Abstract
Posttranslational modifications of histones such as methylation, acetylation, and phosphorylation regulate chromatin structure and gene expression. Here we show that protein kinase C-related kinase 1 (PRK1) phosphorylates histone H3 at threonine 11 (H3T11) upon ligand-dependent recruitment to androgen receptor (AR) target genes. PRK1 is pivotal to AR function since PRK1 knockdown or inhibition impedes AR-dependent transcription. Blocking PRK1 function abrogates androgen-induced H3T11 phosphorylation, and inhibits androgen-induced demethylation of histone H3. Moreover, serine 5-phosphorylated RNA polymerase II is no longer observed at AR target promoters. Phosphorylation of H3T11 by PRK1 accelerates demethylation by the Jumonji C (JmjC) domain-containing protein JMJD2C. Thus, phosphorylation of H3T11 by PRK1 establishes a novel chromatin mark for gene activation, identifying PRK1 as a gatekeeper of AR-dependent transcription. Importantly, levels of PRK1 and phosphorylated H3T11 correlate with Gleason scores of prostate carcinomas. Finally, inhibition of PRK1 blocks AR-induced tumour cell proliferation, making PRK1 a promising therapeutic target.
Keywords: PRK1, androgen receptor, histone phosphorylation, prostate cancer
The N-terminal tails of histones are subject to a plethora of posttranslational modifications such as methylation, acetylation, and phosphorylation by specific chromatin-modifying enzymes1. During gene expression, these modifications influence chromatin structure to facilitate the assembly of the RNA polymerase II transcription machinery1, 2. Androgen receptor (AR)-dependent gene expression is characterized by changes in chromatin modifications such as removal of repressive methyl marks from lysine 9 of histone H3 (H3K9)3, 4. The demethylases Jumonji C (JmjC) domain-containing protein JMJD2C4 and lysine specific demethylase 1 (LSD1)3 co-operate to remove trimethyl and di-and monomethyl marks from H3K9, respectively. In addition, other chromatin modifications such as acetylation of lysines 9 and 14 of histone H3 (H3K9/K14)5, are necessary for AR-dependent transcription. However, little is known about the upstream regulators that govern these histone modifications. Since protein kinase C-related kinase 1 (PRK1) controls AR-dependent gene expression6, we asked whether PRK1 signalling regulates histone modifications at AR target genes.
Results
PRK1 controls AR-dependent gene expression
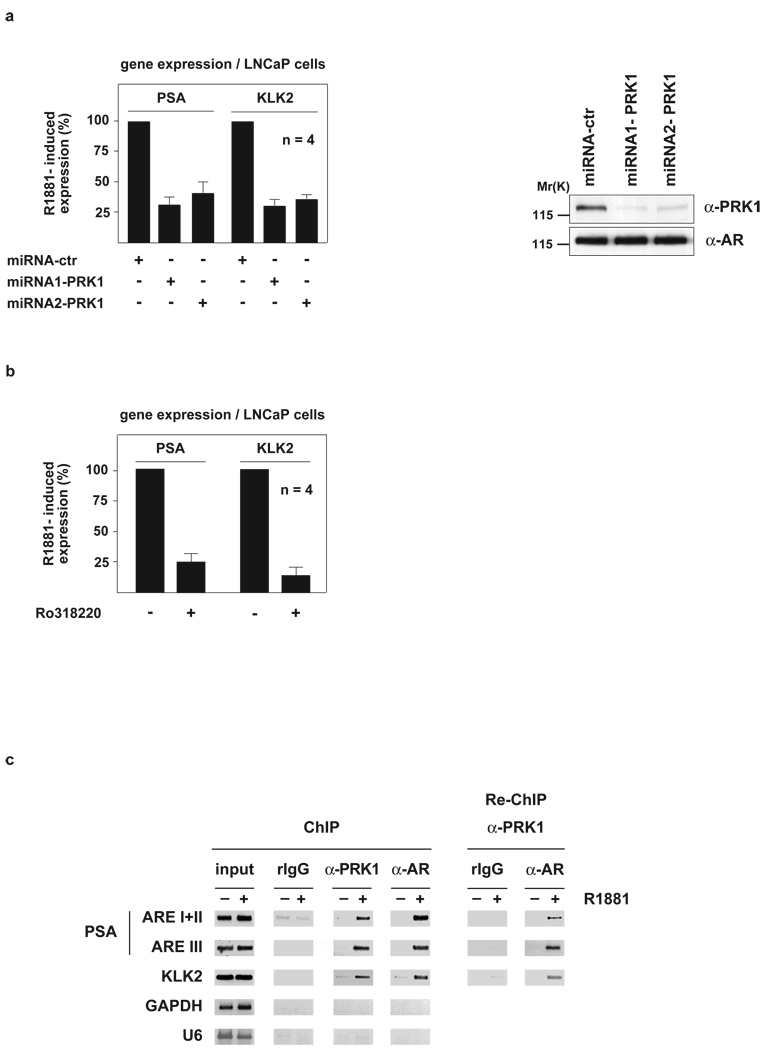
To initiate our study, we analysed the effect of PRK1 knockdown on the expression of endogenous AR target genes. LNCaP prostate tumour cells were transduced with lentiviruses expressing miRNAs directed against PRK1, which results in an efficient and specific down-regulation of endogenous PRK1 (Fig. 1a, right panel). Quantitative RT-PCR analyses demonstrate that the reduction of PRK1 levels strongly impairs androgen-induced expression of endogenous AR target genes such as Prostate Specific Antigen (PSA) or Kallikrein 2 (KLK2), but does not affect unrelated control genes (Fig. 1a, left panel and see Supplementary Information, Fig. S1a). In addition, treatment with the PRK1 inhibitor Ro3182206 severely impedes androgen-induced expression of AR target genes, showing that the kinase activity of PRK1 is essential for AR function (Fig. 1b and see Supplementary Information, Fig. S1b). Similarly, miRNA-mediated knockdown of PRK1 or treatment with Ro318220 results in a strong decrease in ligand-induced expression of various AR-dependent reporters (Supplementary Information Fig. S1c, d).
