Abstract
Human APOBEC3G (A3G) and activation-induced deaminase (AID) belong to a family of DNA-cytosine deaminases. While A3G targets the last C in a run of C’s, AID targets C in the consensus sequence WRC (W is A or T and R is a purine). Guided by the structures of the A3G carboxyl-terminal catalytic domain (A3G-CTD), we identified two potential regions (region 1 and region 2) that may interact with DNA and swapped the corresponding regions between a variant of A3G-CTD and AID. The resulting hybrids were expressed in Escherichia coli and two different genetic assays and a biochemical assay were used to determine the sequence selectivity of the hybrids in promoting C to T mutations. The results show that while the 10 amino acid region 2 of A3G was its principal sequence-specificity determinant, region 1 of A3G enhanced the target cytosine preference conferred by region 2. In contrast, neither of the two regions in AID individually or in combination were sufficient to confer the DNA sequence preference of this protein upon A3G. Instead, introduction of AID sequences in A3G relaxed the sequence-specificity of the latter protein. Our results show that the sequence-selectivity of APOBEC family of enzymes is determined by at least two separate sequence segments and there may be additional regions of the protein involved in DNA sequence recognition.
Keywords: Somatic hypermutations, Class-switch recombination, Mutagenesis, Domain-swaps
1. Introduction
The APOBEC family of genes of higher vertebrates is unusual in that their products act as DNA mutators in performing their normal biological function. One member of the family, activation-induced deaminase (AID), is required to create base substitution mutations (called somatic hypermutations) in immunoglobulin genes during antibody maturation, while another member, APOBEC3G (A3G), causes C to T mutations in the cDNA copy of the HIV-1 genome [1,2]. All members of the APOBEC family for which enzymatic function has been established, are DNA- or RNA-cytosine deaminases and when AID or A3G are expressed in Escherichia coli, they overwhelmingly promote C:G to T:A transitions especially in uracil-DNA glycosylase deficient, ung strains [3,4]. This is explained by the replication of U•G mispairs created due to deamination that escape DNA repair.
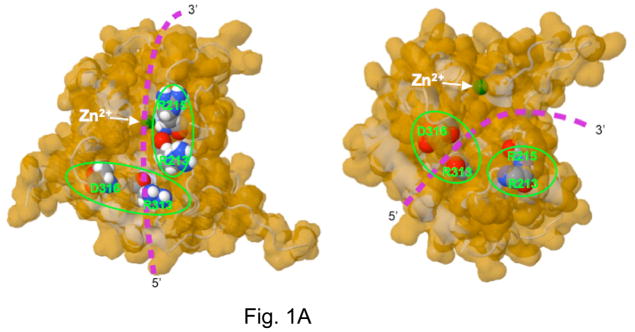
Recent solution structures of APOBEC3G carboxyl terminal domain (A3G CTD) by NMR spectroscopy [5–7] and crystal structure of nearly the same domain by X-ray diffraction methods [8] provide insight into how this class of enzymes interact with DNA and perform catalysis. The active site of A3G CTD contains a Zn2+ ion coordinated by Cys288, Cys291, His257 and a water molecule. Glu259 hydrogen bonds with the water molecule and presumably activates it for an attack at C4 of the target cytosine when single-stranded DNA binds to the enzyme. However, neither structure was solved in the presence of DNA and there are significant differences between the predictions for the path of DNA in the two structures (Fig. 1A).
Figure 1.
Putative DNA-binding regions of APOBEC3G-CTD and construction of hybrids.
A. The structure of the A3G-CTD (2K3A variant) determined by NMR analysis (Left) and of the WT A3G-CTD determined by X-ray crystallography (Right) are shown. The predicted path of DNA in each structure and the position of two amino acid residues in the putative DNA-binding segments are shown to highlight the differences between the two structures. Adapted from a figure in reference [8].
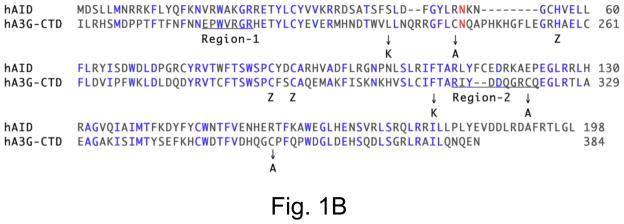
B. Alignment of human AID sequence with carboxyl terminal residues of human APOBEC3G. The residues shared by the two enzymes are shown in blue. Five amino acids replaced to make the 2K3A variant of A3G-CTD are identified by arrows below the sequence and the replaced amino acids placed below the arrows. The putative DNAbinding regions 1 and 2 of the two proteins are underlined.
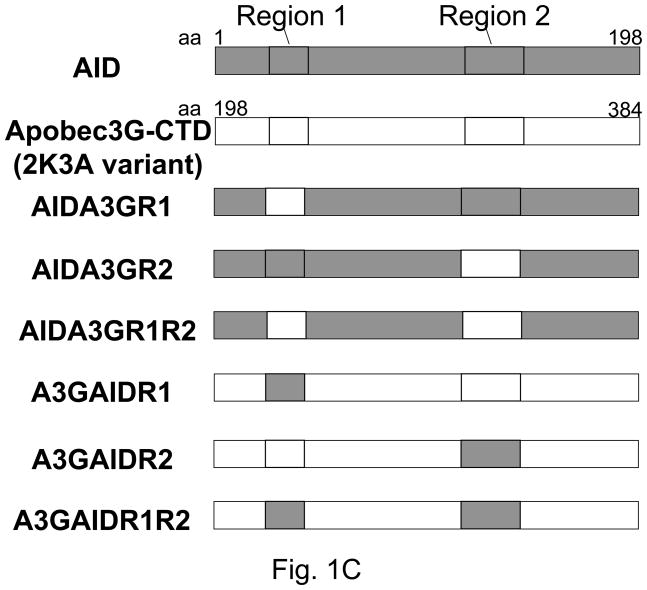
C. Schematic of construction of hybrids between AID and A3G-CTD. AID sequences are shown as grey boxes, while A3G sequences are shown as open boxes.
These differences arise in part because the structures contain loops that appear to acquire different 3D structures in solution (NMR) compared to crystalline form. In particular, the position of the “loop 1” is significantly different in the two structures. The two groups also relied on different criteria to predict where DNA may bind. Chen et al [5] used NMR chemical shift perturbations when 5′-CCT oligomer was added to A3G CTD to identify amino acid residues that may interact with DNA. In contrast, Holden et al [8] relied on the presence of a deep groove in the X-ray structure to identify candidate DNA-binding residues. Both groups performed site-directed mutagenesis and deamination or mutagenesis assays to narrow the list of residues to one (region 1; X-ray structure) or two (region 1 and region 2; NMR structure) putative DNA-binding regions (Fig. 1A and 1B).
