Abstract
Characterizing the biomolecular systems’ properties underpinning prognosis signatures derived from gene expression profiles remains a key clinical and biological challenge. In breast cancer, while different “poor-prognosis” sets of genes have predicted patient survival outcome equally well in independent cohorts, these prognostic signatures have surprisingly little genetic overlap. We examine ten such published expression-based signatures that are predictors or distinct breast cancer phenotypes, uncover their mechanistic interconnectivity through a protein-protein interaction network, and introduce a novel cross-“gene expression signature” analysis method using (i) domain knowledge to constrain multiple comparisons in a mechanistically relevant single-gene network interactions, and (ii) scale-free permutation resampling to statistically control for hubness (SPAN - Single Protein Analysis of Network with constant node degree per protein). At adjusted p-values < 5%, 54 genes thus identified have a significantly greater connectivity than those through meticulous permutation resampling of the context-constrained network. More importantly, eight of ten genetically non-overlapping signatures are connected through well-established mechanisms of breast cancer oncogenesis and progression. Gene Ontology enrichment studies demonstrate common markers of cell cycle regulation. Kaplan-Meier analysis of three independent historical gene expression sets confirms this network-signature’s inherent ability to identify “poor outcome” in ER(+) patients without the requirement of machine learning. We provide a novel demonstration that genetically distinct prognosis signatures, developed from independent clinical datasets, occupy overlapping prognostic space of breast cancer via shared mechanisms that are mediated by genetically different yet mechanistically comparable interactions among proteins of differentially expressed genes in the signatures. This is the first study employing a networks’ approach to aggregate established gene expression signatures in order to develop a phenotype/pathway-based cancer roadmap with the potential for (i) novel drug development applications and for (ii) facilitating the clinical deployment of prognostic gene signatures with improved mechanistic understanding of biological processes and functions associated with gene expression changes. http://www.lussierlab.org/publication/networksignature/
Keywords: systems biology, protein-interaction networks, breast cancer, gene signatures, context-constrained networks
Introduction
Since their conceptual inception in 2002 [1,2], clinical outcome-tied molecular signatures in breast cancer have become a central topic of research. Signatures for poor prognosis [2], recurrence [3], invasiveness [4], and metastasis [5,6] have been experimentally derived from patient groups and biological hypotheses. Despite the proliferation of signatures, genes constituting distinct signatures exhibit poor genetic overlap (share few genes), even though they paradoxically occupy a common prognosis space. They are similarly efficient in predicting bad clinical outcome in new cohorts “raising questions about their biologic relevance, significance and clinical implication” [7,8]. A critical problem to solve for cancer biologists and oncologists is whether these disjoint genetic signatures can “jointly” provide a unified mechanistic insight on their respective scales of biology between gene expression and clinical outcome.
Clearly, the complexity of heterogeneity becomes a chief consideration when trying to compare across signatures. As varied as these gene signatures are, so too are the tissues and methods in which they were derived. A sampling of tissue types evaluated have included ER+ tissue[3], ER+/ER− tumor specimens [6], inflammatory breast carcinoma tissue [9], to cell lines [5]. Detection of gene expression changes have utilized standard commercial gene chip platforms, to commercial customizable chips to in-house cDNA spotted nylon microarrays[10] and RT-PCR assays [3]. Besides the heterogeneity of probe designs, many of the legacy and custom platform provide partial genome assessments in contrast to the contemporary genome-wide arrays.
Aside from differences in molecular derivation, several hypotheses have been postulated to explain the lack of overlap in the genetic makeup of the signature. Arguments have included a case made for inadequate patient sample size in developing the signature, or incomplete genome coverage, and secondly that although the genes are different, they are merely separate aspects of the same groups of molecular pathways or mechanisms. To overcome the sample size restraint to a given original study, investigators have demonstrated that pooling breast cancer data can enhance classification performance in 73% of the cases in one study or generate a signature that was comparable or superior to the prognostic performance of the original signatures [11,12]. Examination of the hypothesis of common molecular pathways hypothesis underlying the genetic heterogeneity of gene signatures has been attempted using straightforward Gene Ontology enrichment and failed to demonstrate the functional overlap. For example, Van Vliet et al., note that there was less than 1% mean overlap of Gene Ontology enrichment in their selection of breast cancer signatures [12]. More recent efforts of pathway analysis have gone beyond the assessment of Gene Ontology concordance. For instance, Pujana et al. implemented a systems biology approach that does not rely on gene expression data,. By selecting four key biologically validated breast cancer genes of distinct cancer-associated pathways, they generated a network of 118 genes and 866 functional associations capable of predicting the association of the hyaluronan-mediated motility receptor gene (HMMR) with an increased risk of breast cancer [13]. The success of this systems biology approach is seen, too, in other methods such as that of genome-scale reverse engineering of direct gene regulatory mechanisms which have been developed using network modeling and successfully applied to mammalian cells [14,15].
Meanwhile, we and other groups have developed comprehensive protein-protein interaction (PPI) networks that have effectively been used by our group and others to analyze protein interactions underpinning share sub-phenotypes among otherwise seemingly disparate diseases [16,17,18], and to characterize the function of a novel tumor suppressor microRNA [17]. In the context of an individual expression signature in breast cancer, PPI networks have been effectively used to reanalyze gene expression data to detect a subnetwork signature of metastatic disease [4] and more recently used to predict prognosis [19]. Such studies demonstrate the power of PPI networks to better understand complex molecular disease processes at a systems level in single studies.
Further, experts in cancer network analyses have also recognized the need to incorporate cancer-domain knowledge (context-constraints) to network modeling [20,21]. We therefore hypothesize that a context-constrained PPI network may be capable of connecting mechanistically heterogeneous signatures. As most of the signatures were designed, in general, to distinguish good versus bad prognosis of clinical outcome or more specifically to predict more aggressive disease progression, we posit that essential pathways such that of cell cycle regulation that is required for oncogenesis as well as progression will correlate with a poorer outcome. Bearing this in mind, in this paper we focus on developing a mechanistically transparent meta-signature of breast cancer. We evaluate ten breast cancer signatures published in leading journals (e.g. New England Journal of Medicine, Table 1) and find their interconnectivity in a cancer-context constrained PPI network, which we hope, could shed light on the underlying shared molecular mechanisms of prognosis to the understanding of genetically distinct gene expression signatures.
