Abstract
The Arabidopsis histone deacetylase HDA6 is required to silence transgenes, transposons, and ribosomal RNA (rRNA) genes subjected to nucleolar dominance in genetic hybrids. In nonhybrid Arabidopsis thaliana, we show that a class of 45S rRNA gene variants that is normally inactivated during development fails to be silenced in hda6 mutants. In these mutants, symmetric cytosine methylation at CG and CHG motifs is reduced, and spurious RNA polymerase II (Pol II) transcription occurs throughout the intergenic spacers. The resulting sense and antisense spacer transcripts facilitate a massive overproduction of siRNAs that, in turn, direct de novo cytosine methylation of corresponding gene sequences. However, the resulting de novo DNA methylation fails to suppress Pol I or Pol II transcription in the absence of HDA6 activity; instead, euchromatic histone modifications typical of active genes accumulate. Collectively, the data reveal a futile cycle of unregulated transcription, siRNA production, and siRNA-directed DNA methylation in the absence of HDA6-mediated histone deacetylation. We propose that spurious Pol II transcription throughout the intergenic spacers in hda6 mutants, combined with losses of histone deacetylase activity and/or maintenance DNA methylation, eliminates repressive chromatin modifications needed for developmental rRNA gene dosage control.
Keywords: Chromatin, histone deacetylation, DNA methylation, epigenetics, RNA polymerase I, ribosomal RNA genes
In eukaryotes, ribosomal RNA (rRNA) genes are repeated head to tail at chromosomal loci known as nucleolus organizer regions (NORs) (see Fig. 1A). In the plant Arabidopsis thaliana, NORs composed of ∼375 rRNA genes abut the telomeres on the northern ends of chromosomes 2 and 4 (Copenhaver et al. 1995; Copenhaver and Pikaard 1996a,b). Each rRNA gene repeat can be transcribed by RNA polymerase I (Pol I) to produce a 45S pre-rRNA primary transcript. The primary transcript is then processed into the 18S, 5.8S, and 25S rRNAs that partner with 5S rRNA, transcribed by Pol III, and ∼80 proteins, encoded by mRNAs transcribed by Pol II, to form the 40S and 60S ribosomal subunits (Korostelev and Noller 2007).
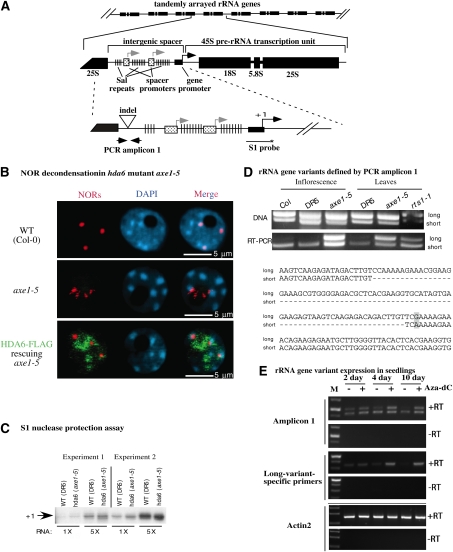
Figure 1.
HDA6 is required for developmentally regulated silencing of 45S rRNA gene variants. (A) rRNA gene organization showing the tandem arrangement of rRNA genes at NORs and the structure of individual rRNA gene repeats. In the middle drawing, the locations of the IGS and the 45S pre-rRNA transcription unit are highlighted, as well as the gene promoter (black arrow, +1), duplicated spacer promoters (gray arrows), and repetitive elements known as Sal repeats. The bottom drawing shows the locations of the promoter S1 probe and PCR amplicon 1, which includes an indel that allows long and short variants to be discriminated. (B) NORs are decondensed in hda6 mutants. Nuclei isolated from wild-type (WT) plants, axe1-5 mutants, or axe1-5 transformed with an HDA6-Flag transgene were subjected to DNA-FISH using a 45S rRNA gene probe (red signals). HDA6-Flag protein was immunolocalized using anti-Flag antibody. Nuclei were counterstained with DAPI (blue signals). Bars, 5 μm. (C) 45S pre-rRNA transcripts initiated at the RNA Pol I start site (+1), detected using S1 nuclease protection of RNA from wild-type (DR5) or hda6 mutant (axe1-5) plants. RNA was isolated from two sets of plants grown independently (experiments 1 and 2) and tested at two input RNA concentrations, showing that the results are semiquantitative. The S1 probe location is shown in A. (D) Long and short rRNA gene variants defined by amplicon 1 are differentially expressed. The top panel shows ethidium bromide-stained amplicon 1 PCR products obtained using genomic DNA from inflorescence or leaves as the template. The bottom panel shows RT–PCR products amplified from cDNA generated by random priming of total RNA. Wild-type Col-0 and DR5 (a Col-0 transgenic line from which axe1 mutants were isolated) controls are compared with the hda6 mutants axe1-5 and rts1-1. Sequences of long and short PCR products in the vicinity of the indel present within amplicon 1 are shown at the bottom of the panel. (E) Silencing of long rRNA gene variants is developmentally regulated. Wild-type Col-0 seedlings were germinated on media containing (+) or lacking (−) 10 mM aza-dC and then harvested 2, 4, or 10 d after germination. Ethidium bromide-stained RT–PCR reaction products, and products of controls in which reverse transcriptase was omitted (−RT), are shown using amplicon 1 primers (top) or primers specific for long rRNA gene variants (middle). The ACTIN2 control shows that equal amounts of RNA were subjected to PCR amplification in all reactions. Size markers (M) are present in the first lane of each gel.
Each rRNA gene is separated from the adjacent rRNA gene by an intergenic spacer (IGS) that contains multiple regulatory elements (Reeder 1989). In A. thaliana, the IGS includes the gene promoter, spacer promoters, and so-called Sal repeats (Fig. 1A; Gruendler et al. 1991). Spacer promoters share >85% similarity with the gene promoter but are only ∼10% as active in transient expression assays (Doelling et al. 1993). The Sal repeats, named for the presence of SalI restriction endonuclease sites, occupy the same positions as enhancer repeats in Xenopus and mice, and can functionally substitute for Xenopus enhancers in frog oocytes (Pikaard et al. 1990; Doelling et al. 1993). Despite their enhancer activity in Xenopus, enhancer activity has not been demonstrated for Sal repeats in Arabidopsis (Wanzenbock et al. 1997).
Only a subset of rRNA genes is active in somatic cells, with distinct post-translational chromatin modifications reflecting the “on” or “off” transcriptional states (Grummt and Pikaard 2003; Preuss and Pikaard 2007; McStay and Grummt 2008). Enzymes that regulate histone acetylation, histone methylation, DNA methylation, and nucleosome positioning are involved in the on/off switch (Sandmeier et al. 2002; Santoro et al. 2002; Lawrence et al. 2004; Earley et al. 2006a; Espada et al. 2007; Preuss et al. 2008). Other mechanisms, including phosphorylation of Pol I transcription factors, regulate the transcriptional capacity of genes in the on state, thereby fine-tuning the dosage control system (Grummt and Pikaard 2003; Russell and Zomerdijk 2006; Moss et al. 2007; McStay and Grummt 2008). As a result of fine-tuning, changes in the number of active genes do not necessarily result in increased rRNA synthesis, due to compensatory changes in the number of transcribing RNA Pol I enzymes per gene (French et al. 2003).
