Summary of recent advances
Expansive compound collections made up of structurally heterogeneous chemicals, the activities of which are largely undefined, present challenging problems for high-throughput screening (HTS). Foremost is differentiating whether the activity for a given compound in an assay is directed against the targeted biology, or is the result of surreptitious compound activity involving the assay detection system. Such compound interference can be especially difficult to identify if it is reproducible and concentration-dependent – characteristics generally attributed to compounds with genuine activity. While reactive chemical groups on compounds were once thought to be the primary source of compound interference in assays used in HTS, recent work suggests that other factors, such as compound aggregation, may play a more significant role in many assay formats. Considerable progress has been made to profile representative compound libraries in an effort to identify chemical classes susceptible to producing compound interference, such as compounds commonly found to inhibit the reporter enzyme firefly luciferase. Such work has also led to the development of practices that have the potential to significantly reduce compound interference, for example, through the addition of non-ionic detergent to assay buffer to reduce aggregation-based inhibition.
Introduction
High-throughput screening (HTS) involves testing hundreds of thousands, if not millions, of chemical compounds for activity in a biological assay. Originally developed as a drug discovery methodology by pharmaceutical companies for identification of chemical entities with potential therapeutic value, HTS has branched into the academic sector as a technique to discover chemical probes, which are applied as research tools to study a biological target or pathway [1–3].
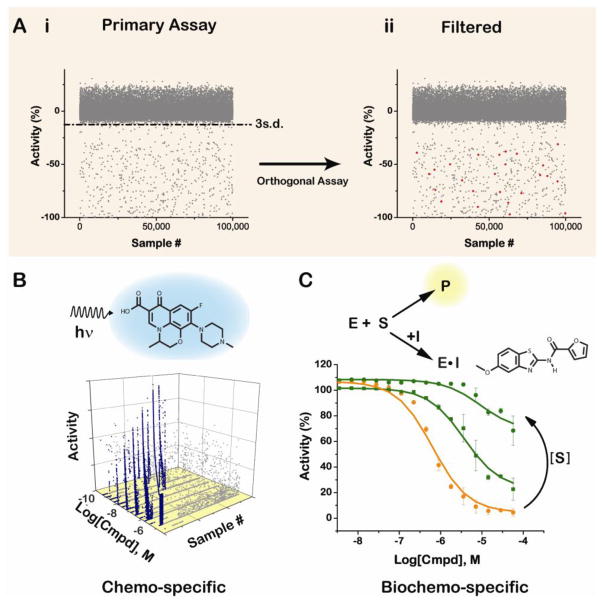
Libraries used for HTS consist of chemicals with a high degree of structural heterogeneity (compared to nucleic acid or peptide libraries, for example) that often have unknown and undefined properties and biological activities. One challenge in HTS is to successfully differentiate between compounds that demonstrate genuine activity against the biological target, target family, or pathway of interest from compounds that interfere with elements of the assay format or technique. Generally, specific activity of a compound against its target is characterized by a high affinity, non-covalent interaction that is reversible [4]. Activity in an assay due to compound interference can result in a “false positive” (see Box 1) [5], and as compounds that are genuinely active against the target are rare (~0.01–0.1% of library), they are easily obscured by a high incidence of false-positives in a screen (Fig. 1A) [6–8]. In this review we will focus on a discussion of compound activity that reproducibly interferes with HTS assays. Such compound interference can arise solely from the compounds themselves [5,9], as is the case for fluorescent compounds (Fig. 1B), or can be the result of their interaction with biological components of the assay system. The latter is the case for compounds that directly inhibit a biological reporter, such as firefly luciferase (Fig. 1C), or compounds that indirectly inhibit or activate proteins in an assay, for example, by undergoing redox cycling to generate hydrogen peroxide. Non-specific chemical reactivity against target proteins, such as covalent binding or metal ion chelation [4,10], is another source of false positives in HTS [11–13], but is out of the scope of this Review and thus will not be discussed in detail here except in the context of redox reactivity.
Box 1. Definition of terms.
Compound interference: For this Review, the activity of a compound in an assay that is reproducible, but may not be related to the biological target of interest. Compound interference contributes to the number of “false positives” in an assay. Common types of compound interference discussed in this Review include compound fluorescence, aggregation, luciferase inhibition, and redox reactivity.
Counter-screen: A screen performed in parallel with or after the primary screen. The assay used in the counter-screen is developed to identify compounds that have the potential to interfere with the assay used in the primary screen (the primary assay). An example of an assay used in a counter-screen would be a biochemical assay to identify compounds that inhibit firefly luciferase. This assay could be used as a counter-screen to a primary screen that utilized the firefly luciferase as a reporter to identify compounds that modulated a biological target/signaling pathway. Counter-screens can also be used to eliminate compounds that possess undesirable properties, for example, a counter-screen for cytotoxicity.
False positive: Generally related to the “specificity” of an assay. In screening, a compound may be active in an assay but inactive toward the biological target of interest. For this Review, this does not include activity due to spurious, non-reproducible activity (such as lint in a sample that causes light-scatter or fluorescence). Compound interference that is reproducible is a common cause of false positives, or target-independent activity.
High-throughput screen (HTS): A large-scale experiment in which collections of compounds are tested for activity against a biological target or pathway. “Screen” for short.
Hits: Slang for putative activity observed during the primary high-throughput screen, usually defined by percent activity relative to control compounds.
Library: A collection of compounds.
Off-target activity: Compound activity that is not directed toward the biological target of interest but can give a positive read-out, and thus be classified as an active in the assay.