Table 1. Ten Breast Cancer Expression Array Signatures Evaluated.
| Phenotype Measured by the Signature |
Tissue used for derivation | No. of genes |
Authors |
|---|---|---|---|
| Invasiveness [22] | CD44+CD24-/low cell lines vs normal breast epithelium |
186 | Liu R, Wang X et al. |
| Poor prognosis [1] (Mammaprint) |
Primary breast tumors | 70 | van ’t Veer LJ, Dai H et al. |
| Metastasis [6] | Primary breast tumors | 76 | Wang Y, Klijn JG et al. |
| PTEN/PIK3 pathway [10] | Primary breast tumors | 246 | Saal LH, Johansson P et al |
| Node (-) disease recurrence [23] |
Primary breast tumors | 16 | Sotiriou C, Wirapati P et al. |
| Lung metastasis [5] | Cell lines | 54 | Minn AJ, Gupta GP et al. |
| Inflammatory breast cancer [9] (IBC_1) |
Primary breast tumors | 109 | Bertucci F, Finetti P et al |
| Inflammatory breast cancer [19] (IBC_2) |
Primary breast tumors | 50 | Van Laere S, Van der Auweral et al. |
| Histologic grade [24] | Primary breast tumors | 264 | Ivshina AV, George J et al. |
| Bone metastasis [25] | Cell lines | 102 | Kang Y, Siegel PM et al. |
Results
Evaluation of breast cancer signatures overlap
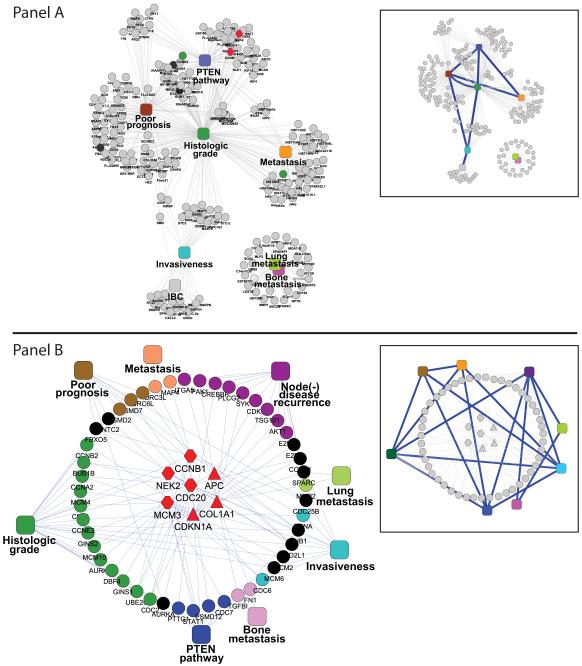
Consistent with findings of previous analyses reported in the literature, we found a slight overlap among various signatures. Out of the systematic evaluation of each pair-wised combination of signature among the ten signatures of this study (45 combinations in total), seven were found to share a few statistically significant genes after adjustment accounting for multiple comparisons [Panel A of Figure 1, Supplemental Figure S1]. However, none of the genes overlapped across all signatures. Thus, as expected, we failed to identify the straightforward genetic overlap to connect different signatures. Additionally, three signatures of metastasis (bone and lung metastasis signature, respectively) formed a separate network aside from that of a tightly nested web of cell cycle regulators, but were not statistically overlapping with the sparsely linked network of 6 other signatures (8 inferred links between signatures in the inset).
Figure 1. Analysis of breast cancer signature overlap using two different methodologies. Panel A. Direct statistical gene signature overlap with no inherent mechanistic meaning.
8 statistically significant inter-signature connections were identified relating 6 of the ten signatures [inset] based on genetic overlap (common genes between signatures) after adjusting for the varying background of genes of the different expression platforms (adjusted p<5%; cumulative hypergeometric distribution). The maximum number of inter-signature connections for a single signature was 3. Signatures for lung and bone metastasis formed a separate unconnected network. Genetic overlap network did not provide inherent mechanistic information. Gene Ontology enrichment identified proliferation pathway markers were strongly associated with increased histologic grade as well as a worsened prognosis. Panel B. Inherent mechanistic and statistical overlap of genetic signature based on “breast cancer context”-constrained molecular interaction networks: 16 statistically significant inter-signature connections were identified relating 8 of the ten signatures [inset] based on “breast cancer context”-driven network genes that mechanistically anchor the signature overlap (red shapes). The Bonferroni adjusted pvalue for the number of relationships to a single gene-network ≤ 0.05 (for the entire network p<0.01). The maximum number of inter-signature connections for a single signature was 5. Noteworthy, the inflammatory breast cancer did not significantly interact or overlap with the rest of the network. Legend: Red circles indicate genes derived from the expression signatures. Triangles represent genes derived from the breast cancer context-driven network. Red hexagons are genes common to both expression signatures and the breast cancer context-driven network. Squares indicate phenotype of gene signatures. Thin grey edges related genes to their respective gene signature (squares, Panels A and B) and indentify any protein interactions in Panel A (no statistics), and only significant gene interactions in Panel B (adjusted pvalues<5%, Methods, SPAN[16,17,18]). Thick blue edges of insets represent “inferred” statistical relationships between genes signatures. Colors represent the following: red indicate genes from the breast cancer context-driven gene set; black indicates genes significantly associated with more than one gene signature (more than one phenotype); the remaining colors indicate separate signatures. Thus, a red hexagon indicates a gene found both in a signature as well as in a breast cancer mechanism gene. Insets contain graphic representations of statistically significant inter-signature connections.
Taking a pure network approach, we previously generated a vast protein-protein interaction (PPI) network with 44,695 protein-protein interactions and 7,321 proteins based on an integration of published databases [17]. The single protein analysis of network (SPAN) [17] of breast cancer signatures were examined based on direct interactions between two expression signatures for each pair of signatures. Although some genes in the signatures did interact directly among signatures, few of these associations reached statistical significance and only four signatures could be directly related significantly after adjustment with false discovery rate for multiple comparisons (data not shown). As expected, when indirect interactions were considered using every intermediary node in the network, only few indirect interactions among signatures reached significance due to the vast number of required adjustments for multiple comparisons. Noteworthy, during our permutation resampling of the network, we maintained the number of partners each protein had consistently in each iteration such that our statistical analysis for the multiple comparisons was particularly stringent for hubs, while it allowed for higher sensitivity in poorly connected nodes than a bootstrap method. In other words, the node degree of each protein is equal to that of the observed distribution in each simulated network. However, while some significant genes were shown to interact more than expected by chance between less than half the breast cancer signatures, this type of analysis was not constrained with biological contexts thus molecular mechanisms derived from such modeling might not be relevant to cancer biology.
