Abstract
Chlorpromazine (CPZ), a potent nicotinic acetylcholine receptor (nAChR) noncompetitive antagonist, binds with higher affinity in the ion channel in the desensitized state than in the closed channel state and with low affinity to additional sites in nAChR-rich membranes. For nAChR equilibrated with agonist, we confirm previous reports that [3H]CPZ occupies a site near the cytoplasmic end of the M2 ion channel domain, photolabeling positions M2-2, M2-6 and/or M2-9 in each subunit. We find that [3H]CPZ also binds at the extracellular end of the channel, photolabeling amino acids at positions M2-16 (α,γ), M2-17 (α,β,δ), and M2-20 (α,β,δ). The photolabeling at the cytoplasmic end of the channel is fully inhibitable by phencyclidine or proadifen, whereas neither drug inhibits [3H]CPZ photolabeling at the extracellular end, establishing that positively charged drugs can bind simultaneously at the cytoplasmic and extracellular ends of the ion channel. [3H]CPZ photolabeling is not detected in the transmembrane domain outside the ion channel, but it photolabels αMet-386 & αSer-393 in the cytoplasmic αMA helix. In the nAChR equilibrated with α-bungarotoxin to stabilize the nAChR in a closed state, [3H]CPZ photolabels amino acids at M2-5 (α), M2-6 (α,β,δ) and M2-9 (β,δ), with no labeling at M2-2. These results provide novel information about the modes of drug binding within the nAChR ion channel and indicate that within the nAChR transmembrane domain, the binding of cationic aromatic amine antagonists can be restricted to the ion channel domain, in contrast to the uncharged, allosteric potentiators and inhibitors that also bind within the δ subunit helix bundle and at subunit interfaces.
The “Cys-loop” superfamily of neurotransmitter-gated ion channels includes the excitatory nicotinic acetylcholine receptors (nAChR1) and serotonin 5-HT3 receptors and the inhibitory GABAA and glycine receptors (1–3). Our knowledge about the three-dimensional structure of these receptors is based upon models of a muscle-type nAChR derived from cryoelectron microscope images of the Torpedo marmorata nAChR (4;5) together with X-ray diffraction models from crystals of molluscan homopentameric acetylcholine binding proteins that are homologous to a nAChR extracellular domain (6;7). The nAChR structure, which was obtained in the absence of agonist and is assumed to represent the nAChR in the closed state, does not have the resolution necessary to accurately identify individual amino acids, but defines the secondary and tertiary structures of the extracellular and transmembrane domains, which are conserved in higher resolution crystal structures of distantly related prokaryotic channels (8–10). The N-terminal half of each subunit contributes to the extracellular domain, containing the neurotransmitter binding sites that are located at subunit interfaces (α-γ and α-δ in the α2βγδ Torpedo nAChR) 30 Å above the level of the membrane. Each subunit’s transmembrane domain is made up of a loose bundle of four α helices (M1-M4), with the amino acids from each M2 helix contributing to the lumen of the ion channel and M4 located most peripheral and in greatest contact with lipid.
A striking feature of the structure of the nAChR transmembrane domain is the presence of pockets within each subunit’s helix bundle and at subunit interfaces that are potential binding sites for allosteric modulators, which contrasts with the compact structure of the transmembrane domain of the prokaryotic channels. This difference in structure may result because the nAChR is in its native lipid environment while the prokaryotic channels were purified in detergent and crystallized in detergent/lipid mixtures, or it may reflect a more fundamental difference between an nAChR which requires cholesterol for channel gating and the prokaryotic channels which function in the absence of cholesterol (11).
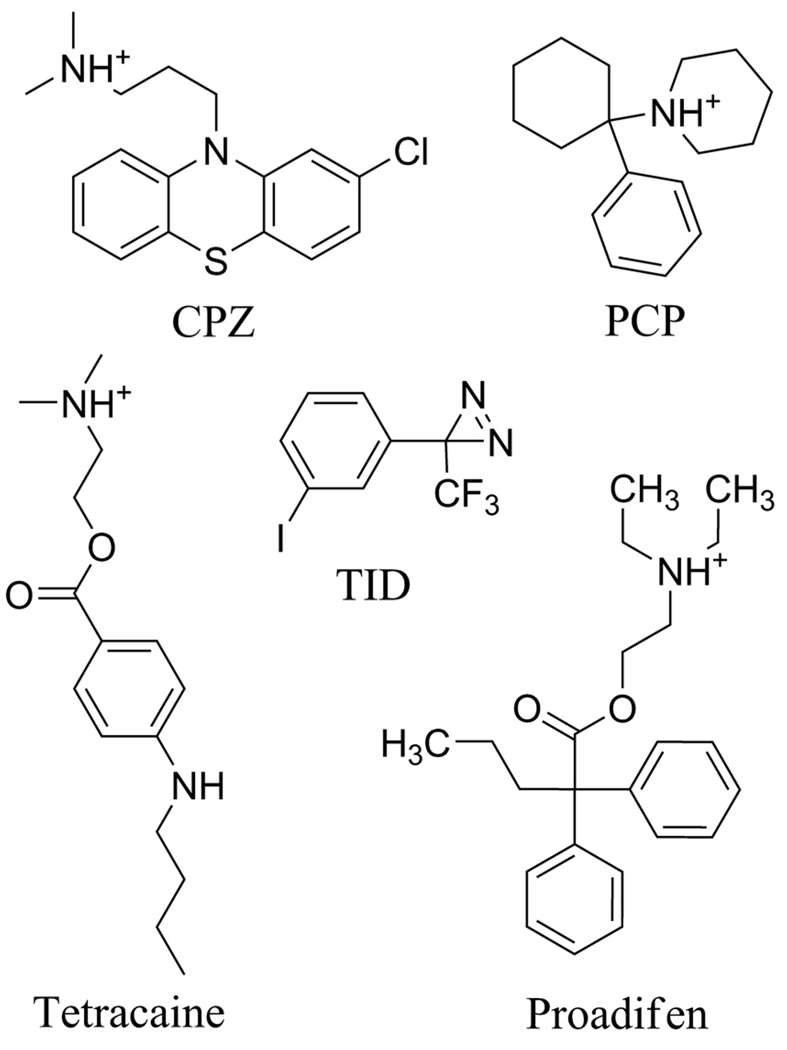
Photoaffinity labeling studies with [3H]chlorpromazine ([3H]CPZ), a phenothiazine tertiary amine (Figure 1) that binds with high affinity to a site in the Torpedo nAChR in the desensitized state (12), provided initial evidence for a drug binding site in the nAChR transmemembrane domain. [3H]CPZ photolabeled amino acids at position M2-6 in each subunit and at M2-2 and M2-9 in some subunits (numbering from the conserved positive charges at the N-terminal (cytoplasmic end) of each M2-helix) (13–16). Its binding site has not been localized in the closed channel state, when it binds with 10-fold lower affinity. Molecular dynamics simulations using the Torpedo nAChR structure predict that CPZ binds near the cytoplasmic end of the closed channel (17), while crystal violet, another aromatic amine, is predicted to bind at the extracellular end (18). Photolabeling with [3H]tetracaine, an aromatic tertiary amine inhibitor that binds preferentially to the channel in the closed state, establishes that it binds at the level of M2-5/6, M2-9 and M2-13 (19). State-dependent binding within the ion channel has been established for [125I]TID, an uncharged, hydrophobic photoreactive drug, that binds at the level of M2-9 and M2-13 in the closed state and at the level of M2-2 and M2-6 in the desensitized state (20;21). Photolabeling studies with uncharged, hydrophobic drugs have also identified drug binding sites in the nAChR transmembrane domain other than the ion channel, including agonist-dependent drug binding at the extracellular end the δ subunit helix bundle (21–23) and a binding site for a positive allosteric modulator at the interface between γ and α subunits (24).
Figure 1. Structures of nAChR channel blockers.
In this report we photolabel the nAChR with [3H]CPZ to identify its binding site in the ion channel in the closed channel state and to determine whether it binds to additional sites in the transmembrane domain of the nAChR in the desensitized state. Photoactivated [3H]CPZ reacts with aliphatic as well as polar amino acid side chains, and this broad reactivity profile is advantageous for the identification of novel binding sites in the hydrophobic nAChR transmembrane domain for inhibitors that are positively charged at physiological pH.
EXPERIMENTAL PROCEDURES
Materials
nAChR-rich membranes were isolated from freshly dissected Torpedo californica electric organs as described (25) and stored in 36 % sucrose, 0.02 % NaN3 at −80 °C until needed. Carbamylcholine chloride (Carb) and the hydrochoride salts of chlorpromazine (CPZ), promazine, promethazine, tetracaine, proadifen and phencyclidine (PCP) were from Sigma-Aldrich. Chlorpromazine sulfoxide (CPZsulfoxide) was from the NIMH Chemical Synthesis and Drug Supply Program. α-Bungarotoxin (αBgTx) was from Biotoxins (St. Cloud, Fl). Staphylococcus aureus endoproteinase Glu-C (V8 protease) was from MP Biomedicals. TPCK-treated trypsin was from Worthington Biochemical Corp. Lysobacter enzymogenes endoproteinase Lys-C (EndoLys-C) was from Roche Diagnostics. [3H]Phencyclidine (27 Ci/mmol) was from Perkin-Elmer Life Science, and [3H]tetracaine (30 Ci/mmol) was from Sibtech (Newington, CT). [3H]Chlorpromazine (16–28 Ci/mmol in ethanol, depending on lot) was from ViTrax (Placentia, CA). The purity of [3H]CPZ was monitored by thin-layer chromatography on a silica gel plate (Kieselgel 60 F254) with the chromatogram developed in a solvent mixture made up of 1-butanol, methanol, water, and acetic acid (1:3:1:0.2). [3H]CPZ migrated as a single spot (Rf = 0.8) well resolved from CPZsulfoxide (Rf = 0.4) or promazine (Rf = 0.6). Stock solutions of [3H]CPZ in ethanol were unstable over time, with [3H]CPZsulfoxide the major degradation product. When needed, oxidized [3H]CPZ (375 µCi) in 0.11 mL methanol/0.01 N HCl was reduced by exposure to metallic zinc (15 mg) for 2 h at room temperature (26), and then [3H]CPZ was purified by thin layer chromatography. For preparative photolabeling experiments leading to the identification of photolabeled amino acids, freshly synthesized [3H]CPZ was used at a purity >95%.
Radioligand Binding Assays
A centrifugation assay (27) was used to measure the effects of CPZ or its analogs on the equilibrium binding of [3H]PCP or [3H]tetracaine to nAChR-rich membranes in phosphate-buffered Torpedo physiological saline (TPS: 250 mM NaCl, 5 mM KCl, 3 mM CaCl2, 2 mM MgCl2, 5 mM sodium phosphate, pH 7.0). For [3H]PCP (6 nM) or [3H]tetracaine (7 nM), 0.2 mL aliquots at 0.7 mg protein/mL (~500 nM ACh binding sites) were incubated with the drugs for 2 h before centrifugation for 40 min at 15,000 rpm in a TOMY TX201 centrifuge. For nAChRs in the desensitized state (+Carb), non-specific binding for [3H]PCP was determined in the presence of 300 µM proadifen, while for nAChRs equilibrated with αBgtx, non-specific binding of [3H]PCP or [3H]tetracaine was measured in the presence of 300 µM tetracaine.
Data Analysis
The concentration dependence of inhibition of [3H]PCP or [3H]tetracaine binding was fit to the following single site binding equation:
where fx is the specific 3H-labeled radioligand bound in the presence of competing drug at concentration x, f0 is the specific radioligand bound in the absence of inhibitor, and IC50 is the total inhibitor concentration associated with 50% inhibition of radioligand binding. Sigmaplot (SPSS) was used for the nonlinear least-squares fit of the data, and the standard errors of the parameter fits are indicated.