APOBEC enzymes target cytosines for deamination in preferred sequence contexts. A3G strongly prefers cytosines in a run of C’s usually targeting the last base in the run [9,10]. AID also shows a significant sequence bias. In uracil excision-defective mice [11] and during incubation of purified enzyme with DNA [12], AID preferentially converts cytosines in WRC sequences to uracil (W is A or T and R is purine). Other APOBECs have been less well-studied but also have DNA distinct sequence preferences such as TC for APOBEC1 [9] and WC for APOBEC3DE [13]. These studies show that the APOBEC family of enzymes have evolved to target cytosines in different sequence contexts and hence their sequence recognition domain(s) may be pliable enough to be changed through genetic manipulations.
To test this possibility we constructed segment swaps between AID and A3G and determined their mutational sequence-specificity. We found that the targeting specificity of both the enzymes could in fact be altered and that both the regions 1 and 2 (R1 and R2) affected the specificities of these enzymes.
2. Materials and Methods
2.1 Strains and plasmids
Escherichia coli K-12 strain BH260 was constructed by introducing ung151::Tn10 allele from BW504 into CC102 (F’ lacI− lacZ461-2 proB+ ara; Δ(lac-proB)XIII) by P1 transduction. Strain BH143 is recA+-derivative of the cloning strain DH10B and has been described before. BL-21 DE3 codon+RIL (Stratagene) is an E. coli B strain with phage λ lysogen containing the RNA polymerase gene and a plasmid containing the tRNA genes for rare codons for arginine, isoleucine and leucine.
The plasmids pGST-AID and pGST-A3G-CTD-2K3A were constructed by respectively cloning the entire AID gene or codons 198 to 384 of the 2K3A variant of A3G [14] into the SmaI and XhoI sites of pGEX-6P-2. A whole plasmid PCR mutagenesis strategy [15] was used to create hybrids between AID and APOBEC3G-CTD-2K3A. The mutagenic primers and reverse primers used for PCR mutagenesis are listed in Supplementary data Table S1. The plasmids were amplified using the mutagenic primer pairs for 18 cycles and the DNA was treated with restriction enzyme DpnI to cleave the parental DNA. The resulting DNA mixture was transformed into E. coli strain BH143 and transformants were selected on plates with carbenicillin. The mutagenesis strategy was designed to introduce new NcoI and BssHII sites in the hybrids and hence plasmid DNA from independent clones was analyzed by restriction digestion with the enzymes NcoI and BssHII and the mutations confirmed by DNA sequencing. For unknown reasons, we could not make AIDA3GR1R2 and the A3GAIDR2 hybrids by this procedure and hence these hybrid genes were synthesized by DNA2.0 (Menlo Park, CA) and cloned into pGEX-6P-2.
2.2 Lac+ papillation assay
The procedure described by Miller [16] was used to perform the Lac+ papillation assay. Briefly, the vector plasmid or plasmids expressing AID/A3G hybrids were introduced into the strain BH260 by transformation. The transformants were grown in glucose minimal media until OD600 reached ~0.3 and dilutions of the cultures were spread on MacConkey-lactose plates containing carbenicillin in such a way that approximately 100–500 colonies grew up on each 150 mm plate. The formation of papillae on each plate was monitored over several days and the papillae appeared as dark red foci within the colonies. The colonies containing one or more papillae were counted as positive and the ratio of such colonies to total number cells on each plate was calculated.
2.3 Rifampicin forward mutation assay
The vector plasmid or plasmids expressing AID/A3G hybrids were introduced into the strain BW504 and independent transformants were selected on LB plates containing 100 μg/mL carbenicillin. Independent transformants were picked and grown overnight in LB media containing carbenicillin. The following day, the cultures were diluted 1000-fold in LB media containing with carbenicillin and grown at 37 °C. When the OD600 reached ~0.03 transcription of AID/A3G genes were induced by the addition of IPTG to 1 mM. The cultures were grown overnight and the following day appropriate dilutions of each culture were spread on LB plates containing carbenicillin to determine the total number of viable cells. The cultures were also spread on LB plates containing 100 μg/mL rifampicin (Sigma-Aldrich) to determine the number of rifampicin-resistant (RifR) colonies. Mutation frequency was obtained by dividing the number of RifR colonies by the number of viable bacteria present in the culture.
2.4 Amplification of rpoB gene and Sequence analysis
A single colony from the center of each rifampicin-containing plate was selected for further analysis. The colony was solely chosen for its location on the plate and not its size to avoid any bias against smaller colonies. The cells from each colony were dispersed in 50 μL distilled water and 5 μL of this mixture was used to perform PCR. A part of the rpoB gene [called cluster 2 in reference [17]] was amplified using the following primers- 5′-CGTCGTATCCGTTCCGTTGG-3′ and 5′-TTCACCCGGATACATCTCGTC-3′, and the products were purified using PCR purification kit (Millipore Corporation). The purified PCR products were sequenced using 5′-CGTGTAGAGCGTGCGGTGAAA-3′ as primer. The sequences of mutants were compared to the wild-type (WT) E. coli K-12 sequence using MacVector software (MacVector, Inc.) and the mutations were identified.
2.5 Purification of AID/A3G hybrid proteins
The plasmid clones containing the GST-tagged hybrids were introduced into the strain BL-21 (DE3 codon+RIL) and the transcription of the fused gene was induced by adding IPTG to a final concentration of 0.2 mM. The cells were grown at 24 °C for four hours and harvested by centrifugation. The cell pellet was resuspended in 30 mL 1X Tris buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.5) along with Complete EDTA-free Protease Inhibitors (Roche Diagnostics, Indianapolis, IN) and lysozyme (1 μg/mL). The cells were broken by sonication and the suspension was cleared by centrifugation. The lysate was passed over a glutathione-sepharose column to bind the GST-containing proteins and the bound proteins were washed with ~ 20 column volumes of 1X Tris buffer followed by elution with 20 mM reduced glutathione [SIGMA-Aldrich; in 50 mM Tris-HCl, pH7.5]. Proteins from different fractions were separated on a 12% SDS-Polyacrylamide gel to identify fractions with hybrid proteins. The fractions containing the protein of interest were pooled; the proteins were dialyzed using slide-A-Lyzer dialysis cassettes (Thermo Scientific, Rockford, IL). The proteins were concentrated using Amicon Ultra centrifugal filter devices (Milipore, Billerica, MA). The proteins were purified further by ion exchange chromatography over a MonoQ column (GE Healthcare). The protein was eluted using 0 to 1 M gradient of NaCl and collected in 1 mL fractions. The hybrid proteins appeared as a prominent peak in the UV absorption profile and the appropriate fractions were pooled, concentrated and equilibrated with the storage buffer [25 mM Tris –HCl, pH 7.5; 1 mM EDTA, 1 mM DTT, 10 % glycerol].
