Abstract
Heterozygous mutations in the ribosomal protein S19 (RPS19) gene are associated with Diamond-Blackfan anemia (DBA). The mechanism by which RPS19 mediates anemia are still unclear, as well as the regulation of RPS19 expression. We show herein that RPS19 binds specifically to the 5′ untranslated region of its own mRNA with an equilibrium binding constant (KD) of 4.1±1.9 nM. We investigated the mRNA binding properties of two mutant RPS19 proteins (W52R and R62W) identified in DBA patients. We observed a significant increase in KD for both proteins (16.1±2.1 and 14.5±4.9 nM, respectively), indicating a reduced RNA binding capability (p<0.05). We suggest that the binding of RPS19 to its mRNA has a regulatory function and hypothesize that the weaker RNA binding of mutant rRPS19 may have implications for the pathophysiological mechanisms in DBA.
Keywords: RPS19, RNA/protein interaction, ribosomal protein S19, 5′TOP sequence, DBA
Introduction
Ribosomal protein S19 (RPS19) belongs to the small 40S subunit of the ribosome, where it presumably binds 18S rRNA [1]. In humans, heterozygous mutations in the RPS19 gene have been identified in approximately 25% of patients with Diamond-Blackfan Anemia (DBA), a congenital bone marrow failure syndrome characterized by a decrease or absence of erythroid precursors [2; 3; 4]. RPS19 is highly conserved among species [5] and missense mutations associated with DBA were recently located using the 3-dimensional structure of an archaeal Rps19 protein [6]. The precise mechanism by which mutations in the ubiquitously expressed RPS19 give rise to the tissue specific phenotype observed in DBA is still unclear [7]. It has been suggested that RPS19 is a target for proteins interacting with the ribosome, or as a factor for the recruitment and translation of particular mRNAs [8; 9]. However, it is believed that perturbed ribosomal biosynthesis and impaired ribosomal function are important contributing mechanisms in DBA [2; 10]. Indeed, RPS19 is required for ribosomal RNA maturation [11; 12], suggesting that RPS19 may have RNA binding properties. Protein-RNA binding may also serve as a way to regulate the levels of ribosomal proteins through the interaction with specific mRNAs. Ribosomal proteins are required in stoichiometric amounts for subunit assembly and reduced amounts of one component involved in this process will be rate limiting [13; 14]. Several ribosomal proteins have been shown to regulate their expression levels through binding to their own mRNA. For example, human ribosomal proteins L30, S13 and S26 interact with their respective mRNA in a feedback mechanism that regulates splicing and ribosomal protein levels [15; 16; 17].
We show herein that rRPS19 binds to the 5′UTR of its own mRNA. We also show that two DBA missense mutations introduced into rRPS19 impair this binding. From our results we suggest that this protein-mRNA interaction has a regulatory function on RPS19 protein levels.
Material and Methods
Recombinant proteins
Recombinant RPS19 (rRPS19) was prepared as described [18]. rRPS19[W52R] and rRPS19[R62W] were kindly provided by T. Buender.
Synthesis of RNA substrates
RNA substrates (wild type 5′UTRs: LONG, SHORT and TOP; 5′UTR mutants: MUT1, MUT2 and MUT3; scrambled control, SCR) were synthesized by in vitro transcription, using T7-RNA polymerase (USB) and PCR products made in standard PCR reactions with Platinum-Taq-DNA Polymerase (GE Healthcare) amplified from human genomic DNA or using two partially complementary primers specific for the construct to be synthesized (sequences available upon request). In vitro transcribed RNA was 5′-end labeled using γ-32P -ATP (Perkin Elmer) and T4-polynucleotide kinase (Fermentas) following manufacturer’s recommendations.
