Abstract
Break-induced replication (BIR) is an efficient homologous recombination (HR) pathway employed to repair a DNA double-strand break (DSB) when homology is restricted to one end. All three major replicative DNA polymerases are required for BIR, including the otherwise nonessential Pol32 subunit. Here we show that BIR requires the replicative DNA helicase (Cdc45, the GINS, and Mcm2–7 proteins) as well as Cdt1. In contrast, both subunits of origin recognition complex (ORC) and Cdc6, which are required to create a prereplication complex (pre-RC), are dispensable. The Cdc7 kinase, required for both initiation of DNA replication and post-replication repair (PRR), is also required for BIR. Ubiquitination and sumoylation of the DNA processivity clamp PCNA play modest roles; in contrast, PCNA alleles that suppress pol32Δ's cold sensitivity fail to suppress its role in BIR, and are by themselves dominant inhibitors of BIR. These results suggest that origin-independent BIR involves cross-talk between normal DNA replication factors and PRR.
Keywords: Break-induced replication (BIR), DNA replication, recombination, Saccharomyces cerevisiae, double-strand break (DSB), post-replication repair
Cells are routinely exposed to potentially lethal DNA double-strand breaks (DSBs) that arise during DNA replication as well as from exogenous exposure to DNA-damaging agents. DSB repair is essential for both cell viability and maintenance of genomic integrity, as aberrant or defective repair of even a single break may result in chromosome loss, genomic instability, or cell death. To counteract these potentiality catastrophic events, cells have evolved multiple mechanisms to repair such lesions. Generally, yeast cells repair a DSB by gene conversion (GC), in which both ends of the break interact with homologous sequences that can be used as a template for repair. These may be found on a sister chromatid, homologous chromosome, or ectopic location (Pâques and Haber 1999; Symington 2002). However, when only one end of the DSB shares homology with a template, another efficient homologous recombination (HR) pathway, known as break-induced replication (BIR), may be used to repair the break. In BIR, recombination is used to establish a unidirectional replication fork that can copy the donor template to the end of the chromosome (McEachern and Haber 2006; Llorente et al. 2008).
BIR is thought to restart stalled or collapsed replication forks during S phase (Haber 1999; Michel 2000). BIR also can elongate budding yeast telomeres in the absence of telomerase in a mechanism that may be analogous to the alternative lengthening of telomeres (ALT), which is seen in many transformed mammalian cell lines and some human cancers (Lydeard et al. 2007). In addition, BIR has been suggested to be a cause of segmental duplications (Payen et al. 2008), and to be the cause of deletions, nonreciprocal translocations, and complex rearrangements seen in a number of human diseases and cancers (Hastings et al. 2009).
Many of the recombination protein requirements for BIR have been well characterized and are common to both GC and BIR. The predominant form of BIR requires the key recombination proteins Rad51 and Rad52, as do all GC events (Davis and Symington 2004; Malkova et al. 2005). Unlike GC, Rad51-dependent BIR requires the nonessential subunit of Polδ, Pol32, to initiate new BIR (Lydeard et al. 2007). BIR also requires both leading and lagging strand replication machinery, although DNA Polɛ is only needed for the elongation of BIR (Lydeard et al. 2007).
Two of the major questions regarding the DNA synthesis steps of BIR are: (1) What DNA helicases are responsible for unwinding the template DNA? (2) How is the replication fork established in the absence of an origin? In eukaryotic S phase, replication is initiated at multiple origins of replication in a highly regulated process so that the entire genome is completely replicated only once (for reviews, see Arias and Walter 2007; Sclafani and Holzen 2007). This regulation is first achieved by regulating the stepwise assembly of the multiprotein prereplication complex (pre-RC) onto DNA at origins of replication. The six-subunit origin recognition complex (ORC) serves to nucleate pre-RC formation at these origins. As the cell transitions from the M to G1 phase, the initiation factor Cdc6 is synthesized and binds to the origin-bound ORC. During G1, another initiation factor, Cdt1, is interdependently imported into the nucleus with the Mcm2–7 DNA helicase and is required for the recruitment of Mcm2–7 to the pre-RC.
In order for the pre-RC to be converted into an active replication fork, the origin DNA needs to be locally denatured by a DNA helicase, so that the replicative DNA polymerases can be loaded onto the origin. These steps are regulated through CDK1-mediated phosphorylation of the essential replication proteins Sld2 and Sld3 that increases their affinity for Dpb11 (Tanaka et al. 2007; Zegerman and Diffley 2007). Formation of this complex stimulates recruitment of Cdc45 and the GINS complex, both of which are required for the pre-RC to be converted into a preinitiation complex (pre-IC) (Kamimura et al. 2001; Takayama et al. 2003). Cdc45 and GINS act in concert with Mcm2–7 to form the functional replicative DNA helicase that travels with the elongating replication machinery away from the origin (Gambus et al. 2006; Moyer et al. 2006; Pacek et al. 2006). The initial recruitment of Cdc45 to form the pre-IC requires an additional protein, Mcm10, which interacts physically with ORC, the Mcm2–7 complex, and Cdc45 (Wohlschlegel et al. 2002; Sawyer et al. 2004).
Recruitment of Cdc45 to form the pre-IC is also dependent on another cell cycle-dependent kinase, Dbf4–Cdc7, which targets many proteins, including Mcm2-7, to allow recruitment of Cdc45 and GINS to the origins and to permit firing of the replication forks after entry into S phase (Sheu and Stillman 2006). Work in mammalian cells indicates that Cdc7/Dbf4-dependent recruitment of Cdc45 to the pre-IC is mediated through Cdt1 (Ballabeni et al. 2009). Following the recruitment of Cdc45 and the GINS complex, the replicative DNA polymerases responsible for copying the leading and lagging strands, the DNA processivity clamp PCNA, as well as the rest of the proteins required to form a stable and functional replisome are recruited to the origin.
