Abstract
The correct levels of deoxyribonucleotide triphosphates and their relative abundance are important to maintain genomic integrity. Ribonucleotide reductase (RNR) regulation is complex and multifaceted. RNR is regulated allosterically by two nucleotide-binding sites, by transcriptional control, and by small inhibitory proteins that associate with the R1 catalytic subunit. In addition, the subcellular localization of the R2 subunit is regulated through the cell cycle and in response to DNA damage. We show that the fission yeast small RNR inhibitor Spd1 is intrinsically disordered and regulates R2 nuclear import, as predicted by its relationship to Saccharomyces cerevisiae Dif1. We demonstrate that Spd1 can interact with both R1 and R2, and show that the major restraint of RNR in vivo by Spd1 is unrelated to R2 subcellular localization. Finally, we identify a new behavior for RNR complexes that potentially provides yet another mechanism to regulate dNTP synthesis via modulation of RNR complex architecture.
Keywords: Ribonucleotide reductase, intrinsically disordered proteins, nuclear import, Cullin 4
Faithful DNA replication, a prerequisite for maintaining genome integrity, requires the maintenance of the correct concentration and the relative ratios of dNTPs (Chabes et al. 2003; Holmberg et al. 2005). dNTPs are formed by ribonucleotide reductase (RNR), which converts ribonucleoside diphosphates into their deoxy forms. Eukaryotes use type Ia RNR complexes comprised of multimers of two subunits: the large (R1) catalytic subunit, and the small (R2) diferric-tyrosyl radical-generating subunit (Stubbe 2003; Nordlund and Reichard 2006). Distinctive for type Ia RNR complexes are two allosteric nucleotide-binding sites on R1 (Reichard 2002; Nordlund and Reichard 2006). The N-terminal “overall activity” site is an ATP cone domain that binds either ATP (stimulatory) or dATP (inhibitory). The specificity site can bind ATP, dATP, dTTP, or dGTP and selects the substrate to be reduced, thus maintaining the appropriate dNTP ratios (Nordlund and Reichard 2006).
RNR is also regulated by a number of further mechanisms. In all eukaryotes studied, RNR protein levels are regulated via transcription. This is particularly obvious in response to genotoxic stress, when DNA repair synthesis requires dNTPs to be present outside of S phase. Work in the budding yeast Saccharomyces cerevisiae identified a further layer of control via the binding of a small RNR inhibitor protein, Sml1, to the R1 subunit (Zhao et al. 1998, 2000; Chabes et al. 1999). Sml1 is degraded as cells enter S phase and in response to genotoxic stress outside of S phase (Zhao et al. 2001). Work in the fission yeast Schizosaccharomyces pombe (Liu et al. 2003) demonstrated that the R2 subunit is localized primarily to the nucleus in non-S-phase cells, and is relocalized to the cytoplasm in response either to S-phase entry or following DNA damage checkpoint activation. Because the majority of the R1 subunit is constitutively cytoplasmic, this relocalization was proposed to promote RNR complex formation and dNTP synthesis. R2 nuclear localization depends on a small RNR regulator, Spd1. In S. cerevisiae, an Spd1-related protein, Dif1, was subsequently shown to promote R2 nuclear import (Lee et al. 2008; Wu and Huang 2008). In cooperation with a nuclear anchor, Wtm1 (Lee and Elledge 2006), Dif1-dependent import results in R2 nuclear accumulation. R2 nuclear accumulation is regulated by S-phase-dependent or genotoxic stress-dependent Dif1 degradation. The reduced Dif1 level decreases nuclear import while nuclear export remains active, promoting a net increase in cytoplasmic R2 (Lee et al. 2008).
S. cerevisiae Sml1 and Dif1 proteins share a domain, the SML box (Fig. 1A; Lee et al. 2008). Synteny analysis suggests that the SML1 and DIF1 genes are derived from the same ancestral locus, diverging when S. cerevisiae underwent genome duplication during its evolution. The SML1 locus subsequently underwent a further direct duplication event so that a related gene, HUG1, is immediately adjacent (Lee et al. 2008). Hug1 and Dif1 also share a sequence motif, the HUG box, that is not apparent in Sml1. Hug1 function is not well defined; its transcript is induced by DNA damage, and it has been proposed to regulate RNR feedback inhibition (Basrai et al. 1999; Benton et al. 2006).
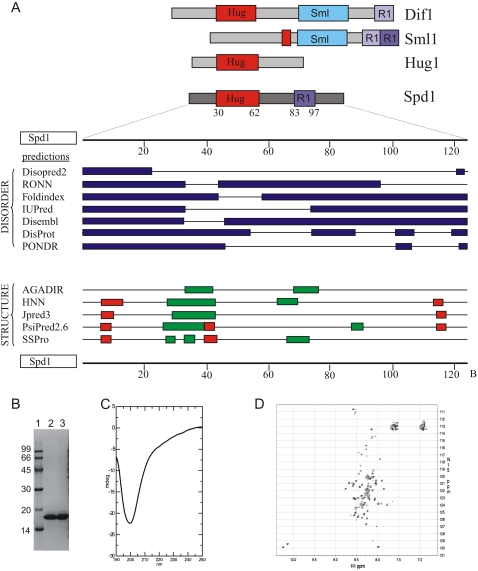
Figure 1.
Spd1 is an intrinsically unfolded protein. (A, top) Cartoon representation of relationship to S. cerevisiae Dif1, Sml1, and Hug1 (data adapted from Lee et al. 2008). (Bottom) Disorder and structure predictions from the indicated software. (Red boxes) Coil; (green boxes) helix. (B) Spd1 was purified to homogeneity and showed a lower electrophoretic mobility than expected (18 kDa vs. 14 kDa). Lane 2 (fraction 6) and lane 3 (fractions 7 and 8) of the MonoQ purification after buffer change to PBS (pH 7.4). (Lane 1) Low-molecular-weight markers; molecular weight is indicated in kilodaltons. (C) A far-UV CD spectrum of Spd1 recorded from 250 to 190 nm on 10 μM Spd1 and 10 mM NaH2PO4 (pH 7.4). A large negative ellipticity with a maximum at 199 nm suggests an unfolded protein with little or no secondary structure. (D) An 15N,1H-HSQC NMR spectrum of 1.0 mM 15N-Spd1 and 10 mM NaH2PO4 (pH 7.4) recorded at 10°C. Very little dispersion of signals is seen, as well as variable peak intensities.
Spd1 in S. pombe and Dif1 in S. cerevisiae regulate R2 nuclear localization. The HUG domain is conserved between Spd1 and Dif1. Dif1 binds R2 via the HUG domain (Lee et al. 2008) to facilitate R2 nuclear import, suggesting that Spd1 may share this function. Spd1 also shares a region of sequence similarity with Sml1, equating to the last half of the suggested Sml1 RNR1-binding domain (Zhao et al. 2000; Lee et al. 2008). This region resides downstream from the C-terminal α-helix region of Sml1, a region with a clear role in RNR1 inhibition (Zhao et al. 2000). Conservation of an R1 interface, albeit limited, would be consistent with the direct association between Spd1 and Cdc22R1 reported to mediate vitro biochemical inhibition of RNR (Hakansson et al. 2006). The sequence conservation between Spd1, Dif1, Sml1, and Hug1 indicates that Spd1 may be the sole S. pombe ortholog of the ancestral gene (summarized in Fig. 1).
Here we provide evidence that Spd1 is an intrinsically disordered protein (IDP) that acts as an import factor for the RNR R2 subunit. Using FRET analysis, we demonstrate that Spd1 controls RNR complex architecture. However, this does not correlate to the formation of active complexes. We suggest it reflects an additional level of RNR regulation beyond formation of canonical RNR tetramers of 2xR1 and 2xR2 subunits (α2β2 tetramer). By structure–function analysis, we separate three roles for Spd1: We identify a mutant defective specifically for Suc22R2 nuclear import (spd1-M2), mutants specifically defective in their ability to restrain RNR function in vivo (i.e., spd1-M12 and spd-M35), and mutants that have specifically lost the ability to promote FRET (i.e., spd1-M1 and spd1-M6). Thus, each of these three roles can be separated from each other, underlining the segmental distribution of function typical of IDPs. Contrary to expectation, we show that the ability of Spd1 to restrain RNR activity in vivo (and thus interfere with S phase) is not a consequence of its ability to sequester Suc22R2 in the nucleus. The restraint of RNR activity in vivo likely reflects the in vitro inhibition of RNR by Spd1 identified by biochemical analysis, although this remains to be established formally.