Figure 1.
PRK1 controls AR-dependent gene expression and associates with chromatin. LNCaP cells were cultivated in the presence or absence of the AR agonist R1881. miRNA-mediated PRK1 knockdown (a) or the PRK1 inhibitor Ro318220 (b) reduce expression of the endogenous PSA and KLK2 genes (a, left panel, b). Western blot analysis (a, right panel) verified the specific miRNA-mediated knockdown of PRK1. Bars represent mean +SD (n=4). ChIP and Re-ChIP (c) using the indicated antibodies demonstrate androgen-dependent association of AR and PRK1 at promoters of AR-regulated genes. The precipitated chromatin was amplified by PCR using primers flanking the AREs in the promoter region of the PSA and KLK2 genes, or the promoters of the unrelated GAPDH and U6 genes.
PRK1 associates with chromatin
To investigate whether PRK1 associates with chromatin in vivo, LNCaP cells were subjected to chromatin immunoprecipitation (ChIP) in the presence or absence of the AR agonist R1881. PRK1 and AR associate with the androgen response elements (AREs) located in the promoters of the PSA and KLK2 genes in a ligand-dependent manner (Fig. 1c, left panel). Recruitment of PRK1 to chromatin is specific, since neither DNA corresponding to the promoters of the unrelated GAPDH and U6 genes (Fig. 1c, left panel), nor to a region between the enhancer and promoter or exon 4 of the PSA gene is enriched (Supplementary Information, Fig. S1e). To show that PRK1 and AR are present in the same complex on the PSA and KLK2 promoters, R1881-treated LNCaP cells were subjected to sequential chromatin immunoprecipitation (Re-ChIP), first with α-AR and then with α-PRK1 antibody. Importantly, the ARE-containing regions are specifically enriched, demonstrating that PRK1 and AR form a complex on chromatin in a ligand-dependent manner (Fig. 1c, right panel and see Supplementary Information, Fig. S1f).
PRK1 phosphorylates H3T11
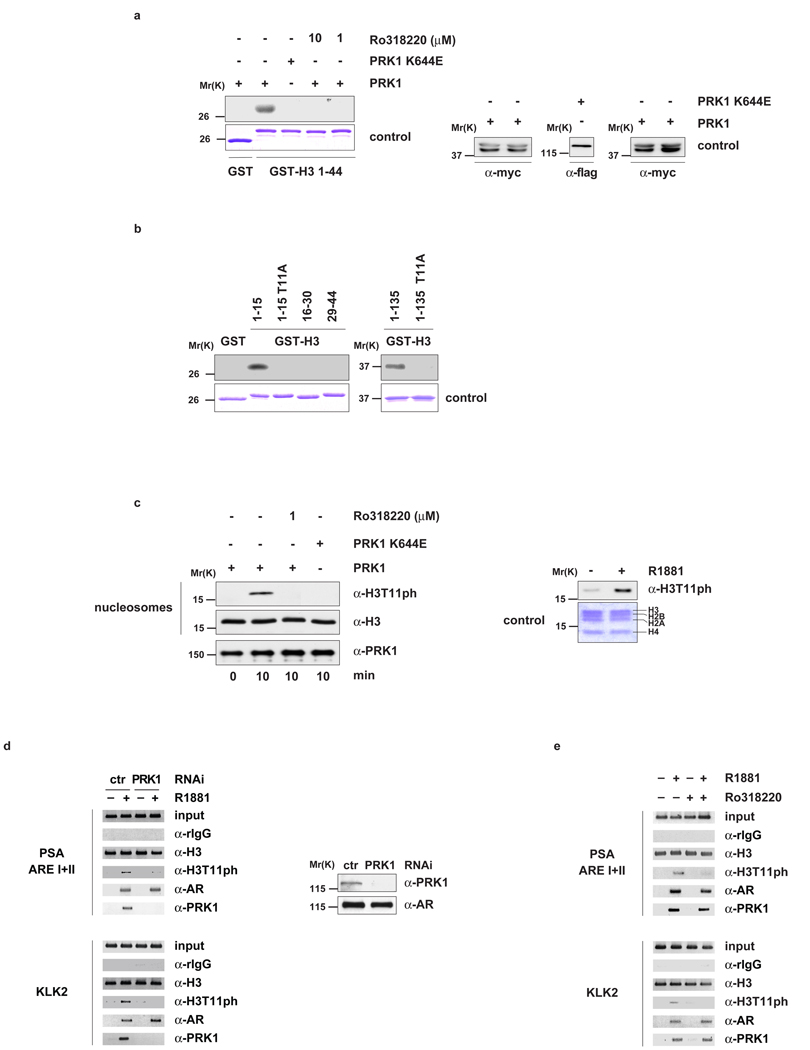
To understand how association of PRK1 and AR with chromatin results in increased gene expression, we tested whether PRK1 directly phosphorylates the N-terminal tail of histone H3. Myc-PRK1 and the flag-tagged kinase dead mutant PRK1 K644E6 were immunoprecipitated from 293 cell lysates with α-myc or α-flag antibody, respectively (Supplementary Information, Fig. S1g, h) and incubated with bacterially expressed and purified GST-H3 1–44 or GST control protein. GST-H3 1–44 is phosphorylated by PRK1 but not by PRK1 K644E (Fig. 2a). The GST control protein is not phosphorylated, thus demonstrating specificity. Furthermore, addition of Ro318220 completely blocks the phosphorylation of GST-H3 1–44 by PRK1 (Fig. 2a). Deletion mapping revealed that only the fragment of histone H3 spanning amino acid residues 1 to 15 (H3 1–15), but not H3 16–30 or H3 29–44, is phosphorylated by purified recombinant PRK1 (Fig. 2b). More importantly, mutation of threonine 11 to alanine in either H3 1–15 (H3 1–15 T11A) or full length H3 (H3 1–135 T11A) abolishes phosphorylation, demonstrating that PRK1 targets histone H3 at threonine 11 (H3T11) (Fig. 2b). In addition, we incubated nucleosomes purified from HeLa cells with recombinant PRK1 in the presence or absence of Ro318220. Western blot analysis, performed with an α-H3T11ph specific antibody (see ref. 7 and Supplementary Information, Fig. S2a–d) demonstrates that PRK1 phosphorylates nucleosomes at H3T11 (Fig. 2c). This phosphorylation is blocked by Ro318220 (Fig. 2c, left panel). Furthermore, nucleosomes purified from LNCaP cells treated with R1881 were phosphorylated at H3T11 demonstrating that phosphorylation at H3T11 occurs in a ligand-dependent manner in vivo (Fig. 2c, right panel and see Supplementary Information, Fig. S2e).
