Abstract
Osteoarthritis (OA), the most prevalent aging-related joint disease, is characterized by insufficient extracellular matrix synthesis and articular cartilage degradation, mediated by several proteinases, including Adamts-5. miR-140 is one of a very limited number of noncoding microRNAs (miRNAs) specifically expressed in cartilage; however, its role in development and/or tissue maintenance is largely uncharacterized. To examine miR-140 function in tissue development and homeostasis, we generated a mouse line through a targeted deletion of miR-140. miR-140−/− mice manifested a mild skeletal phenotype with a short stature, although the structure of the articular joint cartilage appeared grossly normal in 1-mo-old miR-140−/− mice. Interestingly, miR-140−/− mice showed age-related OA-like changes characterized by proteoglycan loss and fibrillation of articular cartilage. Conversely, transgenic (TG) mice overexpressing miR-140 in cartilage were resistant to antigen-induced arthritis. OA-like changes in miR-140-deficient mice can be attributed, in part, to elevated Adamts-5 expression, regulated directly by miR-140. We show that miR-140 regulates cartilage development and homeostasis, and its loss contributes to the development of age-related OA-like changes.
Keywords: MicroRNA, miR-140, osteoarthritis, cartilage, Adamts-5
MicroRNAs (miRNAs) are a class of noncoding RNAs that negatively regulate genes involved in numerous biological processes. Hundreds of miRNAs have been identified in various organisms, and many are evolutionarily conserved. Moreover, an estimated one-third of all mammalian mRNAs are regulated by miRNAs, demonstrating the essential role of miRNAs in controlling gene expression (Lewis et al. 2005). miRNAs are generated as long primary transcripts (pri-miRNAs) that are processed by the enzymes Drosha and Dicer into short ribonucleotides (∼22 nucleotides long). The mature miRNAs are then incorporated into the RNA-induced silencing complex (RISC). The miRNA–RISC complex mediates the degradation of specific mRNA targets and/or the repression of mRNA translation via interactions with the 3′ untranslated regions (UTRs) that are partially sequence-specific (Bartel 2004). Several miRNAs exhibit a tissue-specific or developmental stage-specific expression pattern and have been associated with human diseases such as heart disease (van Rooij et al. 2006) and arthritis (Iliopoulos et al. 2008; Nakasa et al. 2008; Stanczyk et al. 2008; Yamasaki et al. 2009). In addition, mice with limb- or cartilage-specific deletion of the miRNA-processing enzyme Dicer exhibited a severe phenotype with reduced limb size but normal patterning (Harfe et al. 2005; Kobayashi et al. 2008). Dicer is indispensable for mature, functional miRNAs; therefore, this finding suggests that miRNAs play a critical role in skeletal development.
Recent studies have revealed the cartilage-specific expression of miR-140 in mouse embryos and zebrafish (Wienholds et al. 2005; Tuddenham et al. 2006). Using an miRNA microarray, we demonstrated considerable miRNA expression differences between human articular chondrocytes and mesenchymal stem cells. Furthermore, miR-140 exhibited the largest expression difference between the two cell types (Miyaki et al. 2009). The study of miR-140 may thus be the key miRNA to open a new insight of cartilage biology. However, the function of miRNAs in articular cartilage has not been analyzed by targeted deletion.
Previous studies also found reduced miR-140 expression in human osteoarthritis (OA) cartilage (Iliopoulos et al. 2008; Miyaki et al. 2009), which may contribute to the abnormal gene expression pattern characteristic of OA. These findings prompted us to examine the role of miR-140 in cartilage development and homeostasis by generating miR-140-deficient mice and cartilage-specific miR-140 transgenic (TG) mice. The results indicate that miR-140 plays dual roles in both cartilage development and homeostasis, in part via regulating Adamts-5, a major cartilage matrix-degrading protease in OA (Glasson et al. 2005; Stanton et al. 2005).
Results
Targeted deletion of cartilage-specific miR-140
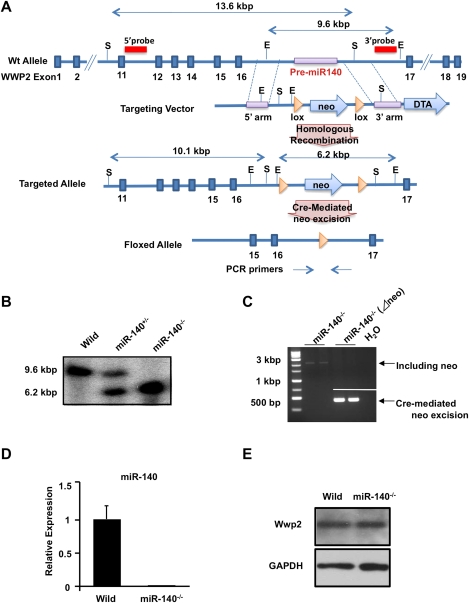
To define the in vivo function of miR-140, we targeted the mouse miR-140 sequence for deletion. We deleted the region containing miR-140 in the intron between exons 16 and 17 of the WW domain-containing E3 ubiquitin protein ligase 2 (Wwp2), and inserted a neomycin (neo) resistance cassette flanked by loxP sites (Fig. 1A,B). The floxed phosphoglycerine kinase (PGK)-neo cassette was removed by crossing with Meox-Cre TG mice, and Cre-mediated neo excision was confirmed by genomic PCR (Fig. 1C). Quantitative real-time PCR (qPCR) analysis of chondrocyte RNA showed a complete absence of miR-140 in miR-140−/− mice (Fig. 1D). We confirmed that the Wwp2 protein levels were unchanged in miR-140−/− mice (Fig. 1E).
Figure 1.
Generation of miR-140-null mice. (A) Targeting of miR-140. The targeting vector was constructed to replace the endogenous pri-miR-140 locus with a PGK-neo cassette by homologous recombination. The 5′ probe and 3′ probe used for Southern blot and PCR primers used for genotyping are indicated. (E) EcoRV, (S) SphI. (B) Southern blot analysis of wild-type (Wild) and miR-140 mutant mice. Genomic DNA was digested with EcoRV and probed with the indicated 3′ probe. (C) Confirmation of Cre-mediated neo excision by genomic PCR. The floxed PGK-neo cassette was removed by crossing with Meox-Cre TG mice. (D) Expression of mature miR-140 in wild-type (Wild) and miR-140−/− mouse embryos at E11.5 as assessed by qPCR. Mature miR-140 was not detected in miR-140−/− mice. Data are expressed as mean ± SEM (n = 3). (E) Western blot analysis of Wwp2. Wwp2 expression was not detectably different in miR-140−/− compared with wild-type mice.