Photolabeling nAChR-Rich Membranes
Photolabeling experiments were carried out with 0.2 mg (analytical) or 10 mg (preparative) of membrane protein for each condition. Unless otherwise noted, nAChR-rich membranes were resuspended at a final concentration of 2 mg protein/mL (~2.5 µM [3H]ACh binding sites) in TPS with oxidized glutathione (1 mM) added to serve as aqueous scavenger. Membrane suspensions were added to a tube containing the appropriate quantity [3H]CPZ (20 µCi for analytical and 150 – 200 µCi for preparative photolabelings) which had been dried under an argon stream. After mixing for 30 min to allow solubilization of [3H]CPZ, the membrane suspensions were aliquoted and incubated on ice with appropriate drugs for an additional 45 min. For analytical photolabeling, aliquots of 100 µL were placed in the wells of a 96-well microtiter plate, whereas 6 cm diameter plastic petri dishes (Falcon 351007) were used for preparative photolabelings. Membrane suspensions on ice were irradiated at 350 nm with a Spectronics EN-16 lamp (40 nm full width for half-maximal transmission) for 30 min for the preparative labeling +Carb ± PCP and in a Rayonet RPR-200 photochemical reactor (Southern New England Ultraviolet Co.) using RPR-3500 bulbs (350 nm, full width for half maximal transmission) for 15 min (+Carb ± proadifen) or 30 min (+αBgTx ± tetracaine). Following irradiation, nAChR-rich membranes were pelleted and resuspended in sample buffer for SDS-PAGE. [3H]CPZ at 1.7 and 0.8 µM was used for preparative photolabeling of nAChRs in closed (+αBgTx) and desensitized (+Carb) states, respectively. Preliminary analytical photolabeling studies, quantified by liquid scintillation counting of excised nAChR subunit bands, revealed differences of <10% in the subunit photolabeling in the presence of oxidized glutathione at 0, 1, and 10 mM, and no difference in the amount of proadifen inhibitable labeling.
Gel Electrophoresis and Proteolytic Digestions
nAChR subunits were separated on 11 cm long, 1.5 mm thick, 8% acrylamide gels (28). Protein bands were visualized using Coomassie blue stain (analytical gels) or GelCode® Blue stain (Pierce). For analytical photolabeling, samples were run in duplicate on two gels, one of which was processed for first fluorography with Amplify (Amersham Biosciences) and exposed to Kodak Biomax XAR x-ray film at −80 °C. 3H incorporation into the nAChR subunits in the gels was quantified by liquid scintillation counting of the excised gel bands (25).
For preparative scale photolabeling, the gel bands containing the nAChR β and δ subunits were excised and passively eluted into 12 mL of elution buffer (100 mM NH4HCO3, 0.1% SDS, and 2 mM dithiothreitol, pH 8.4) over 3 days with gentle agitation. The eluates were filtered and concentrated to < 400 µL in Vivaspin 15 Mr 5,000 centrifugal concentrators (Vivascience), and the concentrates were acetone-precipitated (75 % acetone, −20 °C, overnight) and resuspended in a minimal volume (50 –200 µL) of resuspension buffer (15 mM Tris, 0.1% SDS, and 500 µM EDTA, pH 8.1). The α and γ subunit bands were excised and placed in wells of a 15 cm long, 1.5 mm thick 15 % acrylamide gel with a 4 % stacker (5 cm) for limited “in gel” digestion with V8 protease (28). After electrophoresis, this “mapping” gel was stained with GelCode® Blue stain, and the α subunit proteolytic fragments of 20 kDa (αV8-20), 18 kDa (αV8-18), and 10 kDa (αV8-10), and the 24 kDa γ subunit fragment (γV8-24 (29) were excised, eluted, concentrated, and either filtered before direct reversed-phase high-performance liquid chromatography (rpHPLC) purification or acetone-precipitated and resuspended in digestion buffer. Proteolytic digestions of aliquots of β (trypsin) or δ (EndoLys-C) subunits were separated on 1.5 mm thick, 16.5% T, 6% C Tricine SDS-PAGE gels (29;30). In preliminary experiments, we had found that labeled fragments from EndoLys-C and trypsin digests of [3H]CPZ-photolabeled γ subunit aggregated when fractionated by Tricine SDS-PAGE, while a fragment beginning at the N-terminus of γM2 could be isolated by rpHPLC from a trypsin digest of γV8-24.
Samples of αV8-20 and δ subunit in resuspension buffer were digested with EndoLys-C (0.75 U per sample) at 25 °C for ~2 weeks. Aliquots of β, γ, or δ subunit in resuspension buffer were digested for 3 days with V8 protease (100%, w/w). For trypsin digestion, samples of αV8-10, β subunit, or γV8-24 in resuspension buffer were diluted 5-fold with 0.5 % Genapol C-100 (Calbiochem) in 100 mM NH4HCO3, pH 8.1. After a 10 min incubation, trypsin (~1:1 (w:w) in 20 mM CaCl2 (10 % sample volume)) was added to each sample for a 24 hr digestion at 25 °C.
Reversed-Phase HPLC and N-Terminal Sequence Analysis
Samples were fractionated by rpHPLC as described (24). Material for sequence analysis was isolated from three independent preparative photolabelings: +Carb±PCP; +Carb±proadifen; and +αBgTx±tetracaine. Most samples of interest were loaded onto glass fiber filters (Applied Biosystems # 401111) for sequence analysis. HPLC fractions were slowly drop-loaded onto filters placed on a 45 °C heating block and treatment with Biobrene followed loading to prevent unwanted cleavage at Trp residues (31). Filters loaded with samples containing detergent were treated with gas TFA (5 min), followed by washes with ethyl acetate (4 min) and N-butyl chloride (4 min) to remove excess detergent prior to sequencing. Due to their poor retention on glass fiber filters, HPLC samples containing αM4 or δM1 were loaded onto polyvinylidene fluoride filters using the Prosorb sample preparation cartridge (Applied Biosystems # 401950). Samples were sequenced on an Applied Biosystems Procise 492 automated protein sequencer altered such that 1/6 of each cycle was injected into the amino acid analyzer and 5/6 of each cycle was collected for scintillation counting. The amount of peptide detected was calculated using the equation:
where f(x) is the background-subtracted pmol of the amino acid detected in cycle x (determined by peak height), I0 is the initial amount of peptide, and R is the average repetitive yield. A dotted line representing this calculation is included in the sequencing graphs. Cys, Ser, His, and Trp were not used for this fit due to known problems with their detection and/or quantitation during Edman Degradation. Incorporation into a specific residue (cpm/pmol) was calculated as:
where cpmx is the cpm measured in cycle x.
Molecular Modeling
The Torpedo californica nAChR residues photolabeled by [3H]CPZ were examined in the closed-state Torpedo marmarota nAChR model (PDB # 2BG9) derived from 4 Å cryo-electron microscopy electron density data (5). Forty-seven residue variations exist between the four subunits of the two Torpedo species, sixteen of which are found in the transmembrane helices but only two in the M2 helices: βM2-11 (T. cal., Val; T. mar., Leu) and δM2-6 (T. cal., Ser; T. mar., Cys) and none in the MA helices that are the extensions of the M4 helices into the nAChR cytoplasmic domain. Energy-minimized, protonated models of CPZ, PCP, and tetracaine were constructed and docked to regions of the nAChR model using Discovery Studio Suite (Accelrys) CDOCKER. For channel docking, the ligands were placed at various levels of the M2 helices (M2-2, M2-6, M2-9, M2-13, M2-16/17, M2-20, M2-24, & M2-28), a docking sphere of 12 Å radius was placed around the ligands, and the program identified 150 solutions ranked by interaction energy from 25 randomly chosen initial orientations. CPZ, tetracaine, and PCP were each accommodated sterically within the ion channel from all initial positions, with the most favorable CDOCKER interaction energy for CPZ docked near M2-16/17 with its N+ forming a salt bridge with αGlu-262 (M2-20), an orientation shown in Figures 9 D and E. Docking of CPZ at the level of M2-6 and M2-9, shown in Figures 9 D and F, was less favorable energetically. For tetracaine the strongest interaction energy was also for a molecule docked at the level of M2-16/17 in a similar orientation as CPZ, while for PCP docking at the level of M2-13/16 was favored energetically. These simple calculations, which did not attempt to include phospholipid or energy minimization of the binding site structure (17), were used to identify the locations in the static nAChR that can accommodate CPZ and to visualize our experimental results.
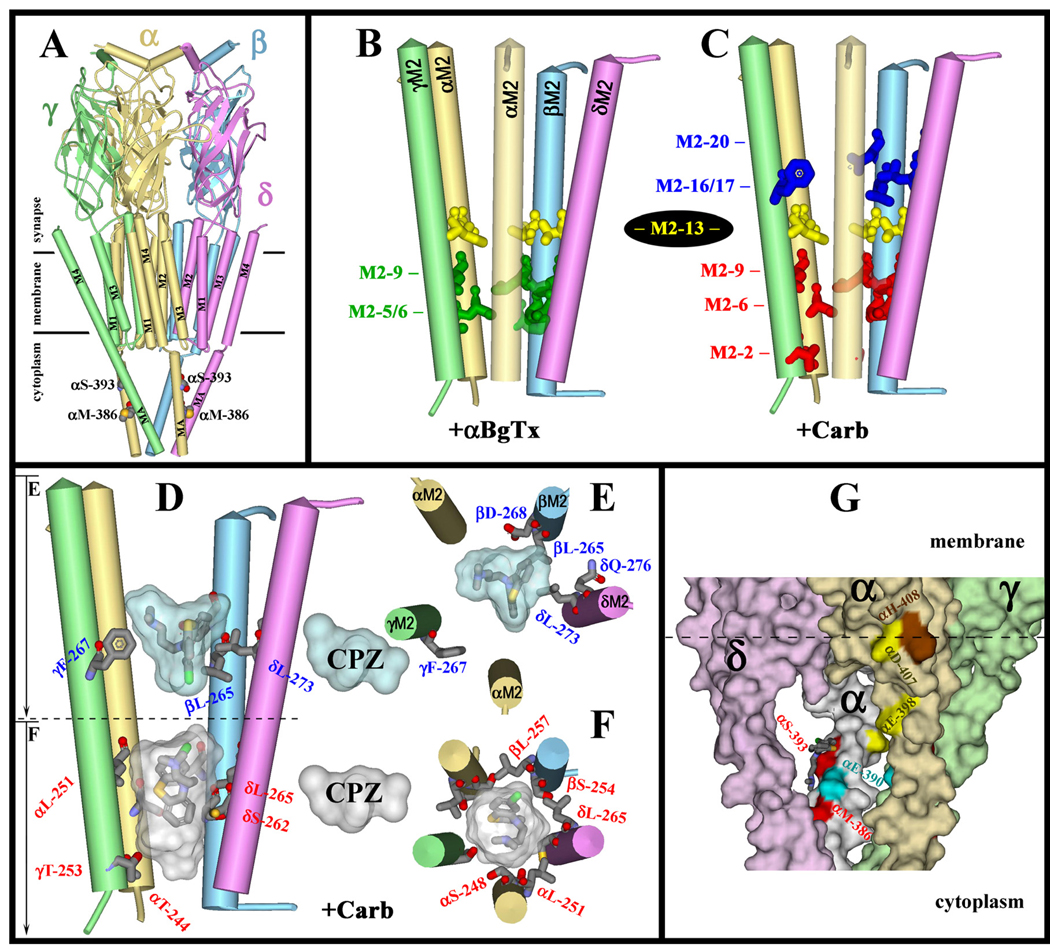
Figure 9. CPZ binding sites within the Torpedo nAChR ion channel.
The model of the Torpedo marmorata nAChR (PDB #2BG9) showing secondary structural elements (α-helices as cylinders, β-sheets as ribbons) color-coded by subunit (α. gold; β, blue; γ, green; & δ, magenta). A, The entire structure is shown for orientation with respect to the membrane. The cytoplasmic residues labeled by [3H]CPZ (αMet-286 & αSer-393) are represented in CPK format. B–F, Images of the M2 helices which form the ion channel domain, with the residues labeled by [3H]CPZ represented in stick format. B & C, The residues labeled by [3H]CPZ in the closed state (B, green) or in the desensitized state (C) at the extracellular (blue, Site17/20) and cytoplasmic (red, Site2/6/9) ends of the channel, along with the ring of Val/Ile at M2-13 (yellow) that are not photolabeled by [3H]CPZ. D–F, The binding pockets for CPZ visualized as Connolly surface representations of the ten lowest energy solutions when CPZ was docked within the ion channel at the level of M2-6 or M2-17 (see Experimental Procedures). In each site, the orientation of the lowest energy docked ligand and the residues labeled by [3H]CPZ are shown in stick format, color coded by atom type (carbon, gray; oxygen, red; nitrogen, blue; sulfur, yellow; & chlorine, green). The ion channel can accommodate two CPZ molecules simultaneously, as viewed from the side (D) or looking down the channel (E, upper binding site, blue pocket; F, lower binding site, white pocket). G, The locations of [3H]CPZ-photolabeled αMet-386 and αSer-393 in the cytoplasmic basket (Connolly surface representation) formed by the MA helices of αγ (beige), γ (pale green), αδ (white), δ (plum), and β (not shown) subunits, with color coding of selected amino acids: [3H]CPZ-labeled αMet-386 & αSer-393 (red); [3H]azietomidate-photolabeled αGlu-390 (cyan) (40); [3H]azioctanol-photolabeled αHis-408 (brown) (37); and [3H]azicholesterol-photolabeled αGlu-398 and αAsp-407 (yellow) (42). The lowest energy solution from docking CPZ near αδSer-393 (150 solutions total) is shown in stick format color-coded by atom type: carbon, gray; nitrogen, blue; sulfur, gold; and chlorine, green.