Orthogonal assay: An assay performed following the primary assay to differentiate between compounds that generate false positives from those compounds that are genuinely active against the target. Conducted on compounds found active in the primary assay, this assay uses a different reporter or assay format in an effort to confirm that activity of the compound is directed toward the biological target of interest. Compounds inactive in an orthogonal assay are removed from further consideration, as a negative result indicates that the original compound activity was most likely assay format-dependent and not specific to the biology of interest.
Primary assay: The assay used for a high-throughput screen.
Secondary assay: An assay used to test the activity of compounds found active in the primary screen (and orthogonal assay) using robust assays of relevant biology. Ideally, these are of at least medium-throughput to allow establishment of structure-activity relationships between the primary and secondary assays and establish a biologically plausible mechanism of action.
Figure 1. Assay interference by compounds can be reproducible and demonstrate concentration dependence, producing false positives in a high-throughput screen (HTS).
(A) False-positives in the primary assay used for HTS are generally eliminated upon testing in an orthogonal assay (see Box 1) conducted on all compounds with greater than average activity. (i and ii) Each dot in the scatter plot represents a compound, plotted based on its percent activity in an assay, in this case, an assay to identify inhibitors. In the example shown, a 3 standard deviation (s.d.) threshold value (dotted line) was used to qualify a compound as active. (i) Results from a primary assay to identify compounds with inhibitor activity, with the percentage of active compounds at 5% of the total screened (found below 3s.d. line). (ii) Following an orthogonal assay, 95% of the active compounds are found to be false positives due to assay interference, leaving only a few genuine actives (red dots; 0.25% of compounds screened). (B) Fluorescent compounds (an example of which is shown) behave much like genuine actives, demonstrating concentration dependence (blue dots in scatter plot) that is reproducible, as shown here for results of a fluorescence-based HTS run in the blue spectral range. Sporadic fluorescence interference is also seen (grey dots), but is not reproducible and does not show concentration dependence (adapted from Simeonov et al., 2008 [14]). (C) Compound interference that directly interacts with an enzymatic reporter, such as firefly luciferase, is shown here. This type of compound interference also demonstrates compound concentration dependence (orange concentration response curve – CRC), and in this specific case, the compound is a competitive inhibitor of firefly luciferase (green CRCs).
Introduction to artifacts typical of light-based detection methods in HTS
Current HTS technologies rely heavily on sensitive light-based detection methods, such as fluorescence or luminescence, to quantify the effect of a compound on a target molecule or signaling pathway [5]. While advantages to light-based detection technologies include a desirable balance between sensitivity and ease of automation for HTS, they are also susceptible to a wide-range of different types of assay interference (see Table 1) [9]. Whereas most light-based assay interference is due to spurious events or occurs only at a high(er) compound concentration(s), compound fluorescence and luciferase inhibition show reproducible concentration dependence, making their identification initially more challenging.
Table 1. Common types of assay interference.
Assay interference described in detail in this Review are underlined. Missing fields in the Table indicate that a substantial amount of work remains to be done to characterize compounds used in HTS and determine the impact of various types of assay interference. Also not included in this Table, due to lack of available information, are the effects of metal ion interference on assays [4,5] and assay interference by compounds that affect cell membrane permeabilization [47].
| Assay Interference | Effect on Assay: | Characteristics of note | Prevention | Prevalence in Library | Percent Enrichment of Actives | Ref |
|---|---|---|---|---|---|---|
| Aggregation |
|
|
|
|
|
[11, 12, 30, 31, 33, 35, 48] |
| Compound Fluorescence |
|
|
|
|
|
[9,11,14,18–20, 49–52] |
| Firefly Luciferase Inhibitors |
|
|
|
|
|
[6,23] |
| Redox Cycling Compounds |
|
|
|
|
|
[37 – 40, 42] |
| Oxidizers |
|
|
|
~20–50% enrichment for some assays | [45, 46, 53] | |
| Covalent reactivity |
|
|
|
|
[4,10,11,13,54] | |
| Cytotoxicity |
|
|
|
[55 – 59] | ||
| Inner filter effect (absorption of light) |
|
|
|
[9,19] | ||
| Light scattering |
|
|
|
[19] | ||
| Quenching |
|
|
[9,19,60] | |||
| Surfactant-like compounds |
|
|
|
[11, 61, 62] |
Compound Fluorescence
Whether or not a compound fluoresces in an assay depends upon its structural properties and the excitation and emission wavelengths used in the experiment. Generally, conjugated bonds within the compound confer fluorescent character, and the greater the degree of conjugation within a compound, the longer the wavelength at which it fluoresces [14]. Compound libraries tend to contain a greater percentage of heterocyclic compounds and compounds with low levels of conjugation [5,15,16], and thus assays that rely on excitation at relatively short wavelengths (λex~ 350 nm) with detection of fluorescence in the blue spectral region (λem= 450–495nm), have a greater likelihood of false positives due to compound fluorescence [14]. An additional consideration is compound purity, as samples with minor, albeit highly fluorescent, impurities are also a source of assay interference.