Skeletal growth in miR-140−/− mice
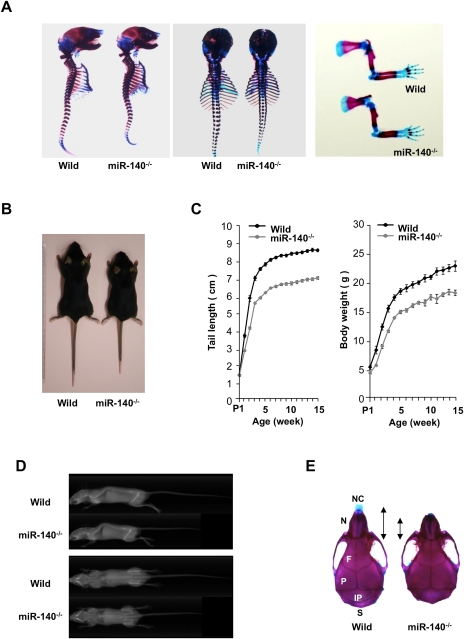
miR-140−/− mice were born in normal Mendelian ratios and were fertile. Skeletal development during embryogenesis in miR-140−/− mice appeared grossly normal (Fig. 2A). Postnatally, miR-140−/− mice manifested a mild skeletal phenotype, with short stature and low body weight (Fig. 2B–D), as well as craniofacial deformities characterized by a short snout and domed skull (Fig. 2E). This craniofacial phenotype in miR-140−/− mice shows partial similarity to that of cartilage-specific Dicer-deficient mice (Kobayashi et al. 2008), supporting the notion that miR-140 is a tissue-specific miRNA important in cartilage development. As miR-140 is expressed in cartilage and not in other tissues by whole-mount in situ hybridization (Wienholds et al. 2005; Tuddenham et al. 2006), reduced skeletal tissues of miR-140−/− mice may account for this weight loss phenotype.
Figure 2.
Growth retardation in miR-140−/− mice. (A) Skeletal preparation with Alcian blue/Alizarin red staining of wild-type (Wild) and miR-140−/− littermates at E17.5. miR-140−/− embryos exhibited no overt abnormalities in skeletal development. (Left) Whole-mount.(Middle) Dorsal view. (Right) Forelimb. (B) Appearance of wild-type and miR-140−/− mice at 4 wk. Growth retardation was observed in miR-140−/− mice. (C) Wild-type and miR-140−/− mice were scored for tail length and body weight at various postnatal stages. miR-140−/− mice showed growth retardation 1 wk after birth (tail length, P < 0.01; body weight, P < 0.05). Data are expressed as mean ± SEM (n = 5–12). (D) No gross skeletal changes were detectable by X-ray in 3-mo-old mice. (E) Skulls of wild-type and miR-140−/− mice stained with Alcian blue/Alizarin red. Dorsal views are shown. miR-140−/− mice exhibited craniofacial bone defects characterized by short nasal bone, short maxilla, and domed skull. (NC) Nasal capsule, (N) nasal bone, (F) frontal bone, (P) parietal bone, (IP) interparietal bone, (S) supraoccipital bone.
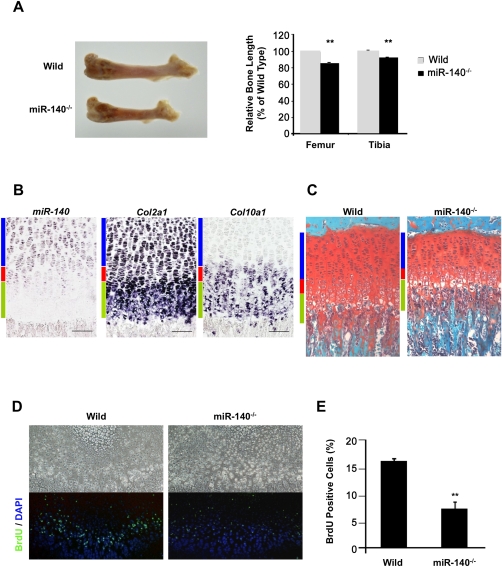
The hind-limb bones were shorter in miR-140−/− mice compared with wild-type mice (Fig. 3A). In the growth plates of wild-type mice, miR-140 was expressed in proliferating chondrocytes but not in hypertrophic chondrocytes that expressed Col10a1 (Fig. 3B). The width of the tibial growth plates in 1-mo-old mice was reduced in miR-140−/− mice compared with wild-type mice (Fig. 3C). The number of proliferating chondrocytes was significantly decreased at postnatal day 10 (P10) in miR-140−/− mouse growth plates (Fig. 3D,E), whereas no significant changes were observed in hypertorophyic zones (Fig. 3C). These findings indicate that the mild skeletal phenotype in miR-140−/− mice with short stature results from a reduction in proliferating chondrocytes.
Figure 3.
Skeletal growth in miR-140−/− mice. (A, left) Length of femur. (Right) Long bones were significantly shorter in the miR-140−/− mice. Data are expressed as mean ± SEM (n = 3). (**) P < 0.01. (B) Expression of pri-miR-140, Col2a1, and Col10a1 in the tibial growth plates of P10 mice. miR-140 was detected in proliferating chondrocytes. (Blue bar) Proliferative zone, (red bar) prehypertrophic zone, (green bar) hypertrophic zone. (C) Safranin O staining of the tibial growth plates in 1-mo-old mice. The proliferating zone was reduced in miR-140−/− mice compared with wild-type mice (Wild). (D,E) BrdU-positive cells were significantly reduced in miR-140−/− mice at P10. BrdU (green) and DAPI (blue) staining indicate nuclei. Data are expressed as mean ± SEM (n = 3). (**) P < 0.01.
OA-like pathology in miR-140−/− mouse knee joints
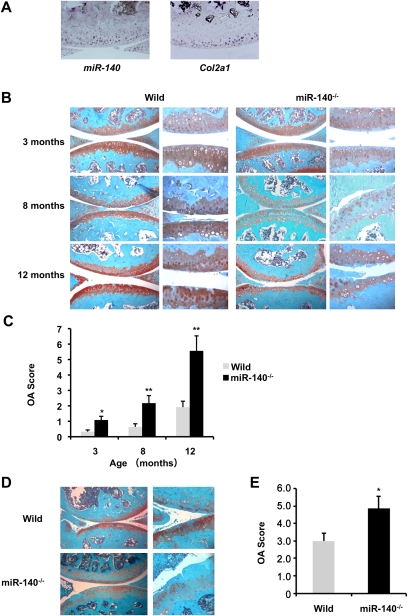
Chondrocytes play a critical role not only in skeletal development, but also in articular cartilage formation and maintenance. Analysis of pri-miR-140 expression in 2-mo-old wild-type mice by in situ hybridization showed miR-140 expression in chondrocytes from the surface to middle zones of articular cartilage and in the menisci, overlapping with expression of the chondrocyte marker Col2a1 (Fig. 4A). Furthermore, the structure and shape of knee joints—including articular cartilage, menisci, and ligaments—were analyzed by Safranin O staining and three-dimensional (3D) computed tomography (CT). They all appeared to be normal in miR-140−/− mice at birth and 1 mo of age (Supplemental Fig. S1).