RESULTS
CPZ and CPZsulfoxide inhibition of [3H]PCP and [3H]tetracaine binding
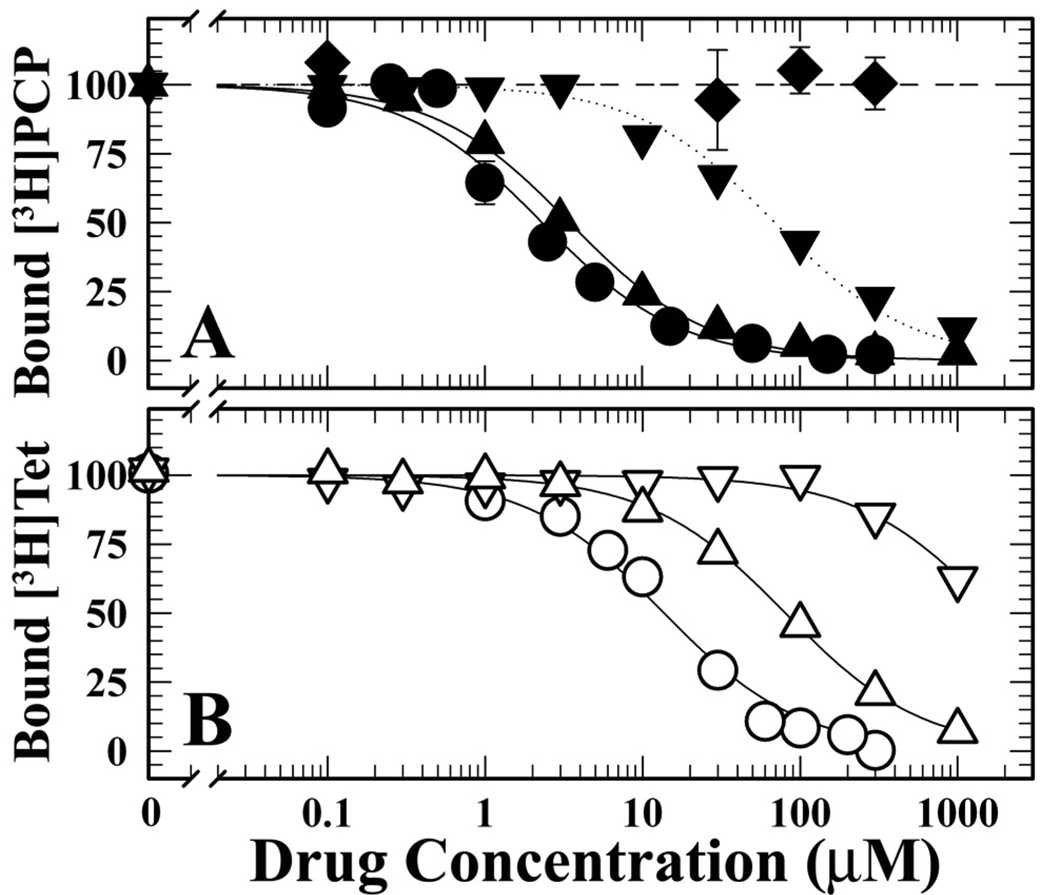
Since [3H]CPZsulfoxide was likely to be a contaminant of even recently synthesized/purified [3H]CPZ, we first characterized its interactions with the nAChR in equilibrium binding assays using drugs that bind preferentially in the nAChR ion channel in the desensitized state ([3H]PCP (32)) or a closed state ([3H]tetracaine (33)). Assays were carried out in the presence of the agonist carbamylcholine (Carb), to stabilize nAChRs in the equilibrium desensitized state, or α-bungarotoxin (αBgTx), a peptide competitive antagonist that occupies the transmitter binding site stabilizing a closed channel state similar to the resting state. Consistent with previous reports (32), CPZ inhibited binding of [3H]PCP (+Carb) and [3H]tetracaine (+αBgTx) with IC50s of 2 µM and 16 µM, respectively. For each nAChR conformation, CPZsulfoxide bound with ~25-fold lower affinity than CPZ (desensitized state, IC50 = 55 µM; closed state, IC50 = ~1 mM) (Figure 2). In parallel assays, we also determined that promazine (deschloro CPZ) inhibited the binding of [3H]PCP (+Carb) and [3H]tetracaine (+α-BgTx) with IC50s of 3 and 80 µM, while phenothiazine itself at concentrations up to 300 µM did not inhibit [3H]PCP binding (+Carb). The low affinity of CPZsulfoxide for a site in the ion channel indicates that the presence of small amounts of this contaminant will be unlikely to contribute to observed photolabeling in the ion channel.
Figure 2. Inhibition of [3H]PCP and [3H]tetracaine binding to nAChR-rich membranes by CPZ and CPZ Analogs.
Equilibrium binding of [3H]PCP (+Carb, closed symbols) (Panel A) and of [3H]tetracaine (+αBgTx, open symbols) (Panel B) was determined by centrifugation in the absence or presence of CPZ (●, IC50 = 2.3 ± 0.4 µM; ○, IC50 = 14 ± 2 µM), CPZsulfoxide (▼, IC50 = 69 ± 6 µM; ▽, IC50 = 1.7 ± 0.2 mM), promazine (▲, IC50 = 3.5 ± 0.2 µM; △, IC50 = 80 ± 3 µM), and phenothiazine (♦). For each competition experiment, the data were normalized to the specific binding in the absence of competitor, which for [3H]PCP (+Carb) was 5530 ± 300 cpm (specific) and 700 ± 200 cpm (non-specific), and for [3H]tetracaine (+αBgTx) was 3500 ± 300 cpm (specific) and 1700 ± 100 cpm (non-specific).
State-dependent [3H]CPZ photoincorporation into the nAChR
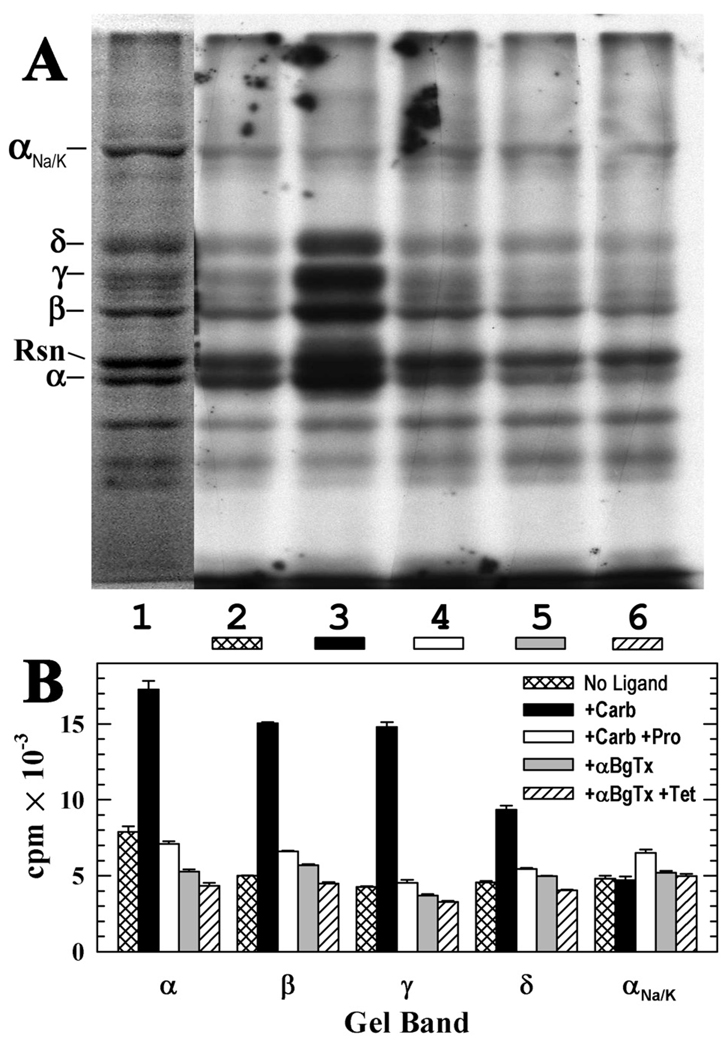
The pharmacological specificity of nAChR photolabeling at the subunit level was determined by SDS-PAGE followed by fluorography and liquid scintillation of excised gel bands (Figure 3). nAChR-rich membranes were photolabeled with 3 µM [3H]CPZ in the absence of other drugs, in the presence of the agonist Carb, or in the presence of αBgTx. Photolabeling of each nAChR subunit was 2–4 fold higher for nAChRs labeled in the desensitized state (+Carb) than in a closed state (+α-BgTx), and for nAChRs in the desensitized state, proadifen, a desensitizing channel blocker (34), inhibited photolabeling of the δ subunit by 40%, the α and β subunits by 60%, and the γ subunit by 70%. For nAChRs photolabeled in the desensitized state, the 17,000 cpm incorporated in the α subunit indicated photolabeling of 0.8% of subunits, and the 15,000 cpm incorporated in the β and γ subunits indicated photolabeling of ~1.4% of subunits. Although subunit photolabeling in the presence of αBgTx was <50% that seen for nAChRs in the desensitized state, at least a portion of that photolabeling was likely to be within the ion channel, since tetracaine inhibited photolabeling of the α, β, and δ subunits by ~20%.
Figure 3. [3H]CPZ photolabeling of the Torpedo nAChR in the closed and desensitized states.