Evidence that histone deacetylation is involved in rRNA gene silencing first came from the study of nucleolar dominance, an epigenetic phenomenon in genetic hybrids that describes the expression of rRNA genes inherited from only one progenitor (McStay 2006; Preuss and Pikaard 2007). Initially, the histone deacetylase (HDAC) inhibitors Trichostatin A (TSA) and sodium butyrate were shown to derepress rRNA genes subjected to nucleolar dominance (Lawrence et al. 2004; Chen 2007). Subsequently, RNAi-mediated knockdown of the 16 predicted HDACs in Arabidopsis suecica, the allotetraploid hybrid of A. thaliana and Arabidopsis arenosa, revealed that HDA6 is required for the preferential silencing of A. thaliana-derived rRNA genes (Earley et al. 2006a). In vitro, HDA6 is a TSA-sensitive enzyme that removes acetyl groups from multiple core histone lysines, suggesting that HDA6 facilitates rRNA gene silencing via regional erasure of histone acetylation (Earley et al. 2006a). However, the rRNA gene intervals where HDA6 exerts its control have not been identified.
In addition to affecting rRNA gene silencing, HDA6 is required for transgene silencing, as revealed by multiple genetic screens, including screens targeting components of the RNA-directed de novo cytosine methylation pathway (Furner et al. 1998; Murfett et al. 2001; Aufsatz et al. 2002; Probst et al. 2004; Matzke and Birchler 2005; He et al. 2009; Matzke et al. 2009). ATCOPIA, ATLINE1-4, ATLANTYS, ATGP1, and Sadhu transposable elements are also suppressed by HDA6, revealing a broad influence of HDA6 throughout the genome (Lippman et al. 2003; May et al. 2005; Rangwala and Richards 2007).
On a chromosomal scale, disruption of nucleolar dominance in allotetraploid A. suecica HDA6-RNAi lines correlates with decondensation of the multimegabase NORs (Earley et al. 2006a). On a genic scale, histone hyperacetylation, increased histone H3 Lys 4 (H3K4) trimethylation, and decreased cytosine methylation occur at rRNA gene promoters in HDA6-RNAi lines (Earley et al. 2006a). In the diploid species A. thaliana, similar chromatin modifications occur in hda6 mutants; likewise, NORs are decondensed (Probst et al. 2004), suggesting an increase in the number of active rRNA genes. However, no change in the abundance of transcripts initiated at rRNA gene promoters could be detected in a previous study of A. thaliana hda6 mutants (Probst et al. 2004), making the basis for NOR decondensation in hda6 mutants unclear.
In this study, we explored the molecular basis for HDA6-mediated repression of rRNA genes in A. thaliana. We show that a major function of HDA6 is to suppress spurious RNA Pol II transcription throughout the IGS. Consequently, sense and antisense IGS transcripts are overproduced in hda6 mutants, giving rise to abundant siRNAs that are diced by DCL3 and DCL4. Resulting siRNAs, in turn, direct de novo cytosine methylation throughout the IGS. Interestingly, losses in maintenance methylation at CG and CHG motifs in hda6 mutants occur in parallel with the gain of de novo asymmetric CHH methylation. Although siRNA-directed cytosine hypermethylation is typically repressive, this is not the case in hda6 mutants. Instead, histone modifications typical of active genes accumulate, and Pol I and Pol II association with rRNA genes increases. Importantly, an abundant class of rRNA gene variants, normally expressed only during early seedling development, fails to be silenced in hda6 mutants, and therefore remains active in mature plants. We propose that, in hda6 mutants, spurious transcription throughout the IGS, combined with losses of maintenance cytosine methylation and HDAC activity, disrupts the chromatin modifications needed to repress rRNA gene promoters during development.
Results
HDA6 suppresses IGS transcription
NORs are dramatically decondensed in A. thaliana hda6 mutants (Fig. 1B; Probst et al. 2004). In diploid A. thaliana, there are four NORs: one pair present on chromosome 2, and the other pair located on chromosome 4. NORs tend to coalesce such that fewer than four DNA-FISH (DNA fluorescent in situ hybridization) signals are typical in interphase nuclei using rRNA gene probes. For example, three NOR signals (red) are detected by DNA-FISH in the wild-type (ecotype Col-0) nucleus shown in Figure 1B. These bright FISH signals correspond to the portions of the NORs where excess, inactive rRNA genes are highly condensed and sequestered in heterochromatin at the external periphery of the nucleolus. In contrast, active rRNA genes are transcribed within the nucleolus, which appears as a black hole in nuclei stained with the fluorescent DNA-binding dye DAPI (blue signals). The filaments of highly decondensed, active rRNA genes do not yield FISH signals bright enough to be detected in Figure 1B.
Unlike wild-type nuclei, whose prominent NOR signals are external to the nucleolus, nuclei of hda6 mutants such as axe1-5 (Murfett et al. 2001) display decondensed NOR signals within the nucleolus (Fig. 1B, middle panel). Transformation of axe1-5 with a transgene encoding HDA6 fused to a Flag epitope tag (HDA6-Flag) rescues the mutation, such that compact NOR signals external to the nucleolus are again observed, as in wild-type nuclei (Fig. 1B, bottom panel). Localization of recombinant HDA6 by virtue of its Flag tag shows that HDA6 localizes within the nucleolus, as well as the nucleoplasm, in agreement with a previous study (Earley et al. 2006a).
NOR decondensation typically correlates with increased rRNA gene transcription. However, using S1 nuclease protection to monitor levels of accurately initiated transcripts at rRNA gene promoters, we observe no change in axe1-5 mutants relative to the nonmutant control DR5 (Fig. 1C), in agreement with prior results (Probst et al. 2004). However, RT–PCR analyses reveal an effect of HDA6 on the differential expression of rRNA gene variants that are polymorphic at their 3′ ends (Fig. 1D). Long and short rRNA gene variants can be discriminated by an 81-base-pair (bp) insertion/deletion (indel) present within PCR amplicon 1 (Fig. 1A, bottom panel). This indel is located within a region of the primary transcript that is removed during pre-rRNA processing (S Tucker, F Pontvianne, and CS Pikaard, unpubl.). PCR amplification of genomic DNA reveals that long and short rRNA gene variants are similar in abundance (Fig. 1D, top panel). However, RT–PCR analysis of total RNA reveals that only short variants are expressed in inflorescence or leaves of mature wild-type (Col-0 and DR5) plants (Fig. 1D, bottom panel). In contrast, both long and short variant transcripts are detected in the hda6 mutants axe1-5 and rts1-1. Collectively, these data indicate that HDA6 is involved in repressing long rRNA gene variants, which represent a significant fraction of the total rRNA gene pool.
During early seedling development in Arabidopsis, condensation of chromocenters and NORs into heterochromatin and silencing of rRNA genes subjected to nucleolar dominance begin 2 to 4 d after germination (Mathieu et al. 2003; Pontes et al. 2007). Consistent with these observations, RT–PCR analysis using amplicon 1 primers, or a primer pair that amplifies only long variants, reveals that long variants are progressively inactivated during early seedling development (Fig. 1E). At 2 d after germination, transcripts of both long and short rRNA gene variants are detected in similar abundance, but, by 10 d, only short variant transcripts are expressed abundantly. If plants are grown on media containing 5-aza-2′-deoxycytidine (aza-dC), an inhibitor of cytosine methylation, suppression of long rRNA gene variants is precluded (Fig. 1E). Taken together, these results indicate that long variant expression in A. thaliana is regulated epigenetically, with the developmental silencing of variants involving HDA6 and cytosine methylation, as is also the case for uniparental rRNA gene silencing in nucleolar dominance in the hybrid A. suecica (Chen and Pikaard 1997; Earley et al. 2006a; Preuss et al. 2008).