Figure 4.
miR-140−/− mice exhibit OA-like pathology. (A) Expression of pri-miR-140 (left) and Col2a1 (right) in articular cartilage of the knee joints in 2-mo-old mice. miR-140 was detected in chondrocytes from the surface to middle zones of articular cartilage. (B) Safranin O staining of the knee joint in 3-mo-old mice (wild-type, n = 6; miR-140−/−, n = 6), 8-mo-old mice (wild-type, n = 15; miR-140−/−, n = 13), and 12-mo-old mice (wild-type, n = 12; miR-140−/−, n = 11). miR-140 deletion caused an early onset of OA-like changes. (C) Histopathologic scores of knee joints. OA scores were significantly increased in miR-140−/− mice. Data are expressed as mean ± SEM. (*) P < 0.05; (**) P < 0.01. (D) Surgical OA was induced in 10-wk-old wild-type and miR-140−/− mice by resecting the MMTL (wild-type mice, n = 8; miR-140−/− mice, n = 11). The knee joints were harvested 8 wk after surgery and stained with Safranin O for histopathologic analysis. (E) miR-140−/− mice showed significantly increased OA scores compared with wild-type mice after surgery. Data are expressed as mean ± SEM. (*) P < 0.05.
Because miR-140 expression was shown to be reduced in human OA cartilage (Iliopoulos et al. 2008; Miyaki et al. 2009), we examined the potential role of miR-140 in cartilage homeostasis. OA initiation and progression is mediated by various stimuli and circumstances, including the following three main factors: age-related changes in homeostatic balance, excessive mechanical stress sometimes triggered by joint injury, and transient inflammation in the articular joint damaging the cartilage matrix. Therefore, to determine the potential role of miR-140 in cartilage homeostasis, we used three different animal models of OA: an aging model, a surgical model, and an antigen-induced arthritis (AIA) model.
First, we tested whether loss of miR-140 affected age-related onset of OA changes, and observed that miR-140−/− mice developed an age-related OA-like pathology. Knee joints from 3-mo-old mice showed reduced Safranin O staining in femoral condyles and tibial plateaus, indicative of proteoglycan loss (Fig. 4B). By 8 mo, overt cartilage degradation was apparent as more severe proteoglycan loss, a roughened articular surface, and fibrillation; these changes were not observed in age-matched wild-type mice (Fig. 4B). By 12 mo, miR-140−/− mice showed severe structural cartilage defects (Fig. 4B) that were not associated with synovial hyperplasia. We also observed OA-like changes in elbow and ankle joint articular cartilage of miR-140−/− mice at 12 mo old, compared with articular cartilage from wild-type mice (Supplemental Fig. S2). To quantify OA-like pathological changes in the articular cartilage, an OA scoring system was used to validate several aspects of the histological changes (Chambers et al. 2001; Glasson et al. 2004). OA scores were significantly higher in miR-140−/− mice compared with wild-type mice (Fig. 4C). These results support the hypothesis that miR-140 is a critical regulator of cartilage homeostasis, and its loss contributes to cartilage degradation characteristic of OA.
Next, we used the surgical arthritis model, in which the articular cartilage was exposed to an excessive mechanical load, due to joint instability caused by surgical resection of the medial meniscotibial ligament (MMTL) (Glasson et al. 2007). Consistent with observations in the aging OA model, the surgical arthritis model also demonstrated that miR-140−/− mice exhibit accelerated proteoglycan loss and fibrillation of articular cartilage in knee joints compared with the wild-type mice at 8 wk after surgery, which is reflected in higher OA scores (Fig. 4D,E).
Cartilage-specific miR-140 TG mice exhibited resistance to AIA
We further examined the role of miR-140 in articular cartilage by using the AIA model, in which transient inflammation was induced by antigen injection. Inflammatory signals such as interleukin-1β (IL-1β) from the chondrocytes and synovial membrane of articular joint spaces mediate OA progression, in part by up-regulating the expression of cartilage matrix degradation enzymes in chondrocytes (Goldring and Goldring 2007). In this model, as well as in human OA pathogenesis, transient joint inflammation causes cartilage damage by the induction of cartilage-degrading enzymes. The AIA model is advantageous in that it leads to cartilage damage in a short time in wild-type mice, which enables us to monitor the beneficial effects of miR-140 against cartilage degradation by a gain-of-function approach.
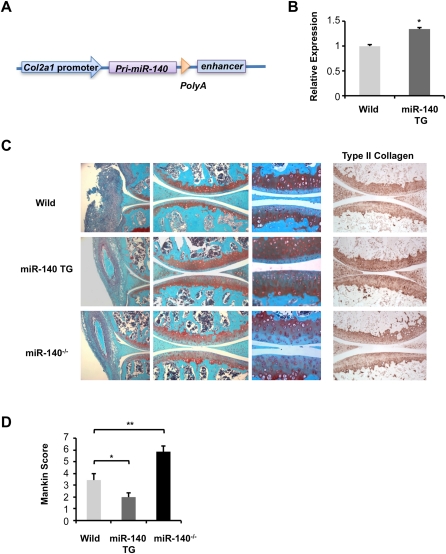
Accordingly, we generated cartilage-specific TG mice in which miR-140 expression was driven by a well-characterized Col2a1 enhancer sequence (Fig. 5A; Zhou et al. 1995; Krebsbach et al. 1996). We obtained three lines of TG mice and observed up-regulation of miR-140 in cartilage of all three lines (Fig. 5B). These TG mice did not show any apparent abnormalities in skeletal development (data not shown). To examine whether the miR-140 level in articular chondrocytes affects cartilage sensitivity to experimental challenge, we assessed AIA in knee joints of miR-140 TG mice, miR-140−/− mice, and wild-type mice. We observed similar levels of synovial hyperplasia among miR-140 TG mice, miR-140−/− mice, and wild-type mice; however, miR-140−/− mice showed reduced Safranin O staining (Fig. 5C). Importantly, miR-140 TG mice were resistant to proteoglycan and type II collagen loss compared with wild-type mice. To quantify the extent of matrix degradation, we used the Mankin scoring system (Mankin 1971; Zemmyo et al. 2003) for proteoglycan loss. Mankin scores were significantly lower in miR-140 TG mice and significantly higher in miR-140−/− mice compared with wild-type mice (Fig. 5D). These findings are consistent with the idea that miR-140 protects against OA progression.
