Cocaine is recognized as the most reinforcing of all drugs of abuse.1,2,3 There is no available anti-cocaine medication. The disastrous medical and social consequences of cocaine addiction have made the development of an effective pharmacological treatment a high priority.4,5,6 An ideal anti-cocaine medication would be to accelerate cocaine metabolism producing biologically inactive metabolites via a route similar to the primary cocaine-metabolizing pathway, i.e. cocaine hydrolysis catalyzed by plasma enzyme butyrylcholinesterase (BChE).5,7,8,9,10,11 However, the native BChE has a low catalytic efficiency against naturally occurring (−)-cocaine.12,13,14,15 (−)-cocaine has a plasma half-life of ~ 45 – 90 min, long enough for manifestation of the central nervous system (CNS) effects which peak in minutes.13,16 Here we report an unconventional computational design leading to discovery of a human BChE mutant with a ~151-fold improved catalytic efficiency, which can be used as an exogenous enzyme in human to prevent (−)-cocaine from reaching CNS. The encouraging outcome not only provides a hopeful anti-cocaine medication, but also demonstrates that a novel general approach of studying enzymatic mechanism and computational drug design is promising.
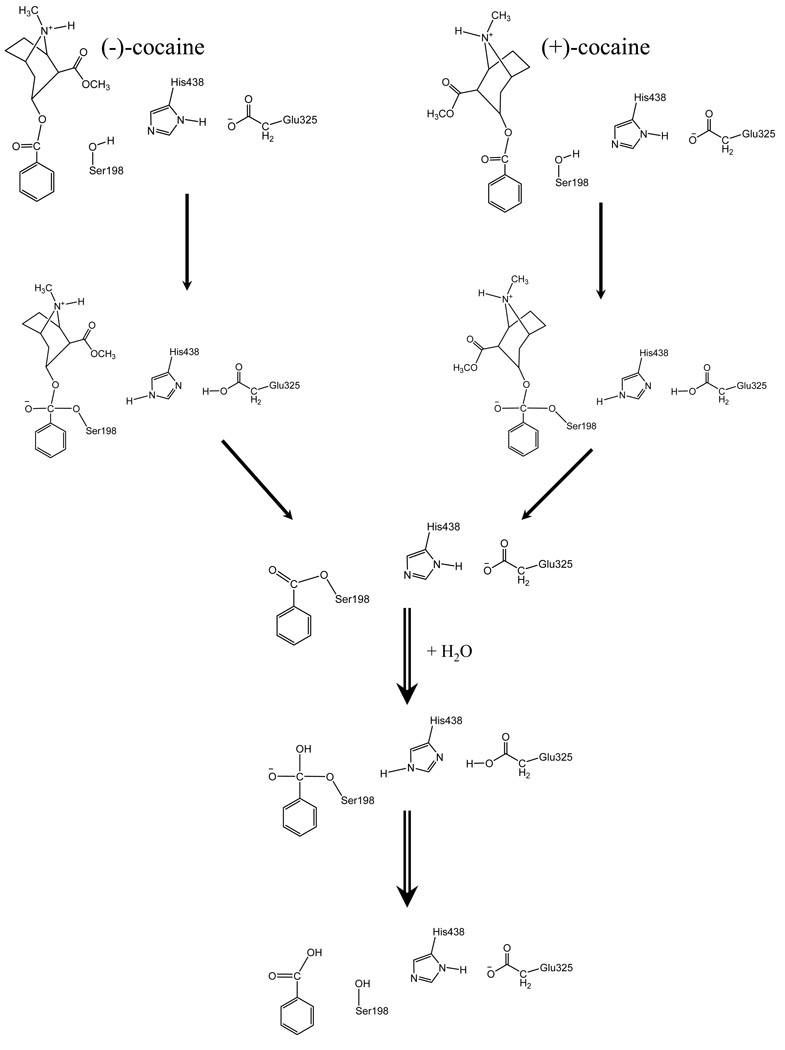
For rational design of a mutant enzyme with a higher catalytic activity for a given substrate, in general, one needs to design a mutation that can accelerate the rate-determining step of the entire catalytic reaction process while the other steps are not slowed down by the mutation. Reported computational modeling and experimental data indicated that the formation of the prereactive BChE-(−)-cocaine complex (ES) is the rate-determining step of (−)-cocaine hydrolysis catalyzed by wild-type BChE,17,18,19,20,21,22,23 whereas the rate-determining step for the faster hydrolysis of the biologically inactive (+)-cocaine enantiomer is the chemical reaction process consisting of four individual reaction steps (see Scheme 1).18 This mechanistic understanding is consistent with the experimental observation17 that the catalytic rate constant of wild-type BChE against (+)-cocaine is pH-dependent, whereas that of the same enzyme against (−)-cocaine is independent of the pH. The pH-dependence of the rate constant for (+)-cocaine hydrolysis is clearly associated with the protonation of H438 residue in the catalytic triad (S198, H438, and E325). For the first and third steps of the reaction process, when H438 is protonated, the catalytic