To determine which components of the replication machinery are required for BIR and how this replication fork is established in the absence of an origin, we took a genetic candidate approach by examining mutations of different replication proteins responsible for both initiation and elongation of DNA replication. We found that BIR requires the Mcm2–7 DNA helicase as well as Cdc45 and the GINS complex. Furthermore, BIR requires Dpb11–Sld3, Mcm10, Ctf4, and Cdt1, but not those components that appear to be exclusively required for formation of the pre-RC: Cdc6 and ORC. BIR also requires the cell cycle-dependent kinase Cdc7 to initiate BIR DNA synthesis, a step that may also involve the post-translational modification of PCNA by Rad18 and Siz1. We also describe novel PCNA alleles that suppress the cold sensitivity of pol32Δ cells, but are themselves dominant-negative for BIR but not GC. Taken together, these findings provide significant insights into the genetic requirements for establishing the BIR DNA replication machinery and its regulation as a replication restart mechanism in eukaryotes.
Results
Assay to study BIR
We showed previously that Rad51-dependent BIR requires both the leading and lagging strand replication machinery (Lydeard et al. 2007). However, it is not known which DNA helicase is responsible for unwinding the template duplex DNA in BIR. To test the importance of various replication proteins, we used the previously described haploid system (Lydeard et al. 2007) to study BIR in Saccharomyces cerevisiae (Fig. 1). In strain JRL346, a HO endonuclease cut site was integrated into the CAN1 gene on the nonessential terminal region of chromosome V (Ch V), such that the 3′ portion of the gene was deleted (designated CA). A 3′ segment of the CAN1 gene (denoted as AN1), sharing 1157-base-pair (bp) homology with CA on Ch V, was introduced in the same orientation into Ch XI, 30 kb from its telomere. Galactose-induced expression of HO endonuclease creates a DSB that must be repaired to maintain cell viability. Nearly all viable cells repair the break by BIR, resulting in a nonreciprocal translocation that restores an intact CAN1 gene—rendering cells canavanine-sensitive—but results in loss of the hygromycin-resistant (HPH) marker. Repair can be monitored by measuring cell viability or by using a PCR assay that detects the first 242 bp of new DNA synthesis (Figs. 1, 2A). To study the role of essential DNA factors in BIR, we used temperature-sensitive (ts) mutations or temperature-inducible degron alleles. Cells were grown at the permissive temperature of 25°C, and then arrested and maintained with nocodazole to ensure that they had completed DNA replication and did not re-enter S phase; this avoids the problem of replication factors being sequestered during DNA replication (Holmes and Haber 1999; Wang et al. 2004). We note that, when the DSB is induced in cycling cells, they experience checkpoint-mediated G2/M arrest long before they initiate new DNA synthesis in BIR (Malkova et al. 2005), indicating that BIR repair normally occurs after replication is complete, although in this case replication factors might have been recruited while cells were still in S phase. The cultures were then split, and either maintained at the permissive temperature or shifted to the restrictive temperature of 37°C; after 3 h, HO endonuclease was induced, and repair was monitored by PCR.
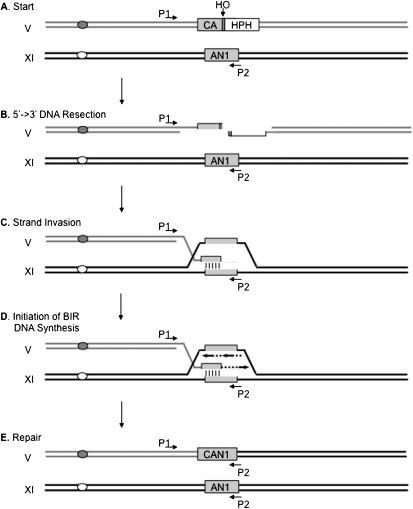
Figure 1.
Experimental system and model of BIR. (A) An HPHMX-marked HO cut site (gray bar) is integrated into the CAN1 gene on Ch V, deleting the 3′ end portion of the gene; the remaining sequences are represented as CA. The AN1 donor sharing 1157-bp homology with CAN1 is integrated into Ch XI. At the start, PCR primers P1 and P2, which are used to monitor the initiation of new DNA synthesis, will not amplify because they sit on different chromosomes. (B) Following HO expression, a DSB is generated and the broken chromosome ends are resected by 5′-to-3′ exonucleases to generate ssDNA. (C) The Rad51 recombinase binds to the 3′-ended single strand and mediates the homology search and strand invasion of the donor template to form a D-loop. (D) The BIR replication machinery is recruited to the recombination intermediate, and the leading and lagging strand coordinately initiate new DNA synthesis. The initiation of new DNA synthesis is detected by PCR when the P2 sequences are covalently linked to the P1 sequences. (E) BIR DNA synthesis extends to the end of the template chromosome, resulting in a nonreciprocal translocation in which the CAN1 gene is restored, while the entire donor chromosome arm is duplicated and sequences distal to the original HO cut site are lost.
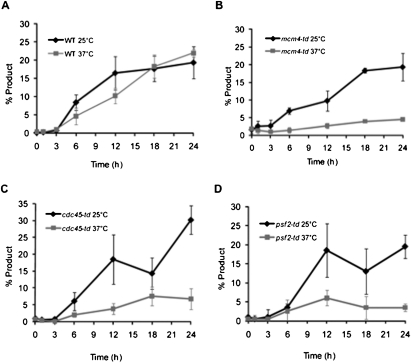
Figure 2.
The Mcm2–7 helicase is required for BIR DNA synthesis. (A–D) Appearance of BIR repair product, as monitored by PCR, in wild-type (WT), mcm4-td, cdc45-td, and psf2-td cells arrested in nocodazole at either 25°C or 37°C. Error bars represent ±SEM for wild type, mcm4-td, and cdc45-td, and the error range for two independent measurements of psf2-td.
The Mcm2–7 helicase is required for the initiation of new BIR DNA synthesis
To examine the role of the Mcm2–7 helicase in BIR, we constructed a temperature-sensitive mcm4-td degron strain (Labib et al. 2000). As measured by the aforementioned PCR assay and shown in Figure 2B, we found that BIR is defective at the restrictive temperature of 37°C in the mcm4-td strain (P < 0.05 at 18 h and later). We then asked if BIR required both Cdc45 and the GINS complex, using strains with degron alleles of cdc45-td (Tercero et al. 2000) and psf2-td (Gambus et al. 2006), which encodes one of the four subunits of the GINS complex. As is the case with mcm4-td, both cdc45-td (P = 0.01 at 24 h) and psf2-td are defective in the initiation of BIR DNA synthesis at 37°C, but are functional at 25°C (Fig. 2). These data clearly demonstrate that all known components of the DNA replicative helicase are required for BIR.