Figure 2.
PRK1 phosphorylates histone H3 at threonine 11 (H3T11). Bacterially expressed GST and GST-H3 (a, b) or nucleosomes from HeLa cells (c, left panel) were incubated for the indicated time with active PRK1 or the kinase dead mutant PRK1 K644E in the presence or absence of the inhibitor Ro318220. Coomassie blue staining (a and b, lower panels) and Western blot analyses (a, right panel) show the amounts of GST fusion proteins and PRK1 used. Western blots were decorated with the indicated antibodies (c, left panel). Nucleosomes purified from LNCaP cells cultivated in the presence or absence of R1881 for 30 minutes were analysed in Western blot (c, right panel). For ChIP (d, e), LNCaP cells were cultivated in the presence or absence of the AR agonist R1881, transfected with either siRNA (d) or treated with or without Ro318220 (e) as indicated, and subjected to ChIP with the indicated antibodies. The precipitated chromatin was amplified by PCR using primers flanking AREs in the promoter region of the PSA and KLK2 genes. Western blot analysis (d, right panel) verified the specific siRNA-mediated knockdown of PRK1.
To determine whether PRK1 controls phosphorylation of H3T11 at promoters of AR-regulated genes in vivo, LNCaP cells were first transfected with either an unrelated control siRNA or a siRNA directed against PRK1, in the presence or absence of R1881, and then subjected to ChIP. Addition of ligand results in phosphorylation of H3T11 at the AREs of the PSA and KLK2 promoters (Fig. 2d, left panel and see Supplementary Information, Fig. S2f). Androgen-induced phosphorylation at H3T11 is PRK1-dependent since it is blocked by knockdown of PRK1. PRK1 depletion is specific and does not affect the levels of endogenous AR (Fig. 2d, right panel). To corroborate that androgen-induced phosphorylation of H3T11 is executed by PRK1, LNCaP cells were cultivated in the presence or absence of Ro318220 and subjected to ChIP. As expected, Ro318220 efficiently blocks ligand-induced phosphorylation of H3T11 (Fig. 2e and see Supplementary Information, Fig. S2g). Taken together, these data demonstrate that PRK1 phosphorylates H3T11. Importantly, the phosphorylation of H3T11 associates with AR-dependent gene expression, thus introducing H3T11ph as a novel chromatin mark for transcriptional activation.
PRK1 controls modifications of histone H3
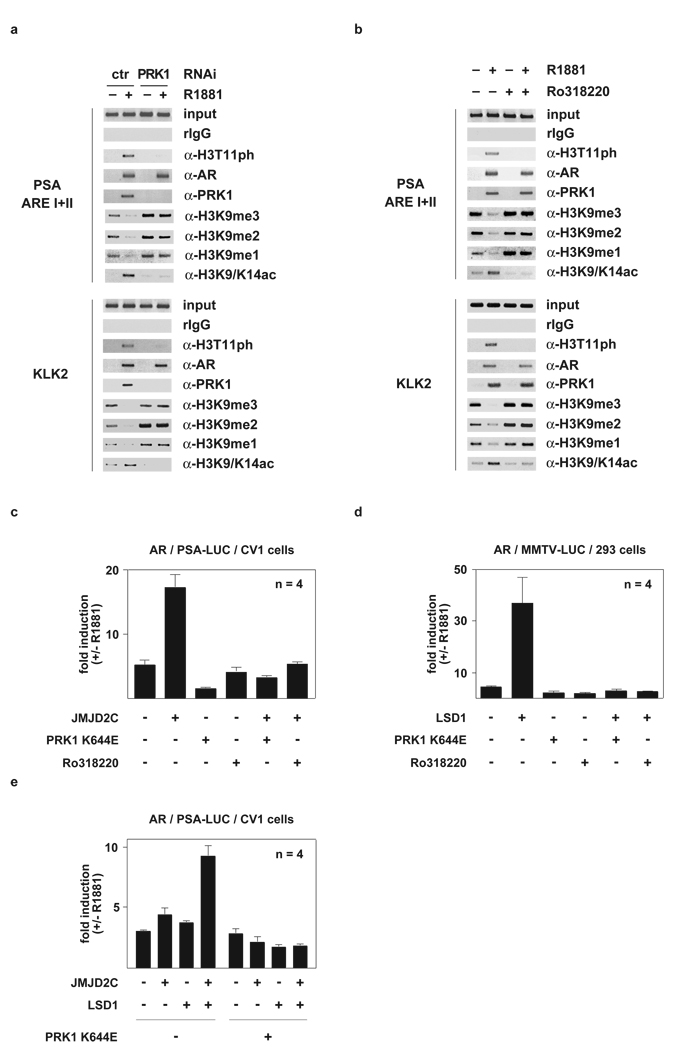
Since ligand-dependent expression of AR target genes demands removal of repressive methyl marks from H3K93, 4 and acetylation of histone H3K9/K145, we analysed whether PRK1 controls changes in these histone marks. Therefore, LNCaP cells cultivated in the presence or absence of R1881 were transfected with either an unrelated control siRNA or a siRNA directed against PRK1 and subjected to ChIP. Ligand-induced demethylation of tri-, di-, and monomethyl