2.6 Deamination Activity Assay
The oligomer CCC-17 (Supplementary data Table S2) was used as substrate for the different AID/A3G hybrids. One pmol of oligomer was labeled at the 5′ end with 33P or 32P and incubated at 37 °C with 2 μg of AID/A3G hybrid proteins in a 10 μL volume in the deamination reaction buffer (25 mM Tris- HCl, pH 7.5; 1 mM DTT; 1 mM EDTA). When the time course of the deamination reactions was studied, the reaction mixtures were scaled up six-fold. At various times, 10 μL aliquots were removed from the reaction tube and the reactions were stopped by the addition of 1,10-phenanthroline (SIGMA-Aldrich) to 5 mM. One unit of E. coli UDG (New England Biolabs) was added to the reactions and incubation was continued 37 °C for 45 min. The reactions were terminated by addition of either NaOH to 0.1 M or piperidine to 10% followed by heating to 95 °C for 7 minutes. The products of the reactions were separated on a 20% sequencing gel. The gel was scanned using Typhoon 9210 scanner. ImageJ software was used to quantify intensities of products -UCC- and -CCU- and of the CCC-17 substrate. For each lane, the intensity of the substrate band was added to intensities for deamination products to obtain total amount of DNA and this numbers was used to calculate the percent of each product in the reaction.
3. Results
3.1 Genetic system to study sequence specificity of AID and A3G
Cupples et al. have described strains designated CC101 through CC111 that can be scored for Lac− to Lac+ reversion and allow detection of mutators that cause specific base substitutions or small addition/deletions [18,19]. In particular, the strain CC102 reverts lacZ allele to lacZ+ through a C:G to T:A transition (G in coding strand; Supplementary Fig. S1). We set out to investigate whether AID and A3G were mutators in this strain because the target cytosine is within a CCC sequence context which is the preferred sequence for A3G [9,10], but disfavored by AID [12]. We expected that A3G should be a good mutator in this genetic system, but not AID. An ung mutation was introduced into CC102 (resulting strain is BH260) to allow all U•G mispairs created by AID or A3G to be replicated to T:A without repair.
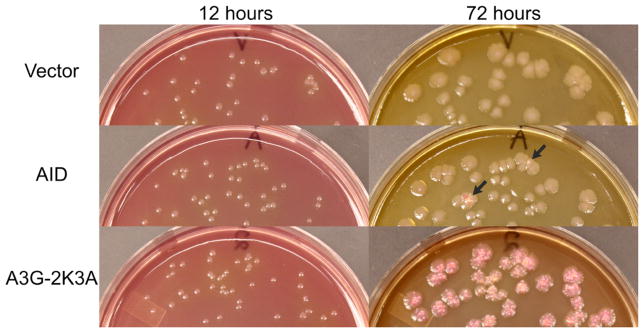
The full-length human AID and a variant of the carboxyl-terminal domain (CTD) of human A3G called 2K3A were fused with GST in the vector pGEX6P2 and expressed in BH260. The 2K3A variant was used here instead of the wild-type A3G-CTD because of greater solubility of the former protein ([5,14] and data not shown) and will be referred to as A3G throughout the manuscript for textual convenience. A3G promoted papillation of colonies within 48 hours and by 72 hours nearly every colony containing A3G displayed one or more papillae (Fig. 2 and Supplementary Fig. S2). In contrast, AID-promoted only a modest amount of papillation (Fig. 2). However, the ability of AID to promote papillation was significantly higher than the vector control (Fig. 3). Representative Lac+ revertants from cells with A3G were sequenced and were found to contain the expected G:C to A:T mutation (data not shown). The strong mutator phenotype of A3G and a much weaker phenotype of AID are consistent with sequence preferences of these proteins, CCC and WRC, respectively [3,9,12]). These results confirmed the expectation that A3G would be a better mutator in this system than AID.
Figure 2.
Lac+ papillation assay.
McConkey-lactose plates with colonies containing vector plasmid, or plasmids containing GST fusions of AID or A3G-CTD (2K3A variant) are shown after 12 or 72 hr growth. The host was BH260 (=CC102 ung::Tn10). Two colonies expressing AID that show p ink papillae after 72 hr are marked with black arrows. All colonies containing A3G show papillation after 72 hr.
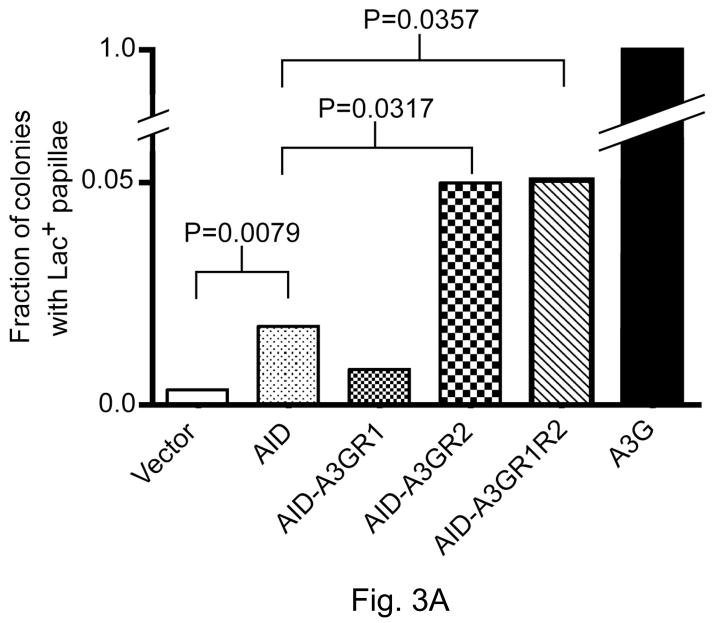
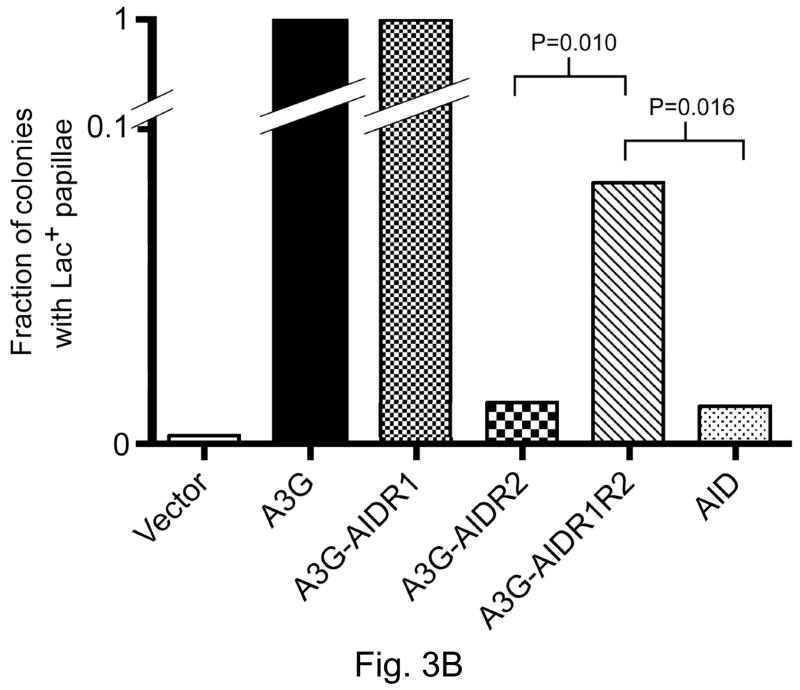
Figure 3.