nAChR-rich membranes (200 µg protein, 280 pmol of ACh binding sites in 100 µL of TPS supplemented with 1 mM oxidized glutathione) were equilibrated with 2.8 µM [3H]CPZ alone (control, lane 2) or with 2 mM Carb (lane 3), 2 mM Carb and 0.1 mM proadifen (lane 4), 10 µM αBgTx (lane 5), or 10 µM αBgTx and 0.1 mM tetracaine (lane 6). After irradiation at 360 nm for 30 min, aliquots of each condition were fractionated by SDS-PAGE on two gels, which were both stained with Coomassie blue with one processed first for fluorography and the stained bands were excised from the second for 3H determination. A, Coomassie blue stain (lane 1) and fluorograph (lanes 2–6; 14 day exposure at −80°C). Indicated on the left are the mobilities of the nAChR subunits, rapsyn (Rsn), and the α subunit of the Na+/K+-ATPase (αNa/K). B, 3H incorporation in the gel bands containing the nAChR subunits and αNa/K, determined by liquid scintillation counting. Plotted are the mean cpm and range determined from bands in the two gels
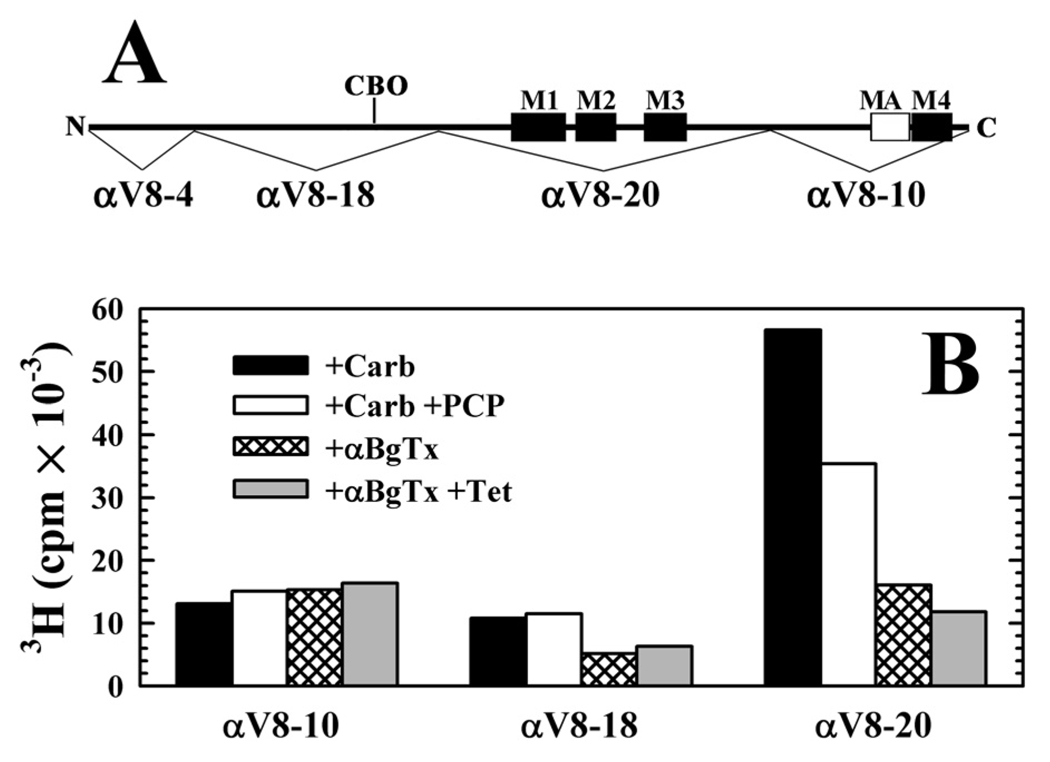
The regions of PCP- or tetracaine-inhibitable photolabeling within the nAChR α subunit were further localized by subjecting the photolabeled subunits to in-gel digestion with V8 protease, which generates large, non-overlapping subunit fragments of 20 kDa (αV8-20, beginning at αSer-173 and containing the M1, M2, and M3 membrane spanning helices as well as the ACh binding site segment C), 18 kDa (αV8-18, beginning at αThr-52 and containing ACh binding site segments A and B), and 10 kDa (αV8-10, beginning at αAsn-338 and containing the cytoplasmic MA helix and the M4 transmembrane helix) (28) (Figure 4). For membranes photolabeled in the presence of agonist, 70% of the 3H was recovered in αV8-20, and PCP reduced that labeling by 40%, with no inhibition of labeling in the gel bands containing αV8-18 or αV8-102. For nAChRs photolabeled in the presence of α-BgTx, 40% of 3H was associated with αV8-20, and tetracaine reduced that labeling by 25%.
Figure 4. Distribution of [3H]CPZ photoincorporation within large fragments of the nAChR α subunit.
A, A schematic indicating the location within the α subunit primary structure of the 4 α subunit fragments produced by in-gel digestion with V8 protease (28): αV8-4, αV8-18, containing most of the extracellular domain and the N-linked carbohydrate (CBO), αV8-20, containing the M1-M3 membrane-spanning helices, and αV8-10, containing M4 and the cytoplasmic amphipathic helix denoted MA. B, [3H]CPZ photoincorporation within the α subunit fragments isolated in separate preparative photolabelings of nAChRs either equilibrated with Carb ± 100 µM PCP (0.8 µM [3H]CPZ) or with αBgTx ± 100 µM tetracaine (1.7 µM [3H]CPZ).
To identify amino acids photolabeled by [3H]CPZ in the nAChR in the desensitized state, nAChR-rich membranes were photolabeled on a preparative scale on two occasions in the presence of agonist, in the absence or presence of PCP or proadifen. To identify amino acids photolabeled in the nAChR in the closed channel state, membranes were equilibrated with αBgTx and photolabeled in the absence or presence of tetracaine. The experimental identification of the photolabeled amino acids are presented by representative sequencing data in the figures, and the average efficiencies of photolabeling at each position, obtained by sequencing multiple samples, are presented in Table 1 in order to compare the relative efficiencies of photolabeling in the different positions.
Table 1.
Pharmacological specificity of [3H]CPZ photoincorporation into residues in the nAChR M2 ion channel domain in the desensitized (+Carb) and closed channel (+αBgTx) statesa.
| +Carb −PCP./+PCP Figure 5 cpm/pmol |
+Carb −proadifen ./+proadifen Figure 6 cpm/pmol |
+αBgTx −tetracaine / +tetracaine Figure 7 cpm/pmol |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| β | δ | α | β | γ | δ | α | β | δ | |
| M2-2 | <0.1 / <0.1 | <0.2 /<0.3 |
3.4 / <0.1 (±1.2) |
0.2 / <0.1 (±0.1) |
52 / <2 | 0.3 / <0.05 | <0.2 / <0.1 | <0.1 / <0.05 | <0.02 / <0.01 |
| M2-5 |
0.3 / <0.1 (±0.1) |
<0.2 / <0.05 |
2.0 / <0.2 (±0.4) |
1.1 / <0.05 (±0.3) |
<1 /<2 | <0.1 / <0.02 |
0.6 / <0.2 (±0.04) |
0.3 / <0.05 |
0.1 / < 0.02 (±0.002) |
| M2-6 |
1.2 / 0.3
(±0.3)/(±0.03) |
0.6 / <0.05 (±0.02) |
12 / <0.2 (±1.4) |
6.2 / 0.2 (±1.2) |
<2 /<2 | 4.9 / 0.2 |
1.0 / <0.2 (±0.04) |
0.2 / <0.05 |
4.4 / 0.1
(±0.2)/(±0.002) |
| M2-9 |
2.7 / 0.4
(±0.3)/(±0.03) |
0.5 / <0.05 (±0.05) |
3.2 /<0.1 (±0.5) |
20 / 0.2
(±4.7) |
<2 /<2 | 2.0 / <0.02 |
1.3 / <0.2 (±0.2) |
8.0 / 0.3 |
0.5 / <0.02 (±0.01) |
| M2-16 |
2.2 / 2.0
(±1.0)/(±0.7) |
1.1 / 1.6
(±0.1)/(±0.2) |
0.6 / 0.5
(±0.5) |
2.7 / 0.5
(±1.2) |
32 / 14 | 1.4 / 0.5 | <0.05 / <0.1 | <0.1 / <0.2 | <0.05 / <0.05 |
| M2-17 |
17 / 23
(± 2)/(± 2) |
3.4 / 5.3
(±0.03)/(±1.7) |
1.5 / 0.7
(±0.3) |
26 / 6.4
(±5.5) |
<5 /<3 | 13 / 3.0 | <0.05 / <0.1 | 0.3 / <0.2 |
0.2 / <0.05 (±0.001) |
| M2-20 |
11 / 11
(±1.2)/(±1.3) |
10 / 14
(± 2)/(±0.5) |
4.1 / 2.1 (±0.5) |
18 / 2.3
(±7.2) |
10 /<5 | 13 / 6.4 |
0.6 / 1.2
(±0.2)/(±0.02) |
0.5 / 1.5 |
0.6 / 0.6
(±0.01)/(±0.005) |
The 3H incorporation in each residue (cpm/pmol of PTH-derivative) was calculated from the observed 3H release and the initial and repetitive yields as described under “Experimental Procedures”. The results are from the three photolabeling experiments, including data from Figs. 5–7. When three or two samples were sequenced, data are presented as a mean (± SEM) (data columns 1, 3, and 7) or as a mean (± range) (data columns 2, 4, and 9), respectively. For the other data, only a single aliquot of each sample was sequenced. The cpm/pmol in bold are the values for positions at which the peak of 3H release was >20% over the background releases of 3H in preceding cycles. The upper limits of photolabeling in the other cycles were determined from the random variation of the background release of 3H in the adjacent sequencing cycles.
[3H]CPZ photoincorporation in the nAChR ion channel in the presence of agonist (desensitized state)
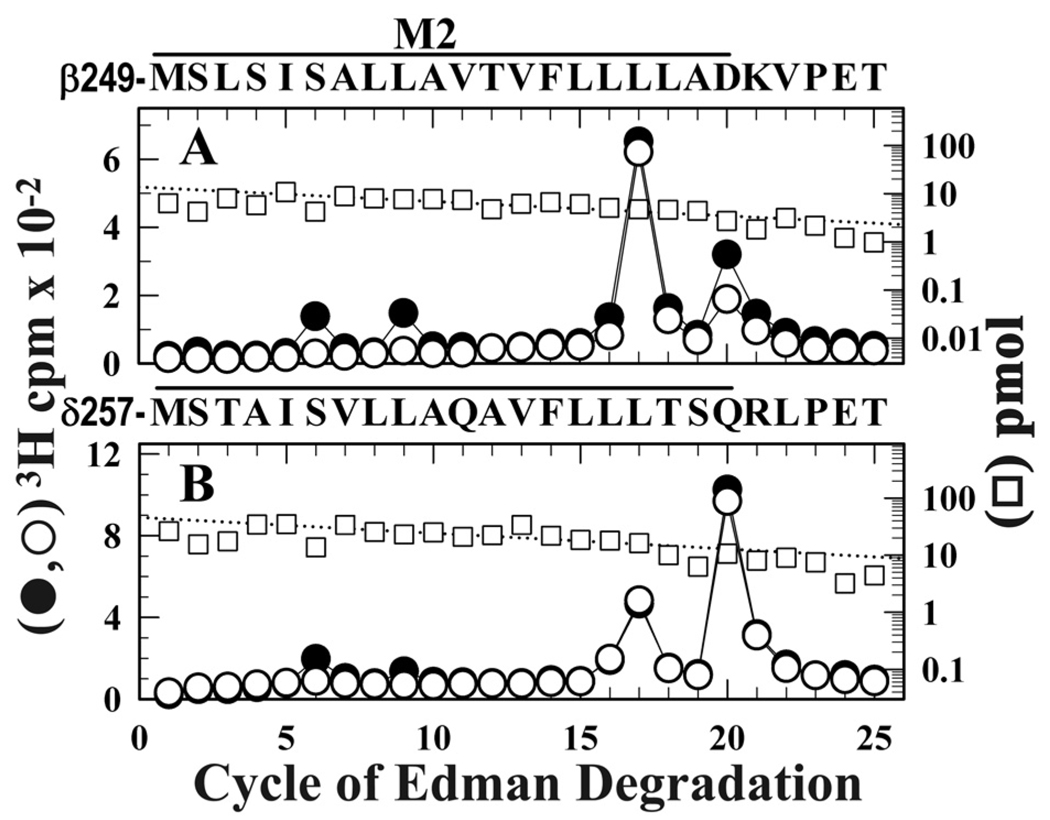
In a first experiment, nAChR-rich membranes equilibrated with agonist were photolabeled with [3H]CPZ ± PCP. Fragments beginning at the N-termini of βM2 (βMet-249) and δM2 (δMet-257) were isolated for sequence analysis by SDS-PAGE and rpHPLC from trypsin digests of β subunit and EndoLys-C digests of δ subunit (Supplemental Figure S1). Within βM2 (Figure 5A), there were peaks of 3H release in cycles 6 and 9 that were fully inhibited by PCP, consistent with labeling of βM2-6 (βSer-254, 2 cpm/pmol) and βM2-9 (βLeu-257, 2 cpm/pmol), but the major peaks of 3H release were in cycles 17 and 20, indicating photolabeling of βM2-17 (βLeu-265, 16 cpm/pmol) and βM2-20 (βAsp-268, 9 cpm/pmol). While PCP inhibited photolabeling of βM2-6 and βM2-9, it did not inhibit photolabeling of βM2-17 and βM2-20 near the extracellular end of the ion channel domain. Similarly, within δM2 (Figure 5B), there was PCP-inhibitable photolabeling of δM2-6 (δSer-262, 0.8 cpm/pmol), but the major peaks of 3H release were in cycles 17 (δLeu-273, 3 cpm/pmol) and 20 (δGln-276, 12 cpm/pmol), neither inhibitable by PCP. While [3H]CPZ photolabeling at the extracellular end of the ion channel has not been previously examined, the PCP-inhibitable [3H]CPZ photolabeling of βM2-6, βM2-9, and δM2-6 is as reported previously by Changeux and coworkers (13;14) in sequence analyses that extended only to M2-15.