PAGE analysis / RNA substrate Refolding / Secondary structure prediction
5′-end labeled RNA substrates were refolded in buffer GS (30 mM Tris/Cl, pH 7.7, 10 mM MgCl2, 1 mM EDTA, 200 mM KCl, 15% Glycerol) by heating 5 minutes to 70°C and subsequent cool-down for 30 minutes at room temperature. Refolded RNA was kept on ice until further use. Approximately 0.2 pmol of refolded RNA were analyzed on a 10% denaturing (8 M Urea) or 10% native polyacrylamide gel (Acrylamind/Bisacrylamide 19:1). Gels were dried and RNA visualized by phosphoimager analysis. Secondary structure predictions were performed using the mfold v3.1 web server at http://www.bioinfo.rpi.edu/applications/mfold [19].
Electrophoretic Mobility Shift Assay
Approximately 0.6 pmol 5′-end labeled substrate RNA were refolded and subsequently incubated for 15 minutes at room temperature with increasing amounts of rRPS19 (5 to 180 nM) in Buffer GS (30 mM Tris/Cl, pH 7.7, 10 mM MgCl2 1 mM EDTA, 200 mM KCl, 15% Glycerol) supplemented with 0,7 μg/μl BSA, 26,7 ng/μl yeast RNA (Ambion), 2,7 ng/μl polycytidylic acid (Poly(C) RNA, GE Healthcare). Mock incubations were run in parallel without any protein or adding BSA. Complexes and free RNA were separated on a native 4% polyacrylamide gel containing 2% glycerol. Dried gels were analyzed by phosphoimager analysis on a BAS 1800 II Bio-Imager (Fujifilm).
Filter Binding Assays
Refolded 5′end labeled substrate RNA (increasing from 0 to 200 nM) was incubated with 80 nM recombinant protein in buffer GS supplemented with 0.7 μg/μl BSA, 26.7 ng/μl yeast RNA, 2.7 ng/μl poly(C) RNA for 15 minutes at room temperature. Reactions were applied to a nitrocellulose filter mounted on a vacuum manifold and equilibrated in GS buffer without glycerol and washed three times with 0.75 ml ice-cold buffer. Nitrocellulose filters were dried and retained RNA determined by Cherenkov counting. The equilibrium binding constant (KD) was obtained by plotting experimental data, and subsequently analyzing the data with non-linear regression (Origin7.0® software, OriginLab Corporation) using the first order binding equation
Obtained KD values for the different mutants were tested for significant difference using student’s two-tailed t-test.
Conservation analysis, alignment and 3D-structure visualization
Conservation of the first exon of human RPS19 was analyzed using the UCSC genome browser (http://www.genome.ucsc.edu/). Sequences of the RNA substrates were aligned using BioEdit Sequence Alignment Editor v7.0.5.3. The 3D structure model of RPS19 was generated using ViewerLite v5.0 and PDB file 2v7f (http://www.rcsb.org/pdb/home/home.do)[6].
Results
RPS19 binds to the 5′UTR of its mRNA
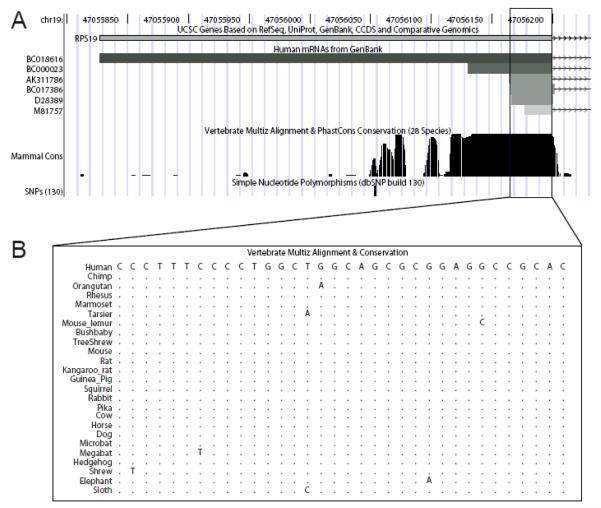
Recent studies have shown that the first exon of RPS19 is almost completely conserved among vertebrates suggesting a functional importance (Figure 1A, [5]). The first exon is non-coding and comprises the 5′ untranslated region only, as the ATG-start codon is located at the immediate beginning of the second exon. Several exon 1 (i.e. 5′UTR) variants have been identified spanning 35 to 372 nucleotides all of which are continuous with genomic DNA (NCBI database at htpp://www.ncbi.nih.gov/ and the UCSC genome browser at http://www.genome.ucsc.edu/; figure 1A). The shortest and capped 5′UTR variant identified in the databases spans 35 nucleotides and contains a so-called 5′terminal oligopyrimidine (5′TOP) sequence [20].