Figure 5.
Cartilage-specific miR-140 TG mice are resistant to AIA. (A) miR-140 TG mice were generated with a construct using the cartilage-specific Col2a1 promoter. (B) Significantly increased expression of miR-140 in chondrocytes of miR-140 TG mice compared with those of wild-type mice was confirmed by qPCR analysis. These data are results from representatives of a single mouse line (n = 3). Data are expressed as mean ± SEM. (*) P < 0.05. (C) Seven days after induction of AIA, Safranin O staining and immunohistochemistry for type II collagen shows higher proteoglycan and collagen levels in miR-140 TG mice and lower levels in miR-140−/− mice compared with wild type. (D) Mankin score for proteoglycan loss in wild-type mice (n = 5), miR-140 TG mice (n = 6), and miR-140−/− mice (n = 6) 7 d after induction of AIA. Data are expressed as mean ± SEM. (*) P < 0.05; (**) P < 0.01.
ADAMTS-5 is a direct target of miR-140 and mediates OA pathogenesis
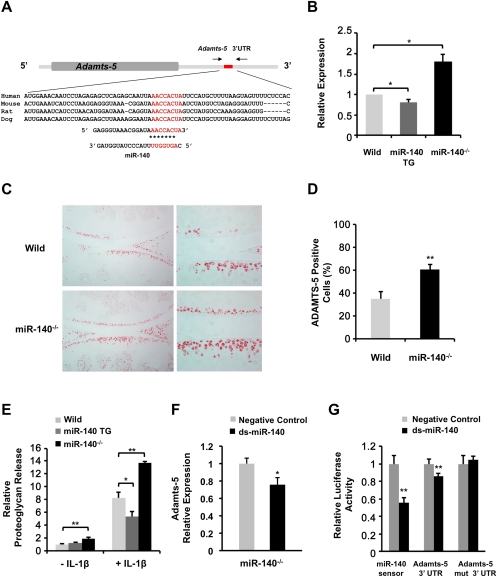
Identification of miR-140 target genes can provide new insights into miR-140 function and OA pathogenesis. To achieve this goal, we applied DNA array analysis to identify miRNA targets (Lim et al. 2005; Valencia-Sanchez et al. 2006). Based on computationally predicted target genes found in the online database TargetScan (http://www.targetscan.org), 17 of the mRNAs in the array that were increased in miR-140−/− chondrocytes exhibited conserved miR-140-binding sites in their 3′ UTRs with 7-mer or 8-mer seeds (Supplemental Table S1). Among the most overexpressed was the gene for ADAMTS-5, which degrades aggrecan and is a critical enzyme for OA pathogenesis (Glasson et al. 2005; Stanton et al. 2005). We analyzed Adamts-5 sequences in humans, mice, rats, and dogs and observed highly conserved miR-140-binding sites with 8-mer seeds in the Adamts-5 3′ UTR (Fig. 6A). Adamts-5 expression was significantly increased in chondrocytes from miR-140−/− mice and significantly decreased in those from miR-140 TG mice compared with wild-type (Fig. 6B). Similarly, increased ADAMTS-5 protein expression in articular cartilage in miR-140−/− mice at 1 and 3 mo old was visualized by immunohistochemistry (Fig. 6C,D; Supplemental Fig. S3). To confirm that miR-140 plays a critical role in aggrecanolysis, we quantified proteoglycan loss from cartilage in wild-type mice, miR-140 TG mice, and miR-140−/− mice. Femoral head cartilage explants were cultured with or without IL-1β, which is a potent catabolic stimulus of cartilage matrix degradation. Cartilage explants from miR-140−/− mice showed significantly increased proteoglycan release compared with wild-type cartilage (Fig. 6E). In contrast, IL-1β-induced proteoglycan release from the cartilage of miR-140 TG mice was significantly lower compared with wild-type cartilage.
Figure 6.
miR-140 regulates Adamts-5 expression. (A) miR-140 targets the Adamts-5 3′ UTR. Sequence alignment of a putative miR-140-binding site in the Adamts-5 3′ UTR shows a high level of sequence conservation and complementarity with miR-140. (B) Differential Adamts-5 expression in rib chondrocytes from wild-type mice, miR-140 TG mice, and miR-140−/− mice was evaluated by qPCR. Data are expressed as fold differences compared with wild-type chondrocytes (mean ± SEM; n = 4–5 per genotype). (*) P < 0.05. (C) Histologic analysis of articular cartilage from 3-mo-old mice. Immunostaining showed an increase in ADAMTS-5-positive chondrocytes in miR-140−/− mice. (D) ADAMTS-5-positive cells were detected in wild-type and miR-140−/− cartilage. Knee joints from 3-mo-old wild-type (n = 3) and miR-140−/− (n = 3) mice were examined. Data are expressed as mean ± SEM. (**) P < 0.01. (E) Femoral head cartilage explants from miR-140−/− mice cultured in DMEM only (n = 13) or DMEM containing IL-1β (n = 6) showed significantly increased proteoglycan release compared with wild-type mice in DMEM only (n = 17) or DMEM containing IL-1β (n = 6). IL-1β-induced proteoglycan release in miR-140 TG mice in medium (n = 12) was significantly lower compared with that of wild-type mice (n = 10). Data are expressed as fold differences compared with wild-type explants in DMEM only (mean ± SEM). (*) P < 0.05; (**) P < 0.01. (F) Relative Adamts-5 expression in miR-140−/− chondrocytes was determined by qPCR after transfection with ds-miR-140 (black). Data are expressed as fold differences relative to negative control cells (gray, mean ± SEM). (*) P < 0.05. (G) Luciferase activity of pLuc2-miR-140 sensor (miR-140 sensor), pLuc2-Adamts-5 3′ UTR (Adamts-5 3′ UTR), and pLuc2-Adamts-5 3′ UTR with mutated miR-140-binding site (Adamts-5 mut 3′ UTR). Luciferase activity was determined in HEK293T cells transfected with miR-140 sensor, Adamts-5 3′ UTR, or Adamts-5 mut 3′ UTR along with a ds-miR-140 (black) or negative control miRNA (gray) (n = 6). Data are expressed as relative luciferase activity (mean ± SEM). (*) P < 0.05; (**) P < 0.01.