Results
Spd1 is an IDP
Sml1 is a member of a group of proteins that are intrinsically disordered (Danielsson et al. 2008). An IDP lacks a well-structured three-dimensional fold (Tompa 2002). However, nuclear magnetic resonance (NMR) studies show that IDPs can adopt transient structure in solution, and that some IDPs fold onto their interaction partner when they associate. This process, coupled folding and binding, can result in protein–protein interactions with relatively low affinity but high specificity (Sugase et al. 2007). The Spd1 sequence has the typical characteristics of an IDP: a high content of charged residues (23%), and a low aliphatic index (52.85; for reference, myoglobin = 95.1). Structural prediction programs such as DrDOS (Ishida and Kinoshita 2007) predict a high probability of disorder, with minor regions that can potentially form intermittent secondary structures. Most consistent are the two helical regions between residues 27–44 and 66–76, plus the possibility of shorter, extended structures between 6–10 and 115–120 (Fig. 1A).
To investigate the disorder characteristics by spectroscopy, recombinant Spd1 was purified to >98% homogeneity (Fig. 1B). Electrophoretic mobility corresponded to ∼18 kDa, higher than the expected 14.2 kDa. This is a general attribute of IDPs (Tompa 2002). A far-UV CD spectrum showed no distinct signs of pronounced secondary structure elements, with very little negative ellipticity in the 210- to 220-nm range (Fig. 1C). Instead, a large negative signal with maximum at 199 nm was evident, highly indicative of an unfolded protein. An 15N,1H- HSQC NMR spectrum recorded at 10°C showed a very narrow distribution of signals in the 1H dimension, also typical of unfolded proteins (Fig. 1D). Importantly, a distribution of both high and low intensities of the NMR signals was observed, which suggests some residues of Spd1 are in intermediate exchange on the NMR time scale, possibly due to sampling of several conformations. Thus, Spd1 possesses all of the hallmarks of an IDP: low electrophoretic mobility, a lack of secondary structure in far-UV CD, and a collapsed NMR spectrum corresponding to an unfolded protein. Similar results have been established previously for Sml1 (Danielsson et al. 2008).
Spd1 regulates R2 nuclear import, but does not act as a nuclear anchor
In S. cerevisiae, two distinct mechanisms contribute to nuclear accumulation of R2: Dif1-dependent nuclear import, and R2 retention by the Wtm1 nuclear anchor. To establish if Spd1 shares the nuclear import function, we examined if Suc22R2 accumulates in the nucleus in spd1-d cells by blocking nuclear export with leptomycin B (LMB), which inhibits Crm1-dependent nuclear export of Suc22R2 (Liu et al. 2003). Suc22R2 did not accumulate in the nucleus in response to LMB treatment (Fig. 2A), indicating that Suc22R2 is no longer transported into the nucleus in the absence of Spd1 and thus cannot accumulate there when export is blocked.
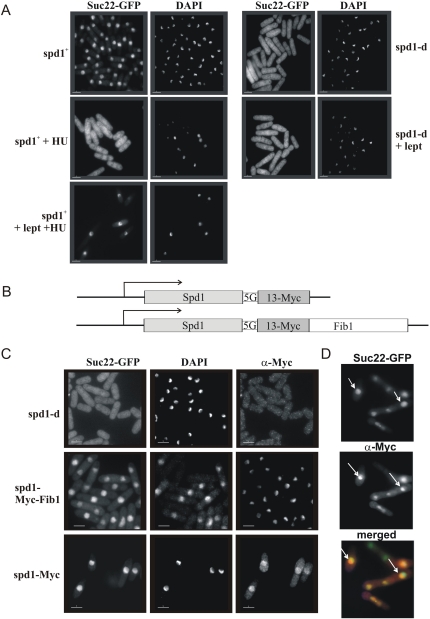
Figure 2.
Spd1 regulates Suc22R2 nuclear import. (A) spd1+ CFP-suc22 and spd1-d CFP-suc22 cultures were treated with HU (20 mM) to arrest cells in S phase and/or leptomycin (100 ng/mL) to block nuclear export. After 2 h, cells were formaldehyde-fixed, and GFP was visualized by epifluorescence and DNA was visualized by DAPI staining. In spd1+ cells treated with HU, Suc22R2 becomes pan-cellular. (Bottom left) Concomitant treatment with leptomycin prevents nuclear export and Suc22R2 redistribution. (Bottom right) In the absence of Spd1, GFP-Suc22 is distributed throughout the cell and does not accumulate in the nucleus when nuclear export is blocked. (B) Cartoon representation of the spd1-Myc and spd1-Myc-Fib1 constructs integrated at the spd1 locus under control of the spd1 promoter. A 5× glycine linker (5G) separates Spd1 from the tags. (C) GFP-Suc22 (epifluorescence), DAPI-stained DNA, and the Myc epitope (indirect immunofluorescence) visualized in fixed cells following logarithmic growth. (D) Merged images of Myc and GFP-Suc22 localization from the spd1-Myc-Fib1 culture. White arrows indicate example Myc staining nucleoli. Bars, 5 μm.
Despite considerable effort, we did not identify a homolog or ortholog of the Wtm1 nuclear anchor mechanism in S. pombe. In addition, spd1 deletion results in complete Suc22R2 delocalization from the nucleus. In S. cerevisiae, Dif1 deletion only partially disrupts R2 nuclear accumulation. The remaining accumulation is Wtm1-dependent (Lee and Elledge 2006). This suggests there is no nuclear anchor for Suc22R2 in S. pombe, but it remains formally possible that Spd1 both contributes to Suc22R2 nuclear import and acts as a canonical Suc22R2 nuclear anchor. If so, forced Spd1 localization to the nucleolus would be predicted to result in concomitant nucleolar Suc22R2. We thus modified the spd1+ locus to express a C-terminal fusion of 13-Myc epitopes followed by the Fib1 fibrillarin-like protein that is known to localize to the nucleolus (Gallagher et al. 1993). The fusion protein (Fig. 2B) expressed by the spd1 promoter retained function, as judged by the correct profile and dynamics of Suc22R2 localization (Fig. 2C; data not shown). In a GFP-Suc22R2 background, we observed that Spd1-13Myc-Fib1 localized to the nucleolus, but Suc22R2 was not enriched in the nucleolus when compared with spd1+ controls (Fig. 2C). Thus, Spd1 shares a nuclear import function with Dif1, but does not contribute to nuclear retention via a nuclear anchor function.
Spd1 is required for FRET between Cdc22R1 and Suc22R2
The increased colocalization of R1 and R2 subunits in the cytoplasm during S phase and in response to genotoxic stress is proposed to allow active RNR complexes to form when dNTPs are required (Liu et al. 2003, 2005; Holmberg et al. 2005; Zhang et al. 2006; Wu and Huang 2008). To explore this possibility, we established a FRET assay (Fig. 3A,B) to examine the Cdc22R1/Suc22R2 interaction. First, we used the fluorescent protein tags to estimate the relative abundance of Cdc22R1 and Suc22R2 in both the nuclear and cytoplasmic compartments in G2- and S-phase cells. The relative fluorescence between Cdc22R1 and Suc22R2 is 3:1 (data not shown). In G2 phase, Suc22R2 fluorescence was approximately threefold more intense in the nucleus than the cytoplasm, with ∼74% of the signal residing in the nucleus (Fig. 3C). In S phase, nuclear Suc22R2 fluorescence is partitioned between two nuclei and comprises ∼26.5% of the GFP-Suc22R2 signal. Individually, each nucleus shows a significant loss of intensity compared with G2 phase. We also see a corresponding gain in intensity (from 26% to 63.5%) in the cytoplasm (Fig. 3C).