Figure 5. PCP modulation of [3H]CPZ photoincorporation in the nAChR ion channel in the presence of agonist.
3H (●, ○) and PTH-amino acids (□) released during sequence analysis through βM2 (A) and δM2 (B) in subunit fragments isolated from nAChR-rich membranes photolabeled with 0.8 µM [3H]CPZ after equilibration with Carb in the absence (●) or presence (○, □) of 100 µM PCP. Trypsin digests of β subunits and EndoLys-C digests of δ subunits were fractionated by Tricine SDS-PAGE and rpHPLC to isolate fragments beginning at βMet-249 or δMet-257 (Supplemental Figure S1). A, The primary sequence began at βMet-249 (□, +PCP, 14 pmol; −PCP, 18 pmol), with any secondary sequence at <0.5 pmol. For nAChRs labeled +Carb (●) the peaks of 3H release in cycles 6, 9, 17, and 20 indicated labeling of βSer-254, βLeu-257, βLeu-265, and βAsp-268 at 2, 2, 16, and 9 cpm/pmol. Addition of PCP (○) reduced labeling of βSer-254 and βLeu-257 to 0.3 cpm/pmol, while labeling of βLeu-265 was increased to 25 cpm/pmol and labeling of βAsp-268 was unchanged (7 cpm/pmol). B, The primary sequence began at δMet-257 (□, +PCP, 46 pmol; −PCP, 40 pmol), with a secondary sequence beginning at δAsn-437 (3 pmol both conditions). For nAChRs labeled +Carb (●) the peaks of 3H release in cycles 6, 9, 17, and 20 indicated photolabeling of δSer-262, δLeu-265, δLeu-273, and δGln-276 at 1, 1, 3, and 12 cpm/pmol. Addition of PCP (○) reduced photolabeling of δSer-262 and δLeu-265 to <0.1 cpm/pmol, while labeling of δLeu-273 (4 cpm/pmol) and δGln-276 (13 cpm/pmol) was not affected.
To characterize the amino acids photolabeled in γM2, we first attempted to fractionate EndoLys-C or trypsin digests of [3H]CPZ-labeled γ subunits by Tricine SDS-PAGE. Since the labeled fragments in these digests migrated as intractable high molecular weight aggregates, as an alternative strategy we took advantage of the fact that V8 protease can be used to cleave the γ subunit at γGlu-208 before γM1, and we sequenced that fragment for 65 cycles to the next Glu after γM2 (Supplemental Figure S2). While sequencing through γM2, there was small peak of 3H release in cycle 59 (20 cpm), consistent with labeling of γM2-16 (γPhe-267), with lower peaks of 3H release in cycles 45 and 52, consistent with labeling of γM2-2 (γThr-253) and γM2-9 (γLeu-260). In addition, there were more prominent peaks of 3H release in earlier cycles of Edman degradation, but each of these could be accounted for by [3H]CPZ photolabeling of γPhe-267 and the presence of an acid-label Gln-Thr bond 5 cycles before that position.
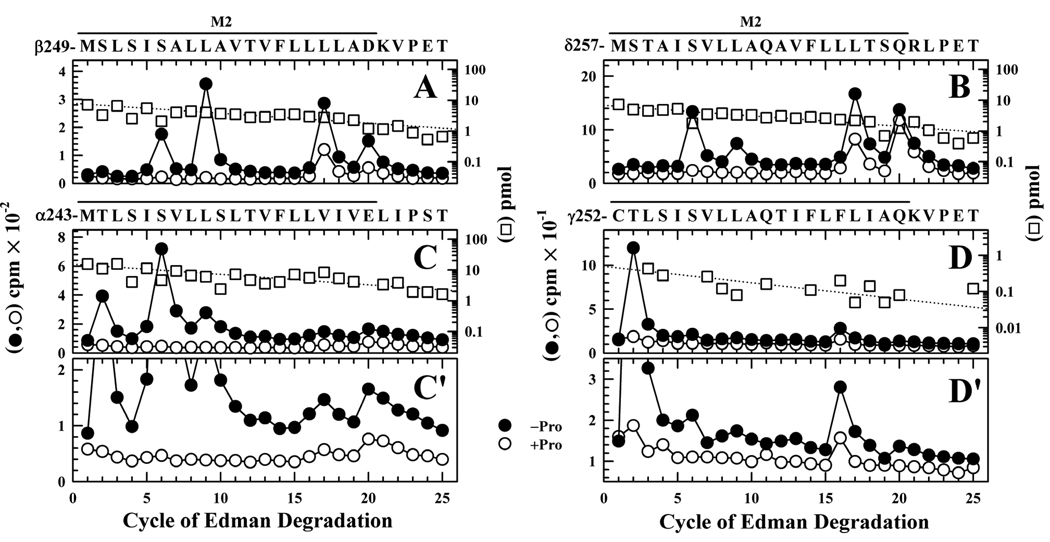
In a second photolabeling experiment, [3H]CPZ photolabeling within each of the M2 segments was characterized when nAChRs equilibrated with Carb were photolabeled ± proadifen (Figure 6). Fragments beginning at the N-termini of βM2 and δM2 were isolated as above, and sequence analysis established that [3H]CPZ photolabeling of positions M2-6 and M2-9 was fully inhibited by proadifen, while photolabeling at M2-17 and M2-20 was partially inhibited (Figures 6A and B). Qualitatively, the photolabeling of positions M2-6 and M2-9 in this experiment was more prominent than in the experiment of Figure 5. Quantitative comparison of the efficiencies of photolabeling (cpm/pmol) at each position indicates that βM2-20 and δM2-20 were each photolabeled with similar efficiency in the two experiments and that it was the photolabeling efficiency at the cytoplasmic end that differed (Table 1). Further studies would be required to identify the origin of this difference3.
Figure 6. Proadifen modulation of [3H]CPZ photoincorporation at the extracellular and cytoplasmic ends of the M2 ion channel domain in the presence of agonist.
3H (●, ○) and PTH-amino acids (□) released during sequence analysis of fragments beginning at the N-termini βM2 (A), δM2 (B), αM2 (C, C’), and γM2 (D, D’), isolated from nAChR-rich membranes photolabeled with 0.8 µM [3H]CPZ after equilibration with Carb in the absence (●) or presence (○, □) of 100 µM proadifen. Photolabeled subunits were isolated by SDS-PAGE, and the βM2 and δM2 fragments were isolated as described in Figure 5, while αV8-20 and γV8-24 were isolated by in gel digestion with V8-protease, and then an EndoLys-C digest of αV8-20 was sequenced directly and the γM2 fragment was isolated by rpHPLC from a trypsin digest of γV8-24 (Supplemental Figure S3). A, The primary sequence began at βMet-249 (I0 = 8 pmol −(□) and +proadifen), with a secondary sequence beginning at βLys-216 (~0.5 pmol)). (10,800 (●) and 2,600 (○) cpm sequenced). B, The only sequence detected began at δMet-257 (I0 = 7 pmol −(□) and 11 pmol +proadifen; 8,000 (●) and 3,300 (○) cpm sequenced). C, The fragment beginning at αMet-243 was the primary sequence (I0 = 16 pmol −(□) and +proadifen), along with α subunit fragments beginning at αSer-173 (10 pmol), αHis-186 (8 pmol), and αTyr-277 (5 pmol), as well as EndoLys-C (Gly-1, 8 pmol) and a V8 protease peptide (Val-180, 20 pmol). (37,000 (●) and 12,200 (○) cpm sequenced). C’ is an enlargement of C to show more clearly the level of labeling at αM2-16, αM-17, and αM2-20. D, While the primary sequence was trypsin (6 pmol), the fragment beginning at γCys-252 was present (I0 = 0.5 pmol −(□) and +proadifen; 1,280 (●) and 510 (○) cpm sequenced). D’ is an enlargement of D included to better show the level of labeling at γM2-16. For all sequencing runs, the efficiencies of photolabeling (cpm/pmol) associated with the peaks of 3H in cycles 2, 6, 9, 16, 17, or 20 are quantitated in Table 1.
Photolabeling within αM2 was characterized by sequencing an EndoLys-C digest of αV8-20 (Figure 6C), which revealed a major peak of 3H release in cycle 6, indicating labeling of αM2-6 (αSer-248, 11 cpm/pmol), with smaller peaks of 3H release in cycles 2 and 9, consistent with labeling of αThr-244 (5 cpm/pmol) and αLeu-251 (3 cpm/pmol). There were also small peaks of 3H release in cycles 16, 17 and 20 (24, 26 & 60 cpm, respectively, expanded ordinate scale, Figure 6C′) indicative of photolabeling of αLeu-258, αVal-259 and αGlu-262 at 1, 1 and 4 cpm/pmol. Photolabeling of αM2-2, αM2-6, and αM2-9 was each reduced by >95% in the presence of proadifen. To characterize photolabeling within γM2 (Figure 6D), the labeled γ subunit was digested “in gel” with V8 protease to produce a 24 kDa fragment (γV8-24, beginning at γ-Ala167 (29)), and a fragment beginning at the γM2 N-terminus (γCys-252) was isolated by rpHPLC at ~55% organic from a trypsin digest of γV8-24 (Supplemental Figure S3). Sequence analysis revealed the presence of the γCys-252 fragment at 0.5 pmol (along with a trypsin fragment at 5 pmol), and a major peak of 3H release in cycle 2 indicating labeling of γThr-253 at 50 cpm/pmol that was reduced >95% by proadifen. A minor peak of 3H release in cycle 16 (Figure 6D′) indicated labeling of γPhe-267 at ~30 cpm/pmol.
[3H]CPZ photoincorporation in the nAChR ion channel in the presence of αBgTx (closed state)
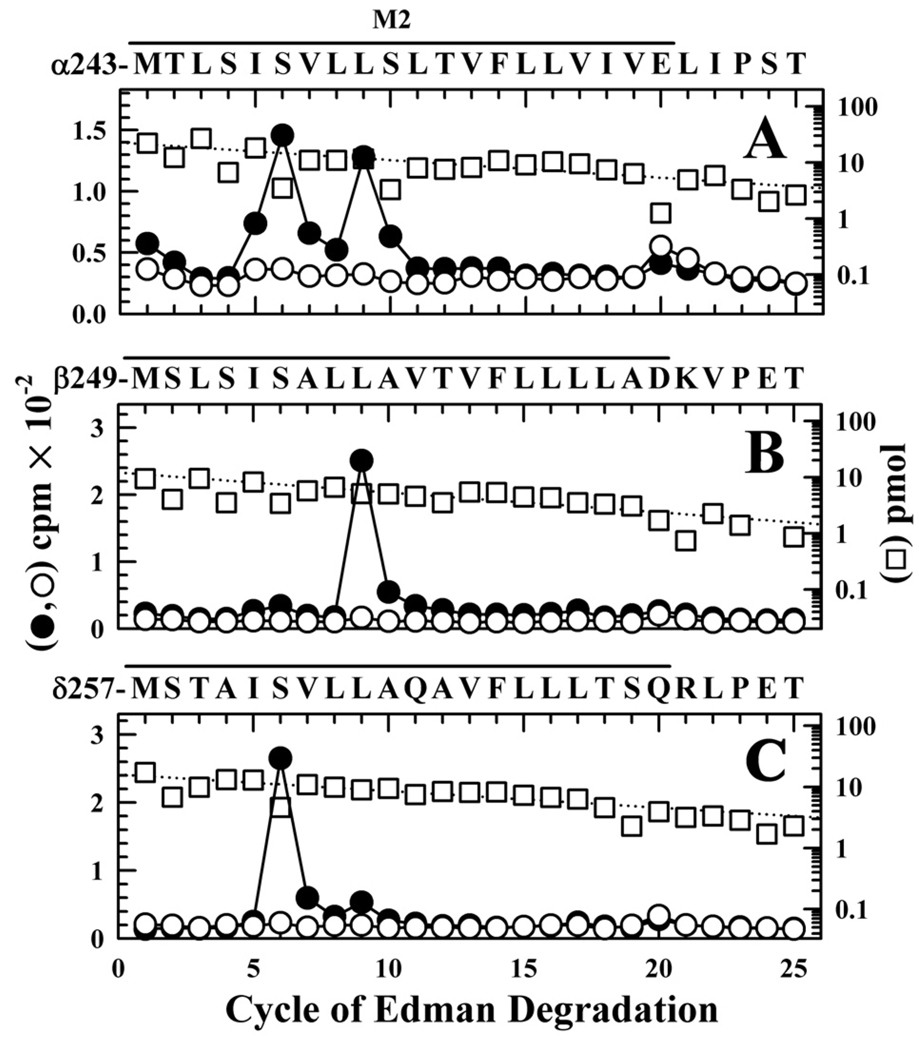
nAChR-rich membranes were equilibrated with αBgTx and then photolabeled on a preparative scale with [3H]CPZ in the absence or presence of tetracaine, a high affinity, closed-state selective channel blocker (19;33). When the 3H incorporation into the polypeptides of the nAChR-rich membrane preparation was determined by analytical SDS-PAGE and fluorography, rapsyn and the nAChR β subunit were the most highly photolabeled polypeptides (Supplemental Figure S4). There was tetracaine-inhibitable labeling within the nAChR α, β and δ subunits, with little, if any, in the γ subunit, in the α subunit of the Na+/K+-ATPase, or in rapsyn. Subunit fragments beginning at the N-termini of α-, β-,and δ-M2 were isolated for sequence analysis by the same procedures used for nAChRs photolabeled in the desensitized state.