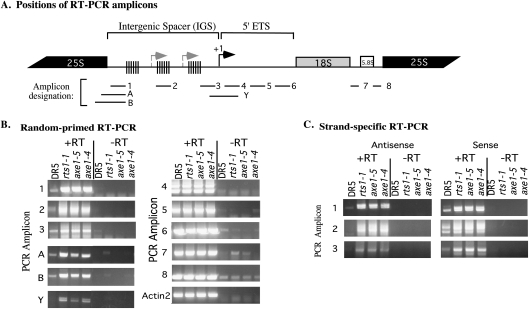
To examine the genic regions where HDA6 exerts its repressive effects, we used RT–PCR to compare transcript levels at various intervals throughout the ∼10-kb rRNA gene repeat (Fig. 2A) in both wild-type (DR5) and hda6 mutant plants (Fig. 2B,C). In axe1-4, axe1-5 (Murfett et al. 2001), or rts1-1 (Aufsatz et al. 2002) mutants, RNA transcript levels are elevated throughout the IGS (Fig. 2B; amplicons 1, 2, 3, A, B, and Y). RT–PCR reactions employing strand-specific primers revealed that these IGS transcripts correspond to both DNA strands, indicative of bidirectional transcription (Fig. 2C). However, downstream from the gene promoter, no significant difference in RNA transcript levels is detected in hda6 mutants compared with the DR5 (wild-type) control (Fig. 2B, amplicons 4–8). These data suggest that HDA6-mediated transcriptional repression is focused on the IGS.
Figure 2.
IGS transcripts are overproduced in hda6 mutants. (A) Diagram showing the positions of RT–PCR amplicons examined in B and C. (B) RT–PCR analysis of RNAs corresponding to the IGS, 5′ ETS, or rRNA coding regions. First-strand cDNA was synthesized from total RNA using random primers and then subjected to PCR using primers specific to the indicated amplicons. Ethidium bromide-stained reaction products derived from RNA of wild-type (DR5) or hda6 mutants (axe1-5, axe1-4, or rts1-1) are compared. Control reactions from which reverse transcriptase was omitted (−RT) reveal background signals attributable to DNA contamination. Amplification of ACTIN2 served as a control to show that equivalent amounts of RNA from each genotype were tested. (C) Strand-specific RT–PCR amplification of IGS transcripts. Sense or antisense strand-specific primers were used to synthesize first-strand cDNA, followed by PCR amplification using amplicon 1, 2, or 3 primers.
Enzymatic activity of HDA6 is required to suppress spurious IGS transcription
HDACs including human HDAC1, Aquifex aeolicus HDAC HDLP (Finnin et al. 1999), Arabidopsis HDA19, and Arabidopsis HDA6 are similar in sequence in the vicinity of their catalytic centers (Fig. 3A). The crystal structure of HDLP in complex with TSA suggests that conserved amino acids corresponding to HDA6 positions 186, 188, 190, 191, and 193 contribute to the active site (Fig. 3A; Finnin et al. 1999). Consistent with this prediction, HDAC1 aspartates circled in Figure 3A, corresponding to D186 and D188 of HDA6, are critical for HDAC activity (Hassig et al. 1998). Using site-directed mutagenesis, we changed each of five amino acids of HDA6 that are invariant among HDAC1, HDLP, HDA19, and HDA6. Mutant and wild-type versions of the HDA6 gene were then transformed into hda6 mutant plants (axe1-5), and resulting HDA6-Flag proteins expressed in the transgenic plants were affinity-purified. Immunoblotting confirmed that similar quantities of recombinant HDA6-Flag were recovered from each transgenic line (Fig. 3B, top panel). HDAC activity of the recovered proteins was tested using core histones acetylated in vitro using recombinant A. thaliana HAC1 and tritiated acetyl-CoA (Earley et al. 2006a, 2007). Whereas wild-type HDA6 deacetylated the histones, causing a loss of signal on the fluorogram (duplicate reactions are shown), HDA6 bearing any of the active site mutations lacks HDAC activity (Fig. 3B, middle panel). Mutant HDA6 proteins also lack HDAC activity on histone H4 N-terminal tail peptides acetylated using human pCAF2 (data not shown).
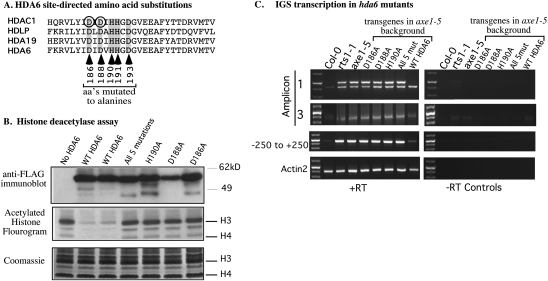
Figure 3.
HDA6 enzymatic activity is required for rRNA gene variant silencing. (A) Multiple alignment of HDA6 and related HDACs in the vicinity of the active site. Circled amino acids are known to be required for human HDAC1 activity. Arrows indicate amino acids mutated to alanines in transgene-expressed recombinant HDA6 tested in B and C. (B) Alanine substitutions of conserved active site amino acids abolish HDA6 enzymatic activity. The top row shows an immunoblot of Flag-tagged HDA6 proteins expressed in the A. thaliana axe1-5 mutant background, affinity-captured on anti-Flag beads, and resolved by SDS-PAGE. Single-alanine substitution mutants (D186A, D188A, or H190A) and an HDA6 mutant bearing alanine substitutions at five positions (186, 188, 190, 191, and 193) are shown alongside wild-type (unmutated) HDA6-Flag and the anti-Flag immunoprecipitate from axe1-5 plants not bearing an HDA6 transgene (no HDA6 control). The middle panel shows a fluorogram of histones that were acetylated in vitro by recombinant A. thaliana HAC1 using radioactive (3H) acetyl-CoA and then incubated with wild-type or mutated HDA6. (Middle panel) Reaction products were resolved by SDS-PAGE and gels were stained with Coomassie Blue, dried, and exposed to X-ray film using a 3H-intensifying screen in order to generate the fluorogram shown. The Coomassie-stained gel revealing the histones is shown in the bottom panel. (C) The HDAC activity of HDA6 is required to suppress long variant expression and IGS transcription. Long and short variant expression (amplicon 1), IGS transcription upstream of the gene promoter (amplicon 3), and transcripts reading through the promoter (−250 to +250 amplicon) were detected using RT–PCR. Total RNA isolated from wild-type Col-0 and hda6 mutants axe1-5 or rts1-1 were analyzed in parallel with RNA isolated from axe1-5 plants transformed with transgenes expressing HDA6 bearing single amino acid mutations of the active site (D186A, D188A, or H190A), the five amino acid mutation of the active site (all 5 mut), or wild-type HDA6 (WT HDA6). Amplification of ACTIN2 cDNA serves as a control to show that equivalent amounts of RNA were tested in all lanes. The right panel shows control reactions from which reverse transcriptase was omitted (−RT).
To determine if the HDAC activity of HDA6 is necessary for rRNA gene variant silencing, we assayed for long and short variant expression in wild-type (Col-0), hda6 (rts1-1 or axe1-5) mutants, axe1-5 mutants rescued by a wild-type HDA6 transgene, or axe1-5 plants expressing HDA6 active site mutant proteins. Only short rRNA gene variants are expressed in vegetative tissues of mature control plants, whereas in hda6 mutants (rts1-1 and axe1-5), both the long and short variants are expressed (Fig. 3C, amplicon 1 results). The wild-type HDA6 transgene rescues the axe1-5 mutant, such that only short variants are expressed, and also restores IGS transcripts to control levels (Fig. 3C, amplicon 3 results). In contrast, none of the HDA6 transgenes bearing active site mutations rescues axe1-5 (Fig. 3C). Collectively, the results indicate that the HDAC activity of HDA6 is required for rRNA gene variant silencing.