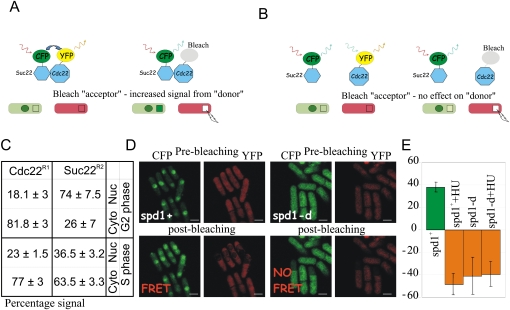
Figure 3.
Spd1 is required for FRET between Cdc22R1 and Suc22R2. (A) Cartoon representation of FRET between CFP- and YFP-tagged RNR subunits. Bleaching the YFP acceptor results in an increased emission signal from the CFP donor. The green circle represents the nucleus (Suc22R2 accumulates in the nucleus). Squares represent the bleached area; in this instance, a region of cytoplasm. (B) Equivalent cartoon showing result when fluorophores are not sufficiently aligned for FRET. Notice that, instead of an increase in the signal from the donor (i.e., evidence of FRET), a moderate decrease is seen (as opposed to no change) due to some overlap in the absorption spectra for CFP and YFP. (C) Percentage of nuclear and cytoplasmic fluorescent intensity of Cdc22-CFP and CFP-Suc22 in single tagged cells visualized under the same microscope slide. (D) Representative images from spd1+ and spd1-d cultures without treatment with HU. (E) Quantification of combined nuclear and cytoplasmic FRET signal. Error bars, SD from mean. Negative FRET (formally no-FRET) results from nominal photobleaching of the donor along with the acceptor.
In spd1+ cells, FRET could be detected in the cytoplasm and the nucleus of cells in both G2 and S phase (Fig. 3D). In this case, FRET was used simply as an indicator of the R1–R2 interaction and not as a quantitative measure of association number or structural proximity. Surprisingly, in spd1-d cells, we did not detect FRET in the cytoplasm or nucleus of either S-phase or G2 cells (Fig. 3E). Treating spd1+ cells with hydroxyurea (HU), an agent that inhibits RNR and synchronizes cells in S phase, resulted in the FRET signal disappearing (Fig. 3E). However, FRET was similarly lost when cdc25-22 cells synchronized in G2 were held in G2 and treated with HU (data not shown), indicating an S-phase-independent effect of HU. Taken together, these data indicate that Spd1 affects the association of Cdc22R1/Suc22R2 subunits, but that this does not correlate with apparent activation of RNR.
Alanine scanning mutagenesis of Spd1
From the available data, we can postulate three possible in vivo functions for Spd1: regulation of Suc22R2 nuclear import, an influence on RNR complex architecture that equates to our FRET analysis, and, finally, an in vivo restraint of RNR activity that correlates with an increased dNTP concentration in the absence of Spd1 in csn1-d and ddb1-d cells (Holmberg et al. 2005) and is possibly equivalent to either the inhibition observed in vitro (Hakansson et al. 2006), the nuclear sequestration of Suc22R2, or a combination of both. To understand how these potential functions relate to each other, and to shed light on the mechanism by which Spd1 inhibits RNR, we created 41 independent spd1 mutants in which each sequential group of three residues were substituted for alanine (Fig. 4A), and examined their influence on Spd1 degradation and Spd1-dependent phenotypes.
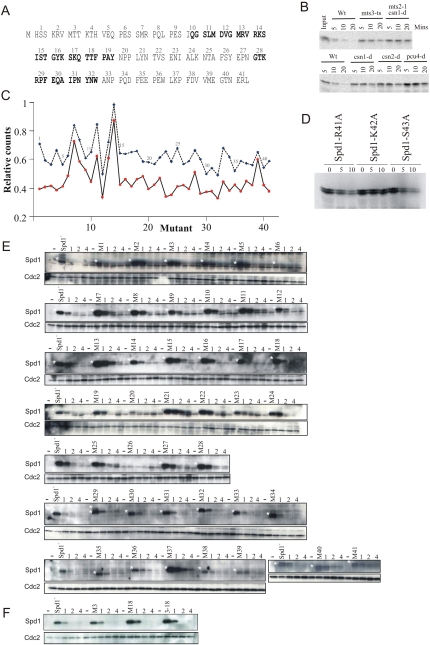
Figure 4.
Stability of Spd1 mutant proteins. (A) The 41 mutants of spd1 created. These are designated as spd1-M1, spd1-M2, etc. Each mutant results in the three indicated amino acids being changed to alanine. Bold indicates conserved HUG and R1 domains (see Fig. 1A). (B) In vitro degradation of 35S-labeled Spd1. In vitro translated Spd1 is incubated for the indicated number of minutes with whole-cell extract derived from either wild type (Wt) or the indicated mutants. mts3-ts and mts2-1 are mutations in genes encoding subunits of the proteasome and show compromised degradation in this assay. As is seen in vivo, Spd1 degradation is dependent on the signalosome subunits Csn1 and Csn2, and on the Cullin 4 homolog Pcu4. (C) Quantification of in vitro degradation assay for the individual mutants. (Blue symbols and dashed line) 10-min incubation; (red symbols and solid line) 20-min incubation. (D) The three amino acids mutated in spd1-M14 were changed individually to alanine and tested for stability in wild-type cell extract. Both bands are Spd1-specific. (E) In vivo stability of Spd1 mutant proteins. Each mutant was integrated separately at the spd1 genomic locus under control of the spd1 promoter. Logarithmically growing cells were incubated with HU (20 mM) for 1, 2, and 4 h; extract was prepared and subjected to SDS-PAGE; and Spd1 was detected by Western analysis with polyclonal α-Spd1 antisera. A white star marks the Spd1-specific bands. Before each mutant, a control lane contains extract from spd1-d cells. Blots were probed in parallel for Cdc2 as a loading control. (F) Identical analysis of spd1-M3, spd1-M18, and the combined spd1-M3-18 mutations that cover the two conserved Cds1Chk2 kinase consensus sites.
Analysis of Spd1 degradation in vitro and in vivo
We established an in vitro assay for Spd1 degradation by incubating 35S methionine-labeled Spd1 in whole-cell extract. Degradation kinetics were monitored by SDS-PAGE and autoradiography (Fig. 4B). Degradation was dependent on the Cullin 4 E3 ubiquitin ligase Pcu4–Ddb1Cdt2 and the signalosome, as predicted (Liu et al. 2003). Each individual mutant protein (Fig. 4C) was 35S methionine-labeled and incubated with degradation-competent extract, and the percentage of protein remaining after 10 and 20 min was quantified. The results did not identify a domain responsible for degradation, but did reveal a single stable mutant, Spd1-M14. Deconvolution of this mutant into the three individual alanine substitutions revealed that a single lysine residue (K42) was required for efficient degradation (Fig. 4D).
We were surprised not to define a degron domain, and were also wary of the observation that a single lysine is required: Usually, disrupting a single lysine residue in vivo results in adjacent lysines acting as alternative ubiquitin acceptor sites. We thus integrated each mutant into the spd1 locus, where they are expressed under control of the spd1 promoter. To assay degradation, each strain was grown to logarithmic phase and treated with 20 mM HU. This induces the Cdt2 targeting subunit of Cul4–Ddb1Cdt2 ubiquitin ligase and promotes Spd1 degradation (Liu et al. 2005). Samples for Western blot analysis were prepared immediately before HU addition and at 1, 2, and 4 h. Again we did not define a specific domain controlling Spd1 degradation (Fig. 4E). The Spd1-M14 and Spd1-K42A mutant proteins identified in the in vitro studies also showed no evidence of stability (Fig. 4E; data not shown). There were several additional observations. First, a double mutant encompassing the two checkpoint kinase consensus sites, Spd1-M(3 + 18), was not stabilized (Fig. 4F). Second, the initial protein levels of many mutants varied significantly. This is a reproducible observation seen with two independent α-Spd1 antibodies, and likely reflects that triple alanine substitution in an IDP is expected to affect intrinsic stability. Third, only two mutant proteins, Spd1-M21 and Spd1-M23, were significantly stabilized.