triad cannot function and, therefore, the enzyme becomes inactive. The lower the pH of the reaction solution is, the higher the concentration of the protonated H438 is, and the lower the concentration of the active enzyme is. Hence, the rate constant was found to decrease with decreasing the pH of the reaction solution for the enzymatic hydrolysis of (+)-cocaine.17 Based on the above mechanistic understanding, the previously reported efforts for rational design of the BChE mutants have focused on how to improve the ES formation process.18,20,24
Scheme 1.
Schematic representation of BChE-catalyzed hydrolysis of (−)- and (+)-cocaine.
Experimental observation18 also indicated that the catalytic rate constant of A328W/Y332A BChE is pH-dependent for both (−)- and (+)-cocaine. The pH-dependence reveals that for both (−)- and (+)-cocaine, the rate-determining step of the hydrolysis catalyzed by A328W/Y332A BChE should be either the first or the third step of the reaction process. Further, if the third step were rate determining, then the catalytic efficiency of the A328W/Y332A mutant against (−)-cocaine should be as high as that of the same mutant against (+)-cocaine because the (−)- and (+)-cocaine hydrolyses share the same third and fourth steps (see Scheme 1). However, it has been observed that the A328W/Y332A mutant only has a ~9-fold improved catalytic efficiency against (−)-cocaine, whereas the A328W/Y332A mutation does not change the high catalytic activity against (+)-cocaine.18 This analysis of the experimental and computational data available in literature clearly shows that the rate-determining step of (−)-cocaine hydrolysis catalyzed by the A328W/Y332A mutant should be the first step of the chemical reaction process. Further, recently reported computational modeling also suggests that the formation of the prereactive BChE-(−)-cocaine complex (ES) is hindered mainly by the bulky side chain of Y332 residue in wild-type BChE, but the hindering can be removed by the Y332A or Y332G mutation.20 Therefore, starting from the A328W/Y332A and A328W/Y332G mutants, our current study for improving the catalytic efficiency of BChE against (−)-cocaine aimed to decrease the energy barrier for the first reaction step without significantly affecting the ES formation and other chemical reaction steps. To achieve this aim, molecular dynamics (MD) simulations25 were performed to simulate the structures of the first transition state (TS1) for (−)-cocaine hydrolysis catalyzed by wild-type BChE and its various mutants.
