developmental disorders due to metal toxicity would be to combine high throughput genetic methods with a prospective, longitudinal birth cohort where exposures have been measured for each developmental window.
Genome wide approach versus candidate gene approach in Gene-environment Interaction Research
We noted previously that geneticists typically do not account for critical developmental windows in their study designs; but environmental scientists also do not adequately address genetic factors in their designs. Very few environmental scientists have addressed the role of genetic interactions with environmental factors prospectively or on a genomic scale. In studies in which environmental factors are measured prospectively, measures of genetic variation in response to a toxic agent typically focus on a few candidate genes. The candidate genes will typically be culled from previous basic science work on that agent. While there are strengths to such an approach, including biologic plausibility and clear a priori hypotheses, there are also limitations to selecting only a few SNPs for a study.[16] The foremost limitation is that such work can only validate previous research and cannot discover new information regarding the environmental factor’s toxicity. Also, given the multifaceted nature of biologic interactions, it is difficult to conceive that one SNP from one gene can fully account for the complexity of an entire biologic pathway. As previously noted, there may be genes or SNPs that have important roles in chemical pathogenesis or neurodevelopment that are not yet known, and a candidate approach would not be able to identify such genes. The selection of SNPs in a candidate approach is therefore always potentially open to bias.
A newer approach is to use genome wide scans, which allow for screening of the genome in an unbiased manner with respect to genetic risks factors.[17] However, such an approach also has strengths and weaknesses. The greatest strength is that it allows new biological relationships to be discovered. The primary weakness is that the vast majority of positive findings are false positives, due to the nature of testing hundreds of thousands of comparisons. Nonetheless, the field of gene-environment interaction research would profit from the use of a genome wide approach to identify genetic factors that modify the effects of environment. Such an approach would be a paradigm shift in environmental health away from hypothesis-driven research to a hypothesis-generating research.
Discovery and Validation / Replication
A primary technique to limit false positive findings in genomics is to plan replication of the results a priori. Using genomic platforms to test for interactions with an environmental factor will generate thousands of false positive interactions. If 1 million SNPs are tested, then 50,000 are likely to be significant merely by chance. How then can researchers determine which significant result is real? Statistical analysis addressing multiple comparisons is a common tool and a critically needed method by which to address this issue, but such methods assume that biological causation correlates with statistical associations. Such an assumption is unlikely to be correct. The simple ranking of gene-environment interactions, would treat a gene that may have greater biological plausibility a priori as equal to all other genes, even genes that may not be expressed in the target tissue. A challenging problem after genome wide genotyping is to balance the statistical evidence of gene-environment interactions with a priori evidence of biological plausibility. To better address this issue of multiple comparisons, genetic epidemiologists typically plan a priori replication studies in independent populations. That is, statistical associations are first “discovered” in one population using genome wide methods and then the most significant associations are then “tested” a 2nd or even 3rd time in separate, independent populations. This will greatly reduce the number of false positives, as results have to be consistent at each stage and in each population. This repeating or validation of findings brings biology into the results as replication is a hallmark of a true biological finding. However, choosing which SNPs to replicate may not be clear and the number of SNPs to genotype in replication may be limited by financial resources. Recently, the incorporation of previously established biological information into the Discovery phase analysis has been proposed as an additional adjunct means to further reduce the false positive rate.[18-20] These methods systematically prioritize SNPs based on known biological plausibility in a genomic statistical analysis. This approach has been shown to reduce the false positive rate of genome wide association studies when compared to standard statistical rankings.[18]
Going from Genome Wide Gene-Environment Interaction to Identifying the Functional Variant Fixed SNP arrays do not directly genotype the causal SNP. Instead the goal is to discover a genetic region where the functional SNP is found. Actually finding the causative genetic variant requires additional work even after the results are replicated in independent populations. The phenomena of linkage disequilibrium allows researchers to go from the results of fixed SNP arrays to potentially functional SNPs. Linkage disequilibrium (LD) refers to non-random associations between polymorphisms at different loci. Two variants with high LD will tend to be inherited together and one variant will effectively “tag” the other. The details of the methodology for this process are beyond the scope of this review, but have been facilitated in large part by the the International HapMap project.[21-23] Finding the true functional genetic variant typically involves fine gene mapping to identify variants that may be in linkage disequilibrium[24] with the markers genotyped in fixed SNP microarrays. In fine mapping, the genes in that region are identified and sequenced to determine their SNPs. These SNPs are then tested for association based on proximity to statistically significant SNPs in the replication phase and their functional properties are ultimately tested.