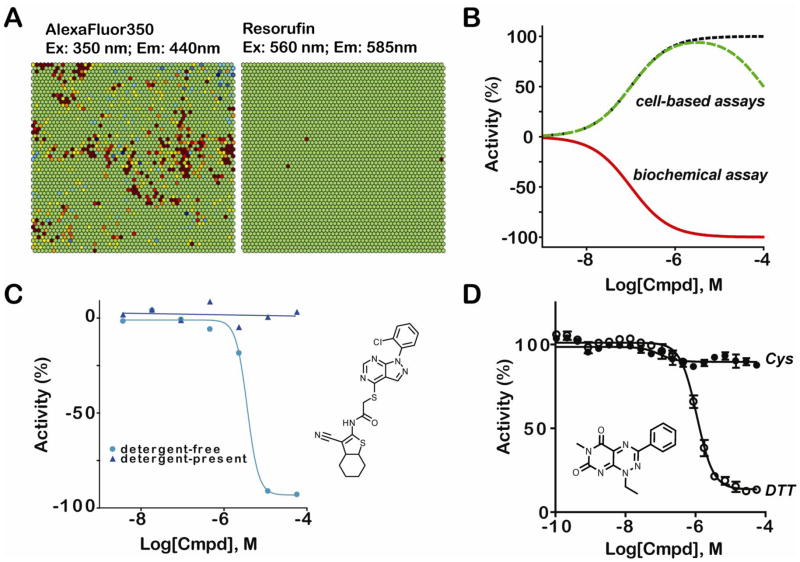
The prevalence of fluorescence interference also depends on the concentration of compound used in the assay relative to the assay fluorophore. This is especially a problem in HTS if fluorophores are used at low concentrations, for example, 1nM or less, in screens with compounds that are commonly tested at concentrations of 10μM or greater, so that if a compound sample is fluorescent, it has a high likelihood of interfering with genuine assay signal [15]. Efforts to profile a representative compound collection (http://pubchem.ncbi.nlm.nih.gov; PubChem Assay IDs – AIDs – 587–594, 709) indicated that ~2–5% of the compounds in the library fluoresced in the blue spectral region (e.g. AlexFluor 350, coumarin-like fluorescence λex~350nm/λem~440nm), whereas with excitation at ~560nm and detection at ~585nm (typical of resorufin, an orange fluorophore), only 0.004–0.01% of the library fluoresced (Fig. 2A and Table 1) [11,14]. Additionally, Simeonov et al. (2008) found that for several fluorescence-based assays involving excitation/emission in the blue spectral region, up to 50% of the actives identified in the screen were actually fluorescent [14].
Figure 2. Different types of assay interference: compound fluorescence, inhibition of the reporter enzyme firefly luciferase (FLuc), aggregation-based inhibition, and redox cycling compounds (RCC).
(A) Self-organizing map (SOM) depicting the relationship between chemical structure and fluorescence for compounds in a representative library whose fluorescence profile overlaps with AlexaFluor 350 or resorufin [14]. Each hexagon represents a cluster of compounds that are structurally related. Clusters shaded red/orange indicate an enrichment of compounds in the structural class that fluoresce (compared to the average), whereas clusters shaded blue indicate a structural class with fewer than average fluorescent compounds. As can be seen, significantly more compound classes fluoresce in the blue-spectral range, compared to those that fluorescence in the orange region. Additionally, compound classes that fluoresce tend to be related, as demonstrated by an enrichment in fluorescent compounds in adjacent clusters [14]. (B) Some compounds that inhibit the FLuc enzyme (red CRC for biochemical assay) appear to be active in cell-based FLuc reporter gene assays (green and black CRCs) due to the interaction of the compound with the FLuc enzyme. Apparent activation in cell-based assays due inhibitor-based FLuc stabilization can generate different types of CRCs, with two example CRCs shown here. If the inhibitory compound is competed off the enzyme by excess FLuc substrates found in the detection mix, or by washing of the cells prior to addition of the detection mix, a CRC similar to the one shown by the black-dotted line can be generated. If inhibition is not relieved at high compound concentrations, then a CRC similar to the one indicated by the green hatched-line is commonly seen. (C) Addition of Triton X-100 to the assay buffer significantly relieves inhibition of a compound that forms aggregates. Aggregation-based inhibition shows concentration dependence and the tendency for a compound to form aggregates is assay-specific. (D) Enzyme assay behavior of an example redox cycling compound (RCC). Data shown are from caspase-1 assays performed in the presence of 10 mM DTT (PubChem AID: 896) or 10 mM Cysteine (PubChem AID: 929). Changing the reduction potential of the buffer from a strong (DTT) to a weak (Cys) redox potential alleviates the apparent inhibition by the compound shown (PubChem CID: 647501), a RCC that generates peroxide upon exposure to DTT (PubChem AIDs: 672, 878, 787, 1234, 2035).
Detection of fluorescence in the blue spectral range is also plagued by spurious “activity”, which is often due to fluorescence of lint or other particulate matter (Fig. 1B, grey dots) [14,17] or scattering of light by precipitates/assay components. Both of these phenomena occur with greater incidence at shorter wavelengths of light [14,15,18,19]. Obtaining fluorescence intensity data at a range of compound concentrations can help identify if the activity seen is spurious or due to compound fluorescence [14,20]. Compound fluorescence is reproducible and concentration dependent, with the degree of fluorescence detected proportional to compound concentration (Fig. 1B, blue dots) [14]. If activity follows a concentration-response, it is important to use an orthogonal assay (ideally using a different output reporter, like bio-, chemi-, or electrochemi-luminescence) to determine if the activity is independent of the assay format/technique [14]. There are a number of ways to significantly reduce the occurrence of fluorescence-based artifacts in an assay, as indicated in Table 1.