Ability of AID/ A3G hybrids to promote Lac+ papillation.
The ratio, number of colonies with one or more papillae divided by the total number of colonies on a plate, is shown on Y-axis. Each bar represents the mean from five independent plates. P-values were calculated at the 95% confidence interval using the Mann-Whitney test.
A. AID hybrids with regions of A3G.
B. A3G hybrids with regions of AID.
3.2 Properties of AID hybrids containing A3G regions in Lac+ papillation assay
To determine whether either of the putative DNA-binding segments from A3G (Fig. 1B) was sufficient to confer the 5′-CCC sequence preference upon AID, we replaced in separate constructs Region-1 and Region-2 of AID with corresponding segments from A3G. In a third constructs both the Region-1 and Region-2 of AID was replaced with the corresponding segments from A3G. The resulting hybrids were respectively referred to as AIDA3GR1, AIDA3GR2 and AIDA3GR1R2, and were tested for their ability promote Lac+ papillae.
AIDA3GR1 and AIDA3GR2 performed significantly differently from each other in the papillation assay. While AID-A3GR1 was a weaker mutator than WT AID in the papillation assay, AID-A3GR2 was a significantly stronger mutator (at 95% confidence level). However, neither was as good a mutator as A3G (Fig. 3). The ability of AIDA3GR1R2 to promote papillation was indistinguishable from that of AIDA3GR2 suggesting that the presence of Region-1 in the former construct did not improve significantly the ability of AID to target CCC sequences.
3.3 Properties of AID hybrids with A3G regions in rifampicin-resistance assay
We used rifampicin-resistance (RifR) forward mutation assay to test the ability of the AID/A3G to target cytosines in many different sequence contexts [17]. When the AID-A3G hybrids were compared using this assay, all three hybrids were more mutagenic than AID, but were weaker mutagens than A3G (Supplementary Fig. S2). It is noteworthy that AIDA3GR1, which is a poor mutator in the Lac+ papillation assay (Fig. 3), is slightly more mutagenic than either AIDA3GR2 or AIDA3GR1R2 in the RifR assay. This suggests that the inability of AIDA3GR1 to revert mutation in lacZ-416 is likely to be due the sequence context of the target of cytosine rather than enzyme activity, protein stability and other biochemical factors.
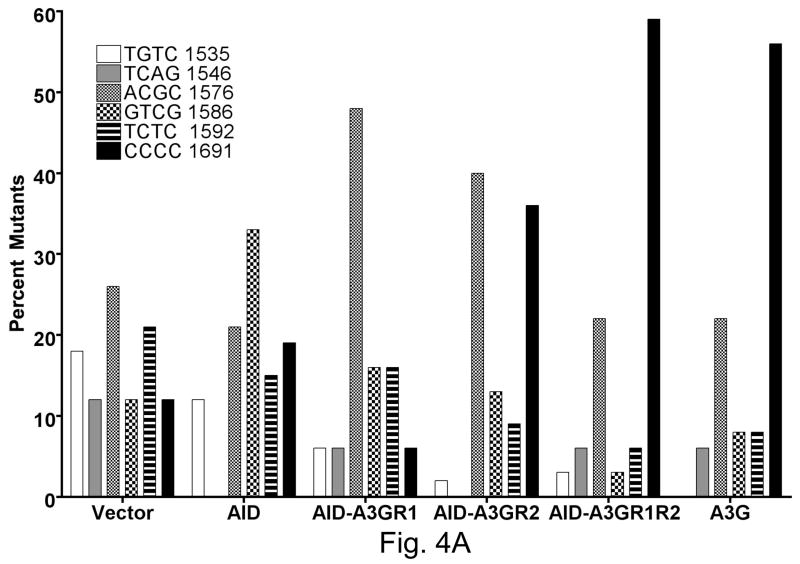
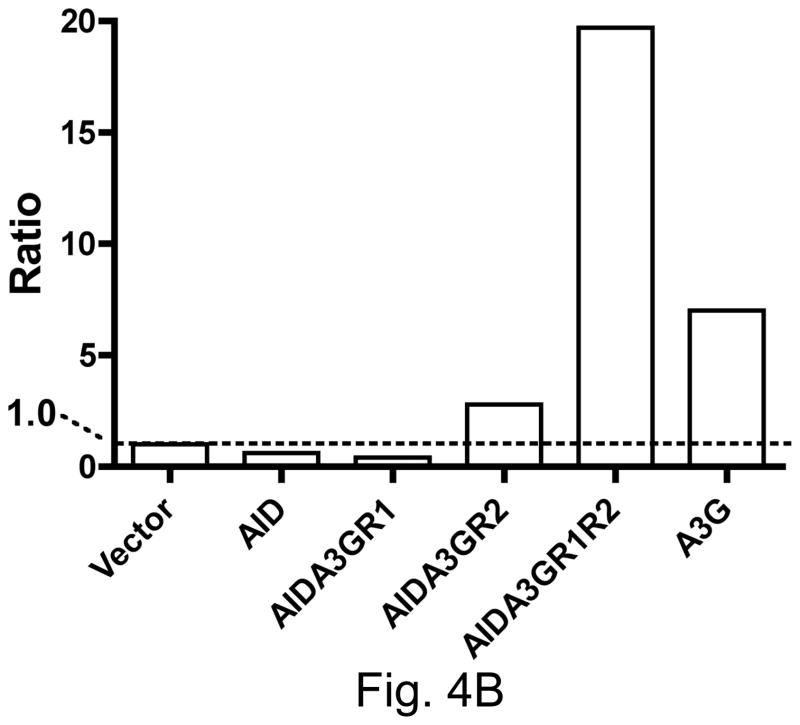
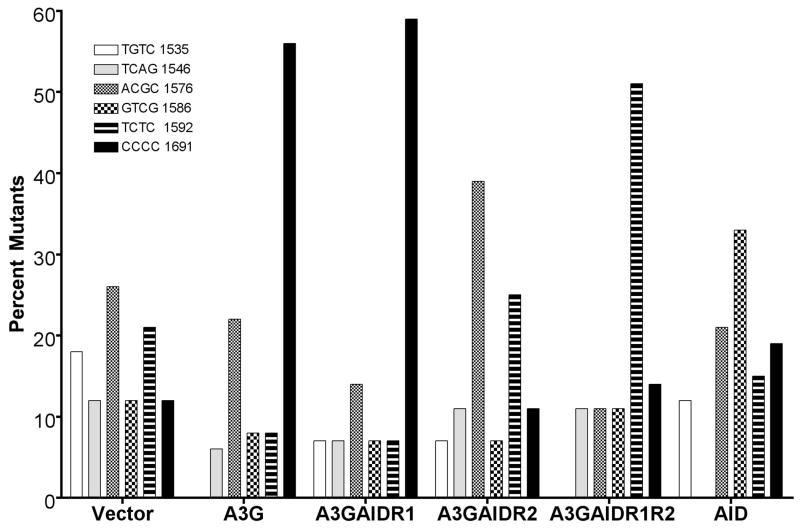
A section of the rpoB was sequenced in a number of independent RifR mutants obtained with each hybrid construct and a C:G to T:A mutation was found in >98% RifR mutants at one of six positions. While AID created the largest fraction of mutations at position 1586 (~ one third of total), A3G created a majority of the mutations at position 1691 (~60%; Fig. 4A). The mutation spectra of the three hybrid constructs were significantly different from each other (Fig. 4A). Overall, while the AIDA3GR1 mutation spectrum resembled the spectrum of AID, the AIDA3GR2 and AIDA3GR1R2 spectra resembled the spectrum of A3G. The most informative sequence contexts for cytosine among these sites are rpoB sequence positions 1586 and 1691. At the former position the cytosine is in the template strand and has WRC sequence context (5′ GTAT/5′ATAC; target cytosine underlined), while it is CCC at the latter position (5′ CCCC). Thus a ratio of the fraction of mutations at 1691 to those at 1586 provides a rough measure of the tendency of a AID-A3G hybrid to have sequence specificity similar to A3G.
Figure 4.
Mutation spectrum of AID hybrids in rpoB gene.
Frequency of mutations at six positions in the rpoB gene is shown. The position of the target cytosine within the gene and the sequence context of this cytosine are indicated. At positions 1546 and 1586 the deaminated cytosine is in the template strand. The sequence shown is in the non-template strand.
A. AID with regions from A3G.
B. Ratio of the frequency of mutations at position 1691 to that at position 1586.
