Abstract
MicroRNAs (miRNAs) have emerged as important modulators in development, tissue homeostasis, and diseases. In this issue of Genes & Development, Miyaki and colleagues (pp. 1173–1185) report that miR-140 is involved in the pathogenesis of osteoarthritis by regulating, at least in part, ADAMTS5. Moreover, mice lacking miR-140 are dwarf as a consequence of impaired chondrocyte proliferation. This study is the first in vivo demonstration that miR-140 has a critical and nonredundant role in cartilage development and homeostasis.
Keywords: MicroRNA, miR-140, osteoarthritis, cartilage, Adamts-5
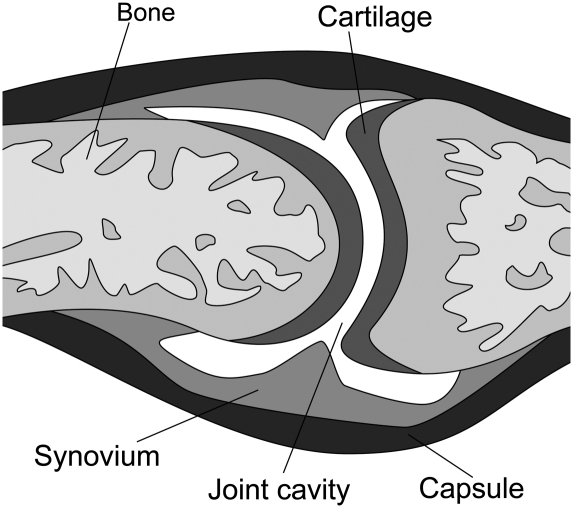
A joint is where two bones come together (Fig. 1; Goldring and Goldring 2005). Synarthroses are immovable joints. In these joints, a thin layer of fibrous connective tissue connects the bones. The sutures in the skull are examples of immovable joints. Amphiarthroses are slightly movable joints. In this type of joint, the bones are connected by hyaline cartilage or fibrocartilage. The ribs connected to the sternum by costal cartilages, the symphysis pubis, and the intervertebral disks are examples of this type. Most joints in the adult body are diarthroses, or freely movable joints. In this type of joint, the ends of the opposing bones are covered with hyaline cartilage, the articular cartilage, and they are separated by a space called the joint cavity. The components of the joints are enclosed in a dense fibrous joint capsule. The inner layer of the capsule is the synovial membrane that secretes synovial fluid into the joint cavity for lubrication. Because all of these joints have a synovial membrane, they are sometimes called synovial joints (Fig. 1).
Figure 1.
Schematic representation of a synovial joint. The diarthrodial joint connects two bones, and it allows their reciprocal movements. Two bony epiphyses are surrounded by a layer of articular hyaline cartilage, and the joint cavity separates them. A tight joint capsule connects the ends of the bone, and its inner layer is made of synovial membrane that secretes synovial fluid into the joint cavity.
Osteoarthritis (OA) is a disease of joints that affects most of the elderly population, the main features of which are damage of the articular cartilage surface, extensive subchondral bone remodeling, inflammation of the synovial membrane with thickening of the joint capsule, and osteophyte (bone spur) formation at the joint margins (Little and Fosang 2010). To this day, there is no treatment for the damaged articular cartilage besides the surgical replacement with an artificial joint.
Molecular mechanisms underlying joint formation are not yet fully elucidated (Kingsley 2001). A well-studied model of synovial joint formation is the digital ray. Metatarsals; proximal, medial, and distal phalanges; and the joints between the same bones arise from a single condensation called the digital ray. The first indication of joint development in the digital ray is the formation of the interzone, a specialized region of higher cell density within the condensation. The interzone evolves into a three-layered interzone, consisting of two regions of higher cell density sandwiching a region of lower cell density. The cells in the zone of lower cell density progressively disappear, possibly by programmed cell death, thereby forming the joint cavity. The two regions of higher cell density are thought to differentiate into the articular cartilage covering the articular surfaces of the adjacent bones. The joint capsule—i.e., ligaments and synovial lining—and tendons develop from condensations located lateral to the digital ray. Therefore, the integration of endochondral bone development and joint development in the digits results in a functional synovial articulation: two bones whose articular surfaces are covered with articular cartilage, separated by a synovial fluid-filled point cavity, and connected by the joint capsule. Soluble proteins such as GDF5, Wnt14, and Noggin, and transcription factors such as Sox5, Sox 6, and Hif-1α are the main regulators of joint development (Brunet et al. 1998; Storm and Kingsley 1999; Hartmann and Tabin 2001; Provot et al. 2007; Dy et al. 2010). However, it is still largely unclear how these different signals are integrated to form articular joints.