Hypothesizing that non-cancer related nodes in our PPI network were generating background interaction noise or simply reducing the power of the statistical analysis due to the multiplicity of comparisons, we improved our network model by developing a “breast cancer context”-constrained PPI network using known literature knowledge about breast cancer. As described in the Methods section, we used, as inter-signature nodes, 250 cancer-related genes curated from the literature that were previously identified by Paik et al [3] in the New England Journal of Medicine. The biological and pathophysiological causal mechanistic role of these 250 breast cancer genes were selected by traditional in vitro and in vivo biological studies. Examining SPAN networks between each mechanistically derived breast cancer gene and each signature, and after correcting for multiplicity and for node degree (Methods on the permutation resampling [17]), 54 genes (what we are defining as the network-signature) were found to be significantly more connected than by empirical distribution (adjusted p < 5%) and were inherently mechanistically anchored (Table 2). Each single protein connectivity of the deduced molecular mechanisms of breast cancer signature was independently tested within each signature and corrected for multiple comparisons. Thus, the observed interconnectivity among signatures arose from shared intrinsic molecular mechanisms rather than from inherent computational/statistical design to connect signatures. In particular, seven breast cancer context genes effectively anchored the inter-signature connections: CCNB1, APC, CDC20, MCM3, CDKN1A, COL1A1, and NEK2 (Panel B of Figure 1, red nodes), and were highly enriched for cell cycle- and cellular movement-dependent involvement in G2/M DNA damage checkpoint regulation along with ATM signaling based on Ingenuity Pathway Analysis [26]. 16 significant inter-signature relationships were identified between eight of the ten signatures, including the two-metastasis signatures that could not be connected using simple statistical enrichment (Panel B of Figure 1). 15 of the 54 network-signature genes were found to connect at least three gene signatures. Consistent with our prior gene overlap method, lung and bone metastasis were connected to one another; but, this time, they also connected to other signatures via their significant interaction with the node(+) disease recurrence signature. Moreover, five signatures were each independently connected to five other signatures demonstrating an extremely tight, intertwined web of interconnectivity (Panel B of Figure 1). Noteworthy, genes of the inflammatory breast cancer signatures IBC-1 and IBC-2 were the only genes that did not interact significantly with the 250 breast cancer-related genes.
Table 2. 54-Gene Composing the Network-Signature.
Breast cancer constrained SPAN network modeling identified 54-genes associated with the breast cancer mechanism. We also evaluated the PPI network property of each gene: H = hub gene, B = bottleneck gene, H/B = hub and bottleneck gene (Methods).
| Statistically Significant Genes of the Network Signature | |||||
|---|---|---|---|---|---|
| APC (H) | CDC2 (B/H) | GINS2 | PCNA (H/B) | CCNE1 (H) | MCM10 (H) |
| AURKA (H/B) | CDC20 (H) | ITGA5 (H) | PSMD12 (H) | CDK7 (H) | MCM3 (H) |
| AURKB (H/B) | CDC25B | MAD2L1 (H) | PSMD2 (H) | COL1A1 (H) | MCM4 (H) |
| BUB1 | CDC6 (H) | MAP4 | PSMD7 (H) | CREBBP (H/B) | MMP2 |
| BUB2 | CDC7 (H) | MCM2 (H) | PTTG1 (H) | DBF4 (H) | ORC3L (H) |
| CCNA2 | CDKN1A (H/B) | MCM6 (H) | STAT1 (H/B) | E2F3 | PAK1 (H/B) |
| CCNB1 (H) | CDT1 (H) | NEK2 | TGFBI (H/B) | E2F5 | PLCG2 (H) |
| CCNB2 | FBXO5 | ORC6L (H) | TSG101 | FN1 (H) | SPARC |
| CCNE2 | GINS1 | PAK3 (H) | AKT1 (H/B) | KNTC2 | SYK (H/B) |
Gene Ontology enrichment studies identified these genes to be predominantly regulators of the cell cycle pathway (Table 3). For example, cell cycle (GO: 0007049) was annotated with 25 of the 54-genes (adjusted p = 0.4 × 10−11). Similarly, cell division (GO:0051301) and mitosis (GO:0007067) ranked highly. Corroborating these observations using separate software, the Ingenuity Pathway Analysis also revealed enrichment of the 54 genes in the network signature represented in multiple canonical pathways involved in cell cycle regulation. In particular, the top 6 Ingenuity pathways identified included: mitotic roles of polo-like kinase, cell cycle G2/M DNA damage regulation, molecular mechanisms of cancer, CHK proteins in cell cycle regulation, G1/S checkpoint regulation, and cell cycle control by BTG family protein.
Table 3. Gene Ontology Enrichment of Network-Signature Genes.
| GO ID | Function Name | Network- signature genes |
GO term genes |
Adjusted P-Value |
|---|---|---|---|---|
| GO:0007067 | Mitosis | 15 | 128 | 3.8E-11 |
| GO:0007049 | cell cycle | 25 | 306 | 3.8E-11 |
| GO:0051301 | cell division | 18 | 160 | 3.8E-11 |
| GO:0006260 | DNA replication | 11 | 102 | 4.5E-11 |
| GO:0006270 | DNA replication initiation | 5 | 19 | 9.6E-07 |
| GO:0007051 | spindle organization and biogenesis | 4 | 7 | 9.6E-07 |
| GO:0048015 | phosphoinositide-mediated signaling | 5 | 26 | 4.6E-06 |
| GO:0006268 | DNA unwinding during replication regulation of cyclin-dependent protein |
3 | 11 | 7.6E-04 |
| GO:0000079 | kinase activity traversing start control point of mitotic |
4 | 37 | 9.4E-04 |
| GO:0007089 | cell cycle | 2 | 5 | 9.8E-03 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 3 | 28 | 1.5E-02 |
The significant biological processes were identified by comparing genes in PPIS and Affymetrix U133a chips using Onto-Express software. The criterion of significance is set at 0.01 with enrichment calculated using the cumulative hypergeometric p-value adjusted for multiple testing.
Network properties of the 54-genes
To establish whether these 54-genes coded for proteins that indeed contained distinct network properties, we identified “hub” proteins versus “bottleneck” proteins as described by the Gerstein research group [27] over the entire PPI. Gerstein et al. defined hubs as proteins that have the 20% highest number of neighbors and bottlenecks as the proteins that are in the top 20% in terms of betweenness (connecting groups of proteins). Hub and bottleneck can occur independently of one another. In contrast, of the 54 genes that we selected, 37 (69%) were hub proteins among which 11 (30%) were also bottleneck proteins. This is far in excess of the baseline, thus indicates the central interacting role that these genes may play in the context of breast cancer networks.
As a separate “signature” validation of our approach, we examined the direct overlap of our 54-gene network-signature with an independent 168-gene “signature of proliferation” based on a cluster of gene associated with in vitro oncogenesis [28]. This cluster of genes centered around p53 and INK4A signaling pathways that had been demonstrated in historic datasets to be associated with poor prognosis – similar to that of the evaluated signatures. Interestingly, in total, 14 out of 168 genes significantly overlapped with the 54-gene network-signature (p value<5%).
Validation of the prognostic potential of the 54-gene from the network-signature
We tested the network-signature in three separate genome-wide microarray datasets comparing breast cancer patients outcome: GSE7390 (198 patients), GSE4922 (249 patients) and GSE2990 (189 patients), were downloaded from NCBI GEO database and analyzed in the same way for independent validation [23,24,29]. Two datasets were used in the generation of the original expression signatures (histologic grade and node-negative recurrence) along with a third independent dataset that had been used as a separate validation for the node-negative signature. The third dataset thus could be used to confirm the validity of the network-signature. Time to recurrence was used for GSE7390 data analysis. Time to distant metastasis was used for the analysis of the remaining two datasets, as the original gene signatures derived from GSE4922 and GSE2290 did not assess this clinical endpoint.