BIR requires Cdt1 but is independent of Cdc6 and ORC
In S-phase replication, the Mcm2–7 helicase and replication machinery are recruited to defined origins of replication; in contrast, BIR is initiated by recombination at apparently any homologous donor sequence. It is possible, however, that the assembly of a BIR replication fork would depend on helicases that were bound at origins, and are then somehow able to slide to the sites of recombination, although BIR can occur in G2/M-arrested cells where there should not be any pre-RCs. We hypothesized that the pre-RC components that are required only to initiate origin-specific replication would be dispensable in BIR. We therefore tested genes required for assembly of the pre-RC for their ability to successfully complete BIR.
ORC is bound to origins of replication throughout the cell cycle, and is essential for forming the pre-RC (Diffley et al. 1994). After sequential recruitment of Cdc6, Cdt1, and an initial Mcm2–7 complex to the origin, additional ORC ATPase and Cdc6 ATPase activity is required for repeated loading of Mcm2–7 onto origins (Randell et al. 2006). To test if Cdc6 is required for BIR, we introduced the well-characterized cdc6-1 conditional allele (Zhou et al. 1989). In G2-arrested cells, at the restrictive temperature of 37°C, cdc6-1 cells have no discernable defect in the ability of BIR to initiate DNA synthesis (Fig. 3A).
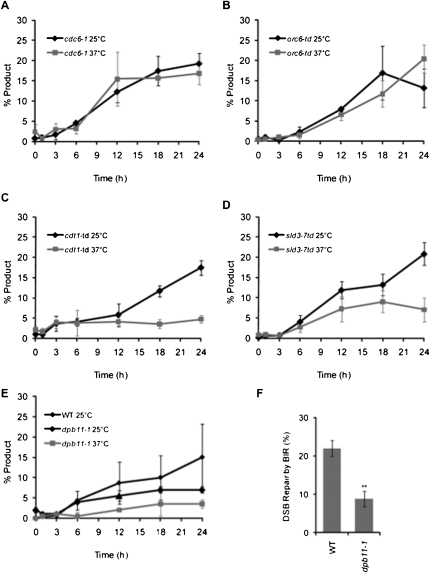
Figure 3.
Cdt1, Dpb11, and Sld3 are required for BIR DNA synthesis. (A–E) Appearance of BIR repair product, as monitored by PCR, in cdc6-1, orc6-td, cdt1-td, dpb11-1, and sld3-7td cells arrested in nocodazole at either 25°C or 37°C. Error bars represent ±SEM for cdc6-1, orc6-td, cdt1-td, and sld3-7td, and the error range for two independent measurements of dpb11-1. (F) Efficiency of BIR in wild type (WT) and dpb11-1 as measured by viability following a DSB at the permissive temperature of 25°C. Error bars represent ±SEM.
Orc6 interacts physically with Cdt1, is essential for Cdt1 and Mcm2–7 recruitment to origins (Chen et al. 2007), and is important in maintaining Mcm2–7 association with chromatin (Semple et al. 2006; Da-Silva and Duncker 2007). We used the conditional orc6-td degron allele (Chen et al. 2007). In nocodazole-arrested cells, there was no difference in the ability of Orc6-td to initiate BIR DNA synthesis at 37°C, compared with repair at the permissive temperature of 25°C (Fig. 3B).
Finally, we tested Cdt1 using the well-characterized Tet-regulated cdt1-td conditional degron allele (Tanaka and Diffley 2002). Under restrictive conditions—in which the cells are maintained at 37°C to degrade the degron allele and 20 μg/mL doxycycline is added to inhibit transcription—cdt1-td cells (P < 0.01 at 18 h and later) cannot initiate new BIR DNA synthesis (Fig. 3C). These results suggest that Cdt1 is essential for establishing a recombination-dependent replication fork that does not depend on either ORC or Cdc6.
BIR requires the Dpb11–Sld2–Sld3 complex for initiation of new DNA synthesis
The Dpb11–Sld2–Sld3 complex is required to recruit Cdc45 and the GINS complex to the pre-RC. To assess its role in BIR, we used the temperature-sensitive dpb11-1 allele. At the restrictive temperature, dpb11-1 cells are proficient in pre-RC formation but defective in initiating DNA synthesis (Kamimura et al. 1998). In the PCR assay, even at the permissive temperature of 25°C, dpb11-1 cells are defective in initiating BIR as compared with wild-type cells at 25°C (Fig. 3E). The defect is even more severe at 37°C when compared with wild type at 37°C or with dpb11-1 at 25°C. Even at the permissive temperature, the viability of these cells was reduced from 20% in wild-type cells to 10% (P = 0.003) (Fig. 3F), a value consistent with the amount of product seen in the PCR assay. A temperature-sensitive degron mutant of Dpb11's binding partner, Sld3, sld3-7td (Kanemaki and Labib 2006), also shows a defect in BIR at 37°C (P = 0.028 at 24 h) (Fig. 3D). We conclude that the Dpb11–Sld3–Sld2 complex is required for the initiation of new BIR synthesis, most likely by recruiting Cdc45 and the GINS complex to the assembling replication machinery at the BIR template.
Mcm10 and Ctf4 are important for BIR
Mcm10 is required for loading of Mcm2–7, Cdc45, and GINS to the origin. Along with another replication factor, Ctf4, Mcm10 also recruits DNA lagging strand polymerase Polα to the origin and maintains its interaction with the traversing replication fork (Ricke and Bielinsky 2004; Zhu et al. 2007). We studied the previously characterized degron allele mcm10-td (Ricke and Bielinsky 2004) and found that mcm10-td cells are defective in initiation of new BIR DNA synthesis at 37°C (Fig. 4A). This result is consistent with the requirement for Mcm2–7, Cdc45, and GINS in BIR, and our previous findings that the lagging strand DNA polymerase Polα–primase complex is required for BIR (Lydeard et al. 2007).
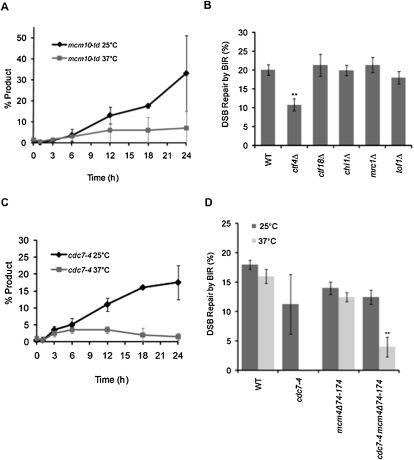
Figure 4.