Separation of Suc22R2 localization and RNR inhibitory regulation
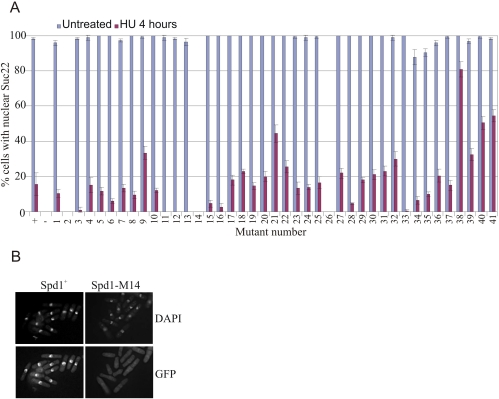
We crossed each spd1 mutant into the GFP-suc22 background and observed Suc22R2 localization by direct fluorescence in untreated cells and cells exposed to 20 mM HU for 4 h (Fig. 5). In spd1+ cells, Suc22R2 was largely nuclear (most asynchronous S. pombe cells are in G2). Following HU treatment (cells arrested in S phase; Spd1-degraded), ∼20% of cells retained weak nuclear GFP fluorescence, and Suc22R2 became pan-cellular in the remaining ∼80%. spd1-m2, spd1-m14, and spd1-m26 were fully defective for Suc22R2 nuclear accumulation. A small but notable effect on nuclear accumulation was observed for spd1-m34, spd1-m35, and spd1-m36. Four mutants—spd1-m11, spd1-m12, spd1-m13, and spd1-m33—showed more dramatic loss of Suc22R2 nuclear accumulation upon HU treatment compared with spd1+ cells. Finally, four mutants—spd1-m21, spd1m-38, spd1-m40, and spd1-m41—displayed robust Suc22R2 nuclear accumulation poorly reversed by HU treatment. No mutant showed a complete inability to delocalize Suc22R2 to the cytoplasm (the phenotype seen when Spd1 is completely stable; i.e., in pcu4-d, cdt2-d, and csn1-d mutants). Notably, the region that corresponds to the conserved HUG domain (spd1-m10 through spd1-m20) was particularly sensitive to mutation in the assay for nuclear localization. In Dif1, this domain is proposed to bind the R2 subunit and promote its nuclear import.
Figure 5.
Subcellular localization of Suc22R2 in the individual spd1 mutants. (A) GFP-Suc22 was visualized by epifluorescence in fixed untreated cells and cells treated for 4 h with HU (20 mM) before fixation to arrest S phase. Controls were spd1+ (+) and spd1-d (−). These are shown on the left. The majority of spd1+ cells show nuclear GFP-Suc22. Following incubation in HU, no evidence of nuclear accumulation of GFP-Suc22 is seen in >80% of cells, the signal being pan-cellular. In the absence of Spd1, nuclear accumulation is not seen in either untreated or treated cells. (B) A representative image: spd1-M14 behaves like a null mutant of spd1 in this assay. GFP-Suc22 was visualized in spd1-M14 cells without HU treatment. DNA was visualized by DAPI staining. Error bars, SD from mean.
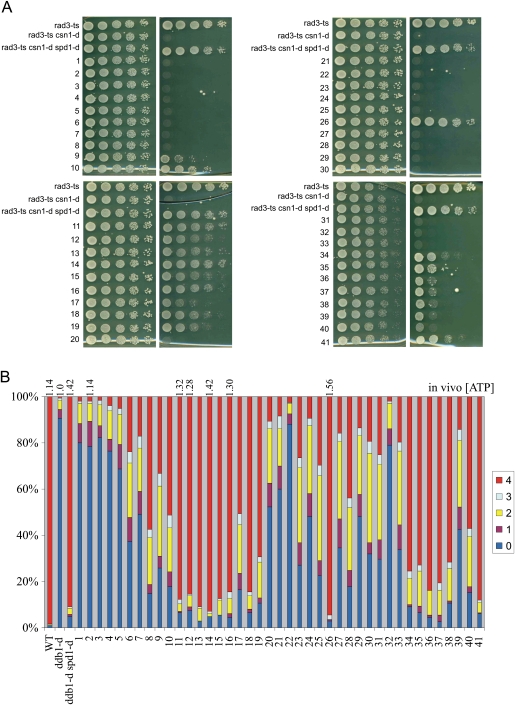
If the mechanisms by which Spd1 restrains RNR function in vivo depend on its ability to localize Suc22R2 in the nucleus, then RNR activation would be at least partially dependent on the loss of nuclear Suc22R2 accumulation in S phase, and spd1 mutants unable to accumulate Suc22R2 in the nucleus should phenocopy the spd1-d null. We thus tested two robust phenotypes for Spd1-dependent restraint of RNR activity. The first is the ability of spd1-null mutants to suppress the synthetic inviability associated with concomitant loss of the csn1 signalosome component and the rad3 checkpoint gene (Liu et al. 2003). Essentially, in csn1-d mutants, Spd1 is stable, and thus RNR activity is restrained in S phase—a situation reminiscent of the inviability of MEC1 deletion and the ability of SML1 deletion to suppress this (Zhao et al. 1998). Mutants in the HUG domain—plus spd1-m26, spd1-M34, spd1-M35, and spd1-M41—rescued rad3-d csn1-d synthetic lethality with significant efficiency, in many cases approaching that of the spd1-d null (Fig. 6A).
Figure 6.
Ability of individual spd1 mutants to restrain RNR function in vivo. Two in vivo assays for RNR function are shown. (A) The ability of spd1 mutants to rescue the synthetic lethality of a rad3-ts csn1-d double mutant at 34°C. A dilution series for each mutant was spotted, and plates were incubated at 34°C (partial loss of checkpoint). Viability is a semiquantitative measure of the loss of the in vivo inhibitory function of Spd1 (Liu et al. 2003). (B) The ability of spd1 mutants to rescue the spore formation defect of homothalic/h90 ddb1-d mutants. The number of spores formed gives a semiquantitative measure of the loss of the in vivo inhibitory function (Holmberg et al. 2005). The in vivo dATP concentration relative to ddb1-d cells were measured in selected mutants and are given above. Loss of inhibitory function correlates with increased dNTP pools. Wild-type (Wt) ddb1-d and ddb1-d spd1-d controls are shown on the left.
The second assay we chose is the ability of spd1 deletion to rescue the spore formation defect evident in the ddb1-d background (Liu et al. 2003; Holmberg et al. 2005). Ddb1 is a component of the Pcu4–Ddb1Cdt2 ubiquitin ligase required for Spd1 degradation, so Spd1 restrains RNR activity in S phase and lowers dNTP pools in ddb1-d mutants, and thus they cannot progress through meiosis and form spores. This phenotype and the low dNTP pools are reversed by deleting spd1. We combined each of the 41 spd1 mutants with h90 ddb1-d and scored the percentage of asci with either zero, one, two, three, or four spores (Fig. 6B). Spore formation was restored (>70% four-spored asci, close to that of ddb1-d spd1-d double-null cells: >90%) in mutants spd1-m11 through spd1-m16, spd1-m18, spd1-m19, spd1-m26, spd1-m34 through spd1-m38, and spd1-m41.
Both of these assays are robust and semiquantitative (Liu et al. 2003; Holmberg et al. 2005). Taking them together, we conclude that the defect in Suc22R2 nuclear import does not correlate with the biological evidence for restraint of RNR activity. Of particular interest are mutants spd1-m2 (import-defective, no defect in restraining RNR) and spd1-m10 plus spd1-m11 (little or no import defect, but significantly unable to restrain RNR activity). The broader trends in the data make clear that HUG domain mutations (spd1-m10 through spd1-m20) influence both import and restraint. Mutations in the C-terminal region (spd1-m34 through spd1-m41) also influence both import and restraint, and likely define a new segment of Spd1. We suggest this is named the Spd1 domain, in keeping with the nomenclature of Lee et al. (2008). Finally, mutations within the putative R1-binding region (of which only three residues are identical between Sml1 and Spd1) do not appear to dramatically influence either import or restraint of RNR activity (spd1-m28 through spd1-m32).