Figure 1.
The 5′ region of RPS19 with known UTR variants and degree of conservation.
(A) Exon 1 and the 5′UTR variants retrieved from the NCBI database (http://www.ncbi.nih.gov/) using the UCSC genome browser (http://www.genome.ucsc.edu/). The most abundant 5′UTR variant (boxed) spans 35 nucleotides and contains a 5′TOP sequence [20]. The sequence conservation between mammals is indicated (for details, see Martinez Barrio et al [5]). No common variants have been reported in the human polymorphism database (dbSNP, http://www.ncbi.nlm.nih.gov/SNP/, build 130). (B) Alignment of the human 35 nt 5′UTR region containing the TOP sequence with orthologous sequences from 24 additional mammalian species. The 5′UTR TOP sequence (D28389) is highly conserved.
We investigated binding of RPS19 to the 5′UTR sequence of its own mRNA by incubating increasing amounts of recombinant RPS19 (rRPS19) with three distinct 5′end labeled RNA substrates followed by an electrophoretic mobility shift assay (data not shown). These substrates correspond to accession numbers BC018616 (375 nt 5′UTR including AUG, termed LONG), BC000023 (72 nt 5′UTR including AUG, termed SHORT) and D28389 (38 nt 5′UTR including AUG, termed TOP), respectively (see figure 1). We observed binding of rRPS19 to all three 5′UTR variants. As the TOP sequence is present in all three variants (figure 1), we reasoned that binding is dependant on this sequence. Therefore, we focused our subsequent analysis on the TOP variant in order to characterize the binding in more detail.
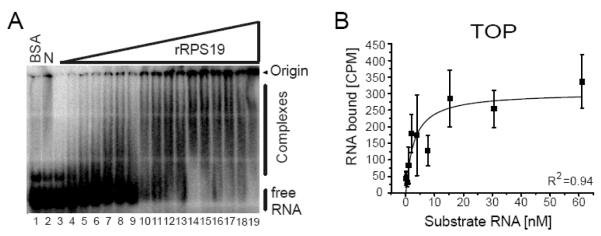
rRPS19 binds to the TOP RNA substrate in a concentration dependent manner, indicating that rRPS19 has RNA binding properties (figure 2A). We then used filter binding assays to quantify the RNA binding of rRPS19 and to clarify whether the observed binding is RNA-sequence specific. The equilibrium binding constant (KD) was measured by incubating rRPS19 with increasing amounts of TOP RNA substrate and the binding constant was determined to KD[TOP]=4.1±1.9 nM (figure 2B, table 1).
Figure 2.
RPS19 binds to the 5′UTR of its own mRNA.
(A) Binding of rRPS19 to the TOP sequence spanning nt −35 to +3 of RPS19 mRNA as analyzed by EMSA. A 32P-labeled TOP RNA substrate was incubated without protein (N: lane 2), with increasing amounts of rRPS19 (lane 3 to 19) or BSA (lane 1). Complexes were separated on a native 4% polyacrylamide gel and visualized by phosphoimager analysis. Binding of rRPS19 to the RNA substrate is demonstrated by retardation of the RNA substrate in the gel (complexes), retention at the top of the gel, and by disappearance of free RNA. (B) A filter binding assay was used to determine the equilibrium binding constant of RPS19 binding to the TOP RNA substrate. The average ±standard deviation of five independent experiments was plotted. The correlation coefficient is indicated (R2).
Table 1.