H3K9 at the AREs of the PSA and KLK2 promoters is severely impaired by PRK1 knockdown (Fig. 3a and see Supplementary Information, Figs. S2h, S3a, S3c–h). Furthermore, ligand-induced acetylation of H3K9/K14 is also blocked (Fig. 3a). Similarly, inhibition of PRK1 activity by Ro318220 results in loss of demethylation of H3K9 and acetylation of H3K9/K14 (Fig. 3b and see Supplementary Information, Figs. S3b, S3i–n), demonstrating that the kinase activity of PRK1 is pivotal in controlling these alterations in histone marks at AR target genes.
Figure 3.
PRK1 controls modifications of histone H3 and AR-dependent gene expression. For ChIP (a, b) and transient transfections (c, d, e), cells were cultivated in the presence or absence of the AR agonist R1881 and the inhibitor Ro318220 (b, c, d), or transfected with siRNA (a). ChIP analyses were performed with the indicated antibodies. The precipitated chromatin was amplified by PCR using primers flanking AREs in the promoter region of the PSA and KLK2 genes. For transient transfections, CV1 (c, e) or 293 (d) cells were co-transfected with AR expression plasmid and AR-dependent reporters. Bars represent mean +SD (n=4).
As previously shown, JMJD2C4 and LSD13 remove repressive methyl marks from H3K9 during AR-dependent transcription. Since PRK1 controls demethylation of H3K9, we investigated the interplay between PRK1 and the demethylases during gene expression in transient transfections. Co-expression of AR with either JMJD2C (Fig. 3c and see ref. 4) or LSD1 (Fig. 3d and see ref. 3) results in a strong ligand-dependent activation of the PSA-LUC or MMTV-LUC reporters. Co-activation by the demethylases is abrogated by PRK1 K644E, acting as a dominant negative mutant, or by treatment with Ro318220. To examine the effect of PRK1 K644E on co-operative stimulation of AR activity by JMJD2C and LSD1, we expressed both demethylases in limited amounts, which alone do not activate AR, but together induce a strong AR superactivation4. As shown in Figure 3e, PRK1 K644E blocks co-operative stimulation of AR activity. The control reporter TK-LUC is not affected by PRK1 (Supplementary Information, Fig. S4a–c). Collectively, these data demonstrate that PRK1 signalling controls transcriptional activation of AR by the demethylases JMJD2C and LSD1.
PRK1 controls transition to the initiation complex
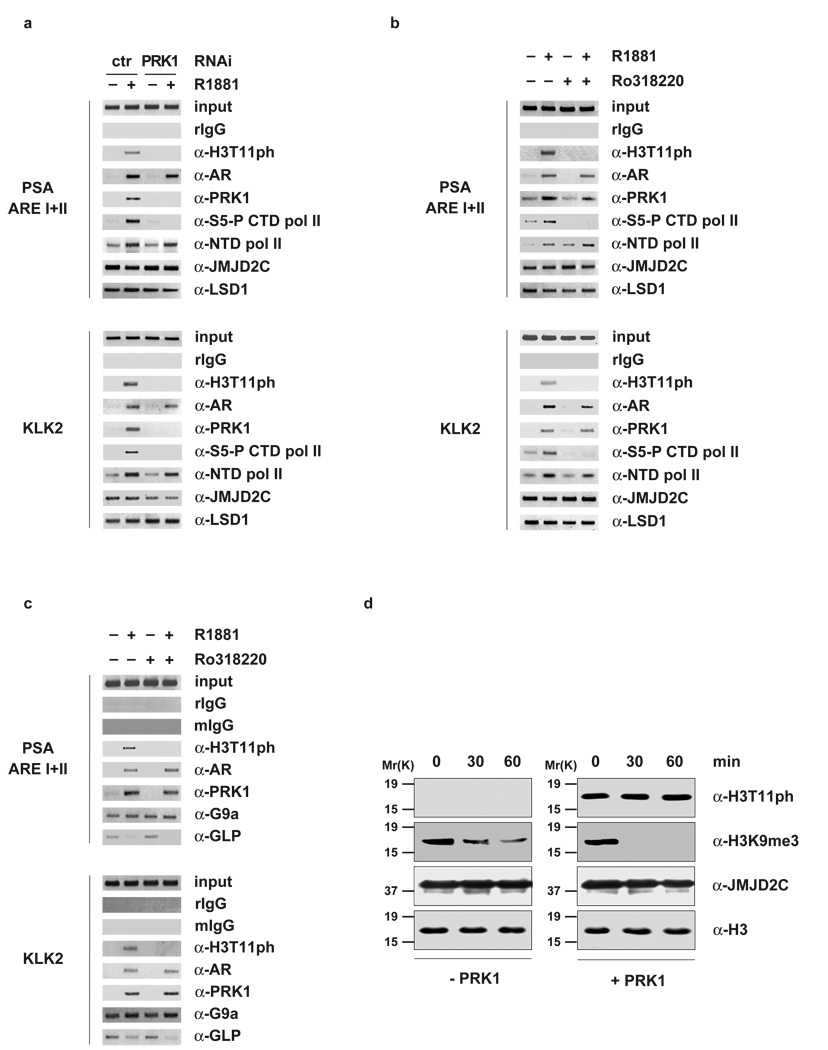
Initiation of transcription requires transition from the pre-initiation to the initiation complex, which is characterized by phosphorylation of RNA polymerase II at serine 5 in the C-terminal repeat domain (S5-P CTD pol II) by the CDK7 component of TFIIH2. To determine whether depletion or inhibition of PRK1 interfered with the formation of the transcriptional initiation complex at AR-regulated promoters, we performed ChIP using an antibody that