We first explored the relationship between the network-signature genes and pathologic parameters using hierarchical clustering and GSEA. The 198 patients in GSE7390 were classified into two clusters. Via chi-square testing, one cluster was found to be enriched with disease advanced and more aggressive tumors including pathological features of ER− (p<0.0001), pathological grade 3 (p<0.0001), and lymph node infiltration stage 3 (p=0.0405). However, there was no significant difference in time to recurrence between the two clusters (Supplemental Figure S2 Logrank p = 0.394). The majority of the network-signature genes demonstrated higher expression levels in ER− than in ER+ samples (Supplemental figures S3, S4, S5). 29 out of the 54 network-signature genes were expressed consistently and significantly (Student’s T-test p-value < 0.05) higher in ER− than ER+ samples across all of the 3 microarray datasets (Supplemental Table S1).
We then examined a GSEA comparison between ER+ versus ER− samples, high versus low grade, poor versus good prognostic samples defined by conventional prognostic tools: Adjuvant!Online, St. Gallen and Nottingham Prognostic Index (NPI). Again, as observed in the datasets described above, GSEA showed that only the network-signature demonstrated an overall upregulation of signature gene expression in ER(−) samples (FDR =0), while none of the conservatively selected control gene sets demonstrated significant upregulation in ER(−) samples. Similarly, in the GSE7390 dataset, overexpression of the network-signature genes was found in the poor prognostic samples predicted by the aforementioned conventional tools (FDR =0) and also the high-grade samples.
These prognostic scores were not available in GSE4922 and GSE2290, and thus cannot be verified in these two datasets. Kaplan-Meier plot revealed that the high and low risk groups determined by network-signature did not show strong significant different outcome in the three datasets (Supplemental Figure S2).
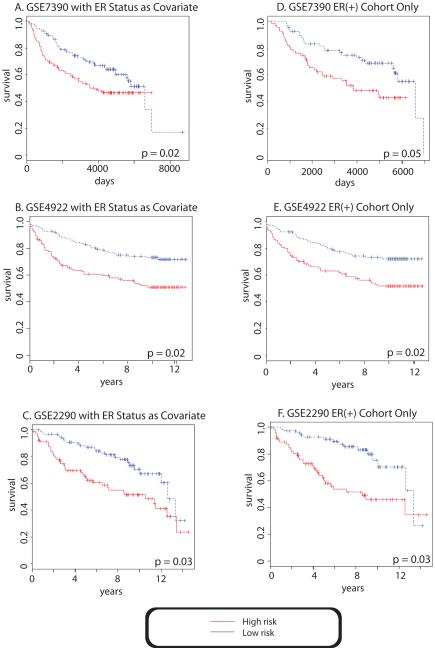
After gaining a better understanding of the clinical correlations, we then examined the utility of the signature for prognostication. We noted that the network-signature alone was not predictive for the clinical outcome in the datasets tested (Logrank p-value 0.22 for GSE7390, 0.05 for GSE4922 and 0.07 for GSE2290). However, multivariate analysis demonstrated that the network-signature with ER status as a covariate, successfully predicted two risk groups that displayed significant different outcome in all of the three datasets (Figures 2, Panels A, B, C). The Logrank p-value was 0.02 for GSE7390, 0.02 for GSE4922 and 0.03 for GSE2290. In contrast, ER status alone failed to provide an independent prediction of clinical outcome (p = 0.22 for GSE7390, p= 0.53 for GSE4922 and p = 0.20 for GSE2990) consistent with clinical observations demonstrating the insufficiency of ER status alone to provide accurate prognosis. However, pathological grade as a covariate with the network-signature failed to predict the outcome (data not shown).
Figure 2. Kaplan-Meier analysis of the 54-gene network signature in three historical GEO datasets.
Endpoints: time to recurrence was used for breast cancer microarray GSE7390[23], time to distant metastasis was used for GSE4922 [24] and GSE2290 [29]. Multivariate analysis with ER status as a covariate successfully predicted two risk groups that displayed significant different outcome in all of the three datasets (Figures 2, Panels A, B, C). The Logrank p-value was 0.02 for GSE7390, 0.02 for GSE4922 and 0.03 for GSE2290. Univariate analysis of ER(+) samples demonstrated a logrank p-value of the ER+ samples was 0.02 for GSE7390, 0.02 for GSE4922 and 0.03 for GSE2990 (Figures 2, Panels D, E and F).
Therefore, multivariate analysis of the network-signature with ER status, but not the network-signature or ER status alone accurately predicted the clinical outcome of the heterogeneous population of breast cancer patients without the requirement of extensive machine learning.
The network-signature was also evaluated for its prognosis of ER+ stratified samples by univariate analysis. The Logrank p-value of the ER+ samples was 0.02 for GSE7390, 0.02 for GSE4922 and 0.03 for GSE2990 (Kaplan-Meier curve shown in Figures 2, Panels D, E and F). Thus, the prognostic power of the network-signature maintained among ER+ patients. Therefore, the network-signature demonstrated the potential to stratify the therapeutically responsive ER positive patients into high and low risk groups that will have distinct clinical outcomes measured by survival. This prognostic stratification potential of the network signature has significant clinical implications since it could help to identify the high-risk patients for additional and more aggressive therapeutic intervention prior to disease progression.
Overlay of the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways
To understand the interplay and biology of the 54-gene network-signature, we calculated the statistical significance of protein-protein interaction (PPI) among these 54 genes using SPAN (Methods and [17]) and organized the interactions according to their connectivity relationship with the respective prognostic signature (Figure 3, Methods and [30]). Thereafter, we conducted functional enrichment analysis of KEGG pathways [31] and overlaid the statistically prioritized four KEGG functions (Cell cycle, p53 signaling, ErbB2 signaling and focal adhesion) onto the PPI gene interaction map (Figure 3, color coding for different KEGG pathway function). The resultant visualization in Cytoscape facilitated more readily an appreciation of the mechanistic underpinning associated with each signature and the overlap we identified via PPI modeling. For example, “focal adhesion” KEGG pathway linked “bone metastasis”, “lung metastasis” signature and “Node(−) disease recurrence” signatures via COL1A1, which is one of the seven inter-signature genes identified via context-driven gene analysis (Figure 1, Panel B). Another example is CDKN1A (p21), which was also identified by context-driven gene analysis and a negative regulator of cell cycle. It was a multifunctional node (cell cycle/ErbB2/focal adhesion, Figure 3) that connected the cluster of three metastasis gene signatures anchored by COL1A1 to the “histologic grade” signature and the “invasiveness” signature. Additionally, we observed a strong overlap of PPI-genes shared between the “poor prognosis” signature and “histologic grade” signature. Such observed molecular pathway overlaps in PPI network provided a mechanistic explanation of a well-documented histologic observation of high incidence of metastasis and poor prognosis associated with high-grade tumors. Therefore, COL1A1, CDKN1A and MCM3 and their direct interacting genes could be further tested and characterized for their potential as functional prognostic markers that can be used for patient stratification for more personalized treatment. Collectively, these observations have demonstrated the significant functional overlaps among the 54 mechanistic network-signature genes and the molecular underpinning of the association between high histologic grade and high incidence of invasiveness or/and with poor prognosis.