The ability to promote R1–R2 FRET does not correlate with RNR inhibition or nuclear import
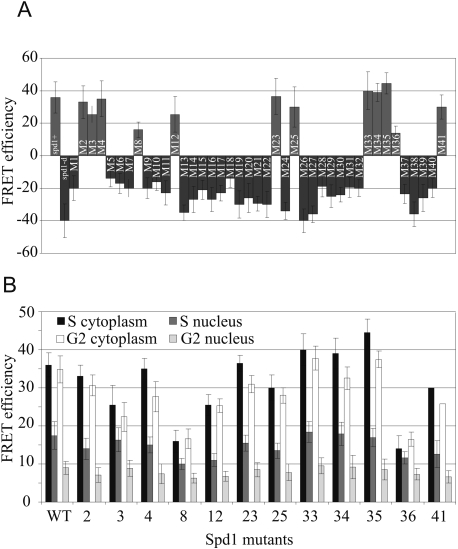
Each of the 41 spd1 mutants was crossed into the cdc22R1-CFP YFP-Suc22R2 strain and tested for their ability to promote Cdc22R1–Suc22R2 FRET. Only 12 of the 41 mutants could promote R1–R2 FRET (Fig. 7A). In most of these 12 cases, the FRET signal was comparable with that seen in spd1+ cells, and was seen in G2 and S phase in both the nuclear and the cytoplasmic compartments (Fig. 7B). The ability to promote R1–R2 FRET did not correlate with either the Suc22R2 nuclear import function of Spd1 or its ability to restrain RNR activity. For example, spd1-M2 keeps the R1–R2 FRET signal but lacks the R2 nuclear localization function. Mutant spd1-M12 similarly promotes R1–R2 FRET but has lost the ability to restrain RNR activity. Mutants demonstrating the converse are also apparent; i.e., spd1-M1 kept both the Suc22R2 nuclear import function and the ability to restrain RNR activity, but has lost the ability to promote FRET.
Figure 7.
Ability of the individual spd1 mutants to promote Cdc22R1-Suc22R2 FRET. Each of the 41 individual mutants was crossed to the cdc22-YFP CFP-suc22 background and tested for FRET signal. (A) Summary of FRET intensities. FRET was examined in both the nucleus and the cytoplasm of both mononuclear (G2) and septating binucleate (S-phase) cells. (Left) Controls are spd1+ and spd1-d. (B) The data for each circumstance for the mutants able to promote FRET. Error bars, SD from mean.
Spd1 can interact with both Cdc22R1 and Suc22R2
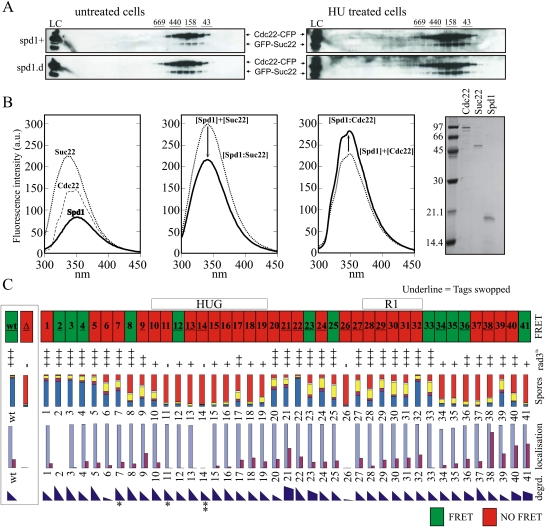
Active RNR in vitro consists of an α2β2 tetramer consisting of 2xR1 and 2xR2 subunits (Nordlund and Reichard 2006). Since RNR is active in spd1-d cells, R1–R2 FRET cannot simply reflect active tetramers. Furthermore, R1–R2 FRET does not decrease when cells enter S phase, so it is unlikely that the FRET signal reflects inactive R1–R2 complexes. Both Escherichia coli (Rofougaran et al. 2008) and mouse (Rofougaran et al. 2006) R1 subunits can be induced to form hexamers (α6) in vitro by either dATP or ATP binding. These subsequently form α6β2 octamers by association with a 2xR2 dimer (β2). α6β2 complexes have been suggested to represent the primary active RNR form because sufficient ATP is available in cells to occupy the majority of activity sites, and a correlation has been observed between α6 hexamer formation and ATP activation (Rofougaran et al. 2008). We used size fractionation to establish whether the presence of Spd1 correlated with the presence of higher-order RNR complexes (Fig. 8A). No evidence for multimer formation was observed in either exponential spd1+ or spd1-d cells. Limited higher-mass complexes were observed following HU treatment, but these were not spd1-dependent. Thus, we believe it unlikely that R1–R2 FRET reflects different higher-order complex formation.
Figure 8.
Spd1 interacts with both Cdc22R1 and Suc22R2. (A) Gel filtration of extracts prepared from cdc22-CFP GFP-suc22 double-tagged spd1+ and spd1-d cells after cross-linking with (right) and without (left) 4-h HU treatment. The α2β2 complex is predicted to be 274.6 kDa. Size markers are given above. (B) Fluorescence quenching assay for interaction between individual RNR subunits and Spd1. (Far left) Fluorescence emission spectra of individual proteins. (Middle left) Suc22R2 + Spd1. Predicted additive fluorescence emission (thin line) and experimental results (thick line) differ, demonstrating an interaction. (Middle right) Equivalent assay for Cdc22R1 + Spd1. (Far right) Aliquots of purified proteins assessed by SDS-PAGE. (C) Summary of data. (From the top) Ability to promote FRET. The region corresponding to the HUG and RNR interaction domains (see Fig. 1) are indicated. Where the mutant number is underlined, FRET data have been verified by reversing the tags (i.e., cdc22-CFP YFP-suc22). The ability of spd1 mutant cells to grow at 34°C in the rad3-ts csn1-d mutant background. Growth is scored semiquantitatively from the data in Figure 6A and several repeats. Spore viability data are reproduced from Figure 6B. Nuclear accumulation of Suc22R2 is reproduced from Figure 5A. The degradation of Spd1 in vivo is semiquantitatively estimated (by eye) from the data in Figure 4E and several repeats for all mutants. Asterisks indicate those mutants with partial (*) or complete (**) resistance to the in vitro degradation assay.
Despite extensive efforts, we were not able to identify conditions where Spd1 can be coprecipitated with Cdc22R1 or Suc22R2 from cell extracts. IDPs can bind multiple substrates with high specificity but low affinity, which is consistent with the low-affinity Spd1–Cdc22R2 interaction reported (Hakansson et al. 2006). We thus used fluorescence quenching to establish if Spd1 could bind either R1 or R2 subunits (Fig. 8B). Recombinant Spd1 can interact with both Cdc22R1 and Suc22R2 in vitro, as predicted by Hakansson et al. (2006) and supported by our identification of a role for Spd1 in Suc22R2 nuclear import.
Discussion
RNR dysregulation results in inappropriate total dNTP levels and/or unbalanced dNTP ratios, which are changes detrimental to genome stability (Chabes et al. 2003). RNR subunit availability is regulated in most organisms via transcriptional control (Elledge et al. 1993; Nordlund and Reichard 2006). In the fission yeast, cdc22R1 and suc22R2 are induced transcriptionally at the transition between G1 and S phase by the Cdc10, Res1, and Res2-containing transcription factor MBF (Fernandez Sarabia et al. 1993). In addition, DNA structure checkpoint activation results in inhibition of the Nrm1 MBF corepressor and subsequent MBF-dependent transcription (de Bruin et al. 2006). However, in cycling cells, the levels of Cdc22R1 and Suc22R2 protein do not change dramatically during the cell cycle or after checkpoint activation (Watson et al. 2004). Most likely, transcriptional regulation ensures that cells entering the cell cycle from stationary phase, or those damaged during stationary phase, have a supply of RNR subunits. A significant proportion of RNR regulation in cycling cells thus occurs post-translationally.
The in vivo inhibitory function(s) of Spd1
In S. cerevisiae, the Sml1 inhibitor binds to the R1 subunit (Zhao et al. 2000; Zhang et al. 2007) and prevents dNTP synthesis by inserting its C-terminal aromatic residue into a cleft usually occupied by the C-terminal residue of R2. Each S phase, Mec1 kinase activates the downstream Dun1 kinase to promote Sml1 degradation (Zhao and Rothstein 2002), likely by phosphorylating serine residues within the Sml1 phospho-degron (SML box) (see Fig. 1). When Mec1 is absent, Sml1 is not degraded, and dNTP synthesis is inhibited during S phase. Since mec1 mutants are also checkpoint-defective, MEC1 is an essential gene: A combination of low dNTP pools and a checkpoint defect results in cell death. Concomitant deletion of SML1 and MEC1 restores cell viability because S phase is no longer inhibited and the checkpoint thus is not required (Zhao et al. 2001).