specifically recognizes S5-P CTD pol II. Importantly, knockdown of PRK1 or treatment of cells with Ro318220 results in the loss of S5-P CTD pol II at the promoters of PSA and KLK2. In contrast, recruitment of RNA polymerase II is not affected, as shown by ChIP using an antibody directed against the N-terminal domain of RNA polymerase II (α-NTD pol II) (Fig. 4a, b and see Supplementary Information, Fig. S4d, e). Taken together, these data show that PRK1 not only controls changes in histone marks, but also regulates the transition from pre-initiation to initiation complex.
Figure 4.
PRK1 controls transition to the initiation complex and accelerates demethylation by JMJD2C. For ChIP (a, b, c), cells were cultivated in the presence or absence of the AR agonist R1881 and the inhibitor Ro318220 (b, c), or transfected with siRNA (a). ChIP analyses were performed with the indicated antibodies. The precipitated chromatin was amplified by PCR using primers flanking AREs in the promoter region of the PSA and KLK2 genes. Untreated nucleosomes (d, left panel) and nucleosomes phosphorylated at H3T11 in vitro (d, right panel) were incubated with 10 µg bacterially expressed and purified His-JMJD2C for the indicated times. Western blots were decorated with the indicated antibodies.
H3T11ph accelerates demethylation
To gain mechanistic insight into the role of H3T11 phosphorylation during transcriptional regulation, we asked whether PRK1 knockdown or inhibition with Ro31220 affects the presence of JMJD2C and LSD1 on the promoters of the PSA and KLK2 genes. As shown in figure 4a and 4b, the presence of JMJD2C and LSD1 is neither influenced by depletion nor inhibition of PRK1 and PRK1 does not phosphorylate JMJD2C, LSD1 or AR (Supplementary Information, Figs. S4f, g and S5a). In addition, ChIP experiments reveal that PRK1 does not regulate the presence of the H3K9 methyltransferases G9a8 and GLP9 or the acetyltransferase Tip6010 on the PSA and KLK2 promoters (Fig. 4c and see Supplementary Information, Fig. S5b). Conversely, inhibition of PRK1 abolishes androgen-dependent recruitment of p30010 (Supplementary Information, Fig. S5b). Moreover, depletion of neither JMJD2C nor LSD1 nor TIF26 impairs ligand-induced phosphorylation of H3T11 (Supplementary Information, Fig. S5c–e).
Next, we asked whether nucleosomes trimethylated at H3K9 and phosphorylated at H3T11 might serve as better substrates for JMJD2C and thereby accelerate demethylation. We phosphorylated nucleosomes at H3T11 in vitro and performed demethylation assay using bacterially expressed and purified JMJD2C. Importantly, nucleosomes phosphorylated by PRK1 at H3T11 are more rapidly demethylated by JMJD2C compared to unmodified nucleosomes (Fig. 4d). Taken together, these data show that H3T11ph activates AR-dependent transcription by enhancing JMJD2C-dependent demethylation.