Population Stratification
A potential problem with genetic epidemiologic studies is that population stratification —systematic differences in ancestry among study subjects—can result in spurious associations or disguise genuine associations.[25, 26] Such effects could also impact gene-environment interactions and are identical to confounding by ethnicity. For example, the prevalence of genetic variants correlates with ethnicity, but most variants have no functional role and only mark one’s ethnic background. A cultural trait that also marks ethnicity, such as diet, might be the true causal factor in a study. If diet and genetics are highly correlated, and diet causes a particular disease, then genetic factors may show associations with that trait, when in fact the dietary factors are the true underlying cause.[27-29] The risk of population stratification can be minimized by including ancestry-informative markers which are SNPs that infer genetic ancestry and correct for stratification. Unless the population contains no variation in ethnicity, all genetic association analyses should incorporate an appropriate adjustment for ancestry.
Sample Size
We did not address the role of sample size in this paper. Interactions always require larger sample sizes than studies of main effects. In addition, the use of cohort designs will limit the ability to study qualitative traits (diabetes, autism) and would favor quantitative traits (serum glucose, IQ/behavioral tests).
Very large studies, such as the proposed National Children’s Study, would allow for some common disease traits to be addressed using this framework, but for many diseases case-control designs will still be the primary design of choice despite their limitations. Nonetheless, there are traits that can be addressed using our proposed framework and we would urge that whenever feasible, prospective designs are used to study childhood health outcomes.
Conclusion
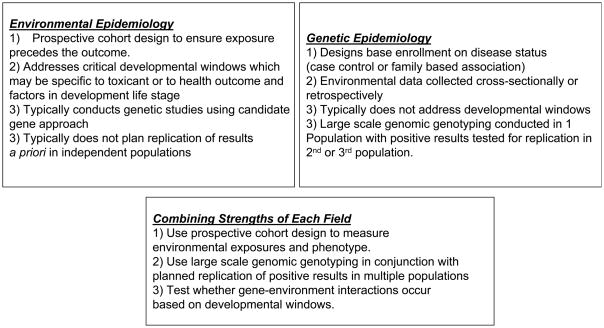
An ideal gene-environment interaction study will combine methods used in genetics (high-density genotyping, planned replication of results) with methods used in environmental epidemiology (prospective-longitudinal cohorts measuring critical developmental windows). (Summarized in Figure 1) Studies of gene-environment interaction must consider critical developmental windows which call for longitudinal designs. This ensures that environmental exposures precede the phenotype and that periods during which critical genes are expressed are captured. The rationale is that a combination of principles unique to each field is needed to effectively study both genetics and the environment simultaneously. A two-stage study design, that is, a discovery phase followed by a validation/replication phase and the collection of longitudinal exposure are the critical design features.
Figure 1. Integration of Principles of Genetic and Environmental Epidemiology.
Summary of common design approaches in Environmental Health and Genetics and summary of proposed combined approach.
Acknowledgments
Funded in part by the National Institutes of Health: R01 ES014930, R01 ES013744, P30 ES00002; R01ES015533.
References
- 1.Rothman K, Greenland S. Causation and causal inference in epidemiology. Am J Public Health. 2005;95(Suppl 1):S144–50. doi: 10.2105/AJPH.2004.059204. [DOI] [PubMed] [Google Scholar]
- 2.Rodier PM. Environmental causes of central nervous system maldevelopment. Pediatrics. 2004 Apr;113(4 Suppl):1076–83. [PubMed] [Google Scholar]
- 3.Faustman EM, Silbernagel SM, Fenske RA, et al. Mechanisms underlying Children's susceptibility to environmental toxicants. Environ Health Perspect. 2000 Mar;108( Suppl 1):13–21. doi: 10.1289/ehp.00108s113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amin-Zaki L, Elhassani S, Majeed MA, et al. Intra-uterine Methylmercury Poisoning in Iraq. Pediatrics. 1974 November 1;54(5):587–95. [PubMed] [Google Scholar]
- 5.Marsh DO, Myers GJ, Clarkson TW, et al. Fetal methylmercury poisoning: Clinical and toxicological data on 29 cases. Annals of Neurology. 1980;7(4):348–53. doi: 10.1002/ana.410070412. [DOI] [PubMed] [Google Scholar]
- 6.Tilson HA, Jacobson JL, Rogan WJ. Polychlorinated biphenyls and the developing nervous system: Cross-species comparisons. Neurotoxicology and Teratology. 1990;12(3):239–48. doi: 10.1016/0892-0362(90)90095-t. [DOI] [PubMed] [Google Scholar]
- 7.Engel SM, Berkowitz GS, Barr DB, et al. Prenatal organophosphate metabolite and organochlorine levels and performance on the Brazelton Neonatal Behavioral Assessment Scale in a multiethnic pregnancy cohort. Am J Epidemiol. 2007 Jun 15;165(12):1397–404. doi: 10.1093/aje/kwm029. [DOI] [PubMed] [Google Scholar]