Figure 3. 54-gene breast cancer network signature overlaid with KEGG pathway.
We evaluated the connectivity of the 54-gene breast cancer network signature to itself through permutation resampling of the PPI controlling for hubness. Nodes which had an empiric p-value of <0.05 were retained. KEGG pathways cell cycle, p53 signaling, ErbB2 signaling and focal adhesion were then overlaid onto the network facilitating a visual “roadmap” among signatures (squares), breast cancer signatures (circles), and breast cancer background genes (triangles). Breast cancer signature genes that were also included in the breast cancer background gene list are represented by hexagons. For example, multifunctional node CDKN1A (cell cycle/ErbB2/focal adhesion) connects the cluster of three metastasis gene signatures anchored by COL1A1 to the “histologic grade” and “invasiveness” signatures.
Discussion
In this study, 10 breast cancer signatures are essentially mapped onto a protein-protein interaction network of known cancer molecules. Although previous papers have noted that these breast cancer signatures appear to be genetically “disjoint” – we have found a significant degree of mathematical overlap among signatures which nonetheless failed to provide mechanistic understanding (Figure 1, Panel A). In contrast, we employed a networks approach in which we regard different signatures not in isolation, but in totality, as a vast interconnected array of causal and non-causal molecules associated with breast cancer state and interrelating through canonical molecular pathways (Figure 1, Panel B; Table 1; Figure 3). The 54 genes are in essence prioritized genes via statistical enrichment of shared biomolecular systems properties of the aggregate molecular signatures that also have phenotypic/prognostic associations. Indeed, we propose that a mechanistic overlap vis-à-vis pathway allows the researcher to “do more with less”. Whereas in the simple statistical overlap method yields only seven gene-mediated significant inter-signature relationships (Figure 1, inset of Panel A), our context-constrained pathway overlap methodology is able to find twice the number of significant relationships between signatures using far fewer genes (Figure 1, inset of Panel B). Also biologically interesting are the gene signatures that did not connect in our SPAN. Inflammatory breast cancer (IBC) histologically, clinically and molecularly behave different than traditional ductal adenocarcinomas [32]. Researchers are beginning to develop targeted treatments for this subtype and our results – given the lack of connectivity of either IBC-1 or IBC-2 signatures to the PPI network – are consistent with the belief that mechanisms of IBC progression are different (Table 1, Figure 1 Panel B).
From an informatics perspective, traversing the generated PPI network deeper than the first interactor generates noise and reduces the ability to find significance. In contrast, a carefully selected first interactor causally associated to breast cancer in the network provides computational relevance and consequently statistical power beyond first interactors between breast cancer signatures. In other words, connecting the gene of a first signature to that of second signature via an intermediate breast cancer interactor corresponds to connecting a second level interactor in the network with a constrained intermediating layer of genes causally associated with breast cancer. As a result, signature genes statistically connected to causal breast cancer genes more than expected by conservative statistical controls thus become explicit interactors of a known breast cancer mechanism. We also recapitulate a well-established issue by bioinformaticians involved in the analysis of protein interaction networks: cellular context matters; unexpressed proteins contribute to noise in a context-independent PPI.
The ability of the 54-gene signature to predict poor outcome in breast cancer reassures the validity of our network approach in understanding gene signatures. Moreover, its significant overlap with 168 mechanistically selected genes in a model of tumorigenesis confirms that indeed that we are capturing underlying pathway deregulation associated with oncogenesis [28]. This prediction is obtained from 54 genes of the signatures that can comprise as many as 250 genes and are often derived from intensive machine learning algorithms in the absence of conducting machine learning of outcome datasets.
However, the network signature required ER status to assist in predicting clinical outcome in the heterogeneous breast cancer populations. It is possible that the published gene signatures are intrinsically biased toward ER(+) tumors because they were generated using heterogeneous patient data which was primarily ER(+). Indeed, ER(+) breast cancer comprises 70 percent of breast cancer patients [33] and ER status alone is known incapable of predicting survival in these populations. Although ER status provides a useful treatment target, ER specific gene markers can be equally found in signatures that correlate with both good and poor prognosis [34].
Another possibility is that this 54-gene network signature and the ER status lacked the statistical power alone to show survival prediction and requires their joint utilization to reach significance in non-stratified heterogeneous populations we analyzed. However, in a stratified population of ER positivity, the mechanistic network-signature has the independent prognostic power to further stratify patients into the high and low risk groups that have distinct clinical outcomes.
Clinically, the proposed 54-gene network-signature and the resulting PPI subnetwork are invaluable for understanding the role of existing gene signatures. In particular, the van’t Veer et al. 70-gene signature (MammaPrint, see “poor prognosis” in Table 1 and Figures 1 and 3) [1] which has been validated in clinical trials as an excellent independent marker of prognosis [35] is more closely related (Figure 3) to the gene signatures of histological grade and invasiveness. While highly enriched in cell cycle genes, MammaPrint (poor prognosis) did not connect to the bone or lung metastasis signatures that appear to be mediated by cellular adhesion as noted by the KEGG pathway enrichment. The conclusion we may draw is that although Mammaprint and these other signatures (some of which have been validated in other tumor types) map to the same prognostic space, they are in part mechanistically complimentary as they poorly overlapped with the genes in our network. Prospective clinical studies are required whether network signature, such as the one we report will be more effective at providing stratified molecular diagnosis or/and prognosis.
From a practical clinical practice standpoint, our methodology elucidates similarities and differences among signatures and points us toward potential biomarkers that may help us determine the choice of treatment. Tantalizing are highly- connected highly genes such as COL1A1 that sits on the intersection of bone metastasis, lung metastasis, and node(−) recurrence. Unsurprisingly, it is tagged with the KEGG pathway as being part of focal adhesion. Previous researchers have noted that this gene is highly overexpressed in a meta-analysis of 13 publications [36]. However, no further research has been performed to evaluate its utility as a prognostic marker. With our network, the KEGG mechanism associated with COL1A1 and its placement in our network makes for a clearer picture of the gene’s phenotype and prevents a convincing case for its potential as a functional biomarker. One may speculate that alteration of the focal adhesion pathway as evidenced by an alteration in COL1A1 expression leads to detachment of the breast cancer cells from the host environment and increase the possibility of distant metastasis. Targeting this gene then, may ultimately maintain regional control.