Both the regions 1 and 2 influenced the ratio of mutations at 1691 and 1586 (Fig. 4B). AIDA3GR2 was significantly more A3G-like than AID in its targeting and AIDA3GR1R2 was even more biased towards mutations at C1691 (Fig. 4B). AIDA3GR1R2 promoted a higher percentage of mutations at C1691 and lower percentage at G1586 than AIDA3GR2. This suggests that when both regions 1 and 2 of A3G are introduced into AID, they cooperate in the resulting protein to create a more A3G-like sequence selectivity. However, AIDA3GR1 does not show A3G-like sequence selectivity and hence region 1 of A3G is not sufficient to confer its preference for run of cytosines in DNA upon AID.
3.4 Genetic Properties of A3G hybrids with AID regions
We wished to determine whether the conclusions based on experiments described above were valid for hybrids with reverse configuration; i.e. for A3G hybrids in which region 1 or 2 (or both) of that protein was replaced with corresponding regions from AID. All three possible hybrids (A3GAIDR1, A3GAIDR2 and A3GAIDR1R2) were made and tested in the Lac+ papillation assay and RifR mutation assay.
The results of the papillation assay with these hybrids were somewhat different than those with AID containing R1, R2 (or both) of A3G. Like the corresponding AID hybrid, when R1 of A3G was replaced with R1 of AID, the papillation frequency remained unchanged. In contrast, but again similar to the corresponding AID hybrid, when R2 of A3G was replaced with R2 of AID, the papillation frequency was reduced to approximately same low level as AID (Fig. 3B). Thus replacement of A3GR2 with AIDR2 resulted in a loss of targeting at CCC sequences. However, when both R1 and R2 of A3G were replaced with corresponding regions of AID, the papillation frequency was intermediate between A3G and AID (Fig. 3B). The observation that A3GAIDR1R2 promotes Lac+ papillation at a frequency significantly higher than A3GAIDR2 suggests that the region R1 of AID does play some role in DNA binding, target selectivity or catalyic activity of the hybrid enzyme.
The A3G hybrids with the putative AID DNA-binding regions also behaved somewhat differently than the AID hybrids in the rifampicin-resistance assay (Fig. 5). While A3GAIDR1 had a mutation spectrum similar to A3G, A3GAIDR2 behaved significantly differently than AID in this assay. The latter hybrid most frequently mutated cytosines at positions 1576 (39%) and 1592 (25%) neither of which is in a WRC sequence context. Strikingly, only 7% of the mutations were found at the WRC sequence at 1586 and the same percentage was found at 1691 (Fig. 5). The low number of mutations found at these positions made presentation of data as ratio of mutations at 1691 and 1586 position not very meaningful. The mutation spectrum of A3GAIDR1R2 was also considerably different than of either A3G or AID. This hybrid promoted most mutations at cytosine at 1592 (51%), and few mutations at 1586 (hotspot for AID) or 1691 (hotspot for A3G). These results suggest that the sequence-specificities of A3GAIDR2 and A3GAIDR1R2 are significantly different than those of either AID or A3G.
Figure 5.
Mutation spectrum of A3G hybrids in rpoB gene. Frequency of mutations at six positions in the rpoB gene is shown. The labeling of the bar graph is similar to that in Fig. 4A.
3.5 Biochemical Properties of A3G Hybrids
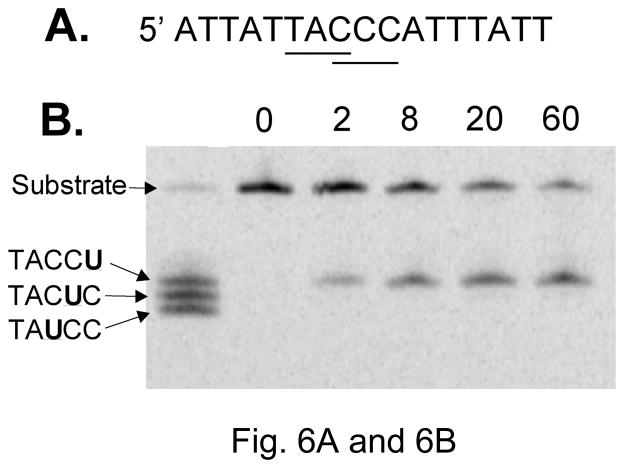
To confirm biochemically that the hybrid deaminases had altered sequence specificities, A3G-CTD variant 2K3A and its hybrids with AID were purified as GST fusions and used in a deaminase activity assay. A Coomasie brilliant blue G-250 stained gel of the proteins and a Western blot of the same using anti-A3G antibodies is shown in Supplementary Fig. S3. The substrate DNA oligomer contained overlapping WRC and CCC sites to allow monitoring of deamination at either site (Fig. 6A). In this assay, A3G readily converted the third cytosine in CCC to uracil but did not convert the first or the second cytosine in the sequence at detectable levels (Fig. 6B and 6C). In fact none of the hybrids of A3G deaminated the second C in the sequence at a significant level (data not shown) and hence deamination of only the outer two cytosines was quantified. The results show that A3G preferentially deaminated the third C over the first C (Fig. 6C).