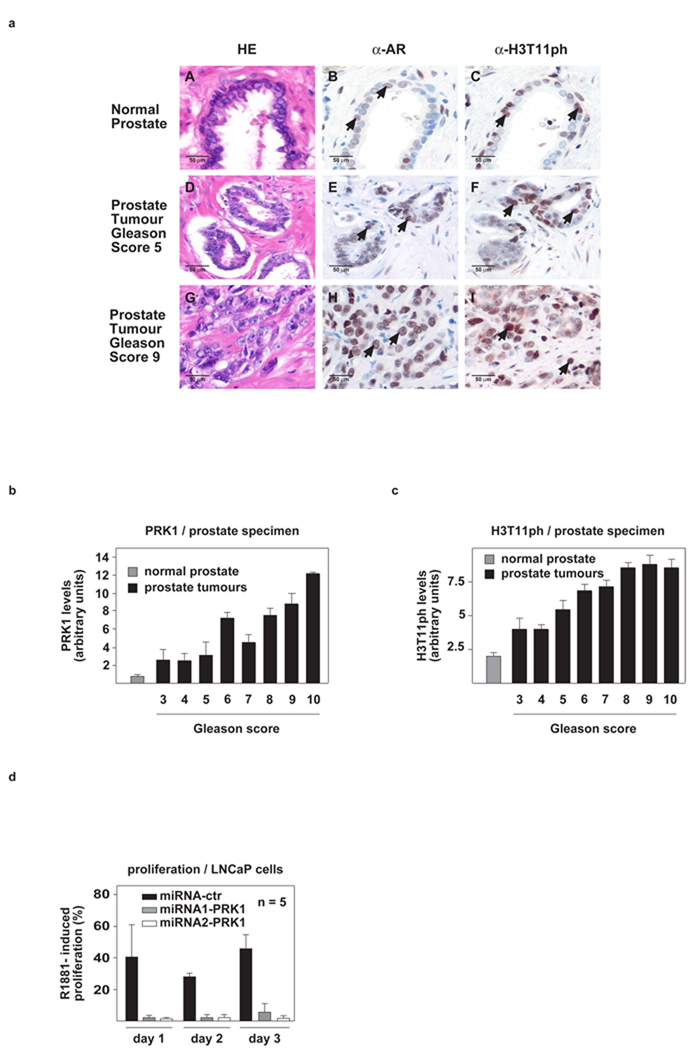
H3T11ph levels correlate with prostate cancer malignancy
To unravel the physiological importance of PRK1, we investigated the levels of PRK1 and H3T11ph in vivo by immunostaining a panel of 20 normal human prostates and 111 prostate carcinomas on tissue microarrays. Quantification of immunoreactivity by scoring staining intensity and percentage of positive carcinoma cells11 reveals that high PRK1 and H3T11ph levels significantly correlate with high Gleason scores and indicate aggressive biology of the tumours (Fig. 5a–c). Furthermore, to examine whether PRK1 regulates tumour cell proliferation, we monitored androgen-dependent cell growth by quantifying proliferation of pLenti6-miRNA-PRK1-infected LNCaP cells. When compared to cells expressing an unrelated control miRNA, androgen-induced proliferation of LNCaP cells is dramatically reduced by PRK1 knockdown (Fig. 5d). Similar results were obtained using the PRK1 inhibitor Ro318220 (Supplementary Information, Fig. S5f), thus underlining the importance of PRK1 in the control of AR-dependent tumour cell growth.
Figure 5.
PRK1 and H3T11ph levels positively correlate with the malignancy of prostate cancer and control tumour cell proliferation. (a), Immunohistochemical staining of AR and H3T11ph in human normal and tumour prostate. AR (B, E, H) and H3T11ph (C, F, I) immunoreactivity is detected in the secretory epithelium of normal prostate (B, C, arrows) and prostate carcinoma cells (E, F, H, I, arrows). Hematoxilin-eosin (HE) stained sections are shown in A, D, and G. All sections were taken from the same radical prostatectomy specimen (magnification: ×250). (b, c) The correlation of elevated PRK1 and H3T11ph levels with high Gleason score in a panel of 111 human prostate carcinomas is highly significant: r=0.499, p<0.001 (b) and r=0.450, p<0.001 (c). Normal prostate specimens (n=20) are included as a control. (d) In LNCaP cells, miRNA-mediated PRK1 knockdown severely reduces R1881-induced cell proliferation. Bars represent mean +SD (n≥4).
Discussion
In this manuscript, we demonstrate that phosphorylated H3T11 is a novel chromatin mark for transcriptional regulation. Phosphorylation of H3T11 is executed by PRK1 in an androgen-dependent manner and enhances demethylation of H3K9 by JMJD2C. By controlling subsequent steps of gene activation such as demethylation of tri-, di-, and monomethyl H3K9, acetylation of H3K9/K14, and the presence of S5-P CTD pol II at target promoters, PRK1 functions as a gatekeeper of AR-regulated gene expression.
At AR target promoters, phosphorylation of T11 by PRK1 represents an activating chromatin mark required for demethylation of H3K9. In vitro, we demonstrate that phosphorylation of H3T11 by PRK1 accelerates demethylation by JMJD2C. Conversely, phosphorylation of the adjacent serine 10 (H3S10) has been shown to block demethylation of trimethylated H3K9 peptides by JMJD2A, a close homologue of JMJD2C12. In full agreement with this report, we did not observe ligand-induced phosphorylation of H3S10 at AR target promoters by PRK1 after 30 minutes (Supplementary Information, Fig. S2e). In contrast, it has been reported that H3S10 phosphorylation is necessary for Myc-dependent transcriptional activation and oncogenic transformation13. However, the mechanisms involved are yet to be fully understood.
PRK1 also regulates the transcriptional activity of other nuclear receptors such as mineralocorticoid and progesterone receptor6, possibly via a mechanism similar to that controlling AR target gene expression. This raises questions whether other transcription factors are regulated by PRK1, and whether there exist other H3T11 kinases that control the activity of specific transcription factors. Recently, Dlk/ZIP has been described to phosphorylate H3T11 during mitosis7. However, we did not find any evidence that Dlk/ZIP can affect the transcriptional activity of AR (Supplementary Information Figure S5g, h). Since the molecular mechanisms of Dlk/ZIP-mediated phosphorylation of H3T11 remain unclear, it will be interesting to investigate whether the kinase associates with other transcription factors.
A recent study14 revealed that histone methylation prevents nuclear receptors and other classes of regulated transcription factors from constitutive gene activation in the absence of stimulating signals. In full agreement with these data, we detect methylated H3K9 at AR target promoters in the absence of ligand, while addition of ligand induces demethylation (Fig. 3a, b). Intriguingly, PRK1 knockdown or inhibition results in increased H3K9 methylation levels, even in the absence of ligand (Fig. 3a, b). While clarification of this observation requires detailed future investigation, we currently hypothesize that this increase could be due to the existence of an AR-independent pathway of PRK1 signalling, which inhibits histone methyltransferases. Blocking PRK1 would thus activate methyltransferases, resulting in increased methylation levels of H3K9. We speculate that the AR-independent targeting of methylases by PRK1 might help to fine-tune the methylation status of specific histone marks and prevent formation of a permanently repressive chromatin environment.