In essence, interaction networks, such as Figure 3, provide the clinicians with a more synthetic and mechanistic visualization to understand gene expression changes associated with breast cancer prognostication, and to facilitate the design of most appropriate combinations for personalized cancer treatment. We view contemporary clinical stratification of patients using ER, PR, and HER2 status as the beginnings of a rudimentary road map, a preamble for individualized drug selection.
In 2007, Massague et al. commented on the necessity to explore the mechanisms of the shared prognosis space between disjoint signatures [8]. Based on our computational modeling and validation using clinical datasets, we propose a model (summarized in Figure 4) that provides, the first systems-based explanation for a subset of signature genes that determines the mechanistic makeup of genetically diverse gene signatures. These observations suggest that each network-based molecular signature is likely associated with one or more aspects of a large protein-protein network. First, they may recapitulate known portions of canonical pathways or identify new significant relationships augmenting the known pathways. The second portion of the signature, that we have been deeming the “noise” may very well contain vital oncogenic pathways that remain to be characterized for their roles in cancer and their relationships with the canonical pathways. And the third aspect is that naturally there may be intermediary/interacting molecules that have yet to be characterized rounding out the “unknown” portion of the molecular signature.
Figure 4. Model for Understanding Molecular Signatures.
Illustrated in this figure is the derivation of the 54-gene network-signature that can be conceived as a subset of the “known mechanisms” of context-constrained networks (top orange component). Molecular signatures can be thought as mapping to different portions of a network. The “known mechanisms” portion overlay well on top of experimentally determined proteininteractions in the laboratory. The “imputed pathways” corresponds to poorly described associations in the PPI network and due to their lack of characterization can be mistaken for background noise of the network. The “unknown pathways” portion of the gene signature refers to molecular interactions that may not be in the network.
Limitations
We are beholden to the data we use as our SPAN constraint as much as we are beholden to the databases we used to generate the SPAN which consisted of both eukaryotic and prokaryotic data. However, our goal was to put forth an extensible method that could grow with increasing data and knowledge. We intend to rerun our analyses at a future time points with more carefully selected gene signatures and a more informed constraint. Indeed, because of the constraint we selected, a clinical limitation of our analysis was that we were not able to include the 21-gene signature that has been commercialized as OncotypeDX [37] which is a popular test available in the United States. These genes 21 genes were not derived from expression analysis but rather from the 250 Paik genes that we used as our contextual genes with causal associations to breast cancer. Consequently, our statistical analysis would have been inherently biased. Future studies using a different context-constraint will be required to evaluate the OncotypeDX and MammaPrint overlap.
A computational limitation of our approach is that there may be other valuable intermediaries, but the multiplicity of comparisons required to evaluate every single protein of the PPI reduces the greatly the statistical power. An alternate approach could have used a metanalysis of expression arrays of breast cancer and normal tissue to identify on a genome-wide basis, which subset of the network should be explored rather than relying on a well-acknowledged set of genes from the review of literature. More complex methods for aggregation of nodes in protein interaction networks can identify patterns beyond the immediate interactor and should be explored in future studies.
Conclusion
We provide a novel demonstration that genetically distinct prognosis signatures, developed from independent clinical datasets, inherently occupy overlapping prognostic space of breast cancer via shared mechanisms that are mediated by genetically different yet mechanistically comparable interactions among proteins of differentially expressed genes in the signatures. This is the first application of a network-based approach to aggregate established gene signatures in developing a phenotype/pathway-based cancer roadmap with a potential (i) for prospective applications in drug development and (ii) for elucidating molecular mechanisms underpinning dissimilar molecular profiles sharing interchangeable prognostic capabilities.
Methods
Signatures
A total of ten gene expression signatures were examined in this study. This included signatures from Minn, et al.[5], Liu, et al.[22], Wang, et al. [6], Bertucci, et al. [9], Ivshina, et al. [24], Kang, et al. [25], Saal, et al. [8], Sotiriou, et al. [9], Van de Vijver, et al. [10], and Van Laere, et al. [11]. Genes comprising each of the signatures were taken from each of the papers or their supplementary materials and translated into a representative set of SwissProt identifiers using the DAVID tool [12]. The original genes, translation tables (where needed), and results are available on http://www.lussierlab.org/publication/networksignature/.
Gene Overlap Analysis Between Two Signatures
The SwissProt translated signatures were analyzed for overlap using a Perl script comparing the accession numbers of the genes in each signature. A similar technique was used to test for connectivity through the combined interaction network. Accession numbers from pairs of signatures were matched to the proteins in the network and analyzed to determine connectivity between proteins.
Equation 1 allows us to formally calculate the statistical significance of overlapping genes between two expression signatures. Variable ‘N’, the common background genes, represents the total number of genes overlapping between the total probes of chips used in two studies. Variables M and n are the number of genes of the two compared expression signatures. Variable m corresponds to the number of genes that are found to overlap between the two compared expression signature (M ∩n). Genes of each array were translated in standard HUGO gene identifiers. Translation of non-standard arrays of the older studies required manual revision of hundred of probes with non-standard probes.
| (Equaton 1) |
Results are adjusted to account for multiple testing using the Dunn-Sidak adjustment, a Bonferroni-like method. In Equation 2, p’ and p represent the corrected and uncorrected p-values, respectively, and n represents the number of independent comparisons in the study. The resulting statistically significant connections were drawn using Cytoscape [30].
| (Equaton 2) |
Generation of the protein-protein interaction (PPI) network from multiple databases
As we described in a previous publication, the protein-protein interaction network was generated by integrating six protein interactions and signaling datasets[16,17,18]. In brief, protein interactions from each dataset were standardized to a two-column list of pairwise interactions and merged into a non-redundant interaction network. Identifiers were converted to a common SwissProt standard coding using translation tables from HUGO and the data sources’ own cross-mappings. We only included interacting pairs that were generated from physical experiments using methods other than the yeast two-hybrid or dosage rescue. Imputed interactions were not used. Datasets included BioGRID 1 [38], Reactome [39] DIP data [40], MINT [41] Human Proteome Reference Database 6 (HPRD) [42],BIND [43].
Single Protein Analysis of Networks (SPAN [17]), a conservative permutation re-sampling of the PPI
Permuted PPI networks were generated using a link randomization approach[44]. Proteins are considered as nodes and interactions between proteins are links. Since biological networks are scale-free rather than random [45], link randomization can create conservative “permuted networks” as controls, from which we can derive an empirical distribution of interactions between a subset of proteins. Furthermore, our implementation of a link-randomization conserves the number of “connections” of each specific protein (node-degree) [45]. Thus the scale free properties of the original distribution are preserved in every permutation as well as the node degree of each specific protein, a distinctive and highly conservative approach that we previously published [17], while the interactions (links) between these proteins vary. Self-interactions, such as those formed by homomultimers, were ignored to avoid introducing bias into the network. Duplicate protein interaction pairs were also excluded in the permutation. 10,000 of these permuted networks were generated from the original amalgamated interaction network consisting of real datasets.