Mcm10 and Cdc7 are required for the initiation of BIR DNA synthesis, and Ctf4 is important for BIR. (A) Appearance of BIR repair product, as monitored by PCR, in mcm10-td cells arrested in nocodazole at either 25°C or 37°C. Error bars represent the error range for two independent measurements. (B) Efficiency of BIR in wild-type (WT), ctf4Δ, ctf18Δ, chl1Δ, mrc1Δ, and tof1Δ cells, as measured by viability following a DSB. Error bars represent ±SEM. (C) Appearance of BIR repair product, as monitored by PCR, in cdc7-4 cells arrested in nocodazole at either 25°C or 37°C. Error bars represent the error range for two independent measurements. (D) Efficiency of BIR in wild-type (WT), cdc7-4, mcm4Δ74-174, and cdc7-4 mcm4Δ74-174 cells, as measured by viability following a DSB at the permissive temperature of 25°C or restrictive temperature of 37°C. Error bars represent ±SEM.
We also tested the role of the nonessential replication factor Ctf4 in BIR. We measured the viability of ctf4Δ cells after inducing a DSB that can only be repaired by BIR (Fig. 4B). There was a twofold decrease in ctf4Δ cells (P = 0.001) compared with wild type, indicating that Ctf4 is also important for BIR. Furthermore, we found that ctf4Δ cells arrested in nocodazole are impaired in the ability to initiate BIR synthesis (Supplemental Fig. S1).
In addition to its role in replication initiation and elongation, Ctf4 is also involved in sister chromatid cohesion (Hanna et al. 2001). We tested other nonessential factors that are involved in both replication fork maintenance and sister chromatid cohesion, including Mrc1, Tof1, Ctf18, and Chl1 (Gambus et al. 2006; Lengronne et al. 2006; Xu et al. 2007). None of these factors affects the viability of BIR in copying a template 30 kb to its telomere (Fig. 4B). Thus, Ctf4, but none of the other well-characterized genes involved in both sister chromatid cohesion and replication, plays a significant role in completing BIR. We suspect that Ctf4 is important for BIR through its function with Mcm10 in recruitment and maintenance of Polα in replication.
Cdc7 is required for BIR
Our results showed that the BIR replication machinery requires most of the same essential replication machinery that is recruited to the pre-IC, but is not initiated in the same way. Activation of normal replication depends on Cdc7/Dbf4. Cdc7 is also involved in post-replication repair (PRR), which is presumably independent of origins, particularly in the error-prone translesion synthesis (TLS) branch of the Rad6 pathway (Pessoa-Brandao and Sclafani 2004). Using the cdc7-4 conditional allele that is inactive at 37°C (Bousset and Diffley 1998), we observed little product in cdc7-4 cells at 37°C, whereas product formation was unaffected at 25°C (Fig. 4C). These data demonstrate that Cdc7 is required for BIR.
Recently, Sheu and Stillman (2010) showed that the essential role of Cdc7 in replication can be bypassed by removing the inhibitory domain in Mcm4, indicating that its phosphorylation is the sole essential function of Cdc7. Introduction of the mcm4Δ74-174 bypass allele into the wild-type strain did not significantly affect viability or the efficiency of BIR (Fig. 4D). We found that mcm4Δ74-174 suppressed the growth lethality of cdc7-4 at 37°C in the absence of a DSB, but only partially suppressed the defect in BIR at 37°C (P < 0.001) (Fig. 4D). These results indicate that the role of Cdc7 in BIR is not limited to phosphorylation of Mcm4.
Ubiquitination and sumoylation of PCNA contribute to the efficiency of BIR
Although the Cdc7 bypass allele of MCM4 suppresses the lethality of cdc7Δ cells, these cells are still sensitive to additional replicative stress, such as that caused by hydroxyurea (Sheu and Stillman 2010), indicating that Cdc7 is also important for S-phase progression, which may involve BIR or PRR. Because mcm4Δ74-174 fails to strongly suppress the BIR defect of cdc7-4, although it suppresses defects in normal DNA replication, it is possible that BIR requires a function involved in PRR. There are several subpathways of PRR that have significant overlap but whose exact substrates are not well characterized. The key regulatory target in PRR is the DNA processivity clamp PCNA (for reviews, see Moldovan et al. 2007; Bergink and Jentsch 2009). PCNA is a homotrimer that can be monoubiquitinated at the highly conserved lysine residue K164 by Rad6–Rad18 in response to DNA damage. The initial ubiquitination of this residue signals the error-prone pathway of TLS that is carried out by the specialized DNA polymerases Polη (Rad30) and Polζ (Rev3–Rev7). This modification may be modified further by Rad5–Ubc13–Mms2-dependent polyubiquitination at K63 to signal an error-free mechanism that is believed to involve HR and template switching of the replication machinery. Alternatively, PCNA can be sumoylated at the same Lys164 and, to a lesser extent, at K127. The sumoylation of PCNA has been thought to inhibit recombination events through its interaction with Srs2, a helicase that disrupts Rad51 filament formation. Recently, Branzei et al. (2008) showed that sumoylation of PCNA is also important for Rad18–Rad5-mediated template switching, and that this repair is coordinated with HR.
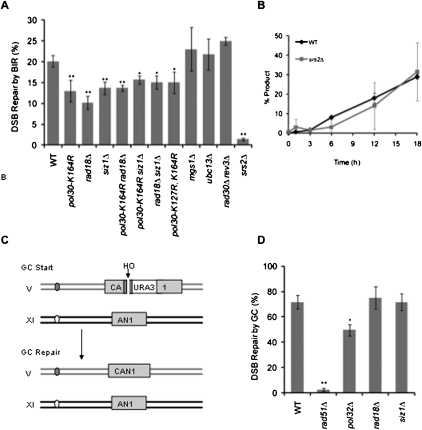
Our previous work showed that PCNA is required for BIR (Lydeard et al. 2007), but the importance of PCNA modifications in BIR is unknown. We first replaced POL30 with the PCNA pol30-K164R allele that cannot be modified by ubiquitination or sumoylation. As shown in Figure 5A, there was a moderate but significant 33% reduction in viability (P = 0.002). To determine if the defect in BIR was due to the lack of ubiquitination at K164, we deleted the E3 ubiquitin ligase responsible for this modification, Rad18. We found that the viability of rad18Δ cells was reduced by half (P = 0.001) compared with wild-type cells, and that the pol30-K164R rad18Δ double mutant was not statistically different from either single mutant. To determine if any of the downstream components of either the Rad18-dependent error-free pathway or TLS pathways are involved, we deleted Ubc13 or both Rev3 and Rad30. We found that neither ubc13Δ nor rad30Δ rev3Δ cells were defective in BIR (Fig. 5A). These data indicate that Rad18-mediated monoubiquitination of PCNA, but not its known downstream effectors, is involved in BIR.
