Abstract
Autophagy is a lysosome-directed membrane trafficking event for the degradation of cytoplasmic components, including organelles. The past few years have seen a great advance in our understanding of the cellular machinery of autophagosome biogenesis, the hallmark of autophagy. However, our global understanding of autophagosome maturity remains relatively poor and fragmented. The topological similarity of autophagosome and endosome delivery to lysosomes suggests that autophagic and endosomal maturation may have evolved to share associated machinery to promote the lysosomal delivery of their cargoes. We have recently discovered that UVRAG, originally identified as a Beclin 1-binding autophagy protein, appears to be an important factor in autophagic and endosomal trafficking through its interaction with the class C Vps tethering complex. Given the ability of UVRAG to bind Beclin 1 and the class C Vps complex in a genetically and functionally separable manner, it may serve as an important regulator for the spatial and/or temporal control of diverse cellular trafficking events. As more non-autophagic functions of UVRAG are unveiled, our understanding of seemingly different cellular processes may move a step further.
Keywords: UVRAG, the class C Vps complex, autophagy, autophagosome maturation, membrane trafficking
Autophagy has been increasingly recognized as essential for cells to maintain homeostasis and is particularly induced in response to various stress stimuli.1-3 Depending on different autophagy modes (selective or non-selective), a cascade of degradative autophagic process is initiated by the engulfment of cytoplasmic cargo into a double-membrane bound autophagosome. Concurrently, the newly formed autophagosomes rapidly acidify as they mature. Fusion with the late endosome and lysosome plays a critical role in this process and results in the delivery of lysosomal hydrolases.4 Defects in autophagosome maturity or autophagosome clearance lead to a piling up of autophagosomes within the cell, which can induce serious disorders including cardiomyopathy, myopathy, neuronal ceroid lipofuscinosis and Danon disease.5-7 Yet despite its importance, little is known about the molecular machinery responsible for autophagosome maturation in mammals. Recently, several proteins involved in endosomal dynamics have been identified to be required for autophagic maturation,7-12 suggesting functional connection and coordinated regulation of two distinct but converged membrane trafficking pathways. Here, we concentrate on very recent findings about mammalian UVRAG, originally recognized as an autophagy protein specifically involved in autophagosome formation, its novel role in modulating autophagic and endosomal maturation.13
UVRAG in Brief
The ultraviolet (UV) radiation resistance-associated gene (UVRAG) was originally identified in a genetic screen for complementing UV sensitivity in xeroderma pigmentosum (XP) cells.14 The name UVRAG was chosen to reflect its role in providing UV resistance. It maps to a tumor-susceptibility locus on human chromosome 11q13 that is frequently implicated in common human cancers, including breast, colorectal and gastric cancers.15-18 UVRAG consists of four major regions: the proline-rich (PR) domain (residues 1–41), the calcium-dependent lipid-binding C2 domain (residues 42–147), the coiled-coil domain (CCD, residues 200–269) and a C-terminal domain (residues 270–699) presumed to be unstructured (Fig. 1A). The PR domain interacts with Bif-1, a Bax activator, which allows Bif-1 to associate with the Beclin 1/PtdIns3-kinase III (PtdIns 3-KC3) complex, thereby inducing autophagy and membrane curvature.19,20 The C2 domain is thought to confer the membrane-lipid association ability of UVRAG.13 The highly hydrophobic CCD domain is the locus for Beclin 1 binding, which is crucial for UVRAG-mediated autophagy activation and tumor suppression in vitro.21,22 Besides its interaction with proteins involved in autophagosome biogenesis, UVRAG was recently shown to bind the class C Vps complex (hereafter referred to as C/Vps),13 a modular protein complex that serves diverse functions in multiple membrane fusion events.23-26 The region of UVRAG that is responsible for this interaction independently maps to the C2 and C-terminal regions. Interestingly, a mutated version of UVRAG that lacks the Beclin 1-binding activity exhibits efficient interaction with the C/Vps complex, whereas a mutated version of UVRAG that lacks the C/Vps complex binding activity maintains efficient Beclin 1 interaction, suggesting that the UVRAG interaction with the C/Vps complex is likely independent from its association with Beclin 1.13
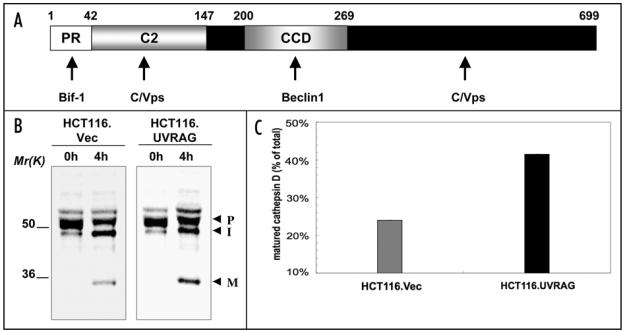
Figure 1.