- 8.Bellinger D, Leviton A, Waternaux C, et al. Longitudinal analyses of prenatal and postnatal lead exposure and early cognitive development. N Engl J Med. 1987 Apr 23;316(17):1037–43. doi: 10.1056/NEJM198704233161701. [DOI] [PubMed] [Google Scholar]
- 9.Schnaas L, Rothenberg SJ, Flores MF, et al. Reduced intellectual development in children with prenatal lead exposure. Environ Health Perspect. 2006 May;114(5):791–7. doi: 10.1289/ehp.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsuang M, Stone W, Faraone S. Genes, environment and schizophrenia. Br J Psychiatry Suppl. 2001;40:18–24. doi: 10.1192/bjp.178.40.s18. [DOI] [PubMed] [Google Scholar]
- 11 *.Collins FS. The case for a US prospective cohort study of genes and environment. Nature. 2004 May 27;429(6990):475–7. doi: 10.1038/nature02628. One of the earliest calls for using prospective data in gene-environment interaction research in place of case-control studies. [DOI] [PubMed] [Google Scholar]
- 12.Manolio TA, Bailey-Wilson JE, Collins FS. Genes, environment and the value of prospective cohort studies. 2006;7(10):812–20. doi: 10.1038/nrg1919. 2006/10//print. [DOI] [PubMed] [Google Scholar]
- 13 *.Davis RL, Khoury MJ. The emergence of biobanks: practical design considerations for large population-based studies of gene-environment interactions. Community Genet. 2007;10(3):181–5. doi: 10.1159/000101760. This paper highlights the potential value of prospective cohort designs in genetics and argues for the merging whenever possible of existing cohorts with biobanks to reduce costs. [DOI] [PubMed] [Google Scholar]
- 14.Manolio TA, Collins FS. Genes, environment, health, and disease: facing up to complexity. Hum Hered. 2007;63(2):63–6. doi: 10.1159/000099178. [DOI] [PubMed] [Google Scholar]
- 15.Yoo KY, Shin HR, Chang SH, et al. Genomic epidemiology cohorts in Korea: present and the future. Asian Pac J Cancer Prev. 2005 Jul-Sep;6(3):238–43. [PubMed] [Google Scholar]
- 16.Rebbeck TR, Spitz M, Wu X. Assessing the function of genetic variants in candidate gene association studies. Nat Rev Genet. 2004 Aug;5(8):589–97. doi: 10.1038/nrg1403. [DOI] [PubMed] [Google Scholar]
- 17.Kronenberg F. Genome-wide association studies in aging-related processes such as diabetes mellitus, atherosclerosis and cancer. Exp Gerontol. 2008 Jan;43(1):39–43. doi: 10.1016/j.exger.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 18 **.Li C, Li M, Lange EM, et al. Prioritized subset analysis: improving power in genome-wide association studies. Hum Hered. 2008;65(3):129–41. doi: 10.1159/000109730. This paper illustrates that the incorporation of prior biological knowledge into genome wide scans will increase the power for replicating results by comparing methods which use purely statistical rankings to methods that employ statistical weighting of SNPs based on prior biological knowledge. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saccone SF, Saccone NL, Swan GE, et al. Systematic biological prioritization after a genome-wide association study: an application to nicotine dependence. Bioinformatics. 2008 Aug 15;24(16):1805–11. doi: 10.1093/bioinformatics/btn315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roeder K, Bacanu SA, Wasserman L, et al. Using linkage genome scans to improve power of association in genome scans. Am J Hum Genet. 2006 Feb;78(2):243–52. doi: 10.1086/500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The International HapMap Project. The International HapMap Project. Nature. 2003 Dec 18;426(6968):789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 22.The International Hapmap Consortium 2005. A haplotype map of the human genome. Nature. 2005 Oct 27;437(7063):1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007 Oct 18;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005 Apr 15;308(5720):419–21. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 25.Campbell CD, Ogburn EL, Lunetta KL, et al. Demonstrating stratification in a European American population. Nat Genet. 2005 Aug;37(8):868–72. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- 26.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006 Aug;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 27.Choudhry S, Coyle NE, Tang H, et al. Population stratification confounds genetic association studies among Latinos. Hum Genet. 2006 Jan;118(5):652–64. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Marignac VL, Valladares A, Cameron E, et al. Admixture in Mexico City: implications for admixture mapping of type 2 diabetes genetic risk factors. Hum Genet. 2007 Feb;120(6):807–19. doi: 10.1007/s00439-006-0273-3. [DOI] [PubMed] [Google Scholar]
- 29.Price AL, Patterson N, Yu F, et al. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007 Jun;80(6):1024–36. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]