Compounds that inhibit firefly luciferase: concentration-dependent inhibition or activation in cell-based assays
Firefly luciferase (FLuc)-based bioluminescence assays are highly favored in HTS due to their sensitivity (extremely low endogenous background signal leads to superior signal to background ratio relative to fluorescence methods) [21] and dynamic response in cell-based reporter gene assays, due to a relatively short FLuc protein half-life [22]. FLuc itself, however, is an enzyme, and is thus susceptible to inhibition by small molecules used in screening. A profiling effort of a 70K compound library determined that at least 3% of the library inhibited the enzymatic activity of FLuc in a concentration-dependent manner (Fig. 1C) [6]. This is actually thought to be an underestimate of the true number of FLuc-active compounds in the library, as weakly competitive FLuc inhibitors were likely not identified using this assay format. Chemical series that include compounds found to inhibit FLuc include quinolines, 1,2,4-oxadiazoles, and benzthiazoles (also benzimidazoles and benzoxazoles) - the latter being structurally similar to the natural FLuc substrate, D-luciferin [6].
Although the short protein half-life of FLuc in cells allows FLuc reporter gene assays to be exquisitely responsive to signal modulation by compounds, it also predisposes the FLuc protein to stabilization by compounds. This phenomenon was originally described by Thompson et al. (1991), in which it was found that incubating cells with certain compounds led to an accumulation of FLuc protein, by interaction with and stabilization of the FLuc protein by the compound, which ultimately led to increased bioluminescent output upon detection (Fig 2B and Supplementary Figure S1) [22]. This apparent activation is due entirely to off-target activity of the compound against the FLuc reporter protein, and also shows concentration-dependence. Retrospective analysis of cell-based FLuc reporter gene assays indicates that as many as 60% of the actives identified for a given screen are actually compounds that act as FLuc inhibitors [23]. In addition, cell-based assays that require long incubation times with compound, or that measure an increase in FLuc activity from an initially low basal level of activity, are especially prone to a significant enrichment of FLuc inhibitors amongst active compounds [23,24]. Further complicating the identification of FLuc inhibitors in cell-based assays is that these compounds can either appear as activators or inhibitors of the cell-based assay depending upon the affinity of the compound for FLuc, the basal FLuc concentration, whether the compound acts as a competitive or non-competitive inhibitor, and what type of detection reagent is used [24,25].
Determining whether a given active is a FLuc inhibitor is simple and highly recommended: test the compound in a purified FLuc assay using KM concentrations of substrates (ATP and D-luciferin). Results from profiling efforts carried out by Auld et al. (2008) are publically available (PubChem AID 411) [6] and have already assisted investigators in the identification of compounds that inhibit FLuc [26]. Additionally, in order to confirm genuine activity against the target, compounds identified in a FLuc assay should be re-tested in an orthogonal assay using an alternate reporter before being tested in secondary assays. It should also be mentioned that it is possible for a compound that inhibits an enzymatic reporter to also have relevant biological activity [27,28].
Promiscuous enzymatic inhibition in biochemical assays: compound aggregation
Compound aggregation was recently discovered to be one of the main causes for promiscuous enzyme inhibition [12,29–32]. Under certain conditions, above certain concentrations, some compounds self-associate to form an aggregate structure, which, at 50–400nm in size, can be visualized by transmission electron microscopy (TEM) [31]. Evidence suggests that enzymes are sequestered on the surface of the aggregate particles, where their function is non-specifically inhibited [30,33]. Prevention of compound aggregation through addition of nonionic detergents, such as Triton X-100, effectively relieves enzyme inhibition by this mechanism [30].
Whether or not a compound is prone to aggregation is dependent upon properties of the compound itself, the assay conditions [11,12] and the protein target [34]. For this reason, and because compounds that aggregate are structurally diverse [31], interference due to aggregate inhibition must be empirically determined for a given assay [5,11]. Generally, though, compounds tend to aggregate at micromolar concentrations, and a compound that aggregates at a higher concentration may have legitimate biological activity at lower concentrations [12,30]. Compound interference by aggregation is relatively easy to identify with a little work, largely because aggregation-based inhibition has hallmark characteristics, as shown in Table 1. While the exact mechanism of how compound aggregates inhibit enzymes is unclear [35], it has been found that addition of 0.01–0.1% Triton X-100 to assay reagents generally allows for significant relief of aggregation-based inhibition (Fig. 2C) [12,30–32], thus making it possible to design a biochemical assay that is less sensitive to this form of inhibition. Significantly, Jadhav et al. (2010) found that a screen run without detergent resulted in 15-times more actives than a screen run in the presence of detergent [11]. A protocol to detect aggregation-based inhibition is published and available [36].
In an effort to estimate the prevalence of aggregation-based inhibition for a typical HTS involving a biochemical assay, investigators have tested various small molecule libraries for enzyme inhibition sensitive to Triton X-100 [12,32]. Interestingly, though the same enzyme (AmpC β-lactamase) was used in two of the assays screened, the percentage of compounds that appeared to be sensitive to detergent-dependent inhibition varied between the two screens, highlighting that compound aggregation is conditional and assay-specific. In a 96-well format assay, Feng et al. (2005) found that 19% of the 1,030 drug-like compounds tested demonstrated detergent-dependent inhibition when screened at 30μM [32]. For a 1536-well assay format, performed against a titration (3nM-30μM) of each of ~70K compounds (quantitative HTS-qHTS), Feng et al. (2007) found that 95% of the actives identified in the screen were detergent-sensitive inhibitors, and consisted of 1.7% of the total library screened (PubChem AIDs 584, 585) [12]. A screen of ~200K compounds targeting the cysteine protease cruzain (PubChem AID 2249) revealed that approximately 1.9% of the library were detergent-sensitive inhibitors, indicating that the prevalence of this type of assay interference is not library specific [11]. In addition, compounds found to be aggregation-based inhibitors of AmpC were not necessarily found to inhibit cruzain, and vice versa, again underscoring the context-dependence of this phenomenon [11]. Results from these screens indicate that aggregation-based inhibition has the potential to significantly inflate the number of apparent actives identified from a screen.