Figure 5.
Rad18-mediated ubiquitination and Siz1-mediated sumoylation of PCNA are important for BIR but not GC. (A) Efficiency of BIR in wild-type (WT), pol30K164R, rad18Δ, siz1Δ, pol30K164R rad18Δ, pol30K164R siz1Δ, rad18Δ siz1Δ, pol30K127R, K164R, mgs1Δ, ubc13Δ, rad30Δ rev3Δ, and srs2Δ cells, as measured by viability following a DSB. Error bars represent ±SEM. (B) Appearance of BIR repair product, as monitored by PCR, in wild-type (WT) and srs2Δ cycling cells. Error bars represent the error range for two independent measurements. (C) The experimental system to study GC at the CAN1 locus (disrupted by URA3 creating a 376-bp gap) on Ch V. The homologous sequences that serve as the donor for repair by GC are found on Ch XI. (D) Efficiency of GC in wild-type (WT), rad51Δ, pol32Δ, rad18Δ, and siz1Δ cells, as measured by viability following a DSB. Error bars represent ±SEM.
To test if the sumoylation of PCNA at K164 was also important for BIR, we deleted the E3 SUMO ligase Siz1 responsible for this modification (Hoege et al. 2002). The viability of siz1Δ cells was decreased by one-third (P = 0.009); moreover, none of the double-mutant combinations of pol30-K164R siz1Δ and rad18Δ siz1Δ (Fig. 5A) were statistically different from either rad18Δ or siz1Δ single mutants. We next tested the pol30-K127R, K164R allele to determine if eliminating the sumoylation of PCNA at K127 in addition to K164 had a greater impairment of BIR. The pol30-K127R, K164R (P = 0.03) was defective in BIR, but was no worse than the pol30-K164R allele (Fig. 5A). We therefore conclude that Siz1-mediated sumoylation of PCNA at K164, but not sumoylation of K127, is important for BIR.
Because sumoylation of PCNA is important in inhibiting HR by recruiting Srs2, we asked if deleting srs2Δ would have an effect on BIR. As seen in Figure 5A, deletion of srs2Δ renders cells inviable following a DSB. However, previous work in our laboratory had shown that Srs2 is also important for turning off the DNA damage checkpoint after a break is repaired; thus, an srs2Δ mutant is unable to resume growth when repair is complete (Vaze et al. 2002). When we monitored the kinetics of repair in cycling srs2Δ cells by the PCR assay (Fig. 5B), we saw no defect compared with wild-type cells. Therefore, the lethality of BIR in srs2Δ cells stems from a failure to turn off the DNA damage checkpoint and not from a failure to repair the break. Finally, we asked if Mgs1, which has been suggested to compete with the Rad6–Rad18 PRR repair pathways (Hishida et al. 2006), was involved in BIR, and found that it also had no effect on viability when deleted (Fig. 5A).
We next tested the kinetics of repair of rad18Δ siz1Δ cells as a representative of the PRR mutants. Although the kinetics of repair were not substantially different from wild type, the reduction in viability in rad18Δ siz1Δ cells was paralleled by a small defect in the final outcome relative to wild type (Supplemental Fig. S2). Our results demonstrate that both Rad18-mediated ubiquitination and Siz1-mediated sumoylation are important for BIR, although the reductions are not as profound as for pol32Δ. To see if Rad18 and Siz1 were also important for other forms of HR, we deleted each gene in a modified strain that can repair the DSB at CAN1 by ectopic GC. For this assay, we introduced into Ch V a 2404-bp region that is homologous to the second end of the DSB (Fig. 5C). When there are two ends of homology with the break, the cells strongly favor GC over BIR (Jain et al. 2009). The viability of the cells with a DSB increases from 20% when there is one end of homology, allowing only BIR, to 70% when GC is used to repair the break (Fig. 5D). Repair is virtually eliminated in the absence of Rad51 (P = 0.002), but deleting Pol32 has only a minor effect on GC (P = 0.04), as shown before (Lydeard et al. 2007; Jain et al. 2009). Deletion of either rad18Δ or siz1Δ had no effect on the efficiency of GC (Fig. 5D), similar to a previous analysis of plasmid gap repair (Moertl et al. 2008). Taken together, these results indicate that both Rad18-dependent ubiquitination and Siz1-mediated sumoylation are involved in BIR but not GC. This suggests that some of the previously described cross-talk seen between PRR and HR (Branzei et al. 2008) may involve BIR events and not GC.
PCNA alleles suppressing pol32Δ cold sensitivity are dominant-negative in BIR
The nonessential Pol32 subunit of Polδ is required for BIR, and its interaction with PCNA is important for BIR (Lydeard et al. 2007). In addition to being defective in BIR, pol32Δ cells are defective for damage-induced mutagenesis, and have an uncharacterized cold sensitivity (Gerik et al. 1998). In a screen of in vitro generated mutations of PCNA (POL30) (Eissenberg et al. 1997), we identified two mutations, pol30-89 (pol30-FF248, 249AA) and pol30-92 (pol30-R80A), that suppressed the growth defect of pol32Δ at 16°C. These alleles were particularly interesting because they had no other discernable phenotype when tested for temperature sensitivity and sensitivity to various DNA-damaging agents, including ultraviolet light, hydroxyurea, and methyl methanesulfonate (Eissenberg et al. 1997; JC Eissenberg and PM Burgers, unpubl.). Based on the requirement of Pol32 and PCNA in BIR, we asked if these PCNA alleles also suppress the requirement of Pol32 in BIR.