Equilibrium binding constants (KD) for the different RNA substrates determined using wild-type rRPS19.
| RNA mutant | KD [nM]a |
|---|---|
| TOP | 4.1±1.9 |
| MUT1 | 5.3±3.5 |
| MUT2 | 7.0±4.9 |
| MUT3 | n.d. |
| SCRAMBLED | n.d. |
Mean ± standard deviation of at least four independent experiments; n.d., not detectable.
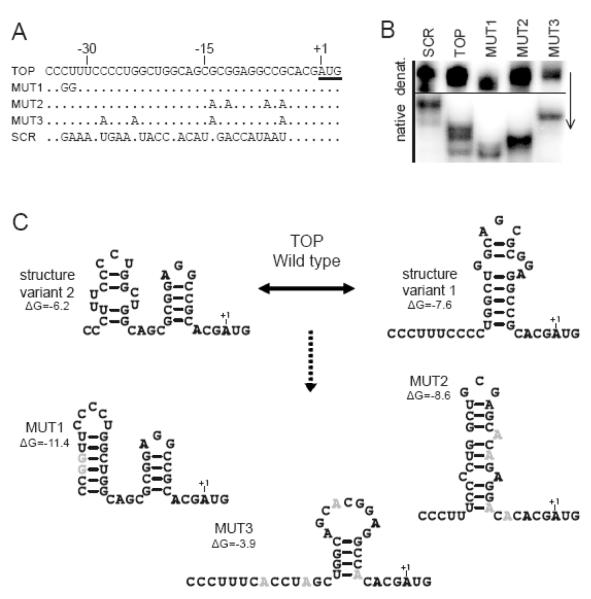
Attempts to map the rRPS19 binding site on the RNA substrate by enzymatic structure probing gave inconsistent results (data not shown). Therefore, we investigated the structural conformation of the wild type TOP RNA substrate by native polyacrylamide gel electrophoresis (PAGE). The wild type TOP RNA substrate appears as multiple bands upon non-denaturing gel electrophoresis suggesting that the RNA is structurally heterogeneous after refolding (figure 3A-B). This may also explain why structure probing was inconsistent.
Figure 3.
Substrate RNA variants used to characterize RNA binding properties of rRPS19.
(A) Sequence alignment of the RNA substrates, showing the wild-type (TOP) and mutated (MUT) variants. MUT: Mutation variants 1 to 3; SCR: Scrambled control. Nucleotides are numbered relative to the RPS19 start codon (underlined). (B) Comparative PAGE analysis of the secondary structure of the five RNA substrates used. The arrow indicates direction of electrophoresis. (C) Predicted secondary structures of the RNA substrates as predicted by Mfold [19]. The calculated folding energy (ΔG) is indicated. Two possible structures with similar folding energy were obtained for the TOP substrate, while only one structure was predicted for variants MUT1 and MUT2. For variant MUT3, the most likely structure with the lowest energy is indicated. No structure was obtained for the scrambled mutant.
We next introduced distinct nucleotide substitutions into the TOP RNA sequence that presumably alter or destabilize the secondary structure of the TOP RNA substrate as predicted by mfold (figure 3A-C). We obtained one distinct structure with similar folding energy for each of the mutant substrates whereas two possible structures were obtained for the wild-type TOP substrate (figure 3C). No secondary structure was obtained for the scrambled RNA substrate used as a negative control. We also analyzed the effects of the introduced mutations on the secondary structure by native PAGE (figure 3B). All TOP mutant substrates are of equal size but display distinct electrophoretic mobility in a native gel. The mutant RNA variants migrate as single bands, indicating that the introduced mutations lock the substrates into distinct conformations (figure 3B-C). This supports the secondary structure prediction of the mutant RNA substrates using mfold (figure 3C, [19]).