Compounds Capable of Redox Cycling and Direct Oxidation
Some compounds, such as certain quinones, undergo redox dependent cycling (redox cycling compounds, or RCCs) in the presence of strong reducing agents such as dithiothreitol (DTT) and tris(2-carboxyethyl)phosphine (TCEP), which results in generation of reactive oxygen species (ROS) [37–40] (Supplemental Figure S2). DTT and other reducing agents are commonly added to buffer systems of enzymatic assays as a means to keep catalytic and key structural amino acids of enzymes in a reduced state, characteristic of a functional protein [40]. The ROS generated by redox-dependent cycling in the presence of DTT or TCEP is hydrogen peroxide (H2O2) [38,40,41], which can, in turn, oxidize cysteine residues of proteins, thus non-specifically and promiscuously inhibiting protein activity [38,39]. In this way, RCC-based interference can lead to exaggerated numbers of apparent actives in HTS assays - Smith et al. [38] noted this as the main artifact responsible for >85% of the active compounds identified in a HTS against caspase-8. Hallmarks of RCC interference can be found in Table 1.
A robust absorbance-based method for H2O2 detection, and thus identification of RCCs, suitable for HTS has been recently described [40]. This assay can detect H2O2 generation (in the 1–100μM range) by RCCs using H2O2-dependent horseradish peroxidase and monitoring oxidation-dependent absorbance changes of phenol red. This assay was used to profile ~200K compounds available in PubChem (AID: 878) [42] for their ability to generate H2O2. Several common scaffolds, including the pyrimidotriazinedione shown in Figure 2D, were found to be among the compounds that generated H2O2 in the presence of 0.8 mM DTT (peroxide generation AC50=0.9μM). Not surprisingly, these compounds were promiscuously active against a number of target proteins [42]. RCC-interference is not just restricted to biochemical assays that employ enzymes where important cysteine, tryptophan or methionine residues can be modified, it also can produce promiscuous effects in cell-based assays [42], since H2O2 is a common cellular messenger [37,39,43]. It is interesting to note that the acute toxicity associated with some RCCs, such as the quinones, has not prevented their use as anti-cancer agents [44].
Compounds can also act directly as an oxidizing agent, without the production of H2O2. In this case, the potency of such a compound is weakened in the presence of reducing reagents (e.g. ≥ 10mM DTT), which act to protect target proteins from either oxidization (as can occur with certain amino-thiophenes) or compound adduct formation (e.g. with p-catechols)[45]. A diagnostic test of target oxidation by a compound is to measure the activity of the protease caspase (which requires a thiol for catalytic activity, thus making it sensitive to oxidation) in the presence of the compound. Increased inhibition of the caspase enzyme in the absence of DTT may be indicative of non-specific cysteine oxidation [46]. It should be noted, however, that the activity of some known drugs is actually through such reactive mechanisms, examples being the sulfur rich disulfiram as well as ethacrynic acid - a drug that contains a Michael acceptor. It is thus essential that reactivity be carefully considered with respect to the desired mechanism of the compound.
Conclusion
Although compound-dependent assay interference in HTS cannot, at this time, be entirely eliminated, it is possible to significantly reduce the probability of its occurrence, in addition to making off-target activity significantly easier to differentiate from activity that is target- and pathway-specific. Acknowledging the potential interference that a given assay may be susceptible to and designing appropriate orthogonal assays to confirm compound activity are the best means of identifying artifactual activity early in the probe or drug discovery process. While assay interference adds an additional challenge to the process of HTS, fortunately, at this time, information on the various types of artifactual activity described in this Review is becoming increasingly available to aid investigators in their chemical biology experiments. Further expansion of publically available data from profiling efforts of representative compound libraries screened for various types of artifactual activity should help identify compounds that may generate false positives in future screens, as well as aid in the design of better assay strategies and systems. The challenge in dealing with assay interference will continue to evolve - as novel assays are developed for HTS (e.g. AlphaScreen®), so, too, will new artifacts specific to these assay systems become evident – and it is the hope that library profiling efforts and cheminformatics will keep pace [13], allowing researchers to focus on genuine chemical probes and avoid diversions caused by artifacts.
Supplementary Material
Acknowledgments
We would like to thank Ruili Huang for preparation and use of the SOM presented in Figure 2. We would also like to sincerely thank Anton Simeonov for data on aggregation-based inhibition used in Figure 2, as well as for critically reading the manuscript. The NIH Chemical Genomics Center is supported by funding from the Molecular Libraries Initiative of the NIH Roadmap for Medical Research.
Footnotes
Conflict of Interest: The authors have no conflicts of interest relating to this publication.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Natasha Thorne, Email: thornen@mail.nih.gov.
Douglas S. Auld, Email: dauld@mail.nih.gov.