IGS siRNAs are overproduced in hda6 mutants
We reasoned that bidirectional IGS transcription in hda6 mutants might give rise to dsRNAs that could be diced into siRNAs. Indeed, RNA blot analyses using probes corresponding to several regions of the IGS reveal a massive accumulation of IGS siRNAs in hda6 mutants compared with wild-type plants (Fig. 4A). Downstream from the transcription start site, within the external transcribed spacer (ETS), no hyperaccumulation of siRNAs is detected, consistent with the RT–PCR evidence that IGS, but not ETS, transcripts accumulate in hda6 mutants (refer to Fig. 2B). Using the IGS2 probe, blot hybridization shows that siRNA overproduction in hda6 is suppressed upon complementation of axe1-5 with wild-type HDA6-Flag (Fig. 4B, two independent transgenic lines are shown). However, HDA6 transgenes bearing D186A, H190A, or five clustered active site mutations fail to rescue the axe1-5 mutation (Fig. 4B). These data indicate that the HDAC activity of HDA6 is required to suppress siRNA overproduction, presumably due to suppressing precursor transcription (see Fig. 2B,C).
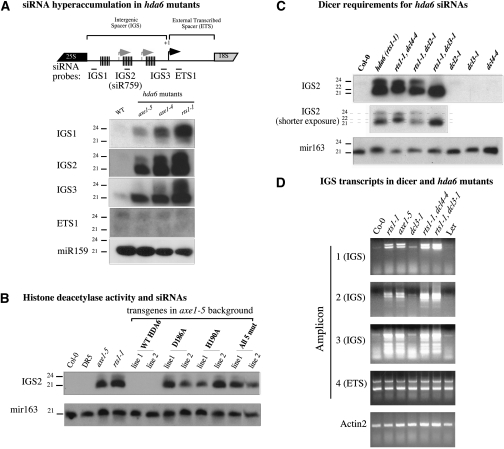
Figure 4.
IGS siRNAs are overproduced in hda6 mutants. (A) IGS and ETS probes were hybridized to Northern blots of small RNAs isolated from wild-type (Col-0) or hda6 mutant plants (axe1-5, axe14, and rts11); see the diagram at the top for IGS and ETS probe locations. Detection of the microRNA miR159 served as an RNA loading control. (B) Small RNAs isolated from wild-type plants (Col-0 and DR5), hda6 mutants (axe1-5 and rts11), or axe1-5 plants expressing transgenes encoding either wild-type HDA6 or an HDA6 mutant bearing the D186A, H190A, or five clustered (all 5 mut) active site mutations were examined by Northern blotting using the IGS2 hybridization probe. Two independent transgenic lines for each construct are shown. Hybridization to a miR163 probe controls for RNA loading. (C) Dicers DCL3 and DCL4 are primarily responsible for IGS siRNAs overproduced in hda6 mutants. Small RNAs isolated from wild-type Col-0, hda6 (rts1-1), hda6 dicer double mutants (for dicers dcl2, dcl3, and dcl4), and dicer single mutants were examined by Northern blotting using the IGS2 probe or mir163 probe; the latter controls for RNA loading. (D) IGS transcripts hyperaccumulate in hda6 dcl4 and hda6 dcl3 double mutants. IGS transcripts were detected by RT–PCR using amplicon 1, 2, or 3 primers; ETS transcripts were detected using amplicon 4 primers (see Fig. 2 for amplicon positions). Total RNA tested was isolated from wild-type Col-0 or Ler plants, hda6 mutants (rts1-1 and axe1-5), dcl3-1, and the double mutants rts1-1 dcl4-4 and rts1-1/dcl3-1.
IGS siRNAs overproduced in hda6 mutants vary in size in different regions of the IGS. The IGS1 probe primarily detects 23- and 24-nucleotide (nt) siRNAs, whereas the IGS2 and IGS3 probes predominantly detect 21-nt siRNAs. To determine which dicers generate IGS siRNAs in hda6 mutants, we assayed siRNA levels in hda6 dcl double mutants. In the rts1-1 dcl4-4 double mutant, 24-nt IGS2 siRNAs are unaffected, but 21-nt siRNAs are lost and are replaced, instead, by 22-nt siRNAs, presumably generated by DCL2 (Fig. 4C). In rts1-1 dcl2-1 double mutants, no change in siRNA size classes occurs, consistent with DCL2 playing a negligible role in IGS dsRNA dicing (unless DCL4 is mutated). In the rts1-1 dcl3-1 double mutant, the 24-nt siRNAs are lost, but 21-nt siRNAs are unaffected. Collectively, the data indicate that DCL4 and DCL3 and are primarily responsible for the 21-nt and 24-nt siRNAs, respectively, that are overproduced in hda6 mutants.
Effects of dicer mutations on the abundance of IGS and ETS transcripts was tested by RT–PCR (Fig. 4D). Controls show that IGS transcripts corresponding to amplicons 1, 2, or 3 are not increased in abundance in either dcl3 (Fig. 4D) or dcl4 mutants (data not shown). However, in rts1 dcl4 and rts1 dcl3 double mutants, IGS transcript abundance is increased above the levels in rts1-1 or axe1-5 single mutants. One explanation of these results could be that IGS transcript stability is increased in the absence of either DCL3 or DCL4-mediated dicing. Alternatively, disrupted siRNA-mediated transcriptional silencing in the absence of dicer activity could potentially result in elevated IGS transcription.
Cytosine methylation gains and losses in hda6 mutants
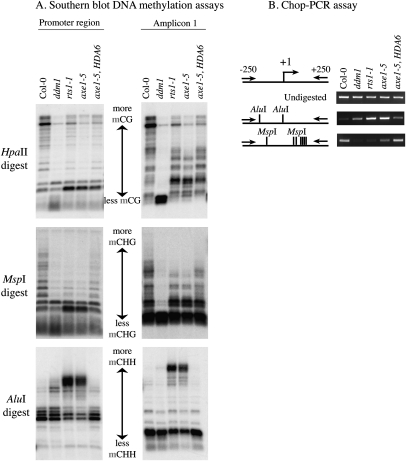
To determine if siRNAs overproduced in hda6 mutants program siRNA-directed DNA methylation of corresponding genomic sequences, we digested genomic DNA using methylation-sensitive restriction endonucleases and performed Southern blot hybridization using a promoter region (−250 to +250) or amplicon 1 probe (Fig. 5). The hda6 mutants axe1-5 and rts1-1 and axe1-5 rescued with the HDA6-Flag transgene were compared with wild-type (Col-0) and ddm1 (decrease in DNA methylation 1) controls. The restriction endonucleases HpaII and MspI recognize the sequence CCGG, but differ in their methylation sensitivities such that MspI digestion provides insight into CHG methylation status (where H denotes A,T, or C), and HpaII provides insight into CG methylation. In ddm1 mutants, methylation is decreased in multiple sequence contexts (Vongs et al. 1993), resulting in increased digestion of IGS sequences by both HpaII and MspI (Fig. 5A). In hda6 mutants, IGS sequences are somewhat more susceptible to HpaII digestion, but not as dramatically as in ddm1. However, using the promoter region probe, similar (but not identical) losses in CHG methylation are apparent in ddm1 and hda6 mutants. Unlike the promoter region, the amplicon 1 probe reveals a more pronounced loss of CHG methylation at MspI sites in ddm1 mutants compared with hda6 mutants. Importantly, CG methylation patterns are not restored to a wild-type pattern in axe1-5 rescued by the HDA6 transgene (Fig. 5A), and CHG methylation is only partially restored, consistent with the fact that maintenance methylation patterns, once lost, are not easily regained (Vongs et al. 1993; Richards 1997).
Figure 5.
Symmetric cytosine methylation decreases, but asymmetric cytosine methylation is gained de novo, in hda6 mutants. (A) DNA methylation assayed using methylation-sensitive restriction endonucleases and Southern blot hybridization. Total genomic DNA was isolated from wild-type Col-0, ddm1, hda6 mutants (rts1-1 and axe1-5), or axe1-5 plants rescued with a wild-type HDA6 transgene. The DNA was digested using HpaII, which reports on methylation in a symmetric CG context; MspI, which reports on methylation in a symmetric CHG context; or AluI, which reports on CHH methylation. Following electrophoresis and blotting, nylon membranes were hybridized to promoter region (−250 to +250) or amplicon 1 large variant-specific probes. (B) Analysis of AluI and MspI site methylation using Chop-PCR. Undigested genomic DNA (top row) or genomic DNA digested with MspI (bottom row) or AluI (midddle row) was subjected to PCR amplification using primers spanning the promoter region.