Like chondrocytes in the developing growth plate, the chondrocytes forming the articular surface of a synovial joint are cells that produce and maintain a unique and abundant extracellular matrix. This cartilaginous matrix has two components: the proteoglycans and the collagens. Collagens provide structure and tensile strength to the matrix (Olsen 1996). Proteoglycans are macromolecules containing a core protein with multiple attached GAGs (glycosoaminoglycans) (Knudson and Knudson 2001). Because of their high content of GAGs, proteoglycans are highly hydrated. One of the most important extracellular proteoglycans is aggrecan, the predominant proteoglycan in articular cartilage. Aggrecan forms large aggregates, which give to cartilage its unique gel-like properties and its resistance to deformation. The central component of these aggregates is a long molecule of hyaluronan (Bastow et al. 2008). Hyaluronan is the only extracellular oligosaccharide that is not covalently linked to a protein, as it is bound to aggrecan in a noncovalent fashion. A link protein that binds to the aggrecan protein and to hyaluronan facilitates this binding. The GAGs attached covalently to aggrecan are keratan sulfate and chondroitin sulfate.
Degradation of aggrecan is an important manifestation of OA (Roughley 2001; Little and Fosang 2010). Because aggrecan loss from cartilage is an early and reversible event, in vitro and in vivo, and because aggrecan can have a protective role in preventing cartilage degradation, a great deal of research has focused on aggrecanolysis at the molecular level. Aggrecanases are the principal proteinases responsible for aggrecan degradation in articular cartilage (Little and Fosang 2010). Other critical enzymes involved in the pathogenesis of OA are matrix metalloproteinases (MMPs) (Little et al. 2009); aggrecanases are active in the early phase of OA, whereas degradation of the collagenous extracellular matrix of cartilage by MMPs is a later event.
ADAMTS-4 and ADAMTS-5 are the most efficient aggrecanases, and generally have been considered to be the most likely candidates for a role in the pathological mechanisms of OA (Ameye and Young 2006; Little and Fosang 2010). ADAMTS-4 and ADAMTS-5 are zinc metalloproteinase members of the “A Disintegrin And Metalloproteinase with Thrombospondin motifs” (ADAMTS) gene family. They consist of a signal sequence, prodomain, catalytic domain, disintegrin-like domain, spacer region, and thrombospondin motifs (TSP), which help to regulate their activity and substrate specificity. ADAMTS-5 is a very small member of the ADAMTS family, having only two TSP motifs, and ADAMTS-4 is the shortest one, with just one TSP motif. How expression of the metalloproteases ADAMTS-4 and ADAMTS-5 is regulated at the transcriptional level is controversial. ADAMTS-5 is the major aggrecanase in mouse cartilage (Glasson et al. 2005; Stanton et al. 2005), whereas it is still unclear whether ADAMTS-5 or ADAMTS-4 is the major aggrecanase in human cartilage (Hardingham 2008; Little and Fosang 2010). Notably, ADAMTS-5-deficient mice (Glasson et al. 2005; Stanton et al. 2005), but not ADAMTS-4-deficient mice, are protected from cartilage erosion in models of experimental arthritis (Glasson et al. 2004; Little and Fosang 2010). Tissue inhibitor of metalloproteinase-3 (TIMP3) is the most significant endogenous extracellular inhibitor of aggrecanases identified in cartilage (Little and Fosang 2010). ADAMTS-5 is expressed in many tissues; however, its function in tissues other than cartilage remains unknown (Little and Fosang 2010).