The levels of PRK1 and H3T11ph directly correlate with Gleason scores of prostate carcinomas. This finding not only suggests a role for PRK1 in tumourigenesis but also indicates that PRK1, similarly to LSD111, may serve as a predictive tumour marker. Of importance is our observation that inhibitors such as Ro318220 control the kinase activity of PRK1 and thereby regulate AR. Thus, specific modulation of PRK1 activity is a promising therapeutic strategy in the treatment of prostate cancer, where AR is pivotal to the control of tumour cell proliferation.
Methods
Plasmids
The following plasmids were described previously: pSG5-AR, GST-AR-NTD, GST-AR-DBD, GST-AR-LBD, pCMX-flag, pCMV-flag-PRK1 K644E, pcDNA3-myc-ΔNPRK1, TK-LUC, MMTV-LUC, Probasin-LUC, and PSA-LUC6; MBP-JMJD2C, His-JMJD2C, and pCMX-flag-JMJD2C4, GST-LSD1 and pCMX-flag-LSD13, GST-H3 1–4415. To construct pLenti6-miRNA1-PRK1, pLenti6-miRNA2-PRK1, pGW-miRNA1-PRK1, and pGW-miRNA2-PRK1, the DNA corresponding to miRNA1-PRK1 (5’-TGCTGATTGCTGTAGGTCTGGATCATGTTTTGGCCACTGACTGACATGATCCACCTACAGCAAT-3’ and 5’-CCTGATTGCTGTAGGTGGATCATGTCAGTCAGTGGCCAAAACATGATCCAGACCTACAGCAATC-3’) and miRNA2-PRK1 (5’-TGCTGTTACTGTCCTGCAACATCTGCGTTTTGGCCACTGACTGACGCAGATGTCAGGACAGTAA-3’ and 5’-CCTGTTACTGTCCTGACATCTGCGTCAGTCAGTGGCCAAAACGCAGATGTTGCAGGACAGTAAC-3’) was cloned into pLenti6/V5-DEST and pcDNA-6.2-GW-EmGFP according to the manufacturer’s instructions (Invitrogen). To construct GST-H3 1–15, GST-H3 1–15 T11A, GST-H3 16–30, and GST-H3 29–44, the corresponding cDNA fragments were cloned into pGEX4T1. To construct GST-H3 1–135 and GST H3 1–135 T11A the corresponding cDNA fragments were cloned into pET-GEX15. Cloning details can be obtained upon request.
Cell culture and transfection
CV1 and LNCaP cells were cultured and transfected as described6. The following amounts were transfected per well: 500 ng of MMTV-LUC, Probasin-LUC, or PSA-LUC; 25 ng of AR expression plasmid; 200 ng (Fig. 3e) or 400 ng (Fig. 3c, d) expression plasmids of LSD1 or JMJD2C, 150 ng PRK1 K644E, 1000 ng expression plasmid of miRNA-control, miRNA1-PRK1, or miRNA2-PRK1 (Supplementary Fig. S1c). Cells were cultivated for 18 hours in the presence or absence of 1 × 10−10 M R1881 (Sigma), 2.5 × 10−6 M (Fig. 3d) or 4.5 × 10−6 M (Fig. 3c) Ro318220 (Roche) as indicated. Luciferase activity was assayed as described6. All experiments were repeated at least four times in duplicate.
Generation of PRK1 antibody
The polyclonal rabbit α-PRK1 antibody was generated according to standard procedures.
Chromatin Immunoprecipitation
ChIP and Re-ChIP experiments were performed as described3, 16. LNCaP cells were cultured for 45 min (Figs. 1c, 2d, e, and 4c) or 210 min (Figs. 3a, 3b, 4a, 4b) in the presence or absence of 1 × 10−8 M R1881 as indicated. Ro318220 (1 × 10−5 M) was added to LNCaP cells (Fig. 3b, 4b, 4c) 60 min before addition of R1881. Three days before harvesting, cells were transfected with siRNA (ctr: 5’-GAACAUGAUCCAGACCUACAGCAAU-3’; PRK1: 5’-GAAAGUCCUAGAUCCACACGCAAAU-3’; TIF2: 5’-CATCCGTTCTCAGACTACTAATGAA-3’; JMJD2C4; Invitrogen, LSD13; Qiagen) using RNAifect (Qiagen) following the manufacturer’s instructions. Immunoprecipitation was performed with specific antibodies (α-H3K9me1 #07-450 lot 24441, α-H3K9me2 #07-441 lot 30309, α-H3K9me3 #07-442 lot 31855, α-H3K9/K14ac #06-599 lot 25233, α-Tip60 #07-038 lot 25012, α-AR #06-680 lot 33529 (Upstate Biotechnology), α-S5-P CTD pol II #ab5131 lot337093, p300 #14984 lot288954, α-H3T11ph #ab5168 lot32710, α-GLP #41969 lot 279169, α-G9a #ab40542 lot 321535, α-H3 #ab1791 lot 172452 (Abcam), α-NTD pol II # sc899 lot A1207 (Santa Cruz), α-TIF2 #T73620 lot 0000068640 (BD Transduction Laboratories), α-LSD15, α-JMJD2C3, and α-PRK1) on protein A-Sepharose 4B (GE-Healthcare). For PCR, 1–5 µl out of 50 µl DNA extract was used. PCR primers for ARE I+II (PSA −459/−121), ARE III (PSA −4288/−3922), KLK2 (−343/−90), GAPDH, U6 and HSP70 were described previously3, 4.