Figure 6.
Sequence selectivity of A3G in cytosine deamination
A. Sequence of the oligonucleotide substrate. The overlapping WRC and CCC sequence within it are underlined.
B. Image of a gel scan of kinetics of cytosine deamination. The numbers above lanes indicate the length of incubation in minutes. The left-most lane contains products from three oligomers with uracils replacing one of three cytosines in CCC.
C. The kinetics of cytosine deamination in the CCC-17 oligomer. Based on quantification of data in Fig. 6B.
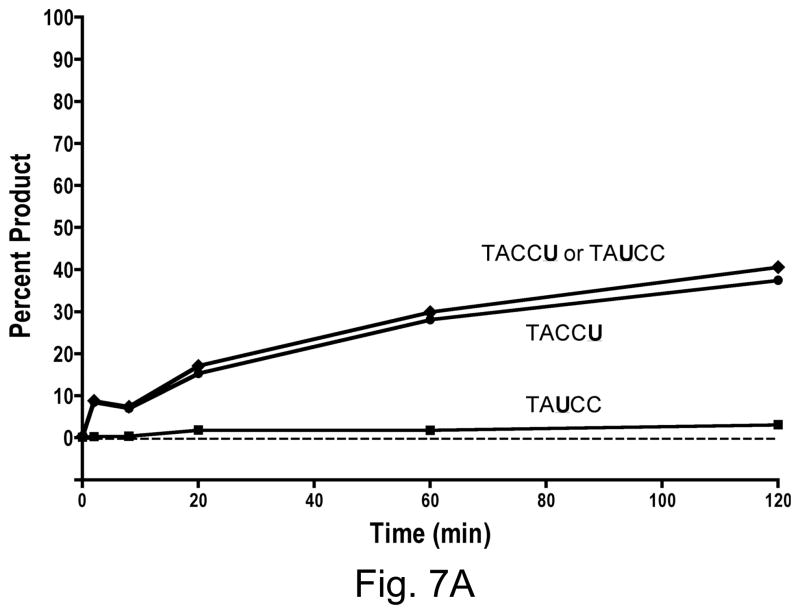
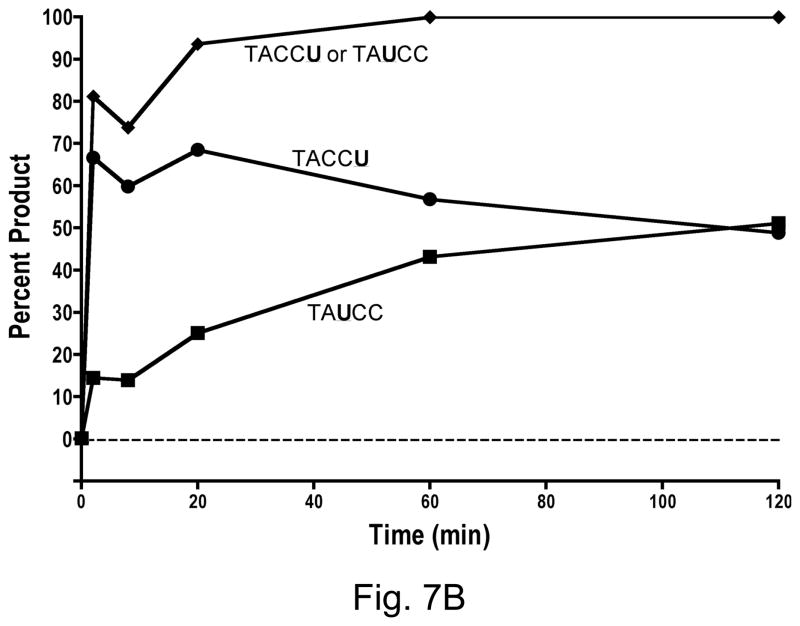
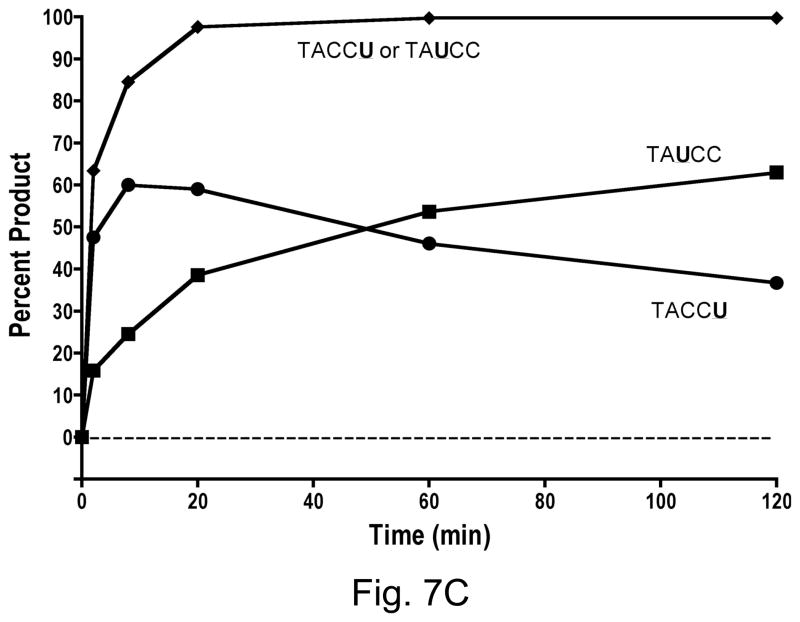
A3GAIDR1 had similarly strong preference for the last cytosine in CCC over the first C (Fig. 7A). It differed from A3G in only one small respect- there was very small but detectable amount of conversion of the first C to a U by this hybrid. In contrast to A3GAIDR1, the A3GAIDR2 and A3GAIDR1R2 hybrids showed strong differences in substrate specificity from A3G. Both these enzymes converted the cytosine in WRC motif (i.e. first C in CCC) to U at significant levels. By 20 min in the reaction both A3GAIDR2 and A3GAIDR1R2 had converted ~90% of the substrate at the first C or the third C (or both) to uracil (Fig. 7B and 7C). Both these hybrids still retained the ability to deaminate the third C in CCC and hence retained the A3G-like specificity. These results show that replacement of R1 and/or R2 regions of A3G with the corresponding regions in AID did not result in a replacement of A3G sequence preference with AID sequence preference, but instead resulted in a broadening of sequence specificity to include WRC in addition to CCC.