Examination of cytosine methylation at AluI restriction sites, which reports on de novo methylation in a CHH sequence context, reveals a dramatic increase in de novo cytosine methylation in hda6 mutants, and a modest increase in ddm1 mutants, as shown using both the promoter region and amplicon 1 probes (Fig. 5A). The de novo cytosine methylation is consistent with the dramatic overproduction of siRNAs in hda6 mutants (see Fig. 4), and a modest increase in siRNA levels in ddm1 mutants (data not shown). In axe1-5 rescued by the HDA6-Flag transgene, CHH methylation is restored to wild-type levels, consistent with the return of siRNA abundance to wild-type levels (refer to Fig. 4B).
As an independent test of DNA methylation status, we used Chop-PCR, an assay in which genomic DNA is subjected to digestion with a methylation-sensitive restriction endonuclease (i.e., it is “chopped”) and then tested as a template for PCR amplification using primers flanking the restriction sites. In wild-type plants, lack of DNA methylation at one or more AluI sites in the promoter region allows AluI to cut the DNA such that the template cannot be amplified by PCR (Fig. 5B; Supplemental Fig. S2). In contrast, the promoter region of wild-type plants is recalcitrant to digestion with MspI, due to CHG methylation; as a result, the region can be PCR-amplified following MspI digestion. In agreement with the Southern blot data, increased de novo methylation at asymmetric sites occurs in the promoter region of ddm1, rts1-1, and axe1-5 mutants, making the promoter region more resistant to AluI cleavage. At the same time, losses of CHG and CG methylation make promoter regions more susceptible to MspI and HpaII digestion, respectively (Fig. 5B; Supplemental Fig. S2). Importantly, wild-type methylation patterns at AluI sites are restored in axe1-5 mutants transformed with the wild-type HDA6-Flag transgene (Fig. 5B). Taken together, the data show that symmetric CHG sites are hypomethylated, but asymmetric CHH sites are hypermethylated, in hda6 mutants. We conclude that HDA6 is not required for de novo CHH methylation, but appears to be required for CG and CHG maintenance methylation. The possibility that HDA6 is also required for de novo methylation at CG and CHG motifs cannot be ruled out.
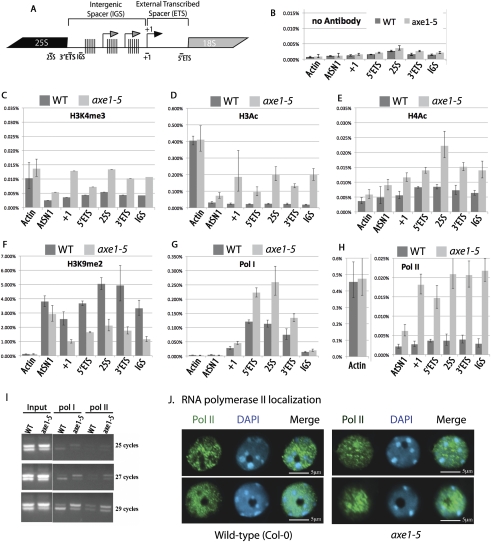
Histone modifications and polymerase occupancy in hda6 mutants
To assay chromatin modifications and polymerase occupancy in rRNA gene IGS and coding sequences (Fig. 6A), we used chromatin immunoprecipitation (ChIP) combined with quantitative PCR (see Fig. 6A for positions of amplicons tested). Mock ChIP reactions conducted in the absence of antibody revealed background signals (Fig. 6B). Antibodies recognizing histone modifications indicative of actively transcribed genes—namely, histone H3 acetylated on Lys 9 and Lys 14 (H3Ac); Histone H4 hyperacetylated on Lys 5, Lys 8, Lys 12, and Lys 16 (H4Ac); or histone H3 trimethylated on Lys 4 (H3K4me3)—yielded greater ChIP signals in hda6 mutants compared with wild-type plants throughout rRNA gene coding and noncoding regions (Fig. 6C–E; Supplemental Fig. S1). Conversely, levels of histone H3 dimethylated on Lys 9 (H3k9me2), a modification characteristic of inactive heterochromatic sequences, are substantially decreased in hda6 mutants relative to wild-type controls (Fig. 6F). At actin genes, which are constitutively transcribed by RNA Pol II, differences in H3K4me3 and H4Ac levels in wild-type versus hda6 mutant plants are not significant. However, at AtSN1 family retrotransposons, which are silenced by HDA6 and RNA-directed DNA methylation, H3K4me3 and histone acetylation levels increase, and H3K9me2 levels decrease in hda6 mutants compared with wild-type plants (Fig. 6). These chromatin changes are consistent with the derepression of AtSN1 elements in hda6 mutants (data not shown).
Figure 6.
Chromatin marks and polymerase occupancy at rRNA genes in wild-type and hda6 mutant plants. (A) Diagram showing the location of IGS, ETS, and coding region amplicons assayed by quantitative PCR following ChIP. (B–H) ChIP using chromatin isolated from shoots of 2-wk-old wild-type (Col-0) or hda6 mutant (axe1-5) plants. Following chemical cross-linking, chromatin was isolated and mock-immunoprecipitated (B) or immunoprecipitated using antibodies recognizing different histone modifications (C–F), RNA Pol I (G), or RNA Pol II (H). ACTIN2 and AtSN1 served as controls for active or (normally) repressed loci, respectively. (I) Association of Pol I and Pol II with long or short rRNA gene variants. Chromatin of wild-type or axe1-5 mutant plants was subjected to ChIP using anti-RNA Pol I or anti-RNA Pol II antibodies. Precipitated DNA was then subjected to PCR amplification using amplicon 1 primers. Reaction products following 25, 27, or 29 cycles of PCR amplification were resolved by agarose gel electrophoresis and stained with ethidium bromide. (J) Immunolocalization of RNA Pol II in interphase nuclei of wild-type or hda6 (axe1-5) mutant plants (green signals). Nuclei were counterstained with DAPI (blue signals). Two representative wild-type and two representative axe1-5 nuclei are shown. Bars, 5 μm.
ChIP using antibodies specific for RNA Pol I revealed Pol I in association with the pre-rRNA transcription unit, including the gene promoter region (+1), the 5′ ETS, the 25S coding sequences, and 3′ ETS sequences that are located immediately adjacent to the 25S coding sequences (Fig. 6G). In each of these intervals, Pol I occupancy is increased nearly twofold in hda6 mutants relative to wild-type controls, consistent with the derepression of long rRNA gene variants, which account for approximately one-half of the total rRNA gene pool. Pol I was not detected at substantial levels in the IGS adjacent to the Sal repeats in either wild-type or hda6 mutant plants (Fig. 6G). At the Actin 2 gene, transcribed by Pol II, and at AtSN1 retrotransposons, thought to be transcribed by Pol III, ChIP signals were essentially undetectable using the anti-Pol I antibody.
ChIP performed using an antibody specific for Pol II revealed that, in wild-type plants, Pol II is not detected in association with rRNA genes at levels above background (Fig. 6, cf. H and B). However, in hda6 mutants, Pol II associates with the rRNA genes throughout the coding and noncoding regions (Fig. 6H). Pol II association with AtSN1 retrotransposons also increases in hda6 mutants, unlike the actin gene control.