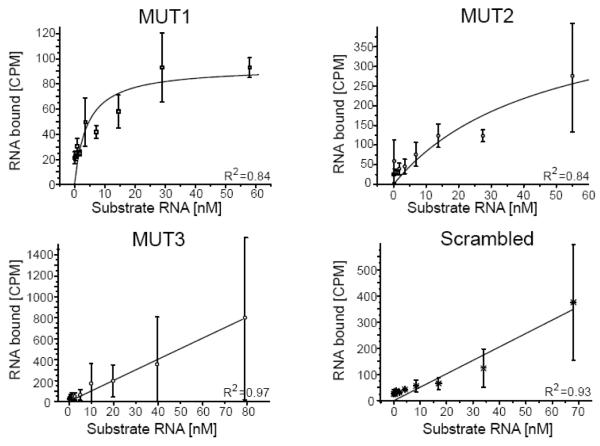
We then measured the equilibrium binding constant (KD) of the interactions between rRPS19 and all mutant RNA variants. A strong interaction was observed between rRPS19 and the variants MUT1 and MUT2, whereas MUT3 and the scrambled variant showed linear relationships indicating unspecific binding (figure 4, table 1). Taken together, the results show that rRPS19 binds specifically to the 5′UTR of its own mRNA and to a sequence corresponding to nt −30 to −5 (figure 3 and figure 4). This binding is presumably dependant on the secondary structure of the RNA.
Figure 4.
Analytic investigation of rRPS19 binding to the 5′UTR variants.
The equilibrium binding constant were determined for the RNA substrate variants (see figure 3) using a filter binding assay. rRPS19 binds to MUT1 and MUT2 with slightly increased KDs compared to wild-type TOP RNA (table 1). The substrate MUT3 and the scrambled control show a linear relationship (indicative of unspecific binding) and KDs could not be obtained (table 1). The average ±standard deviation of three independent experiments was plotted. The correlation coefficients are indicated (R2).
RPS19 mutations associated with DBA alter rRPS19’s RNA binding capacity
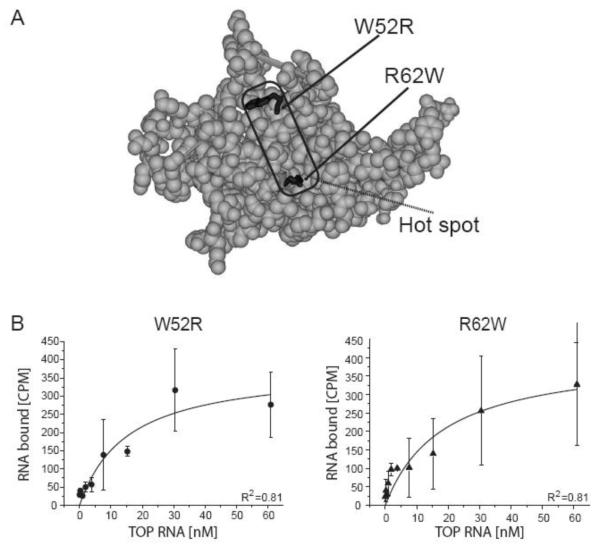
We obtained two mutant rRPS19 proteins (rRPS19[W52R] and rRPS19[R62W]) and analyzed their 5′UTR binding properties. Both missense mutations have been identified in patients with DBA [4]. The two mutations W52R and R62W are located in a central region of the 3-dimensional structure of Rps19 which is compatible with RNA binding (figure 5) [6; 9]. The central region is suggested to be a hot spot region for DBA associated mutations [9]. Analysis of the equilibrium binding constant revealed significantly increased KDs for both mutant proteins compared to wild-type rRPS19 (p<0.05; figure 5, table 2). These data show that the mutant proteins have a significantly reduced binding capacity compared to wild-type RPS19.
Figure 5.
rRPS19 protein variants with two DBA associated missense mutations (W52R, R62W) show reduced binding to the 5′UTR of RPS19 mRNA.
(A) Three-dimensional structure model of RPS19 [6]. The region with the most frequent mutations in DBA (for details see [9]) are indicated and the side chains of the mutations analyzed are highlighted in black. Residues are numbered according to human RPS19. (B) The equilibrium binding constant was determined using filter binding for rRPS19[W52R] and rRPS19[R62W] using the wild type TOP RNA substrate. The average ± standard deviation of three independent experiments was plotted and KDs were calculated using Origin®7.0. Both mutant proteins show a significantly reduced binding capacity compared to wild type protein (p<0.05; see table 2). The correlation coefficients are indicated (R2).