References
- 1.Austin CP, Brady LS, Insel TR, Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 2.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazo JS. Roadmap or roadkill: a pharmacologist’s analysis of the NIH Molecular Libraries Initiative. Mol Interv. 2006;6:240–243. doi: 10.1124/mi.6.5.1. [DOI] [PubMed] [Google Scholar]
- 4.Rishton GM. Nonleadlikeness and leadlikeness in biochemical screening. Drug Discov Today. 2003;8:86–96. doi: 10.1016/s1359644602025722. [DOI] [PubMed] [Google Scholar]
- 5 **.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. A comprehensive review that covers appropriate design and use of HTS assays, especially as pertains to specific biological targets and pathways, and provides a succinct introduction to compound interference in HTS. [DOI] [PubMed] [Google Scholar]
- 6 **.Auld DS, Southall NT, Jadhav A, Johnson RL, Diller DJ, Simeonov A, Austin CP, Inglese J. Characterization of chemical libraries for luciferase inhibitory activity. J Med Chem. 2008;51:2372–2386. doi: 10.1021/jm701302v. This paper includes the first effort to profile a chemical library for compounds with off-target activity against the common reporter firefly luciferase. [DOI] [PubMed] [Google Scholar]
- 7.Inglese J, Auld DS. High throughput screening (HTS) techniques: overview of applications in chemical biology. In: Begley TP, editor. Wiley Encyclopedia of Chemical Biology. Wiley-Interscience; 2009. p. 3188. [Google Scholar]
- 8.Spencer RW. High-throughput screening of historic collections: observations on file size, biological targets, and file diversity. Biotechnol Bioeng. 1998;61:61–67. [PubMed] [Google Scholar]
- 9 *.Shapiro AB, Walkup GK, Keating TA. Correction for interference by test samples in high-throughput assays. J Biomol Screen. 2009;14:1008–1016. doi: 10.1177/1087057109341768. This paper provides an overview of common assay interferences encountered in HTS and provides a novel method for correcting for interference. [DOI] [PubMed] [Google Scholar]
- 10.Rishton GM. Reactive compounds and in vitro false positives in HTS. Drug Discovery Today. 1997;2:382–384. [Google Scholar]
- 11 **.Jadhav A, Ferreira RS, Klumpp C, Mott BT, Austin CP, Inglese J, Thomas CJ, Maloney DJ, Shoichet BK, Simeonov A. Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of thiol protease. Journal of Medicinal Chemistry. 2010;53:37–51. doi: 10.1021/jm901070c. Comprehensive analysis of common assay interferences for a representative 1536-well fluorescence-based HTS assay indicates that aggregation and compound fluorescence were the greatest sources of non-specific activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12 *.Feng BY, Simeonov A, Jadhav A, Babaoglu K, Inglese J, Shoichet BK, Austin CP. A high-throughput screen for aggregation-based inhibition in a large compound library. J Med Chem. 2007;50:2385–2390. doi: 10.1021/jm061317y. This paper describes the first effort to profile a compound library in 1536-well format for detergent-sensitive inhibitors. [DOI] [PubMed] [Google Scholar]
- 13 *.Baell JB, Holloway GA. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J Med Chem. 2010 doi: 10.1021/jm901137j. (in press). In this comprehensive study, cheminformatics was used to identify potentially problematic compound classes found to be promiscuously active in HTS, presumably due to protein reactivity. This paper also identifies compounds that prove problematic specifically for the AlphaScreen® assay format. [DOI] [PubMed] [Google Scholar]
- 14 **.Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, et al. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51:2363–2371. doi: 10.1021/jm701301m. This paper describes the first effort to profile a chemical library for fluorescent compounds, highlighting the prevalence of blue-fluorescing compounds in libraries used for HTS. [DOI] [PubMed] [Google Scholar]
- 15.Vedvik KL, Eliason HC, Hoffman RL, Gibson JR, Kupcho KR, Somberg RL, Vogel KW. Overcoming compound interference in fluorescence polarization-based kinase assays using far-red tracers. Assay Drug Dev Technol. 2004;2:193–203. doi: 10.1089/154065804323056530. [DOI] [PubMed] [Google Scholar]
- 16.Lovering F, Bikker J, Humblet C. Escape from flatland: increasing saturation as an approach to improving clinical success. J Med Chem. 2009;52:6752–6756. doi: 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]
- 17.Oosterom J, van Doornmalen EJ, Lobregt S, Blomenrohr M, Zaman GJ. High-throughput screening using beta-lactamase reporter-gene technology for identification of low-molecular-weight antagonists of the human gonadotropin releasing hormone receptor. Assay Drug Dev Technol. 2005;3:143–154. doi: 10.1089/adt.2005.3.143. [DOI] [PubMed] [Google Scholar]
- 18.Turek-Etienne TC, Small EC, Soh SC, Xin TA, Gaitonde PV, Barrabee EB, Hart RF, Bryant RW. Evaluation of fluorescent compound interference in 4 fluorescence polarization assays: 2 kinases, 1 protease, and 1 phosphatase. J Biomol Screen. 2003;8:176–184. doi: 10.1177/1087057103252304. [DOI] [PubMed] [Google Scholar]
- 19.Comley J. Assay interference: a limiting factor in HTS? Drug Discovery World. 2003 Summer;:91–98. [Google Scholar]
- 20.Imbert PE, Unterreiner V, Siebert D, Gubler H, Parker C, Gabriel D. Recommendations for the reduction of compound artifacts in time-resolved fluorescence resonance energy transfer assays. Assay Drug Dev Technol. 2007;5:363–372. doi: 10.1089/adt.2007.073. [DOI] [PubMed] [Google Scholar]