Direct Gene Interaction Between Connectivity between Two Signatures
This previous network was used for calculating the false discovery rate (FDR) of direct connectivity between each genes of each pair of signatures (based on the number of direct protein interactors between the two signatures). FDR was calculated based on the median number of direct interactions between the two signatures in the empirical distribution divided with the observed number of interactors. First direct interactions were compiled and analyzed, then indirect interactions with one intermediary node as a separate analysis.
Gene Interaction Between an Expression Signatures Genes and Breast Cancer Genes to infer Interaction (Mechanistic Overlap) between Signatures. Context-knowledge constrained PPI network: Single Protein Network Modeling and Prioritized PPI in Cancer
Additionally, we developed a model that estimates the probability of occurrence of an observed Single Protein Network arising from the connectivity of a protein to a list of known proteins derived from a well-established list of 250 breast cancer genes manually curated from the literature and published in the New England Journal of Medicine [3]. The observed number of interactions between the breast cancer signatures and the breast cancer genes can thus be derived and compared to expected distribution from the previously described permutation resampling. The unadjusted p-value of each signature gene connectivity is further adjusted for multiplicity using Bonferroni-type adjustments (the number of genes in each breast cancer signature = the total number of comparisons). A similar procedure was developed to calculate the converse: each single gene in the breast cancer genes was analyzed for its number of interactions with the total lists of genes in each of the breast cancer signatures independently and assigned an adjusted p-value (in this case, controlled for 250 analyses in the gene list). Since breast cancer signatures were independently generated by different authors from distinct cohorts of patients – there were no additional adjustments of p-values for multiplicity between studies of each signature as each signature analysis was considered independent from one another. Thereafter, each breast cancer signature gene that met an adjusted p-value<5% was retained as well as its connected genes from the set of 250 and the converse (since there were two single protein network analyses). The statistically significant single gene networks were simply assembled in a joint network to show shared mechanisms (breast cancer genes known mechanistically to affect the biology of breast cancer). The resulting network was drawn using Cytoscape[30]. It is important to note that in each meticulous permutation we perform, each protein retains a constant node degree. To attain a significant p-value, highly connected proteins are thus required to surpass an equally well-connected protein in the permutation. This non-trivial permutation thus controls well for hubs – and is more sensitive to detect increased connectivity of poorly connected proteins than a straightforward bootstrap.
Permutation Resampling of Networks
Permuted networks were generated with a Perl script that shuffled the connectivity of the nodes in the network while maintaining their node degree as well as the distribution of node degree in the network. 10,000 of these permuted networks were generated from the original amalgamated interaction network. These networks were then used to connect the proteins of the molecular signatures in a permutation resampling. This procedure yielded connectivity distributions at two scales. First, we derived a total connectivity distribution for each signature as a whole when repeatedly reconnected to the set of poor-prognosis candidates. At a smaller, more granular scale, we derived a distribution for each protein in the signatures considered in the study.
Gene expression datasets
Microarray Data source for validation of protein-protein interactions and all breast cancer microarray datasets using the same genome-wide platform (Affymetrix U133A chip) with available .cel files and clinical parameters in NCBI GEO database were downloaded and cell lines were downloaded as referenced prior.
Gene Expression Software
Bioconductor GCRMA package[46], dChip [47], BRB-ArrayTools [48] and GSEA [49] software were used for microarray data analysis. GraphPad prism 4.03 for Windows [50] was used for Chi-square test, Kaplan-Meier plotting and Logrank test. Onto-Express [51] was used for Gene Ontology (GO) analysis.
Identification of biological processes and canonical pathways enriched with the network-signature
The functional profiles of the PPI-signature genes were represented by the biological processes in the Gene Ontology (GO) database [52] or signaling pathways with the number of PPI-signature genes in each GO category or pathway compared to that in the Affymetrix U133A chip to determine the significance function. The analysis of biological processes was performed using Onto-Express, with the default selection of statistical method (hypergeometric distribution followed by Benjamini-Hochberg false discovery rate correction). The lists of the network-signature genes were uploaded into Onto-Express to identify significant biological process (corrected p-value <0.01).
Disregulated genes were uploaded into the Ingenuity Pathway Analysis (IPA) tool from Ingenuity Systems. The genes mapped to corresponding gene objects in the IPA tool are called “focus genes.” The significance of a canonical pathway is controlled by p-value, which is calculated using the right-tailed (referring to the overrepresented pathway) Fisher Exact Test for 2×2 contingency tables. This is done by comparing the number of ‘Focus’ genes that participate in a given pathway, relative to the total number of occurrences of those genes in all pathways stored in the IPKB. The significance threshold of a canonical pathway is set to 2, which is derived by −log10 [adjusted p-value], with the Benjamini-Hochberg corrected p-value ≤ 0.01.
Hierarchical clustering
Unsupervised 2-way hierarchical clustering was performed to associate the expression pattern of the network-signature with clinical parameters, such as ER status, lymph node infiltration, and pathological grade. The default parameter and Pearson correlation in dChip software was used to for hierarchical clustering. Chi-square test was performed to evaluate whether patients with different clinical phenotypes can be classified into different cluster based on the expression pattern of network-signature.
Gene set enrichment analysis (GSEA)
GSEA software was used to quantitatively characterize the expression pattern of network-signature between binary clinical status, such as ER(−) vs. ER(+), high vs. low grade, poor vs. good prognosis. The a priori established gene sets contains the network-signature and 99 gene sets randomly selected from U133A chip, each with the equal number of genes of the network-signature. GSEA is a supervised analysis, which uses the modified nonparametric Kolmogorov-Smirnov test to calculate an enrichment score and thus determine whether a specific gene set is differentially expressed between the binary status of a phenotype. The randomly selected gene sets are used here as controls to determine whether any potential phenotypic association is unique to network-signature.
Survival analysis
BRB-Array Tools were used for time-to-event data analysis, which is referred here as “survival analysis”. The survival data most relevant to the prognosis of BC patients is the time of recurrence or distant metastasis. Two survival analysis tools in the BRB-Array software implemented with Cox’s proportional hazards model were used [53]. The ‘Survival Analysis Prediction Tool’ develops a gene expression based predictor of survival risk group. The survival risk groups were constructed using the supervised principal component method [53]. This method used a Cox proportional hazards model to relate survival time to k “super-gene” expression levels, where k = 2 in the current analysis. The “supergene” expression levels are the first k principal component linear combinations of expression levels of the subset of genes that are univariately correlated with survival. the p value criterion for gene selection is set at 0.999, so that all of the genes in network-signature are used in computing the principle component. To compute a prognostic index for a patient whose expression profile is described by a vector x of log expression level, the following steps were performed. First the components of the vector x corresponding to the genes that were selected for use in computing the principal components are identified. Then the k principal components are computed. These are linear combinations of the components of x, with the weights of each linear combination having been determined from the principal component analysis described above. Finally, the weighted average of these k principal component values is computed, using as weights the regression coefficients derived from the k-variable Cox regression described above. This computation provides a prognostic index for a patient with a log expression profile given by a vector x. A high value of the prognostic index corresponds to a high value of hazard of death, and consequently a relatively poor predicted survival. Two-risk groups are predicted and leave-out-one method was used for cross validation. The prognostic index for the omitted patient was ranked relative to the prognostic index for the patients included in the cross-validated training set. The left-out patient is placed into a risk group based her percentile ranking, and the cut-off percentile is set at 50% for current analysis. This leave-one out analysis was repeated n times (n = sample size), leaving out a different patient each time. It is important to note that the risk group for each case was determined based on a predictor that did not use that case in any way in its construction. Finally, Kaplan-Meier survival curve is plotted for the cases predicted to have above or below average risk. Log rank test is performed by 100 permutations and the criterion for significance is set at p<0.05.