Like S. cerevisiae Sml1, S. pombe Spd1 inhibits RNR in vitro (Hakansson et al. 2006) and restrains RNR activity in vivo when present in S phase (Liu et al. 2003). Importantly, restraint of RNR activity did not correlate with Suc22R2 relocalization. Both of the previously characterized biological assays for in vivo RNR activity gave consistent results and identified several mutants that clearly separate the nuclear import role of Spd1 from its ability to restrain RNR activity in vivo. Specifically, spd1-M12 is unable to restrain RNR activity but is competent for Suc22R2 nuclear import, while mutant spd1-M2 keeps the ability to restrain RNR activity but is, as far as the assay allows us to judge, fully defective for the nuclear import function. The complementary specific loss of function between these two mutants strongly suggests that the major regulatory role of Spd1 on RNR activity is not directly dependent on subcellular distribution of RNR subunits. Most likely, the restraint of RNR activity in vivo correlates with the in vitro inhibition via binding to Cdc22R1 (Hakansson et al. 2006).
We can identify two specific regions of Spd1 that, when mutated, result in decreased restraint of RNR. These two regions are the HUG domain and an additional region, the Spd1 domain, defined by mutants spd1-M34 through spd1-M41. The HUG domain is required for both the nuclear import function and the ability to restrain RNR activity in vivo. This raises the possibility that Spd1 interacts with both R1 and R2 subunits in S. pombe minimally through its HUG domain. We demonstrated that Spd1 can indeed bind both Cdc22R1 and Suc22R2 in vitro, but defining the mechanism of association awaits further biophysical characterization, as the dual specificity of Spd1 likely reflect the potential for low-affinity interactions between IDPs and several substrates.
Spd1 functions to promote nuclear import but is not a nuclear anchor
To establish if Spd1 shares the Dif1 function in nuclear import, we explored whether blocking nuclear export in spd1-deleted cells resulted in nuclear Suc22R2 accumulation: The absence of nuclear export will result in Suc22R2 nuclear accumulation if import is active. In LMB-treated spd1-d cells, Suc22R2 did not accumulate in the nucleus, demonstrating that Spd1 is required for active Suc22R2 nuclear import. A cluster of the mutants analyzed (spd1-M11 through spd1-M16) were either partially or completely defective for Suc22R2 import (data summarized in Fig. 8C). This cluster largely encompasses the conserved HUG domain, suggesting that, like for Dif1, the HUG domain is important for nuclear import and likely defines an R2 interaction surface, a prediction consistent with our identification of a direct association between Suc22R2 and Spd1 in vitro. Two additional mutants, spd1-M2 and spd1-M26, also lost Suc22R2 nuclear accumulation function.
In S. cerevisiae, loss of active import (DIF1 deletion) results in decreased but not absent R2 nuclear accumulation because the Wtm1-dependent nuclear anchor retains R2 subunits (Lee and Elledge 2006). Loss of both Dif1 (active import) and Wtm1 (anchoring) is required for complete loss of R2 nuclear accumulation. In S. pombe, loss of Spd1 alone results in complete loss of Suc22R2 nuclear accumulation. Either there is no R2 nuclear anchor in S. pombe, or Spd1 itself fulfils both import and anchoring roles. However, when Spd1 is localized artificially to the nucleolus, we did not detect additional Suc22R2 in the nucleolus, implying that there is no nuclear anchor for Suc22R2.
Alanine scanning mutagenesis did not define a degron domain
In S. cerevisiae, the SML box of the Sml1 and Dif1 proteins is thought to define a phospho-degron targeted by Mec1ATR-dependent Dun1 kinase activity. In S. pombe, Spd1 degradation is independent of Rad3ATR and the downstream kinase Cds1Chk2 during S phase. While Spd1 degradation becomes dependent on both kinases in response to checkpoint activation, this dependency is known, at least in part, to reflect the checkpoint dependence of cdt2 transcript induction. cdt2 encodes the adaptor for the ubiquitin ligase Pcu4–Ddb1Cdt2 that targets Spd1 for degradation. We did not define a distinct domain required for Spd1 degradation. This is consistent with the lack of evidence for regulation of Spd1 degradation by phosphorylation (Liu et al. 2005). Furthermore, no single serine or threonine mutation prevented degradation. There are no consensus Rad3ATR sites on Spd1, but two potential Cds1Chk2 sites are evident. Mutating both of these sites individually (spd1-M3 and spd1-M18) or together (spd1-M3–18) did not affect Spd1 stability.
Four mutants—spd1-M21, spd1-M23, spd1-M40, and spd1-M41—were partially resistant to degradation. Cells expressing these partially stable proteins were less responsive to loss of nuclear accumulation of Suc22R2 following HU, but none were synthetically lethal with rad3-ts (data not shown). This indicates that, unlike indirect stabilization of Spd1 via deletion of E3 ligase components, these mutant proteins did not keep their ability to fully restrain RNR activity. Intriguingly, the in vitro degradation assay identified a single lysine required for Cul4–Ddb1Cdt2 and signalosome-dependent E3 ubiquitin ligase degradation of Spd1. However, mutating this residue made no difference to in vivo Spd1 stability. One possibility is that K42 is the sole Ub acceptor in vitro, but is likely substituted by other lysines in vivo.
A novel Spd1-dependent level of RNR regulation?
Since we and others have speculated previously that active RNR complexes are formed in the cytoplasm after colocalization of Cdc22R1 and Suc22R2 following Spd1 degradation, we established a FRET assay to examine R1–R2 association in different cellular compartments. Our assumption was that 2xR1–2xR2 tetramer formation (α2β2 complex) would be enhanced in the cytoplasm following Spd1 degradation. While we could clearly visualize FRET between Cdc22-YFP and CFP-Suc22 (and similarly when the tags were reversed) (data not shown), R1–R2 FRET was not enhanced upon loss of Spd1, but was instead completely Spd1-dependent and did not correlate with RNR activity: It was neither increased nor decreased in the cytoplasm of S-phase cells compared with G2 cells. R1–R2 FRET also disappeared when spd1+ cells were treated with HU. However, spd1+ cells held in G2 and treated with HU lost the FRET signal within 30 min (data not shown), despite not progressing into S phase, indicating that HU, a free-radical scavenger, may independently quench the FRET signal.
Of the 41 spd1 mutants tested for R1–R2 FRET, 12 were positive. While there was some variation in the intensity of the FRET signal in these mutants, this was modest, relatively evenly distributed between the nuclear and cytoplasmic compartments, and not specifically different for either G2- or S-phase cells. Interestingly, the distribution of the mutants able to FRET did not correlate with either the nuclear import role or the in vivo restraint of RNR activity. For example, spd1-M2 cells were FRET-competent and had lost nuclear import completely, while spd1-M12 cells were FRET-competent but were unable to restrain RNR activity.
What does the FRET signal represent? In vitro, active RNR complexes consist of 2xR1 and 2xR2 subunits (α2β2). Both ATP (activating) and dATP (inhibitory) binding to the allosteric overall activity site stimulate α2β2 formation, despite their opposite effects on activation. Our gel filtration data indicate the presence of R1–R2 complexes at the size expected for α2β2 complexes and show that these were not dependent on spd1+. Combined with the observation that Spd1 loss leads to RNR activation, we can conclude that the presence of active α2β2 tetramers cannot be the cause of the FRET signal. Recent work using both E. coli (Rofougaran et al. 2008) and mouse (Rofougaran et al. 2006) RNR proteins has suggested that both dATP and ATP induce the formation of R1 hexamers (α6) that can form an α6β2 octamer by association with a dimer (β2) of R2 subunits. However, by gel filtration analysis, we did not observe evidence for Spd1-dependent higher-order RNR complexes. A subfraction of Cdc22R1 and Suc22R2 were seen to migrate at higher molecular weight, but this occurred after treatment with HU and was independent of spd1 status.