(A) Domain organization of UVRAG. The PR, C2, CCD and C-terminal regions are indicated. Amino acid positions are shown at the top. The binding sites for UVRAG-interacting proteins are indicated with arrows. (B) UVRAG promotes cathepsin D maturation in HCT116 cells. HCT116 cells expressing empty vector or wild-type UVRAG were labeled with [35S]-methionine for the indicated time (hours). Cathepsin D was immunoprecipitated and subjected to SDS-PAGE and radiography. The forms of cathepsin D are labeled as follows: P, proform; I, intermediate form; M, mature form. (C) Densitometric quantification of cathepsin D maturation in (B). The percentage of total radiolabeled procathepsin D at time 0 that is present as a mature form after 4 hours in the cells was quantified.
UVRAG in Autophagic Trafficking
The UVRAG-C/Vps interaction connects the C/Vps complex to the mammalian autophagy system—the overexpression of UVRAG induces the association of the C/Vps complex with autophagosomes, whereas a mutated version of UVRAG that lacks C/Vps complex binding fails to do so.13 The C/Vps complex was initially identified in genetic screens for vacuolar protein sorting (vps) mutants.27 One group of vps mutants, called class C, is characterized by the accumulation of multivesicular bodies and the missorting of multiple biosynthetic cargoes, suggesting that it functions at the endosome-lysosome interface.28 Moreover, the C/Vps proteins are evolutionarily conserved and have been identified in many different biological contexts.23,24,29-32 Although it is clear that the C/Vps complex plays a central role in the endolysosomal system, its function in autophagy is less established. Indeed, our findings suggest that the C/Vps complex is a minor participant in the biogenesis of autophagosomes.13 The siRNA-mediated depletion of the C/Vps complex has a minimal effect on UVRAG-mediated autophagosome formation. Instead, our observations strongly support an important role of the UVRAG-C/Vps complex in promoting autophagosome maturation. First, deletion of the C2 domain of UVRAG that disrupts the interaction between UVRAG and the C/Vps complex severely impairs the ability of UVRAG to promote autophagosome fusion with lysosomes, even though it has only marginal effects on UVRAG-mediated autophagosome formation. Second, the siRNA-mediated depletion of the C/Vps complex subunits potently inhibits UVRAG-mediated autophagosome maturation. Finally, the overexpression of wild-type UVRAG, but not the C/Vps binding defective UVRAG mutant, activates Rab7 GTPase activity, a downstream effector of the C/Vps complex involved in both autophagic and endocytic maturation.13
It is important to note that the effects of the UVRAG-Beclin 1 interaction differ in important respects from the UVRAG-C/Vps complex interaction in autophagy regulation. As mentioned above, a mutation in UVRAG that prevents Beclin 1 binding generally has only minor effects on autophagosome maturation, but pronounced effects on autophagosome formation. In contrast, a mutation in UVRAG that prevents C/Vps complex association behaves in an opposite manner. These results imply that mammalian UVRAG functions through two successive activities in autophagy modulation: one involving Beclin 1 in the biogenesis of autophagosomes and the other involving the C/Vps complex in the biogenesis of autolysosomes.
UVRAG in Endosomal Trafficking
Although UVRAG is considered to be autophagosome-related, it is not exclusively present at the autophagosomes. Rather, a large percentage of UVRAG colocalizes with the C/Vps complex at the endosomal compartments carrying the early-endosomal marker Rab5 and EEA1.13 The cellular outcome of this endosome-associated UVRAG was further investigated. Several lines of evidences strongly support the notion that mammalian UVRAG has a major role in promoting endosomal trafficking.13 First, UVRAG is required for receptor-mediated endocytosed epidermal growth factor (EGF) trafficking. Overexpression of UVRAG causes enhanced endocytic trafficking and EGF receptor (EGFR) degradation, whereas depletion of UVRAG is associated with sustained receptor signaling. Notably, UVRAG in mammals is recognized as a putative tumor suppressor.21,22 Tumor cells with enhanced expression of UVRAG show reduced growth rate in culture and in nude mice. Moreover, aberrant levels of UVRAG protein exist in many cancers, although their correlation with carcinogenesis remains to be established.15-18 The tumor suppressor activity of UVRAG has been largely attributed so far to its Beclin 1 binding and the resultant autophagy induction. Whether UVRAG-involved receptor signaling also contributes to malignant transformation is of interest to be investigated. Further support for UVRAG as a trafficking molecule was demonstrated in a DQ-BSA dequenching assay.13 BSA is an endocytosed cargo destined for lysosomal degradation. In UVRAG-overexpressing cells, the dynamics of BSA digestion is significantly increased, which may reflect enhanced trafficking and/or increased lysosome activity. In addition to endocytic trafficking, UVRAG was found to promote the intracellular protein sorting from the TGN to lysosomes as measured by lysosomal protein cathepsin D processing (Fig. 1B and C). Recovery of UVRAG expression in UVRAG-deficient HCT116 cells enhances cathepsin D maturation (Fig. 1B and C). Given the general effects of UVRAG on multiple trafficking routes, it is speculated that UVRAG may regulate trafficking steps at a converged point. Indeed, UVRAG is found to promote endosomal fusion in vitro and in vivo,13 a process that coordinates most of the lysosomal directed membrane dynamics and is also regulated by the C/Vps complex.