Table 2.
Equilibrium binding constants (KD) for two RPS19 protein mutants determined using wild-type TOP RNA substrate.
Mean ± standard deviation of at least three independent experiments
see table 1
Significantly increased compared to wild type rRPS19 protein using student’s two-tailed t-test (p[W52R]=0.0001, p[R62W]=0.020).
Discussion
Diamond-Blackfan anemia is a rare congenital anemia in which 25% of the patients have a mutation in the ribosomal protein S19 (RPS19) gene [3][2; 5]. RPS19 is a ubiquitously expressed protein and the mechanisms mediated by RPS19 gene mutations causing DBA are largely unknown. Recent work has shown that RPS19 is important for rRNA maturation and cell cycle regulation [10; 11; 12; 13; 21; 22]. However, the finding that mutations in other ribosomal protein genes are associated with DBA [23; 24; 25; 26; 27] indicates that the disease is directly related to ribosome function and/or ribosomal biogenesis [2; 28; 29]. Ribosome biosynthesis and subunit assembly are tightly regulated mechanisms both of which are dependant on stoichiometric amounts of ribosomal proteins [13; 14; 30; 31]. The production of ribosomal proteins is controlled at different levels. In this study we show that RPS19 binds to its own mRNA and hypothesize that RPS19 binds to its own mRNA as part of a regulatory mechanism at the translational level. A feed-back mechanism achieved through binding of a ribosomal protein to distinct parts of its mRNA has previously been shown for RPS13, RPS26 and RPL30 [15; 16; 17]. Indeed, our results show that RPS19 binds to a 25 bp motif within its 5′UTR. Furthermore, we show that the binding is reduced when two DBA associated mutations, W52R and R62W, are introduced into rRPS19. In this way, the level of RPS19 in the cell would depend on both the transcriptional activity of the gene (i.e. the amount of mRNA synthesized) and on the translational efficiency of the mRNA (i.e. the amount of RPS19 that binds to the mRNA). Hypothetically, binding of RPS19 to its mRNA could thus be a mechanism to coordinate transcript and protein levels [8].
Sequences similar to the 5′TOP sequence of the RPS19 mRNA are found in 5′UTRs of other ribosomal protein mRNAs [20; 32]. This sequence motif has been shown to play a role in the specific translational regulation of ribosomal proteins and proteins involved in translation [20; 32]. From our findings it may be hypothesized that RPS19 binds mRNAs containing motifs similar to that of the 5′UTR in RPS19. This could be of importance for the previously observed post-transcriptional co-regulation of ribosomal protein levels [13; 14]. Another possibility is that binding of RPS19 to different 5′UTRs is of importance to recruit particular mRNAs to the ribosome and to promote their translation [8; 9]. Several mRNAs would in such a case compete for binding to RPS19, resulting in a potentially tight co-regulation. Our results open up for a possible mechanism underlying RPS19 expression, co-regulation of ribosomal protein levels and translational regulation of specific transcripts mediated by RNA binding properties of RPS19. This regulatory mechanism of RPS19 expression and the possible regulation of other proteins by RPS19 through RNA binding might add to the complexity of DBA.
In summary, we have shown that RPS19 binds to the 5′UTR of its own mRNA. The binding is altered by the structure of the substrate and, notably, by DBA associated mutations in RPS19. We suggest that this mechanism is of importance for the regulation of RPS19 levels and, possibly, for ribosomal subunit biosynthesis with implications for disease mechanisms in DBA.
Acknowledgements
We thank A. Warren for valuable discussions and T. Bünder for RPS19 mutants. This work was supported by National Institutes of Health (5R01-HL079567-04), Marcus Borgström Foundation (to JS), Selanders Foundation, Swedish Research Council, T. and R. Söderbergs Foundation, Children’s Cancer Foundation of Sweden and Uppsala University.