The ChIP data suggest that the high levels of IGS transcription occurring in hda6 mutants is attributable to the derepression of cryptic Pol II transcription units. This raised the possibility that Pol II, rather than Pol I, might be solely responsible for transcripts that include the indel defining the long and short rRNA gene variants. We tested this possibility using anti-Pol I and anti-Pol II antibodies for ChIP, followed by PCR amplification using amplicon 1 primers. In mature wild-type plants, Pol I is preferentially associated with the short variants (Fig. 6I), consistent with the preferential expression of this variant class (refer to Fig. 1). However, in hda6 mutants (axe1-5), Pol I is associated with both long and short variants, in proportion to their relative abundance (see input chromatin controls). Pol II association with rRNA genes also increases in hda6 mutants (Fig. 6I), indicating that Pol II transcription contributes to the synthesis of RNA in the region that defines the long and short variants. Taken together, the data of Figure 6, G and I, suggest that Pol I transcribes both long and short variants, beginning at the gene promoter and including the polymorphic indel region in the 3′ ETS. Superimposed onto this pattern of expression, spurious Pol II transcription occurs throughout the rRNA genes.
RNA Pol II is generally not present in the nucleolus of wild-type Arabidopsis nuclei (Fig. 6J, two representative nuclei are shown). The ChIP data showing that Pol II associates with rRNA genes in hda6 mutants prompted us to investigate whether Pol II localization is altered in the mutants. Indeed, Pol II can be observed throughout the nucleolus in many hda6 nuclei (Fig. 6J, top right panel). In other hda6 nuclei, Pol II is not distributed uniformly throughout nucleoli, but is nonetheless present within the internal periphery of the nucleolus (Fig. 6J, bottom right panel). Collectively, the ChIP and cytological evidence independently reveal the role of HDA6 in suppressing Pol II transcription of nucleolar rRNA genes.
Discussion
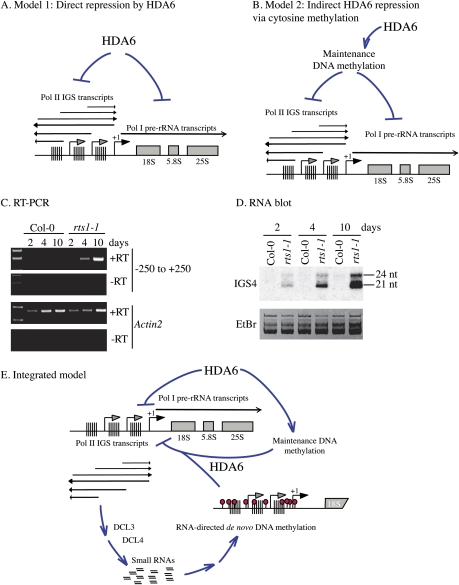
HDA6 is a key regulator of gene silencing that displays a complex interrelationship with DNA methylation. In the absence of HDA6 activity, symmetrical cytosine methylation is decreased at the same time that asymmetric, siRNA-directed de novo cytosine methylation accumulates. siRNA-directed DNA methylation is typically repressive, but this is not the case in hda6 mutants, suggesting that HDA6 acts downstream from de novo DNA methylation. Conversely, the loss of maintenance methylation in hda6 mutants suggests that HDA6 acts upstream of DNA methylation. These observations can be reduced to two simple models, which need not be mutually exclusive. One model is that HDA6-mediated histone modifications silence rRNA genes directly (Fig. 7A). An alternative model is that HDA6 acts indirectly through maintenance DNA methylation, which may be the critical determinant for gene silencing (Fig. 7B). Several observations support the direct action model. First, rescuing an hda6 mutant with a wild-type HDA6 transgene fully restores developmental silencing of long rRNA gene variants (Fig. 3C). However, CG and CHG maintenance methylation patterns are not fully restored in these lines (Fig. 5), arguing that restoration of HDA6 activity, and not maintenance methylation, is key. Second, the ability to clonally inherit maintenance methylation patterns suggests that loss of these patterns early in development in hda6 mutants might yield a relatively constant molecular phenotype. However, IGS transcripts and siRNAs become more and more abundant in hda6 mutants during early development, indicating that HDA6 activity is needed continuously to suppress their expression (Fig. 7C,D). Last, the fact that de novo methylation occurs in hda6 mutants but cannot bring about silencing fits with the hypothesis that HDA6 is critical for silencing in its own right. Based on these considerations, we favor a model in which HDA6 is critical for suppressing Pol II and Pol I transcription, but DNA methylation plays a reinforcing role, potentially helping recruit HDA6 (Fig. 7E).
Figure 7.
Models for the role of HDA6 in rRNA gene variant regulation. (A,B) Models for HDA6 acting directly or through cytosine maintenance methylation. (C) RT–PCR analysis of IGS transcripts in wild-type and hda6 mutants 2, 4, and 10 d after germination. PCR products were subjected to gel electrophoresis and stained with ethidium bromide. (−RT) Controls lacking reverse transcriptase. (D) RNA blot analysis of IGS small RNAs present in total RNA isolated from wild-type and hda6 mutants 2, 4, and 10 d after germination. Ethidium bromide staining shows that similar amounts of RNA were loaded in all lanes. (E) HDA6 suppresses sense and antisense IGS transcription by Pol II. Resulting siRNA production and siRNA-directed de novo cytosine methylation does not repress transcription in the absence of HDA6. Maintenance methylation at CG and CHG motifs is decreased in hda6 mutants, indicating that HDA6 acts upstream of maintenance methylation. Collectively, losses of maintenance cytosine methylation or losses of repressive chromatin marks due to lack of HDA6 activity and rampant IGS transcription prevent the developmental silencing of rRNA gene variants.
In general, the regulation of RNA gene variant expression in nonhybrid A. thaliana shows an overall similarity to uniparental rRNA gene silencing (nucleolar dominance) in hybrid A. suecica. In both cases, silencing is developmentally regulated and requires HDA6 and cytosine methylation. These observations fit with our previous conclusion that nucleolar dominance is a manifestation of rRNA gene dosage control, which operates in all eukaryotes studied to date, including nonhybrids. Interestingly, in A. suecica, nucleolar dominance requires the de novo cytosine methyltransferase DRM2, whereas knocking down the maintenance methyltransferases MET1 or CMT3 does not abrogate rRNA gene silencing (Preuss et al. 2008). However, in the present study, de novo methylation increases in hda6 mutants, yet rRNA genes are derepressed. Although these observations appear contradictory, the loss of CG and CHG methylation at the same time that CHH methylation is gained in hda6 mutants is a complicating factor that precludes a simple interpretation. A combination of de novo methylation and HDA6-dependent maintenance methylation patterns may be important.
The fact that S1 nuclease protection assays do not detect a twofold increase in nascent transcript signals initiating at the rRNA gene promoter when both long and short variants are expressed, as opposed to when only short variants are expressed, is puzzling. This is especially true given that we do observe an approximately twofold increase in Pol I ChIP signals associated with pre-rRNA transcription units in hda6 relative to wild-type plants. One possible explanation for this apparent paradox could be that the half-life of the ETS sequences immediately downstream from the transcription start site becomes shorter as more genes are expressed, perhaps because processing must become more efficient. Further studies will be needed to test this hypothesis.
A surprising aspect of our study is that Pol II transcription is not incompatible with Pol I transcription of rRNA genes in plants. Likewise, a recent study using mammalian cells defective for DNA methyltransferase activity showed that derepression of normally silent rRNA genes correlates with Pol II transcription (Gagnon-Kugler et al. 2009). We speculate that Pol II transcription causes the cotranscriptional replacement of repressive chromatin marks with histone modifications typical of active genes, thereby allowing the Pol I transcription machinery greater access to rRNA gene promoters. Importantly, there has long been evidence for a positive role of IGS transcription in rRNA gene expression in frogs and flies. Xenopus laevis and Xenopus borealis have IGS organizations very similar to the A. thaliana IGS organization depicted in Figure 1A. In Xenopus, full enhancer activity of spacer repeats requires the presence of at least one spacer promoter (DeWinter and Moss 1986). Likewise, at least one spacer promoter is needed in addition to enhancer repeats for X. laevis minigenes to outperform X. borealis minigenes in nucleolar dominance-like competition assays (Caudy and Pikaard 2002). Much of the Drosophila IGS is composed of duplicated spacer promoters that stimulate transcription from the downstream promoter (Grimaldi et al. 1990). Collectively, these observations suggest that spacer transcripts, or possibly the act of spacer transcription, assist in rRNA gene promoter activation.