How might UVRAG regulate endosomal trafficking processes? UVRAG appears to have close association with the endosome machinery. In addition to the C/Vps complex, an interaction between UVRAG and the early-endosomal GTPase Rab5 has been detected (unpublished data). Interestingly, UVRAG preferentially bound to the active (GTP-bound) form of Rab5 rather than the wild-type and inactive (GDP-bound) form of Rab5, which suggests that UVRAG possibly functions as a downstream effector of Rab5. Notably, the UVRAG-associated C/Vps complex mediates Rab5-to-Rab7 conversion, thereby serving as a functional tie between Rab5 and Rab7 in mammalian cells.33 In this context, it is postulated that UVRAG interaction with the C/Vps complex may enhance the ability of the C/Vps complex in Rab conversion, which ultimately promotes endosomal sorting and protein trafficking. In support of this view, disruption of the C/Vps complex binding site prevents UVRAG from activating Rab7 and inducing endosome maturation. However, the detailed molecular function of the C/Vps complex in UVRAG-mediated vesicle transport needs to be further defined. Alternatively or additionally, UVRAG may act in concert with as-yet-undefined factors to assemble specific complexes on endosomal membranes. Together, the studies discussed above indicate that UVRAG constitutes an important component in both autophagic and endosomal membrane trafficking.
Conclusions
In recent years, there has been an explosion of information on the cellular machinery that drives the sorting of autophagic cargoes into autophagosomes and controls the release of these cargos into the lysosomal lumen. Parallel studies have revealed that many endolysosomal-related proteins, including the AAA ATPases SKD1,10 Hrs,12 Vti1,11 and the small GTPases Rab5,34 and Rab7,8,9 have evolved to function in the autophagy pathway and interact with distinct components of the autophagy machinery. One of the key players in both endosomal and autophagosomal pathways is UVRAG, a modular protein that serves diverse functions in the cell ranging from autophagy and tumor suppression to membrane trafficking. Although the role of UVRAG in promoting membrane trafficking awaits further investigation, important insights have been gained through the observation that the binding of UVRAG to the C/Vps complex is required for UVRAG-mediated trafficking activity. An intriguing question is how UVRAG coordinates its distinct membrane-associated (autophagosome and endosome) trafficking activities. How are these complex processes coupled mechanistically? It is also of interest as to how endosomal and autophagic vesicle trafficking destined for the same organelle is differentially regulated. Future studies on these topics will undoubtedly illuminate new views on autophagy as an integral part of the lysosome system, and will, hopefully, suggest new approaches to a number of human diseases associated with compromised membrane trafficking.
Acknowledgements
We would like to thank B. Levine, M.J. Hardwick, S. Virgin, S. Field, T. Yoshimori and Y. Ohsumi for providing reagents. We also thank Steven Lee for proofreading. This work was partly supported by U.S. Public Health Service grants CA82057, CA91819, CA31363, CA106156, RR00168 (J.U.J.). C.L. is a Leukemia & Lymphoma Society Fellow.