In this issue of Genes & Development, Miyaki et al. (2010) use mouse genetics to provide solid and strong evidence that miR-140, a microRNA (miRNA) that is almost uniquely and abundantly expressed in chondrocytes (Tuddenham et al. 2006), has an important role in bone development, at least in part by controlling proliferation. This is the first report that identifies a miRNA with a crucial role in cartilage development and homeostasis in vivo.
miRNAs, development, and diseases
miRNAs are 20- to 23-nucleotide (nt)-long single-stranded noncoding RNA molecules that act as transcriptional repressors by binding to the 3′ untranslated region (UTR) of the target messenger RNA to be down-regulated (Bartel 2004).
miRNAs were first identified in 1993, when two independent research groups showed that the deletion of the gene lin-4 in Caenorhabditis elegans was necessary for correct post-embryonic development (Lee et al. 1993). Surprisingly, this gene did not code for a protein, but for a small 22-nt-long RNA, later named miRNA. Studying this RNA fragment, researchers found that lin-4 was complementary to the gene lin-14 and negatively regulated its expression level. However, the importance of these discoveries was not clear until 2001, when a great number of small RNAs with similar functions as lin-4 were found in both vertebrates and invertebrates (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001).
In recent years, miRNAs have become increasingly popular, as they have been found to be involved in a plethora of biological processes and pathological conditions (Pasquinelli and Ruvkun 2002; Garzon et al. 2009).
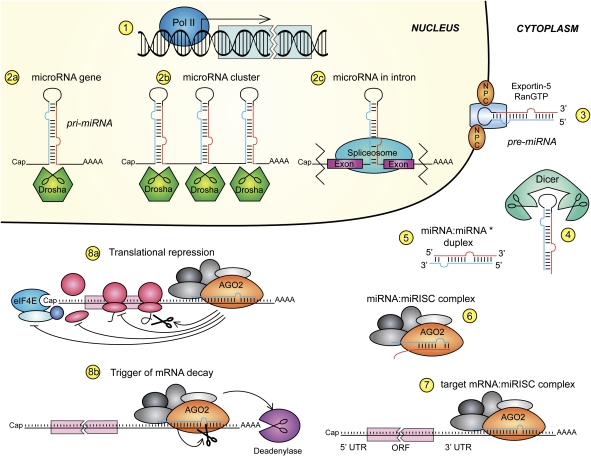
miRNAs are mainly transcribed by RNA polymerase II (Fig. 2, step 1), although transcripts from RNA polymerase III have been reported (Winter et al. 2009). Their classic structure is a small gene located in an intergenic region and with an autonomous promoter (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). The gene is transcribed as a ∼70-nt-long transcript that forms an extended stem–loop structure, shaped like a hairpin, with a partial complementary sequence in the stem region, which harbors the future miRNA: This structure is called pri-miRNA. The maturation of the pri-miRNA takes place in the nucleus with the aid of Drosha. This highly conserved RNase III-type enzyme acts together with DGCR8 (DiGeorge critical region 8) protein (Winter et al. 2009) to endonucleolytically cleave the hairpin and produce what is called a pre-miRNA (Fig. 2, step 2a).
Figure 2.
Biogenesis and action of miRNAs. (Step 1) miRNAs are transcribed mainly by polymerase II. miRNA transcribed from genes (step 2a), polycistronic clusters (step 2b), or the intronic region (step 2c), called pri-miRNAs, and in the first two cases are then processed by the type III RNase Drosha. (Step 3) The newly formed stem–loop structure, pre-miRNA, is recognized by the XPO5, Ran-GTP complex, and is transported to the cytoplasm through the NPC. Dicer cleaves the loop (step 4), leaving a double-strand fragment, the miRNA:miRNA* duplex (step 5). The blue stand is the future mature miRNA, while the red strand is its antisense miRNA*. The duplex is then unwound and loaded into the miRISC complex (step 6) where it recognizes and anneals to its mRNA target 3′ UTR (step 7). The messenger RNA:miRISC complex mediates translational repression (step 8a) and mRNA decay (step 8b). For more details, see the text.