Figure 7.
Kinetics of cytosine deamination by A3G hybrids. Quantification of products created by A3G hybrids. The data points marked “TACCU or TAUCC” are based on the sum of intensities of bands corresponding to TACCU and TAUCC.
A. A3GAIDR1
B. A3GAIDR2
C. A3GAIDR1R2
4. Discussion
We swapped two protein segments between the DNA-cytosine deaminases AID and A3G-CTD (variant 2K3A) and determined the sequence preference of the resulting hybrids for deaminating cytosines using genetic and biochemical assays. Our results show that a 10–12 amino acid segment within A3G (region 2, Fig. 1A) plays a strong role in determining sequence specificity. Both the genetic assays used showed that when the A3G sequences are introduced into the AID sequence framework, the sequence specificity of the resulting hybrid (AIDA3GR2) is significantly more A3G-like (target sequence CCC) than AID-like (target sequence WRC). When region 1 of this hybrid is further replaced with the corresponding region in A3G (hybrid AIDA3GR1R2), the frequency of mutations in rpoB at the CCC site (position 1691) increased from 36% (17 out of 47 mutants) to 59% (19 out of 32; Fig. 4A). Thus A3G region 1 did not change the specificity of the AIDA3GR2 hybrid, only enhanced its preference for the CCC sequence.
These results are consistent with recently published conclusions of Kohli et al. [20] where they described the construction and characterization of a similar AIDA3GR2 hybrid. Like us, these investigators also found that replacement of region 2 of AID with the corresponding region of A3G resulted in a change in rpoB mutation spectrum, increasing mutations at position 1691 and decreasing them at position 1586 (sequence- WRC). Furthermore, they found that purified AIDA3GR2 hybrid had highest activity on a substrate containing two 5-methylcytosines (mC) followed by a cytosine (mCmCC). The mCs were used in this oligomer because some studies have shown [21]- but not others [22]- that they are deaminated by AID at much lower rates than cytosine. Regardless, the hybrid enzyme deaminated the third cytosine in this sequence suggesting that it preferred a CCC sequence context. Kohli et al. [20] did not investigate the role of region 1 of A3G in sequence selectivity. Together with the results presented here we conclude A3G region 2 is a “portable” sequence specificity determinant that may be used to alter the specificity of other members of the APOBEC family of enzymes, but its sequence selectivity is enhanced by a separate 8 amino acid segment in A3G (region 1).
In contrast to A3G region 2, AID region 2 does not represent a portable sequence-specificity determinant. Although the Lac+ papillation assay shows that replacement of region 2 of A3G with corresponding region in AID (hybrid A3GAIDR2) had substantially lower ability to mutate within CCC sequence context, the rpoB mutation spectrum of this hybrid shows that mutations at WRC sequences (position 1586) were not enhanced (Fig. 5A). Instead, this hybrid caused increased mutations at a CGC site (position 1576). The hybrid A3GAIDR1R2 also did not show enhanced mutagenesis at position 1586, but displayed most number of mutations at CTC sequence (position 1592). This apparent relaxation of sequence-specificity of A3G due to region swaps with AID was confirmed by biochemical experiments in which the ability of the purified hybrid proteins to deaminate at WRC and CCC sites were compared. The results show that none of the hybrids had lost their ability to deaminate cytosine in the CCC sequence context. Instead, when A3G region 2 was replaced with corresponding region in AID the hybrid proteins displayed additional ability to deaminate within WRC sequence (Fig. 7B and 7C). Therefore unlike A3G, the region 2 of AID (residues 209 through 216) is not sufficient to switch the specificity of A3G from CCC to WRC.
It is unlikely that the reason why region 2 of AID is unable to act as a portable sequence-specificity domain is because additional amino acids flanking region 2 are required for base sequence recognition. This segment is flanked by four amino acids on either side that are identical between these two proteins (Fig. 1A). Consequently, extending the swaps to flanking residues is unlikely to alter the sequence-specifities of the proteins. Instead, this region of AID may interact with other parts of the protein including region 1 to perform sequence recognition.
We also found evidence for the existence of additional sequence-specificity determinants within A3G. The N-terminus of the A3G-CTD used in our experiments is at residue 198 in the full-length A3G and was based on the observation that residues 198 through 384 formed an active enzyme [14]. A recent report suggested that extending the protein by seven amino acids (i.e. residues 191 through 384) increased the activity of the protein and hence we constructed and tested a similar longer version of protein. We found the purified protein to be more active in biochemical assays as previously reported and promoted RifR mutants at a higher frequency compared to the shorter version ([7]; data not shown). Interestingly, the longer version overwhelmingly promoted mutations at position 1691 of rpoB (22 out of 24; 92%). This is a significantly higher frequency of targeting of CCC than that promoted by the shorter version (56%; Fig. 4A) and is similar to that of the full-length A3G [3].
As discussed above, although A3GAIDR2 and A3GAIDR1R2 hybrids had acquired the ability to deaminate within WRC sequence context, they retained also the ability deaminate the last C in CCC context. What was also noticable was that the CCC to CCU deamination seemed to occur faster than WRC to WRU deamination, peak earlier and then decrease at later times (Fig. 7B and 7C). The eventual decrease of product corresponding to CCU is readily explained by the fact that the oligomer was end labeled at the 5′ end and the WRC sequence is closer to the 5′ end than the CCC. Consequently, when the uracil in WRU is converted to a nick in DNA for gel analysis, the downstream sequence is lost.
However, the apparent targeting of CCC earlier than WRC (Fig. 7B and 7C) is difficult to explain. Several studies have shown that full-length APOBEC3G binds single stranded DNA and processively tracks it in either direction. However, it shows a preference for deamination during 3′ to 5′ tracking and hence substrate sequences nearer the 5′ end are acted upon by the enzyme more frequently than sequences nearer 3′ end [23,24]. Our results show an opposite sequence preference. We do not know whether this is somehow caused by the fact that the WRC and CCC sequence overlap in our oligomer. It is also unclear whether this is caused by our use of the carboxyl terminal domain of A3G, instead of the full length protein. It is not known whether A3G 2K3A variant acts processively in its deamination reactions and has the same directional bias as the full-length protein. Additional experiments are needed to clarify this point.