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–88. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eskelinen EL. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- 7.Saftig P, Beertsen W, Eskelinen EL. LAMP-2: a control step for phagosome and autophago-some maturation. Autophagy. 2008;4:510–2. doi: 10.4161/auto.5724. [DOI] [PubMed] [Google Scholar]
- 8.Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–97. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 9.Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–48. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 10.Nara A, Mizushima N, Yamamoto A, Kabeya Y, Ohsumi Y, Yoshimori T. SKD1 AAA ATPase-dependent endosomal transport is involved in autolysosome formation. Cell Struct Funct. 2002;27:29–37. doi: 10.1247/csf.27.29. [DOI] [PubMed] [Google Scholar]
- 11.Surpin M, Zheng H, Morita MT, Saito C, Avila E, Blakeslee JJ, Bandyopadhyay A, Kovaleva V, Carter D, Murphy A, Tasaka M, Raikhel N. The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell. 2003;15:2885–99. doi: 10.1105/tpc.016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamai K, Tanaka N, Nara A, Yamamoto A, Nakagawa I, Yoshimori T, Ueno Y, Shimosegawa T, Sugamura K. Role of Hrs in maturation of autophagosomes in mammalian cells. Biochem Biophys Res Commun. 2007;360:721–7. doi: 10.1016/j.bbrc.2007.06.105. [DOI] [PubMed] [Google Scholar]
- 13.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008 doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perelman B, Dafni N, Naiman T, Eli D, Yaakov M, Feng TL, Sinha S, Weber G, Khodaei S, Sancar A, Dotan I, Canaani D. Molecular cloning of a novel human gene encoding a 63-kDa protein and its sublocalization within the 11q13 locus. Genomics. 1997;41:397–405. doi: 10.1006/geno.1997.4623. [DOI] [PubMed] [Google Scholar]
- 15.Bekri S, Adelaide J, Merscher S, Grosgeorge J, Caroli-Bosc F, Perucca-Lostanlen D, Kelley PM, Pebusque MJ, Theillet C, Birnbaum D, Gaudray P. Detailed map of a region commonly amplified at 11q13→q14 in human breast carcinoma. Cytogenet Cell Genet. 1997;79:125–31. doi: 10.1159/000134699. [DOI] [PubMed] [Google Scholar]
- 16.Goi T, Kawasaki M, Yamazaki T, Koneri K, Katayama K, Hirose K, Yamaguchi A. Ascending colon cancer with hepatic metastasis and cholecystolithiasis in a patient with situs inversus totalis without any expression of UVRAG mRNA: report of a case. Surg Today. 2003;33:702–6. doi: 10.1007/s00595-002-2567-y. [DOI] [PubMed] [Google Scholar]
- 17.Ionov Y, Nowak N, Perucho M, Markowitz S, Cowell JK. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer Cells with microsatellite instability. Oncogene. 2004;23:639–45. doi: 10.1038/sj.onc.1207178. [DOI] [PubMed] [Google Scholar]
- 18.Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol. 2008 doi: 10.1016/j.humpath.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mul JJ, Pledger WJ, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi Y, Meyerkord CL, Wang HG. BARgaining membranes for autophagosome formation: Regulation of autophagy and tumorigenesis by Bif-1/Endophilin B1. Autophagy. 2008;4:121–4. doi: 10.4161/auto.5265. [DOI] [PubMed] [Google Scholar]
- 21.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 22.Liang C, Feng P, Ku B, Oh BH, Jung JU. UVRAG: a new player in autophagy and tumor cell growth. Autophagy. 2007;3:69–71. doi: 10.4161/auto.3437. [DOI] [PubMed] [Google Scholar]
- 23.Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–27. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato TK, Rehling P, Peterson MR, Emr SD. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol Cell. 2000;6:661–71. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 25.Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci USA. 2000;97:9402–7. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–62. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson MR, Emr SD. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic. 2001;2:476–86. doi: 10.1034/j.1600-0854.2001.20705.x. [DOI] [PubMed] [Google Scholar]
- 29.Kim BY, Kramer H, Yamamoto A, Kominami E, Kohsaka S, Akazawa C. Molecular characterization of mammalian homologues of class C Vps proteins that interact with syntaxin-7. J Biol Chem. 2001;276:29393–402. doi: 10.1074/jbc.M101778200. [DOI] [PubMed] [Google Scholar]
- 30.Kim BY, Ueda M, Kominami E, Akagawa K, Kohsaka S, Akazawa C. Identification of mouse Vps16 and biochemical characterization of mammalian class C Vps complex. Biochem Biophys Res Commun. 2003;311:577–82. doi: 10.1016/j.bbrc.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 31.Pulipparacharuvil S, Akbar MA, Ray S, Sevrioukov EA, Haberman AS, Rohrer J, Kramer H. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J Cell Sci. 2005;118:3663–73. doi: 10.1242/jcs.02502. [DOI] [PubMed] [Google Scholar]
- 32.Richardson SC, Winistorfer SC, Poupon V, Luzio JP, Piper RC. Mammalian late vacuole protein sorting orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol Biol Cell. 2004;15:1197–210. doi: 10.1091/mbc.E03-06-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–49. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 34.Ravikumar B, Imarisio S, Sarkar S, O'Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. 2008;121:1649–60. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]