Our data suggest that Spd1-dependent FRET between R1 and R2 subunits reflects a changed conformation of the RNR α2β2 complex. We hypothesize that Spd1 mediates formation of immature inactive RNR complexes. In these inactive complexes, the fluorophores are appropriately aligned to allow FRET between Cdc22R1 and Suc22R1, reflecting an optimal complex architecture for subsequent activation when Spd1 is degraded. In support of this proposal, the p27 cyclin-dependent kinase inhibitor is an IDP that mediates formation of immature inactive Cdk2–CycA complexes (Russo et al. 1996). p27 folds onto the Cdk2 and CycA subunits, both individually and at the same time. Intriguingly, p27 directly inhibits Cdk2 by altering the conformation of the catalytic cleft and inserting a tyrosine residue as an ATP-mimicking residue. In this context, we note that, in active α2β2 tetramers, the R2 C-terminal residue is buried in a deep cleft of R1, and that interference with optimal RNR complex architecture has been observed using R2 C-terminal-mimicking peptides (Cohen et al. 1986). It has also been suggested that the C-terminal Phe of Sml1 (F104), which is mandatory for full inhibition in S. cerevisiae (Zhao et al. 2000), may also insert into the same deep cleft of R1 that is the binding pocket for the C-terminal aromatic residue of the R2 subunit. The change in fluorescence emission also supports the involvement of tyrosines and/or tryptophans in the interactions.
Once Spd1-dependent inactive complexes are formed, we propose that Spd1 degradation would leave them in the optimal conformation for catalytic activity by removing the direct inhibition and allowing activation via ATP binding to the allosteric “overall activity” site. To explain why spd1-d cells never exhibit FRET but maintain RNR in an active form, we postulate that, in the absence of Spd1, RNR complexes do still form, but with an alternative suboptimal architecture. The fact that these are abundant and not inhibited by Spd1 compensates for the loss of the Spd1-dependent forms.
Conclusion
We showed that Spd1 acts as an import factor for Suc22R2, as predicted from its relationship to Dif1 in S. cerevisiae. Since it also functions to restrain RNR activity in vivo, we asked if these two phenomena were related. Contrary to our expectation, the assays measuring restraint of RNR activity in vivo did not correlate with Suc22R2 import, demonstrating that the major function of Spd1 in regulating dNTP synthesis is unrelated to its role in nuclear sequestration of Suc22R2. We also established a FRET assay that revealed a novel aspect of RNR behavior that has the potential to provide yet another mechanism to regulate RNR, possibly through alterations of tetramer architecture. The precise role for this new effect of Spd1 on RNR and how it relates to the regulation of RNR awaits further analysis. The segmental distribution of Spd1 functionality uncovered here is reminiscent of the hallmarks of IDPs, including the association with multiple partners. Thus, the IDP nature of Spd1 likely explains how this small protein is able to regulate R1–R2 complexes in multiple ways.
Materials and methods
Cloning, expression, stability, purification, and interactions of recombinant proteins
The spd1, suc22, and cdc22 ORFs were PCR-amplified, cloned into appropriate vectors, and verified by sequencing. Individual spd1 mutations were constructed using oligonucleotide-directed mutagenesis, and each was verified by sequencing. Coupled in vitro transcription–translation (Promega) was performed using the manufacturer's instructions to produce 35S-labeled protein. Cell extracts were prepared by resuspending cell pellets in an equal volume of HB buffer (25 mM Tris-HCl at pH 7.5, 15 mM EGTA, 15 mM MgCl2 0.1% NP-40, 1 mM DTT, 0.1 mM NaF) and grinding by mortar and pestle under liquid nitrogen, and were clarified by 10-min centrifugation in a microfuge at 4°C. In vitro degradation reactions were started by adding 35S-labeled protein and were stopped after 10- or 20-min incubation at room temperature by adding SDS sample buffer. Labeled protein was visualized by SDS-PAGE and was quantified using a Storm PhosphorImager. Expression and purification of Cdc22 and Suc22 was as reported previously (Hakansson et al. 2006). For expression of recombinant protein spd1+, ORF was ligated into pET11a and expressed in BL21(DE3) (0.1 M IPTG). For production of 15N-labeled protein, cells were grown in 1 L LB media (100 μg/mL ampicillin) to OD600 0.7–0.8, harvested by centrifugation (25 min at 2000g), resuspended in M9 media (100 μg/mL ampicillin) with (15NH4)2SO4 (1.5 g/L) as the sole nitrogen source, and grown for 1 h before induction. Induction was for 3 h at 37°C. Induced cells were harvested (15 min at 5000g); resuspended in 25 mL of 1× PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), 25% (w/v) sucrose, 5 mM EDTA, and 1% (w/v) Triton X-100; and sonicated three times with intermediate washes in the same buffer. Spd1 was found in the pellets, and this was dissolved in 50 mL of 20 mM Tris (pH 8), 4.5 M urea, and 0.1% (v/v) Triton X-100, and spun 15 min at 20,000g. Supernatant was applied to a Mono Q column equilibrated in buffer A (20 mM Tris at pH 8, 4.5 M urea, 0.1% [v/v] Triton X-100) and eluted in a linear gradient of buffer B (20 mM Tris at pH 8, 4.5 M urea, 0.1% [v/v] Triton X-100, 1 M NaCl). The relevant fractions were identified and dialyzed extensively against 1× PBS (pH 7.4) using a cut-off of 3000 g/mol. Protein was concentrated and stored at −20°C until further use. Gel filtration was performed by loading 2 mg total cross-linked protein on a Superose 6 column following extraction of proteins by grinding in liquid nitrogen. Cells were first incubated with 2 mM homobifunctional cross-linker SDP (Thermo Scientific) for 30 min. Log-phase cells were either treated or not with 20 mM HU for 4 h. Interactions between Cdc22 or Suc22 with Spd1 were assayed by fluorescence quenching spectroscopy using a Perkin-Elmer LS50B and 1 μM protein concentrations alone or in mixture in 10 mM sodium phosphate (pH 7.4). Excitation was at 280 nm at room temperature, averaging five scans and subtracting buffer backgrounds. Theoretical emission spectra for the case of noninteracting proteins were generated by addition of spectra recorded on individual proteins. To assay dATP concentration, small molecule extracts were prepared from 50-mL cultures growing in minimal medium and harvested at 5.0 × 106 cells per milliliter on a 0.45-μm filter, washed once in ice cold water, and resuspended in ice-cold 500-μL 20%TCA/15 mM MgCl2. After three freeze–thaw cycles and final centrifugation, the supernatant was ether-extracted seven times to remove TCA. ATP was measured indirectly using a luciferase-based ATP determination kit from Biaffin GmbH and Co., KG, according to instructions. dATP was determined by a primer extension assay on a dA-specific template as described in Roy et al. (1999). Extended products were quantified on a Storm PhosphorImager.
Spectroscopy
A sample of 10 μM Spd1 was prepared in 10 mM NaH2PO4 adjusted to pH 7.4 using NaOH. A far-UV CD spectrum was recorded at room temperature on a Jasco 810 spectropolarimeter using a light path length of 1 mm. A total of five scans were accumulated from 250 to 190 nm, and buffer background was subtracted. Scanning speed was 20 nm/min, and data pitch was 0.1 nm. The resulting spectrum was smoothed using an FFT filter supplied by the Jasco software. A 15N,1H-HSQC NMR spectrum was recorded at 10°C on a Varian INOVA 750-MHz (1H) spectrometer with 48 transients in the direct dimension and 400 increments in t1. The spectrum was transformed and visualized using NMRPipe (Delaglio et al. 1995). The NMR sample was 1 mM 15N-Spd1 and 10 mM NaH2PO4 (pH 7.4) in 300 μL, which was centrifuged for 5 min at 5000g and transferred to a 5-mm Shigemi NMR tube.