A significant number of miRNAs is instead found in polycistronic units, which are clusters of genomic regions that code for more than one miRNA (Fig. 2, step 2b). These are also formed through Drosha cleavage. A fascinating example is the miR-17-92 cluster, a well-known oncogene but also a potent developmental regulator (Mendell 2008). Last, some miRNAs are generated from introns of messenger RNAs, such as miR-140, through a not fully understood mechanism that involves a spliceosome (Fig. 2, step 2c; Winter et al. 2009).
In the nucleus, the pre-miRNA is recognized by Exportin5 (XPO5), and, in complex with Ran-GTP, is transported to the cytosol through the nuclear pore complex (NPC) (Fig. 2, step 3). The cytoplasmic RNase III Dicer exerts its action on the pre-miRNA (Fig. 2, step 4) by cutting the loop and leaving the miRNA:miRNA* duplex with 2 nt protruding at each 3′ end (Fig. 2, step 5; Winter et al. 2009).
The miRNA action takes place within the miRNA-induced silencing complex (miRISC). Specific ATP-dependent helicases can unwind the miRNA duplex, although helicases are not strictly necessary because the protein Argonaute 2 (Ago2) facilitates uncoiling and loading on the RISC (Fig. 2, step 6; Winter et al. 2009). Theoretically, both strands can generate two mature miRNAs, but usually only the one with the thermodynamically less stable 5′ end is incorporated into RISC, while the other strand is degraded. Once the miRISC:miRNA complex has paired with its target mRNA 3′ UTR (Fig. 2, step 7), it can act as a translational repressor or it can degrade the target mRNA. The crucial nucleotides in the miRNA sequence are usually at positions 2–8, called the miRNA seed, which are almost perfectly complementary to 3′ UTR elements (Bartel 2004).
Translation can be repressed by blocking both the assembly of the translational machinery and the synthesis of nascent polypeptides (Fig. 2, step 8a). The miRISC complex contains Ago2, which competes with the initiation factor eIF4E for binding the mRNA Cap, as well as the anti-association factor eIF6, which inhibits the assembly of the ribosomes on the mRNA (Wu and Belasco 2008). Premature termination of polypeptide synthesis and degradation of the newly formed chain also contribute to translational inhibition, but those processes are not fully understood. Conversely, decay of target mRNA is achieved by endonucleolytic cleavage of mRNA by Ago2—if there is an almost perfect pairing of the miRNA seed:mRNA region—or by RISC-mediated targeting of the 3′ polyA tail to make it susceptible to deadenylase action (Fig. 2, step 8b; Wu and Belasco 2008). The inhibition of translation by miRNA represents a dynamic fine-tuning, as opposed to mRNA decay, which is an irreversible—albeit more efficient—mechanism of post-transcriptional down-regulation.
A detailed description of miRNA biogenesis and post-transcriptional action goes beyond the purpose of this perspective, as excellent reviews on the topic have been published (Bartel 2004; Wu and Belasco 2008; Winter et al. 2009).
miRNAs belong almost exclusively to Metazoan genomes, although it was reported recently that a unicellular green alga, Chlamydomonas reinhardtii, also has miRNAs (Zhao et al. 2007). The number of miRNAs correlates with the complexity of the organism in which they have been identified, and, even among vertebrates, miRNA evolution reflects their taxonomic hierarchy, with an increase in number of miRNAs in primates over other orders (Peterson et al. 2009). According to the microRNA.org data resource (http://www.microrna.org), as of April 2010, 677 miRNAs have been identified in humans and 491 in mice (Betel et al. 2008), while miRBase (http://www.mirbase.org) reports 721 human miRNAs and 579 murine miRNAs (Griffiths-Jones et al. 2008). miRNA genes are generally conserved, continuously added, and almost never lost in the genome. In very primitive organisms, the appearance of new miRNAs correlates well with the evolution of new organs or tissues (Christodoulou et al. 2010). Consistent with this observation, the earlier the miRNA appeared in the genome, the higher it is expressed and the more essential it is for survival of the organism (Peterson et al. 2009).
miRNAs are essential for development and organogenesis. The significance of miRNA regulation in the developing embryo was revealed by the universal knockout of Dicer and Argonaute, respectively, in mice (Bernstein et al. 2003; Liu et al. 2004).