For nAChRs photolabeled in the presence of αBxTx, the peaks of 3H release when sequencing through αM2 indicated photolabeling of αM2-5 (αIle-247), αM2-6 (αSer-248) and αM2-9 (αLeu-251), each at ~1 cpm/pmol, while within βM2 and δM2 the major peaks of 3H release in cycle 9 and 6, respectively, indicated photolabeling of βLeu-257 at 8 cpm/pmol and δSer-262 at 4 cpm/pmol (Figure 7 and Table 1). Photolabeling at each of these positions was reduced by >90% in the presence of tetracaine. In addition to these major peaks of 3H release, in each subunit there was low level release in cycle 20, consistent with labeling of M2-20 at an efficiency ~10% that of position M2-6 or M2-9, and the photolabeling at M2-20 was not inhibited by tetracaine.
Figure 7. [3H]CPZ photolabeling in the M2 ion channel in the presence of α-BgTx.
3H (●, ○) and PTH-amino acids (□) released during sequence analysis of fragments beginning at the N-termini αM2 (A), βM2 (B), and δM2 (C), isolated from nAChR-rich membranes photolabeled with 1.7 µM [3H]CPZ after equilibration with 10 µM αBgTx in the absence (●) or presence (○, □) of 100 µM tetracaine. Labeled subunits were separated by SDS-PAGE and processed as in Figure 5 to isolate peptides beginning at the N-termini of the M2 segments. A, The fragment beginning at αMet-243 was the primary sequence (I0 = 23 pmol −(□) and 17 pmol +tetracaine), along with α subunit fragments beginning at αSer-173 (~12 pmol), αHis-186 (~18 pmol), and αTyr-277 (~2 pmol), as well as EndoLys-C (Gly-1, ~8 pmol) and a V8 protease peptide (Val-180, ~5 pmol). (10,200 (●) and 7,820 (○) cpm sequenced). B, The primary sequence began at βMet-249 (I0 = 12 pmol −(□) and 10 pmol +tetracaine), with any secondary sequence beginning at <10% that level. (1,900 (●) and 960 (○) cpm sequenced). C, The only sequence detected began at δMet-257 (I0 = 15 pmol −(□) and +tetracaine; 5,300 (●) and 2,700 (○) cpm sequenced). For all sequencing runs, the efficiencies of photolabeling (cpm/pmol) associated with the peaks of 3H in cycles 5, 6, 9, 16, 17, or 20 are quantitated in Table 2.
[3H]CPZ photoincorporation outside of the ion channel
Based upon the broad side chain reactivity observed for [3H]CPZ photolabeling in the ion channel, including aliphatic (Val, Ile, Leu), aromatic (Phe) and polar (Ser, Thr, Glu, Asp, and Gln) side chains, we extended our studies to determine whether [3H]CPZ photoincorporated into other regions of the nAChR transmembrane domain. Since CPZ also binds to the agonist sites (12), we characterized photolabeling for nAChRs equilibrated with agonist (desensitized state) to search for binding sites distinct from those sites. Novel binding sites have been identified recently in the nAChR transmembrane domain for uncharged hydrophobic drugs that bind within the ion channel and to sites in the δ subunit helix bundle (TID and benzophenone), or at the interfaces between subunits (benzophenone (γ-α and β-α); TDBzl-etomidate (γ-α) (21–24)).
[3H]CPZ does not bind/photolabel within the δ subunit helix bundle
In the desensitized state, there was no evidence that [3H]CPZ photolabeled δM2-18 or δM2-22 (Figures 5B and 6B), residues that project towards the pocket formed by the δ̣ subunit helix bundle and that are labeled by [125I]TID in the open and desensitized states, along with δIle-288 within the δM2-M3 loop and δPhe-232 and δCys-236 within δM1 (21;22). There was also no evidence of [3H]CPZ photolabeling within δM1, as we found only background 3H in the rpHPLC fractions containing the fragment beginning at δPhe-206 and extending through δM1 and no peaks of 3H release when that fragment was sequenced through δM1 (labeling, if it occurred, was less than 0.4 cpm/pmol or 3% the level of labeling of δM2-20). Also, sequence analysis of rpHPLC fractions from a solution digest of δ subunit with V8 protease, which can be used to identify photolabeling of δIle-288 or amino acids in δM3 (23), established that δIle-288 was not labeled by [3H]CPZ (<0.1 cpm/pmol, data not shown).
[3H]CPZ photolabels within the α subunit MA helix but not in αM4
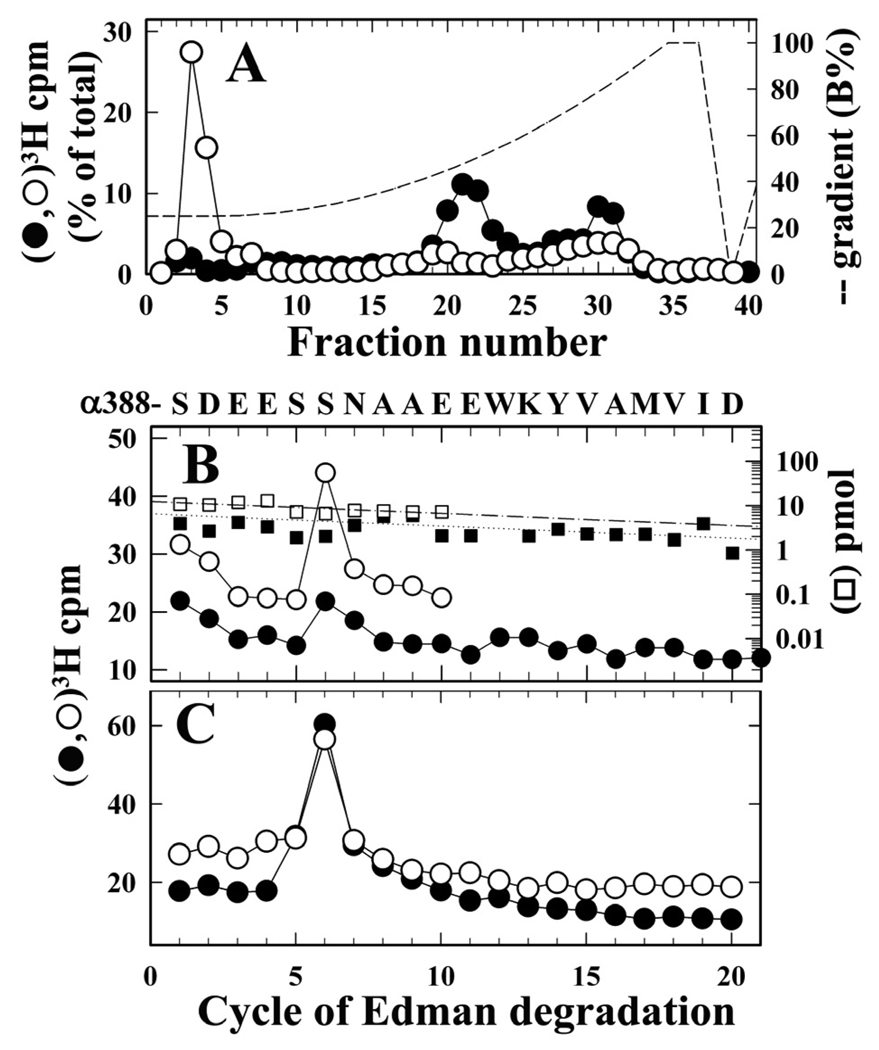
For nAChRs photolabeled in the desensitized state, fragmentation of the α subunit with V8 protease established that there was 3H incorporation in ~10 kDa fragments that include αV8-10, the fragment beginning at αAsn-339 (28) that contains the MA helix from the nAChR cytoplasmic domain (beginning at αIle-376) and the hydrophobic M4 membrane spanning helix beginning at αIle-406 (Figure 4). The amino acids in these fragments photolabeled by [3H]CPZ were characterized by sequence analysis of the peaks of 3H recovered when aliquots of that material were fractionated by rpHPLC before and after trypsin digestion (Figure 8A). Before digestion, 3H was recovered in two broad peaks, one a hydrophobic peak centered at ~80% organic, where αV8-10 elutes (29), and a second hydrophilic peak centered at 50% organic, with <5% of 3H eluting in the column flow-through. Sequence analysis of the hydrophobic peak confirmed the presence of the fragment beginning at αAsn-339 (αV8-10, ~20 pmol), with no peaks of 3H release in 30 cycles of Edman degradation (not shown). When material from the hydrophilic peak was sequenced, there were no peaks of 3H release in 10 cycles of Edman degradation, and the N-terminus of V8 protease was the only identifiable fragment (~2 pmol) (not shown). When the trypsin digest was fractionated by rpHPLC, ~50% of 3H eluted in the column flow-through, whereas 15% was recovered in a broad hydrophobic peak centered at 80% organic (Figure 8A). Sequence analysis of the hydrophobic peak established the presence of fragments beginning at αSer-388 and αTyr-401 (~6 pmol of each), produced by trypsin cleavage at the two lysines N-terminal to M4, and a peak of 3H release in cycle 6 (Figure 8B). In another photolabeling experiment, sequence analysis of the hydrophobic peak from the trypsin digest of αV8-10 established the presence of the αTyr-401 fragment at 5 pmol and <0.3 pmol of the αSer-388 fragment, with no peak of 3H release in cycle 6 (not shown). The simplest interpretation of these results is that the peak of 3H release in cycle 6 (Figure 8B) resulted from the low level labeling of αSer-393 (~0.5 cpm/pmol). When the material in the rpHPLC column flow-through from the tryptic digest was sequenced for 20 cycles, the only peak of 3H release was also in cycle 6 (Figure 8C), though individual fragment sequences were not identifiable. This release in cycle 6 is also likely to result, at least in part, from the photolabeling of αSer-393, in the tryptic fragment extending from αSer-388 to αLys-400 that would be expected to elute in the column flow-through. The 3H in the rpHPLC column flow-through of the tryptic digest (Figure 8A) must also include digestion products of the labeled fragments that eluted in the hydrophilic peak of 3H before trypsin digestion. Further studies are required to identify these photolabeled amino acids, but αSer-393 is one likely candidate, as it could have been contained in a hydrophilic fragment of ~7 kDa produced by V8 protease cleavage of the nAChR α subunit (for example, αAsn-339 to αGlu-391/397).
Figure 8. [3H]CPZ photolabeling in α subunit cytoplasmic (MA) helix.
“In gel” digestion with V8 protease was used to isolate α subunit fragments of ~10 kDa from nAChRs from the photolabeling experiment used in Figure 4 (nAChRs photolabeled +Carb ± PCP). A, rpHPLC fractionation of aliquots of this material, from nAChRs photolabeled −PCP, before (●) and after (○) digestion with trypsin. 3H recovery (%) before and after trypsin digestion: fractions 3–5 (4%, 50%); fractions 20–23 (35%, 6%); fractions 28–32 (23%, 15%). B and C 3H (●,○) and PTH-amino acids (□,■) released during sequence analysis of fractions 28–32 (B) and 3–5 (C) from the rpHPLC fractionations of the tryptic digests of material photolabeled −PCP (●,■) and +PCP (○,□). B, The hydrophobic peak of 3H contained overlapping fragments beginning at αSer-388 (■, I0 = 7 pmol; □, I0 =12 pmol) and αTyr401 (not shown, −PCP, 5 pmol; +PCP, 8 pmol), with 3H release at cycle 6 (4,400 (●) and 3,600 (○) cpm sequenced). For the sample not digested with trypsin, no peak of 3H release was detected in 30 cycles of Edman degradation of fractions 28–32, which contained the fragment beginning at αAsn-339, the N-terminus of αV8-10 (−PCP, 19 pmol), or fractions 20–23, which contained V8 protease (~2 pmol). C, 3H release during sequence analysis of material in fractions 3–5 (11,100 (●) and 8,700 (○) cpm sequenced). Filters were washed before sequencing, as described in Experimental Procedures, to remove excess detergent.