In parallel with evidence that spacer transcription can positively influence rRNA gene expression, there is evidence that IGS transcripts can repress transcription. In mice, structured RNAs that include promoter sequences recruit the NoRC repressor complex (Mayer et al. 2006, 2008). Moreover, our prior studies of nucleolar dominance in A. suecica implicate IGS siRNAs, generated by DCL3, in the RNA-directed repression of rRNA genes. The evidence that spacer transcription can be activating or repressing suggests that not all spacer transcripts are equal, such that their influence on rRNA gene transcription is dependent on the precise transcription unit that is used (Santoro et al. 2010). Deciphering the origins and regulation of IGS transcripts in future studies should yield important new insights into the complex regulation of rRNA genes in both plants and mammals.
Materials and methods
Mutant and control plant lines
DR5, axe1-5, and axe1-4 seeds were provided by J. Murfett and T. Guilfoyle; rts1-1 was provided by W. Aufsatz and M. Matzke; and ddm1 was provided by E. Richards. Plants were grown in growth chambers or a greenhouse.
HDA6 mutagenesis, purification, and assay
The HDA6 gene was PCR-amplified from A. thaliana (ecotype Col-0) genomic DNA using Platinum Pfx (Invitrogen) polymerase and forward (5′-CACCCTTTTACTACTTTACTCTCAAGTCAACCG-3′) and reverse (5′-AGACGATGGAGGATTCACGTCTGG-3′) primers and then was cloned into pENTR-D TOPO (Invitrogen). Site-directed ligase-independent mutagenesis (SLIM) was performed on pENTR-D- HDA6 (Chiu et al. 2004) using primer pairs listed in the Supplemental Material. HDA6 genes in pENTR vectors were recombined into the pEarleyGate 302 destination vector (Earley et al. 2006b) using LR clonase and were transfected into Arabidopsis tumefaciens strain GV3101. Transformation of A. thaliana (Col-0) was by the floral dip method (Clough and Bent 1998). HDA6 purification, histone acetylation, and histone deacetylation assays were performed as described (Earley et al. 2006a, 2007).
Small RNA analyses
Small RNAs were isolated using a mirVana kit (Ambion). Gel electrophoresis and blotting were performed as described (Onodera et al. 2005). Hybridization probes were generated using a mirVana probe construction kit (Ambion), and were purified using Performa DTR Gel Filtration Cartridges, as described previously (Onodera et al. 2005). Oligonucleotide probes are listed in the Supplemental Material.
RT–PCR
Total RNA was isolated using a RNAeasy Plant mini-kit (Qiagen). S1 nuclease protection using a 5′-end-labeled promoter probe was performed as described previously (Lawrence et al. 2004). For RT–PCR assays, RNA was DNase-treated for 60 min using RQ1 DNase (Promega) or 45 min using Turbo DNase (Ambion) according to the manufacturers' instructions. Random-primed cDNA was amplified from 500 ng of total RNA using SuperScript III reverse transcriptase (Invitrogen). PCR amplification conditions were 25–26 cycles of 30 sec at 94°C, 30 sec at 53°C, and 60 sec at 72°C unless noted otherwise. PCR primers are listed in the Supplemental Material.
DNA methylation assays
Genomic DNA was purified from 2-wk-old shoots using a Nucleon Phytopure plant DNA extraction kit (Amersham Biosciences). For Southern blots, 3 g of DNA was digested overnight with 30 U of HpaII, MspI, or AluI. Following 0.8% agarose gel electrophoresis, DNA was depurinated and denatured within the gel and transferred to Hybond-N+ nylon membranes (GE Healthcare). Hybridization was performed overnight at 55°C in 0.25 M sodium phosphate (pH 7.2), 1 mM EDTA, 6.6% SDS, and 1% BSA. Amplicon 1 and promoter (−250 to +250) PCR products were made into probes by random priming (Feinberg and Vogelstein 1983) and labeling with α-[32P]-dCTP. Membranes were washed twice for 5 min with 2× SSC and 0.1% SDS and twice for 20 min in 0.1× SSC and 0.1% SDS and were subjected to phosphorimaging. For Chop-PCR, 100 ng of DNA was digested 60 min with 5 U of HpaII, MspI, or AluI (New England Biolabs) at 37°C. An aliquot of the reaction was then subjected to 20 cycles of PCR amplification, followed by agarose gel electrophoresis and ethidium bromide staining.
ChIP
ChIP was performed as described previously (Wierzbicki et al. 2008). Anti-H3K9me2 (catalog no. ab7312), anti-H3K4me3 (catalog no. ab8580), and anti-H3Ac (catalog no. ab4441) antibodies were purchased from Abcam. Anti-H4Ac (catalog no. 06.598) was obtained from Upstate Biotechnologies. Anti-Pol I and anti-Pol II antibodies were raised against the N-terminal domains of the NRPA2 (At1g29940) or NRPB2 (At4g21710) subunits, respectively. Quantitative PCR was performed on purified immunoprecipitated DNA using 0.5 U of Platinum Taq (Invitrogen), SYBR Green I (Invitrogen), and Internal Reference dye (Sigma). Results were analyzed using the comparative CT method (Livak and Schmittgen 2001) relative to input.
DNA-FISH and protein immunolocalization
DNA-FISH to detect 45S rRNA gene loci (NORs), and protein immunolocalization using anti-Flag antibodies, were performed using isolated nuclei, as described previously (Pontes et al. 2003, 2006). For dual HDA6-Flag/NOR localization, slides were subjected to immunofluorescence and post-fixed in 4% formaldehyde/PBS, followed by DNA-FISH. DNA was counterstained with DAPI (1 g/mL) in Vectashield (Vector Laboratories). Nuclei were examined using a Nikon Eclipse E800i epifluorescence microscope equipped with a Photometrics Coolsnap ES Mono digital camera and a Nikon A1 laser-scanning confocal microscope using 100× objectives. Lasers were 405 nm for DAPI, 488 nm for Alexa 488, and 561 nm for Alexa 594. Confocal and wide-field images were processed using Imaris (Bitplane) and Adobe Photoshop, respectively. For each experiment, 100–200 nuclei were analyzed. Representative nuclei are shown.
Acknowledgments
K.W.E. created all transgenic lines and produced Figures 1, A,C,D; 2, A–C; 3, A,B; 4, B–D; and 6I. F.P. produced Figures 1E, 3C, 5B, and 7 (except part D) and Supplemental Figure S1. A.T.W. produced Figure 6, B–H); S.T. produced Figure 4A; O.P. produced Figures 1B and 6J; and T.B. generated Figures 5A and 7D. P.C.N. first noted accumulation of IGS small RNAs in hda6 mutants. K.W.E., F.P., and C.S.P. wrote the manuscript. We thank Jeremy Haag, Tom Ream, Alexa Vitins, Ek Han Tan, and Sasha Preuss for helpful discussions. This research was supported by NIH grant GM60380 (to C.S.P.). P.C-N. was supported by fellowships SFRH/BD/6520/2001 and SFRH/BPD/30386/2006 from the Fundação para a Ciência e Tecnologia, Portugal. T.B. was supported by a Ruth L. Kirschstein National Research Service Award. O.P. was supported by a grant from the Edward Mallinckrodt Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1914110.