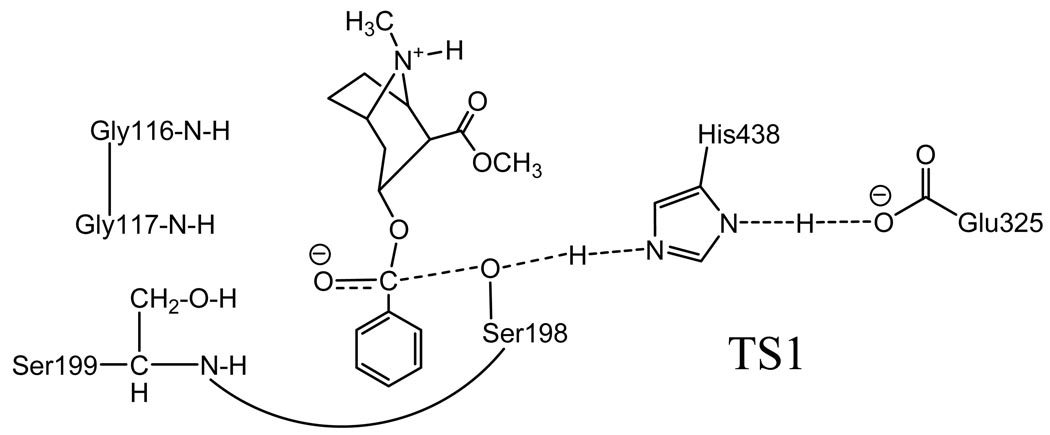
We hoped to predict some possible mutations that can lower the energy of the TS1 structure and, therefore, lower the energy barrier for the first reaction step. Apparently, a mutant associated with the stronger hydrogen bonding between the carbonyl oxygen of (−)-cocaine benzoyl ester and the oxyanion hole of the BChE mutant in the TS1 structure may potentially have a more stable TS1 structure and, therefore, a higher catalytic activity against (−)-cocaine. Hence, the hydrogen bonding with the oxyanion hole in the TS1 structure is a crucial factor affecting the transition state stabilization and the catalytic activity. The possible effects of some mutations on the hydrogen bonding were examined by performing molecular modeling and molecular dynamics (MD) simulations on the TS1 structures for (−)-cocaine hydrolysis catalyzed by wild-type BChE and its various mutants. The initial candidate mutants were chosen by simple geometric consideration of the possible modification of the TS1 structure; only an energy-minimization was carried out in the simple geometric consideration of each possible mutant. Then, the MD simulations were performed only for the candidate mutants whose energy-minimized TS1 structures clearly suggested possibly stronger hydrogen bonding between the carbonyl oxygen of (−)-cocaine and the oxyanion hole of the enzyme.
The MD simulation in water was performed for 1 ns or longer to make sure we obtained a stable MD trajectory for each simulated TS1 structure with the wild-type or mutant BChE. The MD trajectories actually became stable quickly, so were the H˙˙˙O distances involved in the potential hydrogen bonds between the carbonyl oxygen of (−)-cocaine and the oxyanion hole of BChE. The H˙˙˙O distances in the simulated TS1 structures for wild-type BChE and its three mutants are summarized in Table 1 (see supporting information for the key MD trajectory and MD-simulated TS1 structures).
Table 1.
MD-simulated key distances (in Å) and the calculated total hydrogen-bonding energies (HBE, in kcal/mol) between the oxyanion hole and the carbonyl oxygen of (−)-cocaine benzoyl ester in the first transition state (TS1).
| Transition State | Distancea | Total HBEb | ||||
|---|---|---|---|---|---|---|
| D1 | D2 | D3 | D4 | |||
| TS1 structure for (−)-cocaine hydrolysis catalyzed by wild-type BChE |
Average | 4.59 | 2.91 | 1.92 | −5.5 (−4.6) | |
| Maximum | 5.73 | 4.14 | 2.35 | |||
| Minimum | 3.35 | 1.97 | 1.61 | |||
| Fluctuation | 0.35 | 0.35 | 0.12 | |||
| TS1 structure for (−)-cocaine hydrolysis catalyzed by A328W/Y332A mutant of BChE |
Average | 3.62 | 2.35 | 1.95 | −6.2 (−4.9) | |
| Maximum | 4.35 | 3.37 | 3.02 | |||
| Minimum | 2.92 | 1.78 | 1.61 | |||
| Fluctuation | 0.23 | 0.27 | 0.17 | |||
| TS1 structure for (−)-cocaine hydrolysis catalyzed by A328W/Y332G mutant of BChE |
Average | 3.60 | 2.25 | 1.97 | −6.4 (−5.0) | |
| Maximum | 4.24 | 3.17 | 2.76 | |||
| Minimum | 2.89 | 1.77 | 1.62 | |||
| Fluctuation | 0.23 | 0.24 | 0.17 | |||
| TS1 structure for (−)-cocaine hydrolysis catalyzed by F227A/S287G/A328W/Y332M mutant of BChE |
Average | 3.80 | 2.23 | 1.99 | −6.1 (−4.8) | |
| Maximum | 4.61 | 3.08 | 2.88 | |||
| Minimum | 3.16 | 1.75 | 1.67 | |||
| Fluctuation | 0.25 | 0.23 | 0.18 | |||
| TS1 structure for (−)-cocaine hydrolysis catalyzed by A199S/F227A/A328W/Y332G mutant of BChE |
Average | 5.18 | 2.22 | 1.96 | 2.11 | −9.8 (−7.4) |
| Maximum | 5.94 | 3.08 | 2.44 | 3.30 | ||
| Minimum | 4.28 | 1.68 | 1.65 | 1.56 | ||
| Fluctuation | 0.23 | 0.21 | 0.13 | 0.28 | ||
D1, D2, and D3 represent the internuclear distances between the carbonyl oxygen of cocaine benzoyl ester and the NH hydrogen of residues #116 (i.e. G116), #117 (i.e. G117), and #199 (i.e. A199 or S199) of BChE, respectively. D4 is the internuclear distance between the carbonyl oxygen of cocaine benzoyl ester and the hydroxyl hydrogen of S199 side chain in the A199S/F227A/A328W/Y332G mutant.