We further tested whether miR-140 regulates Adamts-5 mRNA in chondrocytes. Treatment of chondrocytes from miR-140−/− mice with ds-miR-140 reduced Adamts-5 expression (Fig. 6F), supporting the notion that miR-140 negatively regulates Adamts-5 mRNA. To determine whether Adamts-5 is a direct miR-140 target in intact cells, the Adamts-5 3′ UTR, which includes a putative miR-140-binding site, was cloned downstream from the luciferase gene in an expression vector driven by the SV-40 promoter (Adamts-5 3′ UTR). Cotransfection of HEK293T cells with ds-miR-140 significantly reduced luciferase activity in cells transfected with Adamts-5 3′ UTR (Fig. 6G). No changes in luciferase activity were observed in cells transfected with the mutated luciferase expression vector (Adamts-5 mut 3′ UTR) in response to ds-miR-140 (Fig. 6G). Taken together, these data indicate that miR-140 directly regulates Adamts-5 expression and proteoglycan loss in articular cartilage.
Discussion
Recent studies have revealed that miRNAs play critical roles in various biological events, including development, disease, and immunity (Stefani and Slack 2008; Xiao and Rajewsky 2009). However, few molecular networks regulated by miRNAs have been characterized in detail, due in part to difficulty in determining the direct targets of each miRNA. Tissue-specific miRNAs targeting mice have been generated to elucidate the role of miRNAs in tissue and organ development, including heart and muscles (van Rooij et al. 2007; Zhao et al. 2007; Liu et al. 2008); however, so far there is very limited information on the function of miRNAs in cartilage/bone development and related diseases.
Transcription factor SRY-box-containing gene 9 (Sox9) is a master regulator of chondrocyte differentiation (de Crombrugghe et al. 2000). The expression of miR-140 shadows Sox9 expression, and the deletion of Sox9 diminishes miR-140 expression during embryogenesis (S Miyaki and H Asahara, unpubl.), indicating that miR-140 is under Sox9 regulation in chondrocytes. Although the disruption of Sox9 in mice results in a complete lack of cartilage development, miR-140−/− mice showed only a mild skeletal phenotype, with short stature and craniofacial changes. Results of studies targeting mice at specific developmental stages show that Sox9 is not only essential for chondrogenesis initiation, but is also important to produce large numbers of proliferating chondrocytes (Akiyama 2008). In the present study, the expression pattern of miR-140 was detected in the proliferation zone during endochondral ossification, and fewer proliferating chondrocytes were observed in miR-140−/− mice. Thus, regulation of proliferating chondrocytes may be one of the Sox9 functions mediated by miR-140.
Mice that are Dicer-deficient in cartilage tissues show a progressive and profound reduction in proliferating growth plate chondrocytes, which leads to severe skeletal growth defects and premature death (Kobayashi et al. 2008); however, this severe phenotype may reflect the effect of reducing all miRNAs, including ubiquitous ones. In this regard, our study provides the first example showing bone and cartilage phenotypes in mice caused by tissue-specific miRNA.
The phenotype of miR-140−/− mice on skeletal framework is not drastic, and the mice can survive for >1 yr; however, this mild phenotype provides us with an opportunity to monitor another aspect of miR-140 function in cartilage. Chondrocytes have two major functions: endochondral ossification for proper skeletal development, and articular cartilage maintenance for joint movement. Although chondrocytes in endochondral ossification disappear after bone development has been completed, articular cartilage chondrocytes persist, secreting the extracellular matrix that protects tissue from damage. The role of miRNAs in tissue homeostasis has not yet been well elucidated, other than a few studies such as those reporting the critical role of miR-143/145 in smooth muscle cell maintenance (Elia et al. 2009).
Articular cartilage is a good tissue for characterizing the molecular network involved in tissue homeostasis. The cartilage framework consists of a cartilage-specific extracellular matrix, which includes type II collagen and proteoglycans, and is maintained by chondrocytes embedded in the matrix. To maintain the integrity of this cartilage framework, the matrix is continuously undergoing remodeling via matrix-degrading enzymes; this degradation is balanced by the secretion of newly synthesized matrix proteins. The balance of these catabolic and anabolic signals in cartilage tissue is critical for cartilage homeostasis; aging, inflammation, or injuries may promote catabolic signals, including ADAMTS-5 and matrix metallopeptidase-13 (MMP-13), which can lead to arthritis changes. OA is the result of this age-related loss of the homeostatic balance between cartilage degradation and repair; it is aggravated by joint inflammation or excessive mechanical load (Goldring and Goldring 2007; Hashimoto et al. 2008; Goldring and Marcu 2009). Treatment options are limited, and new therapeutic targets that regulate the cartilage homeostasis balance should be sought.
To determine the role of miR-140 in articular cartilage, we examined three different arthritis models: age-related, surgical, and inflammatory (AIA). Disruption of miR-140 in vivo induced the early onset of spontaneous OA-like changes in articular cartilage of the age-related model and more severe OA-like changes in the surgical model. Transfection of human chondrocytes with ds-miR-140 down-regulated IL-1β-induced Adamts-5 expression (Miyaki et al. 2009). Consistent with our findings that miR-140 exerts a protective effect in cartilage, miR-140 TG mice were resistant to cartilage matrix degradation in the inflammatory model, suggesting that the regulation of miR-140 has important implications for drug design in OA treatment.
To address the mechanism of enhanced cartilage degradation in miR-140−/− mice, we showed the up-regulation of Adamts-5, a direct target of miR-140. ADAMTS-5 was shown to be a critical cartilage-degrading enzyme, because animals deficient in ADAMTS-5 activity are resistant to cartilage degeneration in the surgical OA model and inflammatory arthritis model (Glasson et al. 2005; Stanton et al. 2005). ADAMTS-5 also appears to be the major enzyme responsible for aggrecan degradation in human OA on the basis of increased mRNA and protein expression in OA cartilage (Malfait et al. 2002). Nevertheless, regulatory mechanisms of Adamts-5 expression have not been clearly elucidated. Two groups reported recently that Runx2 regulates Adamts-5 expression (Thirunavukkarasu et al. 2007), and hedgehog signaling regulates the expression of Adamts-5 via Runx2 (Lin et al. 2009). In addition to these transcriptional regulations, in the present study we demonstrated that Adamts-5 expression was tightly regulated by miR-140 at the post-transcriptional level.