Cell biology and genetics
Yeast strains were constructed using standard methods. spd1 mutants and genomic tagged constructs were created by PCR-amplifying the desired ORF using primers with ∼80-base-pair (bp) homology with the sequences flanking the genomic ORF. The resulting fragment was purified and transformed into an spd1∷ura4 strain where ura4+ had replaced the spd1 ORF. Replacements were selected by growth on 5-FOA, tested by PCR, and verified as correct by sequencing. To assay spore formation, strains were incubated on malt extract agar plates and incubated for 3 d at 25°C, and 200 U were assessed for zygotes/asci with zero to four spores. dATP measurements were performed on purified nucleotides by assessing ability to support primer extension against dNTP standard controls. The dATP level is calculated relative to ATP in the extract; i.e., the primer extension assay was performed on extract volume equal to 75 nmol ATP, as determined by a luciferase-based assay.
For epifluorescence and indirect immunofluorescence, cells were grown in log culture at 30°C (unless otherwise specified) in supplemented yeast extract (YE) media with or without drug as specified. For FRET analysis, cells were harvested, washed with PBS, and air dried on the slide; a drop of mounting medium (50% glycerol, 50% water) and a cover slip were added; and fluorescent proteins were visualized using a laser scanning confocal microscope (LSM 510, Zeiss). Photobleaching of the acceptor was performed by scanning the 514-nm Argon laser across a specific region of interest (ROI) within a cell (cytoplasm or nucleus). Images were processed in ImageJ (NIH). To calculate FRET, images were normalized and intensities were measured for the ROI. IDA indicates intensity of the donor in the presence of acceptor (prebleach), ID indicates intensity of the donor in the absence of acceptor (post-bleach), and IAUTO indicates background intensity (autofluorescence of an untagged control). The FRET efficiency is expressed as a percentage: 1 − [(IDA − IAUTO.)/(ID − IAUTO.)] × 100. We also verified FRET efficiencies using a dedicated ImageJ macro. Levels of complexed and dissociated R1–R2 were unresolved in the present assay: Donor intensities were not corrected for noninteracting R1–R2, rendering FRET efficiencies as a marker of subunit association a quantitative measure of R1–R2 complex concentration or donor–acceptor proximity. Concomitant photobleaching of donor with the acceptor is similarly not accounted for, resulting in negative FRET efficiencies in the absence of R1–R2 interaction and a lower estimate of FRET in cases of positive association.
Acknowledgments
We thank Signe Agernæs Sjørup for assistance with RNR and Spd1 purification. K.N. was supported by CRUK grant C5514/A10722 and MRC grant G0600233. A.-S.S. was supported by EU grant PIOTNGA 215148. C.H. and O.N. acknowledge support from The Danish Cancer Society.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.561910.
References
- Basrai MA, Velculescu VE, Kinzler KW, Hieter P 1999. NORF5/HUG1 is a component of the MEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol Cell Biol 19: 7041–7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MG, Somasundaram S, Glasner JD, Palecek SP 2006. Analyzing the dose-dependence of the Saccharomyces cerevisiae global transcriptional response to methyl methanesulfonate and ionizing radiation. BMC Genomics 7: 305 doi: 10.1186/1471-2164-7-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A, Domkin V, Thelander L 1999. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J Biol Chem 274: 36679–36683 [DOI] [PubMed] [Google Scholar]
- Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391–401 [DOI] [PubMed] [Google Scholar]
- Cohen EA, Gaudreau P, Brazeau P, Langelier Y 1986. Specific inhibition of herpesvirus ribonucleotide reductase by a nonapeptide derived from the carboxy terminus of subunit 2. Nature 321: 441–443 [DOI] [PubMed] [Google Scholar]
- Danielsson J, Liljedahl L, Barany-Wallje E, Sonderby P, Kristensen LH, Martinez-Yamout MA, Dyson HJ, Wright PE, Poulsen FM, Maler L, et al. 2008. The intrinsically disordered RNR inhibitor Sml1 is a dynamic dimer. Biochemistry 47: 13428–13437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin RA, Kalashnikova TI, Chahwan C, McDonald WH, Wohlschlegel J, Yates J 3rd, Russell P, Wittenberg C 2006. Constraining G1-specific transcription to late G1 phase: The MBF-associated corepressor Nrm1 acts via negative feedback. Mol Cell 23: 483–496 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB, Navas TA 1993. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays 15: 333–339 [DOI] [PubMed] [Google Scholar]
- Fernandez Sarabia MJ, McInerny C, Harris P, Gordon C, Fantes P 1993. The cell cycle genes cdc22+ and suc22+ of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol Gen Genet 238: 241–251 [DOI] [PubMed] [Google Scholar]
- Gallagher IM, Alfa CE, Hyams JS 1993. p63cdc13, a B-type cyclin, is associated with both the nucleolar and chromatin domains of the fission yeast nucleus. Mol Biol Cell 4: 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson P, Dahl L, Chilkova O, Domkin V, Thelander L 2006. The Schizosaccharomyces pombe replication inhibitor Spd1 regulates ribonucleotide reductase activity and dNTPs by binding to the large Cdc22 subunit. J Biol Chem 281: 1778–1783 [DOI] [PubMed] [Google Scholar]
- Holmberg C, Fleck O, Hansen HA, Liu C, Slaaby R, Carr AM, Nielsen O 2005. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev 19: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Kinoshita K 2007. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res 35: W460–W464 doi: 10.1093/nar/gkm363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YD, Elledge SJ 2006. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev 20: 334–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YD, Wang J, Stubbe J, Elledge SJ 2008. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell 32: 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T 2003. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev 17: 1130–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Poitelea M, Watson A, Yoshida SH, Shimoda C, Holmberg C, Nielsen O, Carr AM 2005. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4–Ddb1–CSN ubiquitin ligase. EMBO J 24: 3940–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund P, Reichard P 2006. Ribonucleotide reductases. Annu Rev Biochem 75: 681–706 [DOI] [PubMed] [Google Scholar]
- Reichard P 2002. Ribonucleotide reductases: The evolution of allosteric regulation. Arch Biochem Biophys 397: 149–155 [DOI] [PubMed] [Google Scholar]
- Rofougaran R, Vodnala M, Hofer A 2006. Enzymatically active mammalian ribonucleotide reductase exists primarily as an α6β2 octamer. J Biol Chem 281: 27705–27711 [DOI] [PubMed] [Google Scholar]
- Rofougaran R, Crona M, Vodnala M, Sjoberg BM, Hofer A 2008. Oligomerization status directs overall activity regulation of the Escherichia coli class Ia ribonucleotide reductase. J Biol Chem 283: 35310–35318 [DOI] [PubMed] [Google Scholar]
- Roy B, Beuneu C, Roux P, Buc H, Lemaire G, Lepoivre M 1999. Simultaneous determination of pyrimidine or purine deoxyribonucleoside triphosphates using a polymerase assay. Anal Biochem 269: 403–409 [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP 1996. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A–Cdk2 complex. Nature 382: 325–331 [DOI] [PubMed] [Google Scholar]
- Stubbe J 2003. Radicals with a controlled lifestyle. Chem Commun 2511–2513 [DOI] [PubMed] [Google Scholar]
- Sugase K, Dyson HJ, Wright PE 2007. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447: 1021–1025 [DOI] [PubMed] [Google Scholar]
- Tompa P 2002. Intrinsically unstructured proteins. Trends Biochem Sci 27: 527–533 [DOI] [PubMed] [Google Scholar]
- Watson A, Mata J, Bahler J, Carr A, Humphrey T 2004. Global gene expression responses of fission yeast to ionizing radiation. Mol Biol Cell 15: 851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Huang M 2008. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol Cell Biol 28: 7156–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, An X, Yang K, Perlstein DL, Hicks L, Kelleher N, Stubbe J, Huang M 2006. Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc Natl Acad Sci 103: 1422–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yang K, Chen CC, Feser J, Huang M 2007. Role of the C terminus of the ribonucleotide reductase large subunit in enzyme regeneration and its inhibition by Sml1. Proc Natl Acad Sci 104: 2217–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Rothstein R 2002. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci 99: 3746–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 2: 329–340 [DOI] [PubMed] [Google Scholar]
- Zhao X, Georgieva B, Chabes A, Domkin V, Ippel JH, Schleucher J, Wijmenga S, Thelander L, Rothstein R 2000. Mutational and structural analyses of the ribonucleotide reductase inhibitor Sml1 define its Rnr1 interaction domain whose inactivation allows suppression of mec1 and rad53 lethality. Mol Cell Biol 20: 9076–9083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J 20: 3544–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]