Calculated by using the empirical HBE equation implemented in AutoDock 3.0 program suite.27 The total HBE value is the average of the HBE values calculated by using the instantaneous distances in all of the snapshots. The value in parenthesis is the total HBE value calculated by using the MD-simulated average distances.
As seen in Table 1, in the simulated TS1 structures for wild-type, A328W/Y332A, A328W/Y332G, and F227A/S287G/A328W/Y332M BChE’s, the carbonyl oxygen of (−)-cocaine can form up to two N-H˙˙˙O hydrogen bonds with the peptidic NH hydrogen atoms of G117 and A199. The overall strength of the hydrogen bonding between the carbonyl oxygen of (−)-cocaine and the oxyanion hole of the enzyme only slightly increase when wild-type BChE is replaced by the A328W/Y332A or A328W/Y332G or F227A/S287G/A328W/Y332M mutant, as seen from the estimated total hydrogen binding energy (HBE) values in Table 1. In the simulated TS1 structure for A199S/F227A/A328W/Y332G BChE, an O-H˙˙˙O hydrogen bond formed between the hydroxyl group on the side chain of S199 and the carbonyl oxygen of (−)-cocaine, in addition to the two N-H˙˙˙O hydrogen bonds with the peptidic NH of G117 and S199, as seen in Scheme 2 and Table 1. Due to the additional O-H˙˙˙O hydrogen bond, the overall strength of the hydrogen bonding with the modified oxyanion hole of A199S/F227A/A328W/Y332G BChE becomes significantly stronger than that of wild-type, A328W/Y332A, A328W/Y332G, and F227A/S287G/A328W/Y332M BChE’s, as seen from the estimated total HBE values in Table 1. These computational results suggest that the TS1 structure for (−)-cocaine hydrolysis catalyzed by A199S/F227A/A328W/Y332G BChE should be significantly more stable than that by the A328W/Y332A or A328W/Y332G or F227A/S287G/A328W/Y332M mutant, due to the significant increase of the overall strength of hydrogen bonding between the carbonyl oxygen of (−)-cocaine and the oxyanion hole of the enzyme. The aforementioned analysis of the literature17,18,20 also indicates that the first chemical reaction step associated with TS1 should be the rate-determining step of (−)-cocaine hydrolysis catalyzed by a BChE mutant including Y332A or Y332G mutation. Thus, the MD simulations predict that A199S/F227A/A328W/Y332G BChE should have a higher catalytic efficiency than A328W/Y332A or A328W/Y332G or F227A/S287G/A328W/Y332M BChE against (−)-cocaine.
Scheme 2.
Schematic representation of the transition-state structure for first reaction step for (−)-cocaine hydrolysis catalyzed by A199S/F227A/A328W/Y332G BChE.