- 21.Fan F, Wood KV. Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol. 2007;5:127–136. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]
- 22.Thompson JF, Hayes LS, Lloyd DB. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene. 1991;103:171–177. doi: 10.1016/0378-1119(91)90270-l. [DOI] [PubMed] [Google Scholar]
- 23 *.Auld DS, Thorne N, Nguyen DT, Inglese J. A specific mechanism for nonspecific activation in reporter-gene assays. ACS Chem Biol. 2008;3:463–470. doi: 10.1021/cb8000793. This study highlights that compounds that inhibit firefly luciferase can cause apparent activation of firefly luciferase activity in cell-based assays at concentrations commonly used in screening. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Auld DS, Thorne N, Maguire WF, Inglese J. Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression. Proc Natl Acad Sci U S A. 2009;106:3585–3590. doi: 10.1073/pnas.0813345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inglese J, Thorne N, Auld DS. Reply to Peltz et al: Post-translational stabilization of the firefly luciferase reporter by PTC124 (Ataluren) Proceedings of the National Academy of Sciences of the United States of America. 2009;106:E65–E65. doi: 10.1073/pnas.0901936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heitman LH, van Veldhoven JP, Zweemer AM, Ye K, Brussee J, APIJ False positives in a reporter gene assay: identification and synthesis of substituted N-pyridin-2-ylbenzamides as competitive inhibitors of firefly luciferase. J Med Chem. 2008;51:4724–4729. doi: 10.1021/jm8004509. [DOI] [PubMed] [Google Scholar]
- 27.Bakhtiarova A, Taslimi P, Elliman SJ, Kosinski PA, Hubbard B, Kavana M, Kemp DM. Resveratrol inhibits firefly luciferase. Biochem Biophys Res Commun. 2006;351:481–484. doi: 10.1016/j.bbrc.2006.10.057. [DOI] [PubMed] [Google Scholar]
- 28.Herbst KJ, Allen MD, Zhang J. The cAMP-dependent protein kinase inhibitor H-89 attenuates the bioluminescence signal produced by Renilla Luciferase. PLoS One. 2009;4:e5642. doi: 10.1371/journal.pone.0005642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seidler J, McGovern SL, Doman TN, Shoichet BK. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J Med Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- 30 *.McGovern SL, Helfand BT, Feng B, Shoichet BK. A specific mechanism of nonspecific inhibition. J Med Chem. 2003;46:4265–4272. doi: 10.1021/jm030266r. This study demonstrates that aggregation-based inhibition of enzymes can be reversed and prevented by non-ionic detergents. [DOI] [PubMed] [Google Scholar]
- 31.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 32.Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. High-throughput assays for promiscuous inhibitors. Nat Chem Biol. 2005;1:146–148. doi: 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]
- 33.Coan KE, Shoichet BK. Stoichiometry and physical chemistry of promiscuous aggregate-based inhibitors. J Am Chem Soc. 2008;130:9606–9612. doi: 10.1021/ja802977h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giannetti AM, Koch BD, Browner MF. Surface plasmon resonance based assay for the detection and characterization of promiscuous inhibitors. J Med Chem. 2008;51:574–580. doi: 10.1021/jm700952v. [DOI] [PubMed] [Google Scholar]
- 35.Ryan AJ, Gray NM, Lowe PN, Chung CW. Effect of detergent on “promiscuous” inhibitors. J Med Chem. 2003;46:3448–3451. doi: 10.1021/jm0340896. [DOI] [PubMed] [Google Scholar]
- 36.Feng BY, Shoichet BK. A detergent-based assay for the detection of promiscuous inhibitors. Nat Protoc. 2006;1:550–553. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bova MP, Mattson MN, Vasile S, Tam D, Holsinger L, Bremer M, Hui T, McMahon G, Rice A, Fukuto JM. The oxidative mechanism of action of ortho-quinone inhibitors of protein-tyrosine phosphatase alpha is mediated by hydrogen peroxide. Arch Biochem Biophys. 2004;429:30–41. doi: 10.1016/j.abb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Smith GK, Barrett DG, Blackburn K, Cory M, Dallas WS, Davis R, Hassler D, McConnell R, Moyer M, Weaver K. Expression, preparation, and high-throughput screening of caspase-8: discovery of redox-based and steroid diacid inhibition. Arch Biochem Biophys. 2002;399:195–205. doi: 10.1006/abbi.2002.2757. [DOI] [PubMed] [Google Scholar]
- 39.Brisson M, Nguyen T, Wipf P, Joo B, Day BW, Skoko JS, Schreiber EM, Foster C, Bansal P, Lazo JS. Redox regulation of Cdc25B by cell-active quinolinediones. Mol Pharmacol. 2005;68:1810–1820. doi: 10.1124/mol.105.016360. [DOI] [PubMed] [Google Scholar]
- 40 **.Johnston PA, Soares KM, Shinde SN, Foster CA, Shun TY, Takyi HK, Wipf P, Lazo JS. Development of a 384-well colorimetric assay to quantify hydrogen peroxide generated by the redox cycling of compounds in the presence of reducing agents. Assay Drug Dev Technol. 2008;6:505–518. doi: 10.1089/adt.2008.151. This paper describes a simple colorimetric assay to detect hydrogen peroxide generated by compounds in the presence of reducing agents such as dithiothreitol (DTT). It is also the first study to profile a chemical library for redox-cycling activity that leads to enzyme inhibition (via hydrogen peroxide generation) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lal M, Rao R, Fang X, Schuchmann H-P, von Sonntag C. Radical-induced oxidation of dithiothreitol in acidic oxygenated aqueous solution: a chain reaction. Journal of the American Chemical Society. 1997;119:5735–5739. [Google Scholar]