Loss of Dicer in mice results in embryonic lethality at embryonic day 7.5 (E7.5) (Bernstein et al. 2003). Argonaute loss gives rise to severe developmental defects by E10.5 (Liu et al. 2004). The generation of conditional Dicer knockouts in different organs has unveiled the essential developmental roles of miRNAs. An excellent example is the knockout of Dicer in limb mesenchyme (Harfe et al. 2005; Kobayashi et al. 2008).
Embryonic stem (ES) cells, which display a unique set of miRNAs (Houbaviy et al. 2003), do not properly differentiate when the miRNA processing machinery is impaired (Kanellopoulou et al. 2005). For an extensive discussion of miRNAs in development, please see the review by Stefani and Slack (2008).
In the musculoskeletal system, miR-1 and miR-133 are muscule-specific miRNAs (Chen et al. 2006). Many putative targets of miR-1 have been identified, including transcription factors that regulate proliferation versus differentiation, such as histone deacetylase 4 (HDAC4), and the overall result of miR-1 expression is muscular differentiation. miR-133 is transcribed along with miR-1 transcripts, but its expression has an opposite outcome on myogenesis, as it increases overall proliferation and decreases myocyte differentiation, suppressing muscle gene expression through the repression of the serum response factor (SRF) (Chen et al. 2006). The opposite action of those closely related miRNAs has provided important insights about the delicate balance and the profound complexity of this regulatory system.
miR-140
The miRNA-140 gene is located between exons 16 and 17 of the E3 ubiquitin protein ligase gene Wwp2, on murine chromosome 8 and the small arm of chromosome 16 in humans. It was first reported that this miRNA is prevalently expressed in cartilage during both long and flat bone development (Tuddenham et al. 2006). The first study involving miR-140 reported that HDAC4 was down-regulated by this miRNA (Tuddenham et al. 2006). In following studies, it was proven that chemoresistance of osteosarcoma tumor xenografs was mediated in part by the miR-140-dependent suppression of HDAC4, along with induced expression of p52 and p21 (Song et al. 2009). Conversely, in gliomas, miR-140 overexpression correlated with tumor malignant progression (Malzkorn et al. 2009).
miR-140 and OA
In recent years, a putative role for miRNAs in the pathogenesis of OA has emerged (Miyaki et al. 2009; Yamasaki et al. 2009; Akhtar et al. 2010).
In a previously published study, Asahara and colleagues (Miyaki et al. 2009) had shown that normal human articular cartilage expresses miR-140, that this expression is significantly reduced in OA tissue, and that in vitro treatment of chondrocytes with IL-1β, a cytokine classically involved in the pathogenesis of OA, suppresses miR-140 expression. Conversely, transfection of chondrocytes with miR-140 down-regulates IL-1β-induced ADAMTS5 expression (Miyaki et al. 2009).
The same group now reports in this issue of Genes & Development that miR-140 has a critical role in the pathogenesis of OA by a mechanism that could at least partly involve the regulation of ADAMTS5. With a series of elegant in vivo experiments and state-of-the-art mouse genetics, Miyaki et al. (2010) provide solid and clear evidence that, although miR-140 is not required for the formation of the articular surface cartilage, universal knockout of miR-140 predisposes to age-related OA changes, and, conversely, overexpression of miR-140 in chondrocytes protects from OA. The study is important and highly significant for numerous reasons. It demonstrates for the first time that miRNAs control articular cartilage homeostasis in vivo; it thus provides a novel insight into the mechanisms that govern articular cartilage regeneration, and has potentially important therapeutical implications. More generally, it expands our understanding of the role of miRNAs in physiological tissue homeostasis and in pathological conditions. Last, it shows convincing experimental evidence that ADAMTS5 mRNA is a target of miR-140, opening a new window to the still mysterious mechanisms that regulate expression of this aggrecanase. Researchers are now left with the challenge of proving unequivocally that it is indeed the miR-140-dependent regulation of ADAMTS5 mRNA in articular surface chondrocytes that is the main molecular mechanism mediating the critical role of miR-140 in the pathogenesis of OA. In this regard, the study by Miyaki et al. (2010) does not go beyond correlation, although it is still an extremely important and elegant contribution to the field.
miR-140 and growth plate development
In the same study, Miyaki et al. (2010) also report that mir-140 has a nonredundant role in growth plate development.