To identify additional amino acids within αMA located N-terminal to αSer-388, we carried out another [3H]CPZ photolabeling of nAChRs (+Carb) and isolated, as above, αV8-10 for sequence analysis. When the fragment beginning at αAsn-339 (40 pmol) was sequenced for 60 cycles, there was a small peak of 3H release in cycle 55, which confirmed photolabeling of αSer-393 (1 cpm/pmol), and, in addition, there was a larger peak of 3H release in cycle 48, which established that CPZ also photolabels αMet-386 (2 cpm/pmol) (Supplemental Figure S5). Similar to αSer-393, αMet-386 would be contained in a small tryptic fragment extending from αTyr-481 to αLys-387, and labeling of that residue would also contribute to the peak of 3H release seen in cycle 6 of the sequence analysis of the material in the column flow-through from the tryptic digest (Fig. 8B).
DISCUSSION
In this work we use [3H]CPZ to provide new information about the diversity of binding sites in the nAChR transmembrane domain for aromatic amine noncompetitive antagonists. In early photolabeling studies, [3H]CPZ was shown to bind to a site near the cytoplasmic end of the Torpedo nAChR ion channel in the desensitized state, where it photolabeled amino acids within each M2 helix at positions M2-2, M2-6, and/or M2-9, with the [3H]CPZ binding (photolabeling) inhibitable by PCP (13–16). We extended those studies by determining whether [3H]CPZ binds to additional sites in the transmembrane domain of the nAChR in the desensitized state and by identifying its binding site in the nAChR ion channel in the closed state.
For nAChRs in the desensitized state, we find only one additional site for [3H]CPZ in the transmembrane domain: a site at the extracellular end of the channel, identified by the photolabeling of amino acids at positions M2-16 (γ), M2-17 (β, δ), and M2-20 (α, β, and δ), where [3H]CPZ binding/photolabeling is enhanced, rather than inhibited, in the presence of a high concentration of PCP and only partially inhibited by proadifen. We will refer to the two CPZ binding sites as Site2/6/9 and Site17/20, based upon the position of the photolabeled amino acids within the M2 helices. Our results establish that CPZ and PCP (or proadifen) bind in a mutually exclusive manner to Site2/6/9, and that occupancy of that site by PCP does not inhibit binding of CPZ to Site17/204. Thus, the electrostatic constraints within the ion channel are of such a nature that two positively charged ligands can bind simultaneously at the cytoplasmic and extracellular ends of the ion channel. The potentiation of [3H]CPZ photolabeling at Site17/20 seen in the presence of PCP and the partial inhibition of photolabeling seen in the presence of proadifen indicate that the binding of either drug at the cytoplasmic end of the ion channel results in subtle, but different, changes in structure at the extracellular end of the channel.
When nAChRs are stabilized in a closed channel state by the binding of αBgTx at the agonist sites, CPZ binds in the ion channel with 8-fold lower affinity than when the nAChR is in the desensitized state, and the [3H]CPZ photolabeling of amino acids at M2-5 (α), M2-6 (α, β, δ) and M2-9 (β, δ) establishes for the first time that CPZ binds at a similar level in the ion channel in the closed and desensitized states. However, there are differences in the pattern of subunit labeling, such as the favored labeling of αM2-2 and αM2-6 in the desensitized state and of αM2-6 and αM2-9 in the closed state (Table 1), that must result from state-dependent differences in the local structure of the channel.
The nAChR amino acids photolabeled by [3H]CPZ are identified (Figure 9) in the Torpedo nAChR structure (Protein Data Bank code 2BG9), obtained in the absence of agonist and assumed to be the closed state structure. The structural diversity of amino acids photolabeled by CPZ within the ion channel domain, including aliphatic (Leu, Val, Ile), aromatic (Phe) and polar (Ser, Thr, Glu, Asp, Gln ) side chains, is noteworthy and indicates that [3H]CPZ can photoincorporate into any binding site that it occupies. We highlight in Figures 9B and 9C the amino acids photolabeled by [3H]CPZ in the closed channel (+ αBgTx, green) and desensitized (+Carb, red and blue) states, respectively. We also highlight the aliphatic side chains at position M2-13 (yellow) that are not photolabeled by [3H]CPZ in either state but are photolabeled by [3H]TDBzl-etomidate in the desensitized state (24) and contribute, along with the aliphatics at M2-9, to the binding site in the closed channel for the smaller uncharged, hydrophobic drugs [125I]TID and [3H]benzophenone (20;23).
The lack of CPZ photolabeling of the Val/Ile at M2-13 in conjunction with its photolabeling of the Leu at M2-9 and M2-16/17, as well as the fact that CPZ photolabels amino acids at the extracellular end of the channel even when PCP or proadifen occupies the site at the cytoplasmic end, provide strong evidence that CPZ photolabeling at the extracellular end of the ion channel must result from occupancy of a site rather than a diffusional encounter of photoactivated CPZ, which is known to react efficiently with water (35).
Although CPZ only binds at the level of M2-6 and M2-9 in the closed channel, the lumen of the ion channel within the Torpedo nAChR structure can accommodate CPZ at binding sites between the levels of M2-6 and M2-20. We show in Figure 9D a side view of the M2 ion channel domain with one CPZ docked near M2-6 and M2-9, positions labeled in the closed and desensitized states, and a second CPZ docked at the level of M2-17 and M2-20, positions prominently photolabeled only in the desensitized state. While CPZ binding at the level of M2-6 and M2-9 will be in close proximity to side chains from each M2 helix (Figure 9F and Supplemental Figure S6), a view looking down the extracellular half of the channel (Figure 9E) shows that one CPZ can not be in simultaneous contact with all subunits at that level. In the most energetically favored orientation, the CPZ N+ was within 3 Å of αγGlu-262 and its likely photoreactive C-Cl bond (35) was within 5 Å of the photolabeled βLeu-265 and δLeu-273, and 6.5 Å from γPhe-267, consistent with the fact that labeling of αM2-17/20 was at <25% the efficiency of those positions in each other subunit (Table 1). Although positions M2-17 and M2-20 contribute to the lumen of the ion channel in the nAChR structure, it is significant that those residues are labeled only in the desensitized state either by CPZ, by meproadifen mustard (36), or by uncharged, photoreactive anesthetics (37;38). This state dependent labeling establishes that there must be a substantial change in structure at the extracellular end of the ion channel between the closed channel and desensitized states, potentially a reduction in the diameter of the channel lumen to form a more compact binding site.
[3H]CPZ binding at the extracellular end of the ion channel
Our photolabeling results establish that [3H]CPZ binds at equilibrium to ion channel Site17/20 in the nAChR in the desensitized state, and the lack of inhibition by PCP establishes that this site has distinct pharmacological properties from ion channel Site2/6/9. However, the photolabeling data at a single [3H]CPZ concentration are insufficient to determine the CPZ affinity for either site. Since PCP inhibits [3H]CPZ photolabeling at the Site2/6/9, CPZ must bind at that site with a KD of ~1 µM, based upon its inhibition of [3H]PCP binding. Since [3H]CPZ photolabels amino acids in Site17/20 at the same or higher efficiency as at Site2/6/9, it is possible that CPZ binds to the two sites with similar affinities. However, it is also possible that Site17/20 is one of the ~10 low affinity sites/nAChR detected by [3H]CPZ equilibrium binding assays with Torpedo nAChR-rich membranes (12).
Additional [3H]CPZ photolabeling studies will be required to identify drugs that compete with [3H]CPZ binding at the site at the extracellular end of the ion channel. Meproadifen, a quaternary ammonium derivative of proadifen, is a likely candidate, since it partially inhibits photolabeling of αM2-20 by two uncharged drugs, [3H]azioctanol and [3H]azietomidate (37;38), and [3H]meproadifen mustard, its reactive aziridinium analog, reacts selectively with αM2-20 (36).
When [3H]CPZ photolabels Site2/6/9 and Site17/20 in nAChRs equilibrated with agonist, it is most likely that all nAChRs are in the equilibrium desensitized state. However, [3H]CPZ binds most rapidly to a state stabilized only transiently by agonist (39), either the open channel state or a transient desensitized state, and the use of time-resolved photolabeling techniques (22;40) will allow the determination of which site is occupied most rapidly when nAChRs are exposed transiently to agonist.
[3H] CPZ binding sites outside the ion channel
We found no evidence that [3H]CPZ bound in the nAChR transmembrane domain in the δ subunit helix bundle, where [125I]TID and [3H]benzophenone bind in an agonist dependent manner (22;23), and we found no evidence of labeling at the αM4 lipid interface. However, [3H]CPZ photolabeling of αMet-386 and αSer-393 establishes that it binds in the nAChR cytoplasmic domain in the basket formed by the MA helices that are the extensions of the M4 helices (Figure 9G). Photolabeling of αSer-393 was not inhibited by PCP, and, though not examined, we assume that photolabeling of αMet-386 will also be unaffected by PCP. However, the photolabeling of these residues is unlikely to be a result of a random collisional encounter of photactivated [3H]CPZ, since it did not photolabel other serines in αMA (αSer-388, αSer-392) or of any of the acidic side chains, including αGlu-390, which is adjacent to αSer-393 on the MA helix and photolabeled by the uncharged noncompetitive antagonist [3H]azietomidate (40). As the current nAChR structure provides only a very limited definition of the structure of the nAChR cytoplasmic domain, and no definition of the nAChR regions interacting with the coiled-coil domain of cytoplasmic scaffolding protein rapsyn (41), insufficient structural information is available to interpret why CPZ and azietomidate each interact with the same region of nAChR, which may be a drug binding site at the interface between the nAChR and rapsyn (which was also photolabeled by [3H]CPZ).
Supplementary Material
Footnotes
This research was supported in part by US Public Health Service Grant GM-58448 (J.B.C.) and by an award to Harvard Medical School from the Howard Hughes Biomedical Research Support Program for Medical Schools
Abbreviations: nAChR, nicotinic acetylcholine receptor; CPZ, chlorpromazine HCl; CPZsulfoxide, chlorpromazine sulfoxide; Carb, carbamylcholine; αBgTx, α-bungarotoxin; PCP, phencyclidine; Azietomidate, 2-(3-methyl-3H-diaziren-3-yl)ethyl 1-(phenylethyl)-1H-imidazole-5-carboxylate; TDBzl-Etomidate, 4-[3-(trifluoromethyl)-3H-diazirin-3-yl]benzyl-1-(1-phenylethyl)-1H-imidazole-5-carboxylate; TID, 3-(trifluoromethyl)-3-(m-iodophenyl)diazirine; SDS, sodium dodecyl sulfate; PAGE, polyacrylamide gel electrophoresis; rpHPLC, reversed-phase high-performance liquid chromatography; V8 protease, Staphylococcus aureus endoproteinase Glu-C; EndoLys-C, Lysobacter enzymogenes endoproteinase Lys-C.
Further purification by rpHPLC established that only ~20% of the 3H in the gel band containing αV8-18 was associated with that fragment, which eluted as a sharp peak at ~45% organic, well resolved from the major peak of 3H eluting at ~80% organic (not shown). Similarly, rpHPLC purification of the material in the gel bands containing αV8-10 from nAChRs photolabeled either +Carb or +αBgTx established that only ~50% of 3H was associated with αV8-10, which elutes as a broad hydrophobic peak centered at 80 % organic, with the remainder eluting as a hydrophilic peak at ~50% organic (see Figure 8).