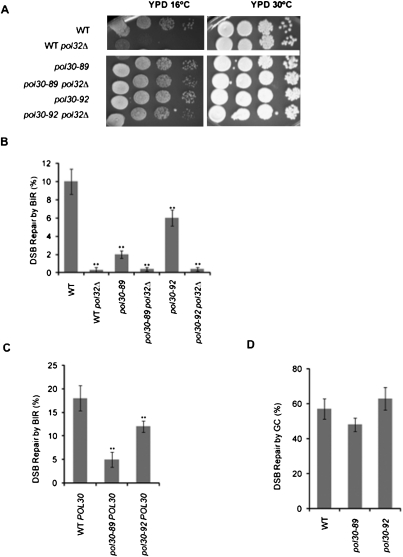
We created strains carrying only pol30-89 and pol30-92, as described in the Materials and Methods. Next, we deleted pol32Δ and confirmed that pol30-89 and pol30-92, but not the wild-type control, suppressed the cold sensitivity of pol32Δ cells (Fig. 6A). Surprisingly, these alleles did not suppress pol32Δ in BIR (P < 0.01 in all cases) (Fig. 6B). Moreover, pol30-89 and pol30-92 by themselves were also defective for BIR, even with a wild-type POL32. Cells carrying the pol30-89 allele were only 20% (P = 0.002) as viable as wild type, whereas those with pol30-92 showed 50% of wild-type viability (P = 0.01).
Figure 6.
Novel PCNA alleles are required for BIR. (A) PCNA mutant alleles pol30-89 and pol30-92 suppress pol32Δ cold sensitivity. Tenfold serial dilutions of the indicated strains incubated for 6 d at 18°C or 3 d at 30°C. (B) PCNA mutant alleles pol30-89 and pol30-92 are defective in BIR. Efficiency of BIR in wild-type (WT), pol32Δ, pol30-89, pol30-89 pol32Δ, pol30-92, and pol30-92 pol32Δ cells, as measured by viability following a DSB. Error bars represent ±SEM. (C) PCNA mutant alleles pol30-89 and pol30-92 are dominant-negative in BIR. Efficiency of BIR in wild-type (WT) (POL30), pol30-89 POL30, and pol30-92 POL30 cells, as measured by viability following a DSB. Error bars represent ±SEM. (D) PCNA mutant alleles pol30-89 and pol30-92 are not required for GC. Efficiency of GC in wild-type (WT) (pol30-0), pol30-89, and pol30-92 cells, as measured by viability following a DSB. Error bars represent ±SEM.
In fact, these pol30 alleles are dominant-negative in BIR, as seen when we tested the viability of cells containing both the endogenous POL30 allele and the mutant allele (or wild-type gene) integrated at the LEU2 locus. Viability of pol30-89 POL30 was decreased nearly fourfold, while pol30-92 POL30 was decreased twofold (P < 0.01 in both) compared with the control carrying POL30 at both locations (Fig. 6C). These decreases are similar to those without the endogenous POL30 gene; therefore, both pol30-89 and pol30-92 alleles are dominant-negative for BIR. We confirmed that these defects were reflected in the level of new DNA synthesis, as monitored by PCR (Supplemental Fig. S3). This assay shows that pol30-89 is dominant-negative; a qualitatively similar decrease was seen with pol30-92 cells, although it was not statistically significant. When tested for the ability of pol30-89 and pol30-92 cells lacking POL30 to successfully complete GC (Fig. 6D), neither allele displayed a defect; thus, these alleles are uniquely required for BIR.
Discussion
Our previous studies had shown that all three major DNA polymerases are required for BIR, but how this replication machinery is recruited to the strand-invaded template and the DNA helicase(s) responsible for unwinding the template DNA had not been addressed. Here we show that Mcm2–7, Cdc45, and the GINS complex are all required to initiate BIR synthesis. Cdt1, the protein responsible for recruiting Mcm2–7 to form the pre-RC, is also essential, whereas other proteins required for pre-RC formation—Cdc6 or ORC—are not necessary for BIR. We also find that the essential initiation and elongation factor Mcm10, along with its cofactor Ctf4, is important for BIR, as are the key replication initiation factors Dpb11 and Sld3. Moreover, we show that the Cdc7 kinase is required for BIR, even when its S-phase replication requirement is suppressed by mutations in Mcm4. In addition, the downstream PRR proteins Rad18 and Siz1 are important for BIR through both the ubiquitination and sumoylation of the DNA processivity clamp PCNA. In contrast, GC does not require Mcm2–7, Cdc45 (and, presumably, GINS), Cdc7, Pol32, or the lagging strand Polα–primase proteins. GC does, however, require Dpb11 (M Yamaguchi and JE Haber, unpubl.).
The finding that the Mcm2–7 helicase and the rest of the essential replicative machinery are required for BIR gives further support to the notion that BIR can function as a bona fide replication restart mechanism in eukaryotes, but it is clear that BIR can also occur outside of S phase, in the G2 phase of the cell cycle. One striking feature of BIR, as we studied it, is its slow kinetics relative to GC. This is true whether the amount of homology shared by the recombining end of the DSB is a few kilobases, in an ectopic recombination event, or 100 kb, when the donor is a homologous chromosome (Malkova et al. 2005). The cell actively regulates the choice of which HR pathway is employed to repair the break through the recombination execution checkpoint (REC), which signals whether both ends of a DSB are engaged with the same template (Jain et al. 2009). REC apparently delays BIR after the Rad51-mediated strand invasion of the donor template but before new DNA synthesis, so that cells are checkpoint-arrested in G2 before the initiation of new DNA synthesis.
We do not yet understand how the replication machinery is established outside of normal S phase, when some important replication factors such as the Mcm2–7 complex and Cdt1 are largely exported from the nucleus or degraded to prevent reinitiation of normal replication (Arias and Walter 2007). CDK1-mediated exclusion of both Mcm2–7 and Cdt1 is an obstacle that BIR must overcome to initiate new synthesis in G2-arrested cells, and may be a key regulatory step of initiating BIR. Presumably, enough Mcm2–7 and Cdt1 are retained or cycle back into the nucleus to cope with a single recombination-induced replication fork. We do know that adding an SV40 nuclear localization sequence to Mcm5, which keeps both Mcm2–7 and Cdt1 in the nucleus (Nguyen et al. 2000), does not accelerate the onset of BIR (M Yamaguchi and JE Haber, unpubl.). In BIR, the invading Rad51 filament serves to denature the duplex DNA, which might allow the MCM helicase to load independently of ORC, but how it is recruited to the donor template is still unclear. Because Cdc6 and ORC are not required for BIR, the proteolytic destruction of Cdc6 and hyperphosphorylation of ORC in G2 have no consequences. Our results also show that the proscription against rereplication involving ARS-dependent, ORC-dependent, and Cdc6-dependent normal replication does not prevent BIR from occurring, either in cells where the DNA damage checkpoint arrests cells in G2/M prior to BIR, or when cells are arrested with nocodazole. Interestingly, Drosophila endoreplication is also independent of ORC, but requires Cdt1 and Mcm2–7 (Park and Asano 2008).