Skeletal development depends on two mechanisms: intramembranous and endochondral (Karsenty and Wagner 2002). The first, in which mesenchymal cells develop directly into osteoblasts, is involved in the formation of the flat bones of the skull. The second, accounting for the development of most other bones, involves a two-stage mechanism whereby chondrocytes form a matrix template—the growth plate—that is then replaced by bone. During endochondral bone development, growth plate chondrocytes undergo well-ordered and controlled phases of cell proliferation, maturation, and death. Proliferative chondrocytes synthesize collagen type II and form a columnar layer; they then stop proliferating, and differentiate into post-mitotic hypertrophic cells. Hypertrophic chondrocytes predominantly express collagen type X and mineralize their surrounding matrix. This unique differentiation process is followed by the death of hypertrophic chondrocytes, blood vessel invasion, and finally replacement of the cartilaginous matrix with bone (Kronenberg 2003).
It has been shown already that miRNAs are critically involved in limb development. In particular, conditional deletion of Dicer in limb bud mesenchyme at early stages of embryonic development leads to formation of a much smaller limb as a consequence of massive cells death (Harfe et al. 2005), whereas lack of Dicer exclusively in chondrocytes impairs chondrocyte proliferation and accelerates their differentiation in fetal growth plates (Kobayashi et al. 2008). Miyaki et al. (2010) now show that universal knockout of mir-140 leads to mild dwarfism, probably as a result of impaired proliferation. The phenotype—although definitively mild—is still significant. Interestingly, lack of miR-140 does not recapitulate lack of Dicer in chondrocytes, suggesting that other molecules other than mir-140 could be critical downstream targets of Dicer in regulating chondrocyte biology. Intriguingly, overexpression of miR-140 does not appear to affect cartilage development in any way. How mir-140 deficiency even modestly impacts chondrocyte proliferation is yet to be resolved.
Acknowledgments
We thank Dr. Tatsuya Kobayashi for helpful discussions. This paper was supported by NIH RO1 AR048191-06 to E.S.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1939310.
References
- Akhtar N, Rasheed Z, Ramamurthy S, Anbazhagan AN, Voss FR, Haqqi TM 2010. MicroRNA-27b regulates the expression of MMP-13 in human osteoarthritis chondrocytes. Arthritis Rheum 62: 1361–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameye LG, Young MF 2006. Animal models of osteoarthritis: Lessons learned while seeking the ‘Holy Grail.’ Curr Opin Rheumatol 18: 537–547 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bastow ER, Byers S, Golub SB, Clarkin CE, Pitsillides AA, Fosang AJ 2008. Hyaluronan synthesis and degradation in cartilage and bone. Cell Mol Life Sci 65: 395–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ 2003. Dicer is essential for mouse development. Nat Genet 35: 215–217 [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C 2008. The microRNA.org resource: Targets and expression. Nucleic Acids Res 36: D149–D153 doi: 10.1093/nar/gkm995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet LJ, McMahon JA, McMahon AP, Harland RM 1998. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 280: 1455–1457 [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38: 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, Klaus S, Snyman H, Hannon GJ, Bork P, Arendt D 2010. Ancient animal microRNAs and the evolution of tissue identity. Nature 463: 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy P, Smits P, Silvester A, Penzo-Mendez A, Dumitriu B, Han Y, la Motte CA, Kingsley DM, Lefebvre V 2010. Synovial joint morphogenesis requires the chondrogenic action of Sox5 and Sox6 in growth plate and articular cartilage. Dev Biol 341: 346–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM 2009. MicroRNAs in cancer. Annu Rev Med 60: 167–179 [DOI] [PubMed] [Google Scholar]
- Glasson SS, Askew R, Sheppard B, Carito BA, Blanchet T, Ma HL, Flannery CR, Kanki K, Wang E, Peluso D, et al. 2004. Characterization of and osteoarthritis susceptibility in ADAMTS-4-knockout mice. Arthritis Rheum 50: 2547–2558 [DOI] [PubMed] [Google Scholar]
- Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, Flannery CR, Peluso D, Kanki K, Yang Z, et al. 2005. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature 434: 644–648 [DOI] [PubMed] [Google Scholar]