- 42.Soares KM, Blackmon N, Shun TY, Shinde SN, Takyi HK, Wipf P, Lazo JS, Johnston PA. Profiling the NIH small molecule repository for compounds that generate H2O2 by redox cycling in reducing environments. Assay Drug Dev Technol. 2010 doi: 10.1089/adt.2009.0247. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2009;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolton JL, Trush MA, Penning TM, Dryhurst G, Monks TJ. Role of quinones in toxicology. Chem Res Toxicol. 2000;13:135–160. doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- 45.Huth JR, Mendoza R, Olejniczak ET, Johnson RW, Cothron DA, Liu Y, Lerner CG, Chen J, Hajduk PJ. ALARM NMR: a rapid and robust experimental method to detect reactive false positives in biochemical screens. J Am Chem Soc. 2005;127:217–224. doi: 10.1021/ja0455547. [DOI] [PubMed] [Google Scholar]
- 46.Crowe A, Huang W, Ballatore C, Johnson RL, Hogan AM, Huang R, Wichterman J, McCoy J, Huryn D, Auld DS, et al. Identification of aminothienopyridazine inhibitors of tau assembly by quantitative high-throughput screening. Biochemistry. 2009;48:7732–7745. doi: 10.1021/bi9006435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnston PA, Johnston PA. Cellular platforms for HTS: three case studies. Drug Discov Today. 2002;7:353–363. doi: 10.1016/s1359-6446(01)02140-7. [DOI] [PubMed] [Google Scholar]
- 48.Feng BY, Toyama BH, Wille H, Colby DW, Collins SR, May BC, Prusiner SB, Weissman J, Shoichet BK. Small-molecule aggregates inhibit amyloid polymerization. Nat Chem Biol. 2008;4:197–199. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson RL, Huang R, Jadhav A, Southall N, Wichterman J, MacArthur R, Xia M, Bi K, Printen J, Austin CP, et al. A quantitative high-throughput screen for modulators of IL-6 signaling: a model for interrogating biological networks using chemical libraries. Mol Biosyst. 2009;5:1039–1050. doi: 10.1039/b902021g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson RL, Huang W, Jadhav A, Austin CP, Inglese J, Martinez ED. A quantitative high-throughput screen identifies potential epigenetic modulators of gene expression. Anal Biochem. 2008;375:237–248. doi: 10.1016/j.ab.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shukla SJ, Nguyen DT, Macarthur R, Simeonov A, Frazee WJ, Hallis TM, Marks BD, Singh U, Eliason HC, Printen J, et al. Identification of pregnane X receptor ligands using time-resolved fluorescence resonance energy transfer and quantitative high-throughput screening. Assay Drug Dev Technol. 2009;7:143–169. doi: 10.1089/adt.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ljosa V, Carpenter AE. High-throughput screens for fluorescent dye discovery. Trends Biotechnol. 2008;26:527–530. doi: 10.1016/j.tibtech.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 53.Epps DE, Taylor BM. A competitive fluorescence assay to measure the reactivity of compounds. Anal Biochem. 2001;295:101–106. doi: 10.1006/abio.2001.5173. [DOI] [PubMed] [Google Scholar]
- 54.Zhou S, Chan E, Duan W, Huang M, Chen YZ. Drug bioactivation, covalent binding to target proteins and toxicity relevance. Drug Metab Rev. 2005;37:41–213. doi: 10.1081/dmr-200028812. [DOI] [PubMed] [Google Scholar]
- 55.Slater K. Cytotoxicity tests for high-throughput drug discovery. Curr Opin Biotechnol. 2001;12:70–74. doi: 10.1016/s0958-1669(00)00177-4. [DOI] [PubMed] [Google Scholar]
- 56.Xia M, Huang R, Witt KL, Southall N, Fostel J, Cho MH, Jadhav A, Smith CS, Inglese J, Portier CJ, et al. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ Health Perspect. 2008;116:284–291. doi: 10.1289/ehp.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho MH, Niles A, Huang R, Inglese J, Austin CP, Riss T, Xia M. A bioluminescent cytotoxicity assay for assessment of membrane integrity using a proteolytic biomarker. Toxicol In Vitro. 2008;22:1099–1106. doi: 10.1016/j.tiv.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crisman TJ, Parker CN, Jenkins JL, Scheiber J, Thoma M, Kang ZB, Kim R, Bender A, Nettles JH, Davies JW, et al. Understanding false positives in reporter gene assays: in silico chemogenomics approaches to prioritize cell-based HTS data. J Chem Inf Model. 2007;47:1319–1327. doi: 10.1021/ci6005504. [DOI] [PubMed] [Google Scholar]
- 59.Crouch SPM, Slater KJ. High-throughput cytotoxicity screening: hit and miss. Drug Discovery World. 2001;6:S48–S53. [Google Scholar]
- 60.Melhuish WH. Molecular luminescence spectroscopy. Pure and Applied Chemistry. 1984;56:231–245. [Google Scholar]
- 61.Goode DR, Totten RK, Heeres JT, Hergenrother PJ. Identification of promiscuous small molecule activators in high-throughput enzyme activation screens. J Med Chem. 2008;51:2346–2349. doi: 10.1021/jm701583b. [DOI] [PubMed] [Google Scholar]
- 62.D’Auria S, Di Cesare N, Gryczynski I, Rossi M, Lakowicz JR. On the effect of sodium dodecyl sulfate on the structure of beta-galactosidase from Escherichia coli. A fluorescence study. J Biochem. 2001;130:13–18. doi: 10.1093/oxfordjournals.jbchem.a002951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.