Although the critical role of miR-140 in cartilage maintenance may be explained largely by identifying Adamts-5 as its direct target, miRNAs are believed to regulate multiple target mRNAs; therefore, the role of miR-140 in cartilage homeostasis may involve the regulation of additional genes, as listed in Supplemental Table S1. Accordingly, gene expression analysis by microarray and qPCR revealed an up-regulation of other cartilage-related catabolic factors, such as matrix degradation enzymes, and a down-regulation of cartilage matrix genes in chondrocytes of miR-140−/− mice as early as P3 (Supplemental Fig. S2). These results suggest that miR-140 may suppress pathways other than Adamts-5 expression, thus regulating the overall balance of cartilage matrix synthesis and degradation. Histone deacetylase 4 (HDAC4), which inhibits hypertrophic differentiation of chondrocytes (Vega et al. 2004), is thought to be an miR-140 target gene (Tuddenham et al. 2006); however, we did not observe significant changes in HDAC4 mRNA or protein expression in the endochondral plate or articular cartilage in the present study. The miR-140−/− and miR-140 TG lines may be useful in identifying other targets associated with cartilage development and homeostasis.
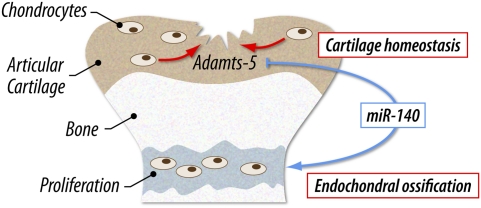
Taken together, our findings demonstrate that miR-140 is required for skeletal development and cartilage homeostasis, and protects against OA-like pathology via Adamts-5 regulation (Fig. 7). We conclude that miR-140 is a novel regulator of cartilage homeostasis, and changes in its expression and function play an important role in diseases associated with cartilage destruction, including OA and inflammatory arthropathies.
Figure 7.
Adamts-5 regulation via miR-140 is required for skeletal development and cartilage homeostasis. miR-140 has dual roles in both endochondral ossification and articular cartilage homeostasis. During endochondral ossification, miR-140 is important for chondrocyte proliferation, although its targets are unclear. miR-140 also plays a critical role in articular cartilage homeostasis by repressing Adamts-5 expression.
Materials and methods
Generation of miR-140-null mice and miR-140 TG mice
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at The Scripps Research Institute and National Institute for Child Health and Development. A vector was constructed to replace the endogenous miR-140 locus with a PGK-neo cassette by homologous recombination in embryonic stem (ES) cells. The 5′ and 3′ sequences flanking the endogenous miR-140 locus were amplified by PCR from a C57BL/6 genomic BAC clone (BACPAC Resource Center). These homologous arms were cloned into a vector incorporating both a neomycin resistance cassette for positive selection and a diphtheria toxin (DTA) gene for negative selection. The targeting vector was linearized and electroporated into TT2F mouse ES cells. Recombinant ES clones were isolated after culture in medium containing G418 antibiotic. Clones were then screened for proper integration by Southern blot analysis with the 5′ probe, 3′ probe, and neomycin resistance cassette sequence indicated in Figure 1. After proper integration was validated by genomic sequencing, two clones were chosen for microinjection into eight-cell-stage embryos. The resulting chimeric offspring were crossed with C57BL/6 mice, and germline transmission was confirmed by Southern blot and PCR. The floxed PGK-neo cassette was removed by crossing with Meox-Cre TG mice; Cre-mediated neo excision was confirmed by genomic PCR.
To generate cartilage-specific miR-140 TG mice, a pri-miR-140 fragment was PCR-amplified from mouse chondrocyte cDNA with the primers 5′-TGGTGTGTGGTTCTATGCCAGC-3′ and 5′-AGCCTCAAGCCAGAATTCAGG-3′.
pri-miR-140 was cloned into the NotI site of a Col2a1-based expression vector (Ueta et al. 2001), which contained the promoter and enhancer of the mouse Col2a1 gene. TG mice were generated by pronuclear injection of the transgene into the B6 strain, and were backcrossed to a C57BL/6 background. Genomic DNA isolated from the tail was analyzed by PCR using specific primers or transgene probes (forward primer, 5′-CAGGCTGTGCTTGGTGGGCATCTG-3′; reverse primer, 5′-CTAGCCTAGGCTCGAGAAGCTTGCT-3′).
qPCR
qPCR was performed using TaqMan Gene Expression Assay probes for Adamts-5 (Mm01344182_m1) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Mm 99999915_g1; Applied Biosystems), and qPCR for miR-140 was performed using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer's protocol. GAPDH or sno202 was used as an internal control to normalize sample differences.
Western blot analysis
Total protein extracts of wild-type and miR-140−/− chondrocytes were prepared for Western blot analysis. The Wwp2 protein, which is encoded by the host gene of miR-140, was detected by the AIP2 antibody (dilution, 1:2000; sc-11896, Santa Cruz Biotechnologies).
Radiologic imaging
X-ray and micro-CT imaging were performed using LaTheta LCT-200 (Aloka) according to the manufacturer's instructions. Three-dimensional CT images of the knee joint were recreated by using VGStudio MAX 2.0 software (Nihon Visual Science).
Bromodeoxyuridine (BrdU) labeling, RNA in situ hybridization, and immunohistochemistry
Proliferating cells were detected by BrdU incorporation using the In Situ Cell Proliferation kit, FLUOS (Roche); 4′,6-diamidino-2-phenylindole (DAPI) served as a counterstain. In situ hybridization of miR-140 was performed as described previously (Yokoyama et al. 2009). We used a probe specific for the primary region of miR-140. The sections of knee joints were immunostained using antibodies against ADAMTS-5 (ab13976, Abcam) and type II collagen (II-II6B3, Hybridoma Bank).
Histopathologic assessment
The miR-140−/− mice and wild-type littermates were obtained from the intercross of miR-140+/− maintained in a C57BL/6 background. Whole-mount Alcian blue and Alizarin red S staining of skeletons were performed on wild-type and miR-140−/− embryos at embryonic day 17.5 (E17.5). Mice were euthanized, and knee joints were harvested from 3-, 8-, and 12-mo-old mice. The knee joints were fixed in 10% zinc-buffered formalin (Z-Fix; Anatech) and decalcified in decalcifier (TBD-2; Shandon). For each animal, 4-μm sagittal sections through the central weight-bearing region of the medial femorotibial joint were stained with Safranin O and Fast Green for analysis of histopathologic differences between wild-type and miR-140−/− mice. The proteoglycan content of articular cartilage was scored using the previously reported Mankin scoring system (Mankin 1971; Zemmyo et al. 2003). OA scores indicating the severity of cartilage degeneration were evaluated for wild-type (n = 6) and miR-140−/− (n = 6) 3-mo-old mice, wild-type (n = 15) and miR-140−/− (n = 13) 8-mo-old mice, and wild-type (n = 12) and miR-140−/− (n = 11) 12-mo-old mice. At least four sections per sample were analyzed microscopically and scored using previously reported semiquantitative scoring systems (Chambers et al. 2001; Glasson et al. 2004).