Supplementary Material
Acknowledgements
This work was supported in part by the NIH/NLM/NCI National Center for Multiscale Analyses of Genomic and Cellular Networks (MAGNET, 1U54CA121852), the NIH/NCRR Clinical & Translational Science Awards (1U54 RR023560-01A1), The Cancer Research Foundation, The University of Chicago Cancer Research Center, and The Ludwig Center for Metastasis Research. We thank Xinan Holly Yang for her help with the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.van ’t Veer LJ, Dai H, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 2.van de Vijver MJ, He YD, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 3.Paik S, Shak S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 4.Chuang HY, Lee E, et al. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minn AJ, Gupta GP, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Klijn JG, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 7.Fan C, Oh DS, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 8.Massague J. Sorting out breast-cancer gene signatures. N Engl J Med. 2007;356:294–297. doi: 10.1056/NEJMe068292. [DOI] [PubMed] [Google Scholar]
- 9.Bertucci F, Finetti P, et al. Gene expression profiling for molecular characterization of inflammatory breast cancer and prediction of response to chemotherapy. Cancer Res. 2004;64:8558–8565. doi: 10.1158/0008-5472.CAN-04-2696. [DOI] [PubMed] [Google Scholar]
- 10.Saal LH, Johansson P, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen R, Ghosh D, et al. Prognostic meta-signature of breast cancer developed by two-stage mixture modeling of microarray data. BMC Genomics. 2004;5:94. doi: 10.1186/1471-2164-5-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Vliet MH, Reyal F, et al. Pooling breast cancer datasets has a synergetic effect on classification performance and improves signature stability. BMC Genomics. 2008;9:375. doi: 10.1186/1471-2164-9-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pujana MA, Han JD, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 14.Margolin AA, Califano A. Theory and limitations of genetic network inference from microarray data. Ann N Y Acad Sci. 2007;1115:51–72. doi: 10.1196/annals.1407.019. [DOI] [PubMed] [Google Scholar]
- 15.Margolin AA, Nemenman I, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lage K, Karlberg EO, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y YX, Huang Y, Fan H, Zhang Q, Wu Y, Li J, Hasina R, Cheng C, Lingen MW, Gerstein M, Weichselbaum RR, Xing HR, Lussier YA. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Computational Biology. 2010 doi: 10.1371/journal.pcbi.1000730. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sam L, Liu Y, et al. Discovery of protein interaction networks shared by diseases. Pac Symp Biocomput. 2007:76–87. [PMC free article] [PubMed] [Google Scholar]
- 19.Van Laere S, Van der Auwera I, et al. Distinct molecular signature of inflammatory breast cancer by cDNA microarray analysis. Breast Cancer Res Treat. 2005;93:237–246. doi: 10.1007/s10549-005-5157-z. [DOI] [PubMed] [Google Scholar]
- 20.Wang K, Alvarez MJ, et al. Dissecting the interface between signaling and transcriptional regulation in human B cells. Pac Symp Biocomput. 2009:264–275. [PMC free article] [PubMed] [Google Scholar]
- 21.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu R, Wang X, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 23.Sotiriou C, Wirapati P, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 24.Ivshina AV, George J, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 25.Kang Y, Siegel PM, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 26.Ingenuity Systems©.
- 27.Yu H, Kim PM, et al. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 2007;3:e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabach Y, Milyavsky M, et al. The promoters of human cell cycle genes integrate signals from two tumor suppressive pathways during cellular transformation. Mol Syst Biol. 2005;1:0022. doi: 10.1038/msb4100030. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desmedt C, Piette F, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 30.Shannon P, Markiel A, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S, et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamauchi H, Cristofanilli M, et al. Molecular targets for treatment of inflammatory breast cancer. Nat Rev Clin Oncol. 2009;6:387–394. doi: 10.1038/nrclinonc.2009.73. [DOI] [PubMed] [Google Scholar]
- 33.Swain SM, Wilson JW, et al. Estrogen receptor status of primary breast cancer is predictive of estrogen receptor status of contralateral breast cancer. J Natl Cancer Inst. 2004;96:516–523. doi: 10.1093/jnci/djh097. [DOI] [PubMed] [Google Scholar]
- 34.Cunliffe HE, Ringner M, et al. The gene expression response of breast cancer to growth regulators: patterns and correlation with tumor expression profiles. Cancer Res. 2003;63:7158–7166. [PubMed] [Google Scholar]
- 35.Wittner BS, Sgroi DC, et al. Analysis of the MammaPrint Breast Cancer Assay in a Predominantly Postmenopausal Cohort. Clinical Cancer Research. 2008;14:2988–2993. doi: 10.1158/1078-0432.CCR-07-4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srour N, Reymond MA, et al. Lost in translation? A systematic database of gene expression in breast cancer. Pathobiology. 2008;75:112–118. doi: 10.1159/000123849. [DOI] [PubMed] [Google Scholar]
- 37.Albain KS, Barlow WE, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 11:55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.BioGRID http://www.thebiogrid.org/downloads.php.
- 39.Reactome http://reactome.org/download/index.html.
- 40.Database of Interacting Proteins http://dip.doe-mbi.ucla.edu/dip/Download.cgi.
- 41.Domino: a domain peptide interactions database http://mint.bio.uniroma2.it/mint/download.do.
- 42.Human Proteome Reference Database http://www.hprd.org/download.
- 43.Biological Objects Network Databank http://bond.unleashedinformatics.com.
- 44.Farkas IJ, Wu C, et al. Topological basis of signal integration in the transcriptional-regulatory network of the yeast, Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:478. doi: 10.1186/1471-2105-7-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 46.Team RDC . R:. A language and environmental for statistical computing. 2005. [Google Scholar]
- 47.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon R LA, Li MC, Ngan M, Menenzes S, Zhao YD. Analysis of Gene Expression Data Using BRB-Array Tools. Cancer Informatics. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, Tamayo P, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.GraphPad www.graphpad.com.
- 51.Onto-Express www.geneontology.org/GO.tools.microarray.shtml#onto-e.
- 52.Ashburner M, Ball CA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox D. Regression models and life tables. JRoyal Stat Soc B. 1972;34:187–202. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.