Photoexcitation of CPZ at 330 nm results almost exclusively in dechlorination and the formation of a reactive radical intermediate (35), while photoexcitation at 280 nm may result in preferential loss of an electron and the formation of a radical cation intermediate ((43), but see (35)). Since the experiments of Figures 5 and 6 used two different Hg lamps, each with filters for optimal excitation at 350 nm, but with different half-widths, it is possible that subtle differences in the wavelength of photoexcitation may determine whether Site17/20 is photolabeled at higher efficiency than Site2/6/9 (Figure 5) or at similar overall efficiency (Figure 6).
[3H]CPZ photolabeling at the extracellular end of the ion channel was not characterized by Changeux and coworkers, since most samples containing M2 segments were sequenced for fewer than 15 cycles of Edman degradation (13–15). However, the γ subunit samples were sequenced for 30 cycles (16), and in addition to peaks of PCP-inhibitable 3H release in cycles 2, 6, and 9, the focus of the report, the data do include a peak in cycle 16, not inhibited by PCP.
SUPPORTING INFORMATION AVAILABLE Five figures as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Lynch JW. Molecular structure and function of the glycine receptor chloride channel. Physiol Rev. 2004;84:1051–1095. doi: 10.1152/physrev.00042.2003. [DOI] [PubMed] [Google Scholar]
- 2.Sine SM, Engel AG. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440:448–455. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 3.Albuquerque EX, Pereira EFR, Alkondon M, Rogers SW. Mammalian nicotinic acetylcholine receptors: From structure to function. Physiol. Rev. 2009;89:73–120. doi: 10.1152/physrev.00015.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–958. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 5.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J. Mol. Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Celie PHN, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 7.Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, Bourne Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO Journal. 2005;24:3635–3646. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilf RJC, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 9.Hilf RJC, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–119. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 10.Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux J-P, Delarue M, Corringer P-J. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 11.Brannigan G, Henin J, Law R, Eckenhoff R, Klein ML. Embedded cholesterol in the nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA. 2008;105:14418–14423. doi: 10.1073/pnas.0803029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidmann T, Oswald RE, Changeux J-P. Multiple sites of action for noncompetitive blockers on acetylcholine receptor rich membrane fragments from Torpedo marmorata. Biochemistry. 1983;22:3112–3127. doi: 10.1021/bi00282a014. [DOI] [PubMed] [Google Scholar]
- 13.Giraudat J, Dennis M, Heidmann T, Chang J-Y, Changeux J-P. Structure of the high-affinity binding site for noncompetitive blockers of the acetylcholine receptor: Serine-262 of the δ subunit is labeled by [3H]chlorpromazine. Proc. Natl. Acad. Sci. USA. 1986;83:2719–2723. doi: 10.1073/pnas.83.8.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giraudat J, Dennis M, Heidmann T, Haumont P-Y, Lederer F, Changeux J-P. Structure of the high-affinity binding site for noncompetitive blockers of the acetylcholine receptor: [3H]chlorpromazine labels homologous residues in the β and δ chains. Biochemistry. 1987;26:2410–2418. doi: 10.1021/bi00383a003. [DOI] [PubMed] [Google Scholar]
- 15.Giraudat J, Gali J-Z, Revah F, Changeux J-P, Haumont P-Y, Lederer F. The noncompetitive blocker [3H]chlorpromazine labels segment M2 but not segment M1 of the nicotinic acetylcholine receptor α-subunit. FEBS Letts. 1989;253:190–198. doi: 10.1016/0014-5793(89)80957-3. [DOI] [PubMed] [Google Scholar]
- 16.Revah F, Galzi JL, Giraudat J, Haumont P-Y, Lederer F, Changeux J-P. The noncompetitive blocker [3H]chlorpromazine labels three amino acids of the acetylcholine receptor γ subunit: Implications for the α-helical organization of regions MII and for the structure of the ion channel. Proc. Natl. Acad. Sci. USA. 1990;87:4675–4679. doi: 10.1073/pnas.87.12.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu YC, Barrantes FJ, Shen JH, Luo XM, Zhu WL, Chen KX, Jiang HL. Blocking of the nicotinic acetylcholine receptor ion channel by chlorpromazine, a noncompetitive inhibitor: A molecular dynamics simulation study. J. Phys. Chem. B. 2006;110:20640–20648. doi: 10.1021/jp0604591. [DOI] [PubMed] [Google Scholar]
- 18.Arias HR, Bhumireddy P, Spitzmaul G, Trudell JR, Bouzat C. Molecular mechanisms and binding site location for the noncompetitive antagonist crystal violet on nicotinic acetylcholine receptors. Biochemistry. 2006;45:2014–2026. doi: 10.1021/bi051752e. [DOI] [PubMed] [Google Scholar]
- 19.Gallagher MJ, Cohen JB. Identification of amino acids of the Torpedo nicotinic acetylcholine receptor contributing to the binding site for the noncompetitive antagonist [3H]tetracaine. Mol. Pharmacol. 1999;56:300–307. doi: 10.1124/mol.56.2.300. [DOI] [PubMed] [Google Scholar]
- 20.White BH, Cohen JB. Agonist-induced changes in the structure of the acetylcholine receptor M2 regions revealed by photoincorporation of an uncharged nicotinic non-competitive antagonist. J. Biol. Chem. 1992;267:15770–15783. [PubMed] [Google Scholar]
- 21.Hamouda AK, Chiara DC, Blanton MP, Cohen JB. Probing the structure of the affinity-purified and lipid-reconstituted Torpedo nicotinic acetylcholine receptor. Biochemistry. 2008;47:12787–12794. doi: 10.1021/bi801476j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arevalo E, Chiara DC, Forman SA, Cohen JB, Miller KW. Gating-enhanced accessibility of hydrophobic sites within the transmembrane region of the nicotinic acetylcholine receptor's δ-subunit - A time-resolved photolabeling study. J. Biol. Chem. 2005;280:13631–13640. doi: 10.1074/jbc.M413911200. [DOI] [PubMed] [Google Scholar]
- 23.Garcia GI, Chiara DC, Nirthanan S, Hamouda AK, Stewart DS, Cohen JB. [3H]Benzophenone photolabeling identifies state-dependent changes in nicotinic acetylcholine receptor structure. Biochemistry. 2007;46:10296–10307. doi: 10.1021/bi7008163. [DOI] [PubMed] [Google Scholar]
- 24.Nirthanan S, Garcia GI, Chiara DC, Husain SS, Cohen JB. Identification of binding sites in the nicotinic acetylcholine receptor for TDBzl-etomidate, a photoreactive positive allosteric effector. J.Biol.Chem. 2008;283:22051–22062. doi: 10.1074/jbc.M801332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Middleton RE, Cohen JB. Mapping of the acetylcholine binding site of the nicotinic acetylcholine receptor: [3H]-nicotine as an agonist photoaffinity label. Biochemistry. 1991;30:6987–6997. doi: 10.1021/bi00242a026. [DOI] [PubMed] [Google Scholar]
- 26.Korczak-Fabierkiewicz C, Robinson DW, Lucas GHW. Conversion of chlorpromazine sulfoxide to chlorpromazine by use of metals in acid solution. J. Chromatogr. 1967;31:539–544. doi: 10.1016/s0021-9673(01)86108-2. [DOI] [PubMed] [Google Scholar]
- 27.White BH, Howard S, Cohen SG, Cohen JB. The hydrophobic photoreagent 3-(trifluoromethyl)-3-(m-[125I]iodophenyl)diazirine is a novel noncompetitive antagonist of the nicotinic acetylcholine receptor. J. Biol. Chem. 1991;266:21595–21607. [PubMed] [Google Scholar]
- 28.White BH, Cohen JB. Photolabeling of membrane-bound Torpedo nicotinic acetylcholine receptor with the hydrophobic probe 3-trifluoromethyl-3-(m-[125I]iodophenyl)diazirine. Biochemistry. 1988;27:8741–8751. doi: 10.1021/bi00424a009. [DOI] [PubMed] [Google Scholar]
- 29.Blanton MP, Cohen JB. Identifying the lipid-protein interface of the Torpedo nicotinic acetylcholine receptor: secondary structure implications. Biochemistry. 1994;33:2859–2872. doi: 10.1021/bi00176a016. [DOI] [PubMed] [Google Scholar]
- 30.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 31.Chiara DC, Trinidad JC, Wang D, Ziebell MR, Sullivan D, Cohen JB. Identification of amino acids in the nicotinic acetylcholine receptor agonist binding site and ion channel photolabeled by 4-[(3-trifluoromethyl)-3H-Diazirin-3-yl]Benzoylcholine, a novel photoaffinity antagonist. Biochemistry. 2003;42:271–283. doi: 10.1021/bi0269815. [DOI] [PubMed] [Google Scholar]
- 32.Oswald RE, Heidmann T, Changeux J-P. Multiple affinity sites for noncompetitive blockers revealed by [3H]phencyclidine binding to acetylcholine receptor rich membrane fragments from Torpedo marmorata. Biochemistry. 1983;22:3128–3136. doi: 10.1021/bi00282a015. [DOI] [PubMed] [Google Scholar]
- 33.Middleton RE, Strnad NP, Cohen JB. Photoaffinity labeling the torpedo nicotinic acetylcholine receptor with [3H]tetracaine, a nondesensitizing noncompetitive antagonist. Mol. Pharmacol. 1999;56:290–299. doi: 10.1124/mol.56.2.290. [DOI] [PubMed] [Google Scholar]
- 34.Boyd ND, Cohen JB. Desensitization of membrane-bound Torpedo acetylcholine receptor by amine noncompetitive antagonists and aliphatic alcohols: studies of [3H]-acetylcholine binding and 22Na+ ion fluxes. Biochemistry. 1984;23:4023–4033. doi: 10.1021/bi00313a003. [DOI] [PubMed] [Google Scholar]
- 35.van den Broeke LT, Ouijja EH, Bojarski J, Beyersgergen van Henegouwen GMJ. In vitro photodegradation of chlorpromazine. Photochem. Photobiol. 1994;59:140–144. [Google Scholar]
- 36.Pedersen SE, Sharp SD, Liu W-S, Cohen JB. Structure of the noncompetitive antagonist binding site in the Torpedo nicotinic acetylcholine receptor: [3H]Meproadifen mustard reacts selectively with α-subunit Glu-262. J. Biol. Chem. 1992;267:10489–10499. [PubMed] [Google Scholar]
- 37.Pratt MB, Husain SS, Miller KW, Cohen JB. Identification of sites of incorporation in the nicotinic acetylcholine receptor of a photoactivatible general anesthetic. J. Biol. Chem. 2000;275:29441–29451. doi: 10.1074/jbc.M004710200. [DOI] [PubMed] [Google Scholar]
- 38.Ziebell MR, Nirthanan S, Husain SS, Miller KW, Cohen JB. Identification of binding sites in the nicotinic acetylcholine receptor for [3H]azietomidate, a photoactivatable general anesthetic. J. Biol. Chem. 2004;279:17640–17649. doi: 10.1074/jbc.M313886200. [DOI] [PubMed] [Google Scholar]
- 39.Heidmann T, Changeux J-P. Time-resolved photolabeling by the noncompetitive blocker chlorpromazine of the acetylcholine receptor in its transiently open and closed ion channel conformations. Proc. Natl. Acad. Sci. USA. 1984;81:1897–1901. doi: 10.1073/pnas.81.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiara DC, Hong FH, Arevalo E, Husain SS, Miller KW, Forman SA, Cohen JB. Time-resolved photolabeling of the nicotinic acetylcholine receptor by [3H]Azietomidate, an open-state inhibitor. Mol. Pharmacol. 2009;75:1084–1095. doi: 10.1124/mol.108.054353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartoli M, Ramarao MK, Cohen JB. Interactions of the rapsyn RING-H2 domain with dystroglycan. J. Biol. Chem. 2001;276:24911–24917. doi: 10.1074/jbc.M103258200. [DOI] [PubMed] [Google Scholar]
- 42.Hamouda AK, Chiara DC, Sauls D, Cohen JB, Blanton MP. Cholesterol interacts with transmembrane alpha-helices M1, M3, and M4 of the Torpedo nicotinic acetylcholine receptor: Photolabeling studies using [3H]azicholesterol. Biochemistry. 2006;45:976–986. doi: 10.1021/bi051978h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motten AG, Buettner GR, Chignell CF. Spectroscopic studies of cutaneous photosensitizing agents-VII. A spin-trapping study of light induced free radicals from chlorpromazine and promazine. Photochem. Photobiol. 1985;42:9–15. doi: 10.1111/j.1751-1097.1985.tb03540.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.