Mouse model of surgically induced OA
Surgical OA was induced in wild-type and miR-140−/− mice at the age of 10 wk by resecting the MMTL in the right knee joint (Glasson et al. 2007). Pathological changes in medial tibial plateau and medial femoral condyle of the joint were evaluated after 8 wk on safranin-O-stained sections by OA score.
Mouse model of AIA
Inflammatory arthritis was induced in knee joints of 10-wk-old wild-type (n = 5), miR-140−/− (n = 6), and miR-140 TG (n = 6) mice by intra-articular injection of methylated bovine serum albumin (mBSA; Sigma catalog no. A1009) in mice preimmunized with mBSA. On day 1, mice were preimmunized by a 100-μL intradermal injection at the base of the tail with an emulsion containing 100 μg of mBSA (in 0.9% saline) and an equal volume of Freund's complete adjuvant (Sigma catalog no. F5881). On day 10, animals were treated with intra-articular mBSA (10 μL of 20 mg/mL mBSA in 0.9% sterile saline or vehicle alone) into the left and right knee joints. On day 17, the knee joints were harvested and processed using standard procedures.
DNA microarray analysis
DNA microarray analysis was performed using the Affymetrix mouse genome 430 2.0 array. RNA samples were collected from cultured rib chondrocytes of wild-type and miR-140−/− mice at P3. Microarray data were summarized by the Robust Multichip Average (RMA) method, and statistical analysis was performed using National Institute on Aging (NIA) Array Analysis (http://Igsun.grc.nia.nih.gov/ANOVA). The microarray data were deposited in the Gene Expression Omnibus (GEO) repository under accession number GSE16007.
Bioinformatics and miR-140 target analysis
We performed searches on the TargetScan prediction program (http://targetscan.org) for the predicted targets of miR-140.
Quantification of proteoglycan
Femoral heads were harvested from 4-wk-old wild-type and miR-140−/− mice. The explants were cultured for 3 d with or without IL-1β (5 ng/mL) in Dulbecco's modified Eagle's medium (DMEM) containing 10 mM HEPES and 1% penicillin/streptomycin. At the end of the culture, the cartilage explants were digested with protease K solution (Tris-EDTA at pH 8.0, 30 μg/mL protease K, 0.5% Tween 20) overnight at 50°C. Proteoglycan content in the protease K digests and conditioned medium were determined using the Blyscan Glycosaminoglycan Assay kit (Biocolor).
Cell culture and transfection of ds-miR-140 assay
Mouse chondrocytes were prepared from rib cartilage of P3 mice by digestion with collagenase. Mouse chondrocytes were cultured in DMEM with 10% fetal bovine serum (FBS) at 37°C. dsRNA oligonucleotides (5 nM) representing mature sequences that mimic endogenous miR-140 and Silencer Negative Control siRNA #1 (Ambion) were transfected into chondrocytes with Lipofectamine 2000 (Invitrogen). The synthesized RNA oligonucleotides 5′-CAGUGGUUUUACCCUAUGGUAG-3′ and 5′-ACCACAGGGUAGAACCACGGAC-3′ were annealed to obtain ds-miR-140. Silencer Negative Control siRNA #1 (siNega) was used at the same concentration as the specific miR-140 ds RNA in each experiment.
Luciferase assay
To create the pLuc2 reporter vector, a luciferase 2 gene (Promega) was incorporated into a modified pGL3-control plasmid with HindIII and EcoRI. To create the pGL3-miR-140 sensor vector (miR-140 sensor), the following chemically synthesized miR-140 multiple target sites were annealed and inserted between EcoRI and XhoI sites downstream from the firefly luciferase coding region in a pGL3 luciferase reporter plasmid with a modified 3′ UTR sequence (sense strand, 5′-AATTCTACCATAGGGTAAAACCACTGCTACCATAGGGTAAAACCACTGCTACCATAGGGTAAAACCACTGCTACCATAGGGTAAAACCACTG-3′; antisense strand, 5′-TCGACAGTGGTTTTACCCTATGGTAGCAGTGGTTTTACCCTATGGTAGCAGTGGTTTTACCCTATGGTAGCAGTGGTTTTACCCTATGGTAG-3′). To create the pLuc2-Adamts-5 3′ UTR vector (Adamts-5 3′ UTR), a fragment of the 3′ UTR of Adamts-5 gene including the predicted miR-140-binding site was PCR-amplified using the primer set 5′-TGAATTCCTACTCTGCTTCCCTCTATGATC-3′ and 5′-TTCTCGAGCTGTGTTTCTTTCCTCAGGAG-3′, and then cloned downstream from the luciferase 2 gene with EcoRI and XhoI. To create the reporter vector with mutated miR-140-binding site (Adamts-5 mut 3′ UTR), the 3′ UTR of Adamts-5 was amplified with the primer set 5′-TGAATTCCTACTCTGCTTCCCTCTATGATC-3′ and 5′-CCCTCTAGACATAGATTTGACGAATATCCGTTTACCCTCCTTAGG-3′, and then cloned into a Adamts5 3′ UTR-containing vector with EcoRI and XbaI. ds-miR-140 and the scrambled siRNA sequences (AGUUGCACGCGCAAUUAdCdC and UAAUUGCGCGUGCAACUdTdT; final concentration, 50 nM) were reverse-transfected with Lipofectamine RNAiMAX (Invitrogen) into HEK293T cells (2.0 × 104 cells per well in a 96-well plate). After 24 h of transfection, culture medium was changed, and the firefly luciferase reporter plasmid (20 ng) and Renilla luciferase control plasmid pRL-SV40 (20 ng) were transfected with Lipofectamine 2000 (Invitrogen). Luciferase activity was determined with the Dual-Glo Luciferase Assay System (Promega).
Statistical analysis
Two-tailed independent Student's t-test and the nonparametric Wilcoxon signed-rank test were used for statistical analysis. Asterisks indicate differences with statistical significance at P < 0.05 (*) and P < 0.01 (**).