- Goldring MB, Goldring SR 2005. Biology of the normal joint. In Kelley's textbook of theumatology, 7th ed (ed. Firestein GS et al. ), pp. 1–34 Saunders Elsevier, Philadelphia [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 doi: 10.1093/nar/gkm952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham T 2008. Extracellular matrix and pathogenic mechanisms in osteoarthritis. Curr Rheumatol Rep 10: 30–36 [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ 2005. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci 102: 10898–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Tabin C 2001. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell 104: 341–351 [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA 2003. Embryonic stem cell-specific microRNAs. Dev Cell 5: 351–358 [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G, Wagner E 2002. Reaching a genetic and molecular understanding of skeletal development. Dev Cell 2: 389–406 [DOI] [PubMed] [Google Scholar]
- Kingsley D 2001. Genetic control of bone and joint formation. Novartis Found Symp 232: 213–222 [DOI] [PubMed] [Google Scholar]
- Knudson CB, Knudson W 2001. Cartilage proteoglycans. Semin Cell Dev Biol 12: 69–78 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, Merkenschlager M, Kronenberg HM 2008. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci 105: 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg H 2003. Developmental regulation of the growth plate. Nature 423: 332–336 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864 [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Little CB, Fosang AJ 2010. Is cartilage matrix breakdown an appropriate therapeutic target in osteoarthritis—Insights from studies of aggrecan and collagen proteolysis? Curr Drug Targets 11: 561–575 [DOI] [PubMed] [Google Scholar]
- Little CB, Barai A, Burkhardt D, Smith SM, Fosang AJ, Werb Z, Shah M, Thompson EW 2009. Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis Rheum 60: 3723–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stuhler K, Meyer HE, Reifenberger G 2009. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol 20: 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT 2008. miRiad roles for the miR-17-92 cluster in development and disease. Cell 133: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaki S, Nakasa T, Otsuki S, Grogan SP, Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK, Asahara H 2009. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum 60: 2723–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, et al. 2010. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev (this issue). doi: 10.1101/gad.1915510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen BR 1996. Role of cartilage collagens in formation of the skeleton. Ann N Y Acad Sci 785: 124–130 [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Ruvkun G 2002. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol 18: 495–513 [DOI] [PubMed] [Google Scholar]
- Peterson KJ, Dietrich MR, McPeek MA 2009. MicroRNAs and metazoan macroevolution: Insights into canalization, complexity, and the Cambrian explosion. Bioessays 31: 736–747 [DOI] [PubMed] [Google Scholar]
- Provot S, Zinyk D, Gunes Y, Khatri R, Le Q, Kronenberg H, Johnson R, Longaker M, Giaccia A, Schipani E 2007. Hif-1α regulates differentiation of limb bud mesenchyme and joint development. J Cell Biol 177: 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughley PJ 2001. Articular cartilage and changes in arthritis: Noncollagenous proteins and proteoglycans in the extracellular matrix of cartilage. Arthritis Res 3: 342–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, et al. 2009. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene 28: 4065–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton H, Rogerson FM, East CJ, Golub SB, Lawlor KE, Meeker CT, Little CB, Last K, Farmer PJ, Campbell IK, et al. 2005. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature 434: 648–652 [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ 2008. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9: 219–230 [DOI] [PubMed] [Google Scholar]
- Storm E, Kingsley D 1999. GDF5 coordinates bone and joint formation during digit development. Dev Biol 209: 11–27 [DOI] [PubMed] [Google Scholar]
- Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T 2006. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett 580: 4214–4217 [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S 2009. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234 [DOI] [PubMed] [Google Scholar]
- Wu L, Belasco JG 2008. Let me count the ways: Mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell 29: 1–7 [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Nakasa T, Miyaki S, Ishikawa M, Deie M, Adachi N, Yasunaga Y, Asahara H, Ochi M 2009. Expression of microRNA-146a in osteoarthritis cartilage. Arthritis Rheum 60: 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y 2007. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes & Dev 21: 1190–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]