Recently, Wang et al. [25] used an E. coli lacZ reversion assay similar to the one described above to screen for AID mutants with increased ability to deaminate cytosines in DNA (“up mutants”). The mutants were obtained from a pool of random mutants covering the entire AID gene and were identified based on increased ability to cause Lac+ papillation. A number of such mutants containing multiple mutations were identified and characterized in terms of their catalytic activity, targeting within the rpoB gene and in terms of their effects on antibody diversification and chromosome translocations. None of the AID up mutants characterized in this study showed altered sequence specificity and hence their increased ability to cause Lac+ papillation was attributed to the increased catalytic activity [25]. The results of Wang et al. differed from those described above in one important respect. Unlike our finding that A3G was a stronger mutator in the Lac+ papillation assay, these investigators did not find significant differences between the abilities of A3G and AID in promoting papillation [25].
This difference could be due to differences in the experimental strategies and criteria used to define Lac+ papillated colonies. First, we used an ung derivative of CC102 for our assay, while Wang et al. used the ung+ host CC102. Repair of U:G mispairs in the latter strain may have decreased the differences between AID and A3G enzymes. Second, we scored the Lac+ papillated colonies at 72 hr (3 days) post-plating while Wang et al. [25] may have waited as long as 7 days before scoring the colonies. The longer wait may have helped cells expressing AID to “catch-up” with A3G in terms of papillation. Finally, Wang et al. [25] counted the number of papillae within each colony and reported the average number of papillae per colony. We found this procedure to be unreliable as papillaes within a colony often merged with each other making the counting procedure inaccurate. Consequently, we scored all colonies that had at least one red outgrowing colony (ie. papilla) as positive.
It is not known whether the sequence-specificities of AID, APOBEC3G and other enzymes in this family are somehow related to their biological function. It is interesting to note that all characterized AID enzymes target WRC(Y) or a closely related sequence. Both murine and human AIDs target 5′-WRC(Y) sequences [26]. This seems also to be true in chickens (in the SHM mode; [27]). Zebra fish AID deaminates cytosines in WRCY sequences in vitro [28] and can complement chicken AID−/− mutant for GC/SHM [29], suggesting a sequence specificity that overlaps with or is identical to WRC(Y). Furthermore, analysis of catfish heavy chain hypermutations shows that they occur predominantly in AGCT and AGCA sequences which are closely related to the consensus WRC(Y) [30]. We have shown here that the sequence specificity of AID can be altered. It would be interesting to determine whether AID with such altered sequence specificity would perform SHM and CSR efficiently. Similarly, it would also be interesting to determine whether APOBEC3G constructs containing AID sequences that display a broadened sequence specificity are as effective in their anti-viral effects as wild-type APOBEC3G. It should possible to use hybrids between AID and APOBEC3G to determine such relationships between the biological functions of these enzymes and their DNA sequence specificities.
Supplementary Material
Acknowledgments
Some of the initial experiments reported here were performed by Mala Samaranayake (New England Biolabs, Ipswitch, MA). This work was supported by grants from National Institutes of Health (GM 57200 and CA 97899).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Navaratnam N, Sarwar R. An overview of cytidine deaminases. Int J Hematol. 2006;83:195–200. doi: 10.1532/IJH97.06032. [DOI] [PubMed] [Google Scholar]
- 2.Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- 3.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 4.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 5.Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 6.Furukawa A, Nagata T, Matsugami A, Habu Y, Sugiyama R, Hayashi F, Kobayashi N, Yokoyama S, Takaku H, Katahira M. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO J. 2009;28:440–451. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harjes E, Gross PJ, Chen KM, Lu Y, Shindo K, Nowarski R, Gross JD, Kotler M, Harris RS, Matsuo H. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J Mol Biol. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol. 2004;337:585–596. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 10.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 11.Xue K, Rada C, Neuberger MS. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J Exp Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 13.Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen KM, Martemyanova N, Lu Y, Shindo K, Matsuo H, Harris RS. Extensive mutagenesis experiments corroborate a structural model for the DNA deaminase domain of APOBEC3G. FEBS Lett. 2007;581:4761–4766. doi: 10.1016/j.febslet.2007.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiner MP, Costa GL. Rapid PCR site-directed mutagenesis. PCR Methods Appl. 1994;4:S131–136. doi: 10.1101/gr.4.3.s131. [DOI] [PubMed] [Google Scholar]
- 16.Miller JH. A Short Course in Bacterial Genetics. Cold Spring harbor Laboratory; Cold Spring Harbor, NY: 1992. [Google Scholar]
- 17.Garibyan L, Huang T, Kim M, Wolff E, Nguyen A, Nguyen T, Diep A, Hu K, Iverson A, Yang H, Miller JH. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amst) 2003;2:593–608. doi: 10.1016/s1568-7864(03)00024-7. [DOI] [PubMed] [Google Scholar]
- 18.Cupples CG, Cabrera M, Cruz C, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics. 1990;125:275–280. doi: 10.1093/genetics/125.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cupples CG, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci U S A. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohli RM, Abrams SR, Gajula KS, Maul RW, Gearhart PJ, Stivers JT. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J Biol Chem. 2009;284:22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 23.Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′ --> 5′ on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 24.Coker HA, Petersen-Mahrt SK. The nuclear DNA deaminase AID functions distributively whereas cytoplasmic APOBEC3G has a processive mode of action. DNA Repair (Amst) 2007;6:235–243. doi: 10.1016/j.dnarep.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Wang M, Yang Z, Rada C, Neuberger MS. AID upmutants isolated using a high-throughput screen highlight the immunity/cancer balance limiting DNA deaminase activity. Nat Struct Mol Biol. 2009;16:769–776. doi: 10.1038/nsmb.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 27.Rogozin IB, Sredneva NE, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. III. Somatic mutations in the chicken light chain locus. Biochim Biophys Acta. 1996;1306:171–178. doi: 10.1016/0167-4781(95)00241-3. [DOI] [PubMed] [Google Scholar]
- 28.Basu U, Wang Y, Alt FW. Evolution of phosphorylation-dependent regulation of activation-induced cytidine deaminase. Mol Cell. 2008;32:285–291. doi: 10.1016/j.molcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatterji M, Unniraman S, McBride KM, Schatz DG. Role of activation-induced deaminase protein kinase A phosphorylation sites in Ig gene conversion and somatic hypermutation. J Immunol. 2007;179:5274–5280. doi: 10.4049/jimmunol.179.8.5274. [DOI] [PubMed] [Google Scholar]
- 30.Yang F, Waldbieser GC, Lobb CJ. The nucleotide targets of somatic mutation and the role of selection in immunoglobulin heavy chains of a teleost fish. J Immunol. 2006;176:1655–1667. doi: 10.4049/jimmunol.176.3.1655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.