Acknowledgments
We thank E. Kim for her excellent technical support, and extend our gratitude to all other members in the laboratory. We also thank Y. Takahashi and M. Asada for their bioinformatics assistance, and E. Lamar for critical reading of the manuscript and discussion. T.S. and H.A. are associate scientists of Tokyo Medical and Dental University. This project was supported by NIH AR050631 (to H.A.), AR056120 (to H.A.), AG007996 (to M.K.L.), and AG033409 (to M.K.L.); the Arthritis National Research Foundation (S.M.); the Arthritis Foundation (S.O.); grants from the Ministry of Health, Labour, and Welfare; the Genome Network Project (MEXT); grants-in-aid for Scientific Research (MEXT); and grants from National Institute of Biomedical Innovation, Research on Child Health and Development, and The Japan Health Sciences Foundation.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1915510.
Supplemental material is available at http://www.genesdev.org.
References
- Akiyama H 2008. Control of chondrogenesis by the transcription factor Sox9. Mod Rheumatol 18: 213–219 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Chambers MG, Cox L, Chong L, Suri N, Cover P, Bayliss MT, Mason RM 2001. Matrix metalloproteinases and aggrecanases cleave aggrecan in different zones of normal cartilage but colocalize in the development of osteoarthritic lesions in STR/ort mice. Arthritis Rheum 44: 1455–1465 [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B, Lefebvre V, Behringer RR, Bi W, Murakami S, Huang W 2000. Transcriptional mechanism of chondrocyte differentiation. Matrix Biol 19: 389–394 [DOI] [PubMed] [Google Scholar]
- Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J, et al. 2009. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: Correlates with human disease. Cell Death Differ 16: 1590–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasson SS, Askew R, Sheppard B, Carito BA, Blanchet T, Ma HL, Flannery CR, Kanki K, Wang E, Peluso D, et al. 2004. Characterization of and osteoarthritis susceptibility in ADAMTS-4-knockout mice. Arthritis Rheum 50: 2547–2558 [DOI] [PubMed] [Google Scholar]
- Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, Flannery CR, Peluso D, Kanki K, Yang Z, et al. 2005. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature 434: 644–648 [DOI] [PubMed] [Google Scholar]
- Glasson SS, Blanchet TJ, Morris EA 2007. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage 15: 1061–1069 [DOI] [PubMed] [Google Scholar]
- Goldring MB, Goldring SR 2007. Osteoarthritis. J Cell Physiol 213: 626–634 [DOI] [PubMed] [Google Scholar]
- Goldring MB, Marcu KB 2009. Cartilage homeostasis in health and rheumatic diseases. Arthritis Res Ther 11: 224 doi: 10.1186/ar2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ 2005. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci 102: 10898–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Nakasa T, Hikata T, Asahara H 2008. Molecular network of cartilage homeostasis and osteoarthritis. Med Res Rev 28: 464–481 [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A 2008. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One 3: e3740 doi: 10.1371/journal.pone.0003740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, Merkenschlager M, Kronenberg HM 2008. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci 105: 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebsbach PH, Nakata K, Bernier SM, Hatano O, Miyashita T, Rhodes CS, Yamada Y 1996. Identification of a minimum enhancer sequence for the type II collagen gene reveals several core sequence motifs in common with the link protein gene. J Biol Chem 271: 4298–4303 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Lin AC, Seeto BL, Bartoszko JM, Khoury MA, Whetstone H, Ho L, Hsu C, Ali AS, Alman BA 2009. Modulating hedgehog signaling can attenuate the severity of osteoarthritis. Nat Med 15: 1421–1425 [DOI] [PubMed] [Google Scholar]
- Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN 2008. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev 22: 3242–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfait AM, Liu RQ, Ijiri K, Komiya S, Tortorella MD 2002. Inhibition of ADAM-TS4 and ADAM-TS5 prevents aggrecan degradation in osteoarthritic cartilage. J Biol Chem 277: 22201–22208 [DOI] [PubMed] [Google Scholar]
- Mankin HJ 1971. Biochemical and metabolic aspects of osteoarthritis. Orthop Clin North Am 2: 19–31 [PubMed] [Google Scholar]
- Miyaki S, Nakasa T, Otsuki S, Grogan SP, Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK, Asahara H 2009. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum 60: 2723–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, Asahara H 2008. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum 58: 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, Detmar M, Gay S, Kyburz D 2008. Altered expression of microRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum 58: 1001–1009 [DOI] [PubMed] [Google Scholar]
- Stanton H, Rogerson FM, East CJ, Golub SB, Lawlor KE, Meeker CT, Little CB, Last K, Farmer PJ, Campbell IK, et al. 2005. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature 434: 648–652 [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ 2008. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9: 219–230 [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu K, Pei Y, Wei T 2007. Characterization of the human ADAMTS-5 (aggrecanase-2) gene promoter. Mol Biol Rep 34: 225–231 [DOI] [PubMed] [Google Scholar]
- Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T 2006. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett 580: 4214–4217 [DOI] [PubMed] [Google Scholar]
- Ueta C, Iwamoto M, Kanatani N, Yoshida C, Liu Y, Enomoto-Iwamoto M, Ohmori T, Enomoto H, Nakata K, Takada K, et al. 2001. Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J Cell Biol 153: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524 [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN 2006. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci 103: 18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN 2007. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316: 575–579 [DOI] [PubMed] [Google Scholar]
- Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, et al. 2004. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119: 555–566 [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH 2005. MicroRNA expression in zebrafish embryonic development. Science 309: 310–311 [DOI] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K 2009. MicroRNA control in the immune system: Basic principles. Cell 136: 26–36 [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Nakasa T, Miyaki S, Ishikawa M, Deie M, Adachi N, Yasunaga Y, Asahara H, Ochi M 2009. Expression of microRNA-146a in osteoarthritis cartilage. Arthritis Rheum 60: 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Ito Y, Ueno-Kudoh H, Shimizu H, Uchibe K, Albini S, Mitsuoka K, Miyaki S, Kiso M, Nagai A, et al. 2009. A systems approach reveals that the myogenesis genome network is regulated by the transcriptional repressor RP58. Dev Cell 17: 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmyo M, Meharra EJ, Kuhn K, Creighton-Achermann L, Lotz M 2003. Accelerated, aging-dependent development of osteoarthritis in α1 integrin-deficient mice. Arthritis Rheum 48: 2873–2880 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D 2007. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129: 303–317 [DOI] [PubMed] [Google Scholar]
- Zhou G, Garofalo S, Mukhopadhyay K, Lefebvre V, Smith CN, Eberspaecher H, de Crombrugghe B 1995. A 182 bp fragment of the mouse pro α 1(II) collagen gene is sufficient to direct chondrocyte expression in transgenic mice. J Cell Sci 108: 3677–3684 [DOI] [PubMed] [Google Scholar]