Unlike GC, BIR requires the second cell cycle-dependent kinase required for replication initiation, Cdc7/Dbf4. Cdc7 either may be required directly in the recruitment of DNA replication proteins to the D-loop or may regulate a critical step in the REC. Our genetic results indicate that the mcm4Δ74-174 bypass allele partially suppresses the requirement of Cdc7 in BIR. Therefore, as in S phase, phosphorylation of Mcm4 by Cdc7 is required to initiate the BIR replication fork, but there must be additional Cdc7 targets to allow efficient BIR.
Another potential Cdc7 target is Cdt1; in human cells, Cdc7 associates with and regulates the abundance of Cdt1 during S phase (Ballabeni et al. 2009). Additionally, Cdt1 promotes Cdc45 recruitment to chromatin in human cells (Ballabeni et al. 2009), and is also present during replication elongation in Drosophila (Claycomb et al. 2002). Although a similar relationship may exist in yeast, these results are not directly comparable because the N-terminal domain of Cdt1 required for its interaction with Cdc7 is not present in yeast. In any case, we believe that the primary role of Cdt1 is to load Mcm2–7 onto the BIR template DNA. It will be important to perform biochemical studies, in particular chromatin immunoprecipitation assays, to determine the temporal order of recruitment of the replication machinery to the BIR template.
In addition to its requirement in replication and BIR, Cdc7 is involved in PRR. In this study, we find that, like Cdc7, Siz1-mediated sumoylation and Rad18-mediated ubiquitination of PCNA at K164 are both involved in BIR, but not GC. The fact that both ubiquitination and sumoylation are important for BIR indicates that either the individual monomers of the PCNA homotrimer are differently modified, or the modifications occur sequentially. Since sumoylated PCNA interacts with Srs2 to prevent recombination during S phase, the BIR defect in siz1Δ and pol30-K164R cells may possibly be linked to Srs2. However, srs2Δ had no effect on completing BIR, although it has a recovery defect, as documented previously. Moreover, because neither rad18Δ nor siz1Δ affected ectopic GC where srs2Δ also has a recovery defect, we believe this finding rules out a role for Rad18 or Siz1 in recovery. Recently, Branzei et al. (2008) found that Siz1 is required for Rad18-dependent template switch events. Unlike BIR, these events are dependent on Rad5–Ubc13–Mms2 polyubiquitination of PCNA. Thus, although there appears to be similar requirements between PRR and BIR, BIR differs from the PRR events as seen by others because it does not require the downstream polyubiquitination of PCNA.
Finally, we identify two PCNA alleles that suppress pol32Δ cold sensitivity but do not suppress the requirement of POL32 in BIR. These results imply that the cold sensitivity of pol32Δ cells is not due exclusively to defective BIR. Second, both pol30-89 and pol30-92 are dominant-negative in BIR but have no impact on GC. As PCNA is a homotrimer, it is possible that inclusion of one pol30-89 or pol30-92 subunit is sufficient to impair BIR but not normal replication. Although the PRR proteins and PRR-related modification of PCNA mutations play a small role in BIR, it is not nearly as profound as the novel pol30-89 and pol30-92 alleles. The mutations of the pol30-89 (pol30-FF248, 249AA) and pol30-92 (pol30-R80A) alleles are far apart within each monomer and on the PCNA homotrimer, as shown in Supplemental Figure S4, and are far from the site of ubiquitination/sumoylation at K164, so the basis of their defects are not evident.
Our understanding of the genetic requirements of BIR is expanding (Supplemental Fig. S5). We uncovered factors that are required specifically for BIR (Pol32), novel requirements for replication factors in DNA damage repair (Cdt1, Mcm10, and Ctf4), factors common to BIR and GC (Rad51 and Dpb11), factors common to replication and BIR but not GC (Cdc7, Mcm2-7, Cdc45, GINS, and some PCNA alleles), and factors common to PRR and BIR but not GC (Cdc7, Rad18, and Siz1). Understanding the interplay of these factors during replication damage repair and uncovering the molecular details of signaling between them will be an area of much future research.
Materials and methods
Strains
All strains used to study BIR are derived from JRL092 (Lydeard et al. 2007) and are described in more detail in the Supplemental Material.
Viability measurements
Logarithmically growing cells grown in YEP + 2% Raffinose were plated on either YEPD or YEP-Gal and grown into colonies. Colonies were counted and then replica-plated onto plates containing either canavanine or hygromycin to confirm repair that occurred by BIR. Experiments were performed at least five times for each strain unless otherwise indicated. To determine the statistical significance between strains, the Student's t-test was used (paired, two-tailed, n ≥ 3 for all strains). Values marked with asterisks in graphs are significantly different: P < 0.05 (*) and P < 0.01 (**).
HO induction and measurement of kinetics of DSB repair
Strains were grown in YEP + 2% Raffinose, and time-course experiments were performed as described previously (Lydeard et al. 2007).
DNA analysis
PCR analysis of BIR was performed as described previously (Lydeard et al. 2007). Experiments of wild-type, cdc45-td, mcm4-td, cdc6-1, orc6-td, cdt1-td, and sld3-7td strains were done three times, while experiments with all other strains were done twice. After DNA isolation, three or more PCR reactions were performed independently for each experiment. For experiments performed three times, the data are graphed as the mean plus the standard error of the mean among the independent experiments, while the data are graphed as the mean plus the data range between experiments for experiments performed twice.
Acknowledgments
We are grateful to Stephen P. Bell, Anja Bielinsky, John Diffley, Enrique Herrero, Karim Labib, Masayuki Seki, and Helle Ulrich for strains, plasmids, and helpful suggestions. We thank Sue Lovett and Paul Miller for thoughtful discussion and advice. This work was supported by NIH grants GM20056 and GM76020 to J.E.H., and GM32431 to P.M.B. J.R.L. was supported by NIH Genetics Training Grant GM07122.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1922610.