Supplemental material is available at http://www.genesdev.org.
References
- Aufsatz W, Mette MF, van der Winden J, Matzke AJ, Matzke M 2002. RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci 99: 16499–16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy AA, Pikaard CS 2002. Xenopus ribosomal RNA gene intergenic spacer elements conferring transcriptional enhancement and nucleolar dominance-like competition in oocytes. J Biol Chem 277: 31577–31584 [DOI] [PubMed] [Google Scholar]
- Chen ZJ 2007. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Pikaard CS 1997. Transcriptional analysis of nucleolar dominance in polyploid plants: Biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc Natl Acad Sci 94: 3442–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu J, March PE, Lee R, Tillett D 2004. Site-directed, ligase-Independent mutagenesis (SLIM): A single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res 32: e174 doi: 10.1093/nar/gnh172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Pikaard CS 1996a. RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. Plant J 9: 259–272 [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Pikaard CS 1996b. Two-dimensional RFLP analyses reveal megabase-sized clusters of rRNA gene variants in Arabidopsis thaliana, suggesting local spreading of variants as the mode for gene homogenization during concerted evolution. Plant J 9: 273–282 [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Doelling JH, Gens JS, Pikaard CS 1995. Use of RFLPs larger than 100 kbp to map the position and internal organization of the nucleolus organizer region on chromosome 2 in Arabidopsis thaliana. Plant J 7: 273–286 [DOI] [PubMed] [Google Scholar]
- DeWinter R, Moss T 1986. Spacer promoters are essential for efficient enhancement of X. laevis ribosomal transcription. Cell 44: 313–318 [DOI] [PubMed] [Google Scholar]
- Doelling JH, Gaudino RJ, Pikaard CS 1993. Functional analysis of Arabidopsis thaliana rRNA gene and spacer promoters in vivo and by transient expression. Proc Natl Acad Sci 90: 7528–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS 2006a. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes & Dev 20: 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS 2006b. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS 2007. In vitro specificities of Arabidopsis co-activator histone acetyltransferases: Implications for histone hyperacetylation in gene activation. Plant J 52: 615–626 [DOI] [PubMed] [Google Scholar]
- Espada J, Ballestar E, Santoro R, Fraga MF, Villar-Garea A, Nemeth A, Lopez-Serra L, Ropero S, Aranda A, Orozco H, et al. 2007. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res 35: 2191–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP 1999. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401: 188–193 [DOI] [PubMed] [Google Scholar]
- French SL, Osheim YN, Cioci F, Nomura M, Beyer AL 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol 23: 1558–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furner IJ, Sheikh MA, Collett CE 1998. Gene silencing and homology-dependent gene silencing in Arabidopsis: Genetic modifiers and DNA methylation. Genetics 149: 651–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon-Kugler T, Langlois F, Stefanovsky V, Lessard F, Moss T 2009. Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol Cell 35: 414–425 [DOI] [PubMed] [Google Scholar]
- Grimaldi G, Fiorentini P, Di Nocera P 1990. Spacer promoters are orientation-dependent activators of pre-rRNA transcription in Drosophila melanogaster. Mol Cell Biol 10: 4667–4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruendler P, Unfried I, Pascher K, Schweizer D 1991. rDNA intergenic region from Arabidopsis thaliana. Structural analysis, intraspecific variation and functional implications. J Mol Biol 221: 1209–1222 [DOI] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS 2003. Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol 4: 641–649 [DOI] [PubMed] [Google Scholar]
- Hassig CA, Tong JK, Fleischer TC, Owa T, Grable PG, Ayer DE, Schreiber SL 1998. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci 95: 3519–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK 2009. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes & Dev 23: 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Noller HF 2007. The ribosome in focus: New structures bring new insights. Trends Biochem Sci 32: 434–441 [DOI] [PubMed] [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS 2004. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell 13: 599–609 [DOI] [PubMed] [Google Scholar]
- Lippman Z, May B, Yordan C, Singer T, Martienssen R 2003. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol 1: E67 doi: 10.1371/journal.pbio.0000067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mathieu O, Jasencakova Z, Vaillant I, Gendrel AV, Colot V, Schubert I, Tourmente S 2003. Changes in 5S rDNA chromatin organization and transcription during heterochromatin establishment in Arabidopsis. Plant Cell 15: 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA 2005. RNAi-mediated pathways in the nucleus. Nat Rev Genet 6: 24–35 [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ 2009. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21: 367–376 [DOI] [PubMed] [Google Scholar]
- May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA 2005. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet 1: e79 doi: 10.1371/journal.pgen.0010079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R 2006. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22: 351–361 [DOI] [PubMed] [Google Scholar]
- Mayer C, Neubert M, Grummt I 2008. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep 8: 774–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B 2006. Nucleolar dominance: A model for rRNA gene silencing. Genes & Dev 20: 1207–1214 [DOI] [PubMed] [Google Scholar]
- McStay B, Grummt I 2008. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol 24: 131–157 [DOI] [PubMed] [Google Scholar]
- Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V 2007. A housekeeper with power of attorney: The rRNA genes in ribosome biogenesis. Cell Mol Life Sci 64: 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfett J, Wang XJ, Hagen G, Guilfoyle TJ 2001. Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13: 1047–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS 2005. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Pikaard CS, Pape LK, Henderson SL, Ryan K, Paalman MH, Lopata MA, Reeder RH, Sollner-Webb B 1990. Enhancers for RNA polymerase I in mouse ribosomal DNA. Mol Cell Biol 10: 4816–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Lawrence RJ, Neves N, Silva M, Lee JH, Chen ZJ, Viegas W, Pikaard CS 2003. Natural variation in nucleolar dominance reveals the relationship between nucleolus organizer chromatin topology and rRNA gene transcription in Arabidopsis. Proc Natl Acad Sci 100: 11418–11423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS 2006. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126: 79–92 [DOI] [PubMed] [Google Scholar]
- Pontes O, Lawrence RJ, Silva M, Preuss S, Costa-Nunes P, Earley K, Neves N, Viegas W, Pikaard CS 2007. Postembryonic establishment of megabase-scale gene silencing in nucleolar dominance. PLoS One 2: e1157 doi: 10.1371/journal.pone.000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss S, Pikaard CS 2007. rRNA gene silencing and nucleolar dominance: Insights into a chromosome-scale epigenetic on/off switch. Biochim Biophys Acta 1769: 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss SB, Costa-Nunes P, Tucker S, Pontes O, Lawrence RJ, Mosher R, Kasschau KD, Carrington JC, Baulcombe DC, Viegas W, et al. 2008. Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol Cell 32: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Fagard M, Proux F, Mourrain P, Boutet S, Earley K, Lawrence RJ, Pikaard CS, Murfett J, Furner I, et al. 2004. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala SH, Richards EJ 2007. Differential epigenetic regulation within an Arabidopsis retroposon family. Genetics 176: 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder RH 1989. Regulatory elements of the generic ribosomal gene. Curr Opin Cell Biol 1: 466–474 [DOI] [PubMed] [Google Scholar]
- Richards EJ 1997. DNA methylation and plant development. Trends Genet 13: 319–323 [DOI] [PubMed] [Google Scholar]
- Russell J, Zomerdijk JC 2006. The RNA polymerase I transcription machinery. Biochem Soc Symp 73: 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier JJ, French S, Osheim Y, Cheung WL, Gallo CM, Beyer AL, Smith JS 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J 21: 4959–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32: 393–396 [DOI] [PubMed] [Google Scholar]
- Santoro R, Schmitz KM, Sandoval J, Grummt I 2010. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep 11: 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ 1993. Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Wanzenbock EM, Schofer C, Schweizer D, Bachmair A 1997. Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana. Plant J 11: 1007–1016 [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS 2008. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]