The catalytic efficiency (kcat/KM) of A328W/Y332A BChE against (−)-cocaine was reported to be ~8.6 × 106 M min−1,18 which is ~9.4 times of the kcat/KM value (~9.1 × 105 M min−1) of wild-type BChE against (−)-cocaine. The catalytic efficiency of A328W/Y332G BChE was found to be slightly higher than that of A328W/Y332A BChE against (−)-cocaine.20 To examine our theoretical prediction of the higher catalytic activity for A199S/F227A/A328W/Y332G BChE, we produced the A328W/Y332A and A199S/F227A/A328W/Y332G mutants of BChE through site-directed mutagenesis.26 To minimize the possible systematic experimental errors of the kinetic data, we performed kinetic studies with the two mutants and wild-type BChE under the same condition and compared the catalytic efficiency of A328W/Y332A and A199S/F227A/A328W/Y332G BChE’s to that of the wild-type for (−)-cocaine hydrolysis at benzoyl ester group. Based on the kinetic analysis of the measured time-dependent radiometric data and the ELISA data, the ratio of the kcat/KM value of A328W/Y332A BChE to the kcat/KM value of wild-type BChE against (−)-cocaine was determined to be ~8.6. The determined catalytic efficiency ratio of ~8.6 is in good agreement with the ratio of ~9.4 determined by Sun et al.17c Further, by using the same experimental protocol, the ratio of the kcat/KM value of A199S/F227A/A328W/Y332G BChE to the kcat/KM value of A328W/Y332A BChE against (−)-cocaine was determined to be ~16.8. These data indicate that A199S/F227A/A328W/Y332G BChE has a ~(151 ± 14)-fold improved catalytic efficiency against (−)-cocaine compared to the wild-type, or A199S/F227A/A328W/Y332G BChE has a kcat/KM value of ~(1.37 ± 0.13) × 108 M min−1 against (−)-cocaine. The catalytic efficiency of A199S/F227A/A328W/Y332G BChE against (−)-cocaine is significantly higher than that of AME-359 (i.e. F227A/S287G/A328W/Y332M BChE, kcat/KM = 3.1 × 107 M min−1, whose catalytic efficiency against (−)-cocaine is the highest within all of the BChE mutants reported so far by other labs)24 which has a ~34-fold improved catalytic efficiency against (−)-cocaine compared to wild-type BChE. By using our designed A199S/F227A/A328W/Y332G BChE as an exogenous enzyme in human, when the concentration of this mutant is kept the same as that of the wild-type BChE in plasma, the half-life time of (−)-cocaine in plasma should be reduced from the ~ 45 – 90 min to only ~18 – 36 seconds.
In summary, this is a study that an enzyme mutant is designed based on the transition-state simulation and the designed BChE mutant has a significantly improved catalytic efficiency against (−)-cocaine, demonstrating that the transition-state simulation is a promising approach for rational enzyme redesign and drug discovery.
Supplementary Material
Acknowledgments
This work was supported by a research grant from NIH/NIDA (R01 DA013930 to C.-G. Zhan). The authors also acknowledge the Center for Computational Sciences (CCS) at University of Kentucky for supercomputing time on Superdome (a shared-memory supercomputer with 256 processors).