Supplemental material is available at http://www.genesdev.org.
References
- Arias EE, Walter JC 2007. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 [DOI] [PubMed] [Google Scholar]
- Ballabeni A, Zamponi R, Caprara G, Melixetian M, Bossi S, Masiero L, Helin K 2009. Human CDT1 associates with CDC7 and recruits CDC45 to chromatin during S phase. J Biol Chem 284: 3028–3036 [DOI] [PubMed] [Google Scholar]
- Bergink S, Jentsch S 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458: 461–467 [DOI] [PubMed] [Google Scholar]
- Bousset K, Diffley JF 1998. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev 12: 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M 2008. SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920 [DOI] [PubMed] [Google Scholar]
- Chen S, de Vries MA, Bell SP 2007. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes Dev 21: 2897–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, MacAlpine DM, Evans JG, Bell SP, Orr-Weaver TL 2002. Visualization of replication initiation and elongation in Drosophila. J Cell Biol 159: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da-Silva LF, Duncker BP 2007. ORC function in late G1: Maintaining the license for DNA replication. Cell Cycle 6: 128–130 [DOI] [PubMed] [Google Scholar]
- Davis AP, Symington LS 2004. RAD51-dependent break-induced replication in yeast. Mol Cell Biol 24: 2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A 1994. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78: 303–316 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Ayyagari R, Gomes XV, Burgers PM 1997. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase δ and DNA polymerase ɛ. Mol Cell Biol 17: 6367–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366 [DOI] [PubMed] [Google Scholar]
- Gerik KJ, Li X, Pautz A, Burgers PM 1998. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J Biol Chem 273: 19747–19755 [DOI] [PubMed] [Google Scholar]
- Haber JE 1999. DNA recombination: The replication connection. Trends Biochem Sci 24: 271–275 [DOI] [PubMed] [Google Scholar]
- Hanna JS, Kroll ES, Lundblad V, Spencer FA 2001. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 21: 3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327 doi: 10.1371/journal.pgen.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishida T, Ohya T, Kubota Y, Kamada Y, Shinagawa H 2006. Functional and physical interaction of yeast Mgs1 with PCNA: Impact on RAD6-dependent DNA damage tolerance. Mol Cell Biol 26: 5509–5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Holmes AM, Haber JE 1999. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96: 415–424 [DOI] [PubMed] [Google Scholar]
- Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N, Haber JE 2009. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev 23: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Masumoto H, Sugino A, Araki H 1998. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol 18: 6102–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Tak YS, Sugino A, Araki H 2001. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J 20: 2097–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Labib K 2006. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J 25: 1753–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JF 2000. Uninterrupted MCM2–7 function required for DNA replication fork progression. Science 288: 1643–1647 [DOI] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F 2006. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 23: 787–799 [DOI] [PubMed] [Google Scholar]
- Llorente B, Smith CE, Symington LS 2008. Break-induced replication: What is it and what is it for? Cell Cycle 7: 859–864 [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE 2007. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448: 820–823 [DOI] [PubMed] [Google Scholar]
- Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE 2005. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol 25: 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Haber JE 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75: 111–135 [DOI] [PubMed] [Google Scholar]
- Michel B 2000. Replication fork arrest and DNA recombination. Trends Biochem Sci 25: 173–178 [DOI] [PubMed] [Google Scholar]
- Moertl S, Karras GI, Wismuller T, Ahne F, Eckardt-Schupp F 2008. Regulation of double-stranded DNA gap repair by the RAD6 pathway. DNA Repair 7: 1893–1906 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S 2007. PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR 2006. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci 103: 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Irie K, Li JJ 2000. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2–7. Curr Biol 10: 195–205 [DOI] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC 2006. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587 [DOI] [PubMed] [Google Scholar]
- Pâques F, Haber JE 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Asano M 2008. The origin recognition complex is dispensable for endoreplication in Drosophila. Proc Natl Acad Sci 105: 12343–12348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet 4: e1000175 doi: 10.1371/journal.pgen.1000175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessoa-Brandao L, Sclafani RA 2004. CDC7/DBF4 functions in the translesion synthesis branch of the RAD6 epistasis group in Saccharomyces cerevisiae. Genetics 167: 1597–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JC, Bowers JL, Rodriguez HK, Bell SP 2006. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol Cell 21: 29–39 [DOI] [PubMed] [Google Scholar]
- Ricke RM, Bielinsky AK 2004. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol Cell 16: 173–185 [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Cheng IH, Chai W, Tye BK 2004. Mcm10 and Cdc45 cooperate in origin activation in Saccharomyces cerevisiae. J Mol Biol 340: 195–202 [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM 2007. Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple JW, Da-Silva LF, Jervis EJ, Ah-Kee J, Al-Attar H, Kummer L, Heikkila JJ, Pasero P, Duncker BP 2006. An essential role for Orc6 in DNA replication through maintenance of pre-replicative complexes. EMBO J 25: 5150–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B 2006. Cdc7–Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell 24: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B 2010. The Dbf4–Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev 66: 630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H 2003. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev 17: 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Diffley JF 2002. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat Cell Biol 4: 198–207 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Labib K, Diffley JF 2000. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J 19: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell 10: 373–385 [DOI] [PubMed] [Google Scholar]
- Wang X, Ira G, Tercero JA, Holmes AM, Diffley JF, Haber JE 2004. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol 24: 6891–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC 2002. Xenopus Mcm10 binds to origins of DNA replication after Mcm2–7 and stimulates origin binding of Cdc45. Mol Cell 9: 233–240 [DOI] [PubMed] [Google Scholar]
- Xu H, Boone C, Brown GW 2007. Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics 176: 1417–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]
- Zhou C, Huang SH, Jong AY 1989. Molecular cloning of Saccharomyces cerevisiae CDC6 gene. Isolation, identification, and sequence analysis. J Biol Chem 264: 9022–9029 [PubMed] [Google Scholar]
- Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, Nutt LK, Kornbluth S, Dutta A 2007. Mcm10 and And-1/CTF4 recruit DNA polymerase α to chromatin for initiation of DNA replication. Genes Dev 21: 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]