Western blot analysis
Experiments were performed as described6. Western blots were decorated as indicated.
Cell proliferation assay
Experiments were performed as described5. pLenti6-miRNA-control, pLenti6-miRNA1-PRK1, and pLenti6-miRNA2-PRK1 were used to produce recombinant lentiviruses to infect LNCaP cells as described17. The infected cells were cultured for 72 hours in medium containing 10% double-stripped FCS. 1 × 104 cells were plated in a 96-well plate in the presence or absence of 1 × 10−9 M R1881. The cell proliferation Elisa BrdU Colorimetric Assay (Roche) was performed according to the manufacturer’s instructions. The figure shows the percentage increase of proliferation in the presence versus absence of R1881. The experiments were performed in quintuplicate.
Quantitative RT-PCR and statistical analysis
Quantitative RT-PCR and statistical analysis were performed as described3. The primers for GAPDH, PSA, and KLK2 were described previously3.
In vitro kinase assay
The kinase assays were performed as described18. 10 µg of either GST-H3, GST-LSD1, GST-AR mutants, or MBP-JMJD2C proteins, or 1 µg of nucleosomes purified from HeLa cells19 were incubated with immunoprecipitated PRK1 proteins (myc-ΔNPRK1 or flag-PRK1 K644E) or 1 µg purified recombinant GST tagged PRK1 (ProQinase GmbH, Freiburg) at 30°C in kinase buffer containing 20 mM Tris-HCl pH 7.5, 20 µM ATP, 8 mM MgCl2, and 5 µCi (γ-32P) ATP. The reaction mixture was analysed by SDS-PAGE followed by autoradiography or Western blotting using antibodies as indicated.
In vitro demethylation assays
Nucleosomes purified from HeLa cells were incubated for 45 min in the presence or absence of purified recombinant PRK1 at 30°C in kinase buffer containing 20 mM Tris-HCl pH 7.5, 20 µM ATP, 8 mM MgCl2. Following dialysis in demethylation buffer, demethylation reactions were performed for the indicated times using bacterially expressed and purified JMJD2C essentially as described4,.
Immunohistochemistry
Stainings were performed using a protocol11 for antigen retrieval and indirect immunoperoxidase. α-AR 441 (Santa Cruz), α-H3T11ph7 antibodies were used at a dilution of 1:75 and 1:500 respectively. Rabbit IgG was used as secondary antibody (1:500; Dako). Immunoreactions were visualized with the ABC-complex diluted 1:50 in PBS (Vectastain, Vector).
Statistical analysis of tissue microarrays
Clinical data of patients and procedures for generating the tissue microarrays were described previously11. Statistical analysis was performed with the Mann-Whitney U-Test using the SPSS 12.0 program (SPSS Inc.) and by calculating the two-tailed Spearman Rank correlation coefficient. The number of cases (n) analysed per Gleason score (Gs) were : Gs 3 (n=5); Gs 4 (n=12); Gs 5 (n=11); Gs 6 (n=25); Gs 7 (n=16); Gs 8 (n=23), Gs 9 (n=10); Gs 10 (n=9). Normal prostate specimen (n=20).
Supplementary Material
Acknowledgements
We thank C. Beisenherz-Huss, C. Schächtele (ProQinase GmbH, Freiburg), and R. Schneider for generously providing reagents. We are obliged to T. Günther, H. Greschik, J. M. Müller and S. Naumovitz for helpful discussions. We thank Franziska Klott for excellent technical assistance. This work was supported by grants of the National Institutes of Health (R01 GM074210) to J.M.G.H., the Deutsche Forschungsgemeinschaft (SFB 746/P2 and Schu 688/9-1), the Dr. Hans Messner Stiftung, and Deutsche Krebshilfe (10-2019-Bu II) to R.S.
Footnotes
Competing interest statement The authors declare that they have no competing financial interests.
Literature
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 3.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 4.Wissmann M, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell. Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 5.Kang Z, Pirskanen A, Janne OA, Palvimo JJ. Involvement of proteasome in the dynamic assembly of the androgen receptor transcription complex. J. Biol. Chem. 2002;277:48366–48371. doi: 10.1074/jbc.M209074200. [DOI] [PubMed] [Google Scholar]
- 6.Metzger E, et al. A novel inducible transactivation domain in the androgen receptor: implications for PRK in prostate cancer. EMBO J. 2003;22:270–280. doi: 10.1093/emboj/cdg023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Preuss U, Landsberg G, Scheidtmann KH. Novel mitosis-specifc phosphorylation of histone H3 at Thr11 mediated by Dlk/ZIP kinase. Nucleic Acids Res. 2003;31:878–885. doi: 10.1093/nar/gkg176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. SET Domain-containing Protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 9.Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 10.Baek SH, et al. Ligand-specific allosteric regulation of coactivator functions of androgen receptor in prostate cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3100–3105. doi: 10.1073/pnas.0510842103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahl P, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 12.Ng SS, et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- 13.Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat. Cell. Biol. 2007;9:932–944. doi: 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Bassets I, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai J, Sultan S, Taylor SS, Higgins JM. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19:472–488. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol. Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 17.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong LQ, et al. Phosphorylation of protein kinase N by phosphoinositide-dependent protein kinase-1 mediates insulin signals to the actin cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5089–5094. doi: 10.1073/pnas.090491897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Neill TE, Roberge M, Bradbury EM. Nucleosome arrays inhibit both initiation and elongation of transcripts by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1992;223:67–78. doi: 10.1016/0022-2836(92)90716-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.