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References and Notes
- 1.Mendelson JH, Mello NK. New Engl. J. Med. 1996;334:965. doi: 10.1056/NEJM199604113341507. [DOI] [PubMed] [Google Scholar]
- 2.Singh S. Chem. Rev. 2000;100:925. doi: 10.1021/cr9700538. [DOI] [PubMed] [Google Scholar]
- 3.Paula S, Tabet MR, Farr CD, Norman AB, Ball WJ., Jr J. Med. Chem. 2004;47:133. doi: 10.1021/jm030351z. [DOI] [PubMed] [Google Scholar]
- 4.Sparenborg S, Vocci F, Zukin S. Drug Alcohol Depend. 1997;48:149. doi: 10.1016/s0376-8716(97)00072-0. [DOI] [PubMed] [Google Scholar]
- 5.Gorelick DA. Drug Alcohol Depend. 1997;48:159. doi: 10.1016/s0376-8716(97)00119-1. [DOI] [PubMed] [Google Scholar]
- 6.Redish AD. Science. 2004;306:1944. doi: 10.1126/science.1102384. [DOI] [PubMed] [Google Scholar]
- 7.Meijler MM, Kaufmann GF, Qi LW, Mee JM, Coyle AR, Moss JA, Wirsching P, Matsushita M, Janda KD. J. Am. Chem. Soc. 2005;127:2477. doi: 10.1021/ja043935e. [DOI] [PubMed] [Google Scholar]
- 8.Carrera MRA, Kaufmann GF, Mee JM, Meijler MM, Koob GF, Janda KD. Proc. Natl. Acad. Sci. US A. 2004;101:10416. doi: 10.1073/pnas.0403795101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landry DW, Zhao K, Yang GX-Q, Glickman M, Georgiadis TM. Science. 1993;259:1899. doi: 10.1126/science.8456315. [DOI] [PubMed] [Google Scholar]
- 10.Zhan C-G, Deng S-X, Skiba JG, Hayes BA, Tschampel SM, Shields GC, Landry DW. J. Comput. Chem. 2005;26:980. doi: 10.1002/jcc.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamendulis LM, Brzezinski MR, Pindel EV, Bosron WF, Dean RA. J. Pharmacol. Exp. Ther. 1996;279:713. [PubMed] [Google Scholar]
- 12.Gateley SJ. Biochem. Pharmacol. 1991;41:1249. doi: 10.1016/0006-2952(91)90665-r. [DOI] [PubMed] [Google Scholar]
- 13.Gatley SJ, MacGregor RR, Fowler JS, Wolf AP, Dewey SL, Schlyer DJ. J. Neurochem. 1990;54:720. doi: 10.1111/j.1471-4159.1990.tb01933.x. [DOI] [PubMed] [Google Scholar]
- 14.Darvesh S, Hopkins DA, Geula C. Nature Rev. Neurosci. 2003;4:131. doi: 10.1038/nrn1035. [DOI] [PubMed] [Google Scholar]
- 15.Giacobini E, editor. Butyrylcholinesterase: Its Function and Inhibitors. Great Britain: Dunitz Martin Ltd; 2003. [Google Scholar]
- 16.Gateley SJ. Biochem. Pharmacol. 1991;41:1249. doi: 10.1016/0006-2952(91)90665-r. [DOI] [PubMed] [Google Scholar]
- 17.Sun H, Yazal JE, Lockridge O, Schopfer LM, Brimijoin S, Pang YP. J. Biol. Chem. 2001;276:9330. doi: 10.1074/jbc.M006676200. [DOI] [PubMed] [Google Scholar]
- 18.Sun H, Pang Y-P, Lockridge O, Brimijoin S. Mol. Pharmacol. 2002;62:220. doi: 10.1124/mol.62.2.220. [DOI] [PubMed] [Google Scholar]
- 19.Zhan C-G, Zheng F, Landry DW. J. Am. Chem. Soc. 2003;125:2462. doi: 10.1021/ja020850+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamza A, Cho H, Tai H-H, Zhan C-G. J. Phys. Chem. B. 2005;109:4776. doi: 10.1021/jp0447136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao D, Zhan C-G. Proteins. 2005 in press. [Google Scholar]
- 22.Zhan C-G, Gao D. Biophys. J. 2005 in press. [Google Scholar]
- 23.Gao D, Zhan C-G. J. Phys. Chem. B. 2005 doi: 10.1021/jp053736x. in press. [DOI] [PubMed] [Google Scholar]
- 24.Gao Y, Atanasova E, Sui N, Pancook JD, Watkins JD, Brimijoin S. Mol. Pharmacol. 2005;67:204. doi: 10.1124/mol.104.006924. [DOI] [PubMed] [Google Scholar]
- 25.See supporting information concerning why classical MD can be employed to simulate the transition state in a way and how the MD simulations were performed.
- 26.See supporting information for the experimental materials and methods.
- 27. Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. J. Comput. Chem. 1998;19:1639. b) Based on the general HBE equation, we have HBE(r) ≈ 5εr012/r12 – 6εr010/r10, in which r is the H˙˙˙O distance in the considered hydrogen bond and r0 is the minimum value of the H˙˙˙O distance for which the HBE equation can be used. The ε value was determined by using the condition that HBE(r) = −5.0 kcal/mol when r = 1.90 Å.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.