Footnotes
Potential Referees:
1. Kathleen Sakamoto, Department of Pediatrics, Hematology/Oncology, Johnsson Comprehensive Cancer Centre, David Geffen School of Medicine at UCLA, 10833 Le Conte Avenue, Los Angeles, California, USA, kms@ucla.edu
2. Radek Cmejla, Department of Cell Biology, Hematology and Blood Transfusion, P.O. Box 74, U Nemocnice 1, 128 20, Prague, Czech Republic, cmejla@uhkt.cz
3. Irma Dianzani, Dipartimento di Scienze Medichi, Universitá di Piemonte Orientale, Via Solaroli 17, 28100 Novara, Italy, irmadia@med.unipmn.it
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lutsch G, Stahl J, Kargel HJ, Noll F, Bielka H. Immunoelectron microscopic studies on the location of ribosomal proteins on the surface of the 40S ribosomal subunit from rat liver. Eur J Cell Biol. 1990;51:140–50. [PubMed] [Google Scholar]
- [2].Ellis SR, Lipton JM. Chapter 8 diamond blackfan anemia: a disorder of red blood cell development. Curr Top Dev Biol. 2008;82:217–41. doi: 10.1016/S0070-2153(07)00008-7. [DOI] [PubMed] [Google Scholar]
- [3].Vlachos A, Ball S, Dahl N, Alter BP, Sheth S, Ramenghi U, Meerpohl J, Karlsson S, Liu JM, Leblanc T, Paley C, Kang EM, Leder EJ, Atsidaftos E, Shimamura A, Bessler M, Glader B, Lipton JM. Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol. 2008 doi: 10.1111/j.1365-2141.2008.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Campagnoli MF, Ramenghi U, Armiraglio M, Quarello P, Garelli E, Carando A, Avondo F, Pavesi E, Fribourg S, Gleizes PE, Loreni F, Dianzani I. RPS19 mutations in patients with Diamond-Blackfan anemia. Hum Mutat. 2008 doi: 10.1002/humu.20752. [DOI] [PubMed] [Google Scholar]
- [5].Barrio A. Martinez, Eriksson O, Badhai J, Frojmark AS, Bongcam-Rudloff E, Dahl N, Schuster J. Targeted resequencing and analysis of the Diamond-Blackfan anemia disease locus RPS19. PLoS One. 2009;4:e6172. doi: 10.1371/journal.pone.0006172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gregory LA, Aguissa-Toure AH, Pinaud N, Legrand P, Gleizes PE, Fribourg S. Molecular basis of Diamond Blackfan anemia: structure and function analysis of RPS19. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Morimoto K, Lin S, Sakamoto K. The functions of RPS19 and their relationship to Diamond-Blackfan anemia: A review. Mol Genet Metab. 2007;90:358–362. doi: 10.1016/j.ymgme.2006.11.004. [DOI] [PubMed] [Google Scholar]
- [8].Angelini M, Cannata S, Mercaldo V, Gibello L, Santoro C, Dianzani I, Loreni F. Missense mutations associated with Diamond-Blackfan anemia affect the assembly of ribosomal protein S19 into the ribosome. Hum Mol Genet. 2007;16:1720–7. doi: 10.1093/hmg/ddm120. [DOI] [PubMed] [Google Scholar]
- [9].Flygare J, Karlsson S. Diamond-Blackfan anemia: erythropoiesis lost in translation. Blood. 2007;109:3152–4. doi: 10.1182/blood-2006-09-001222. [DOI] [PubMed] [Google Scholar]
- [10].Badhai J, Frojmark AS, Davey E, Schuster J, Dahl N. Ribosomal protein S19 and S24 insufficiency cause distinct cell cycle defects in Diamond-Blackfan anemia. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbadis.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Da Costa L, Gleizes PE. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–83. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, Ellis SR. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109:980–6. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Badhai J, Frojmark AS, Razzaghian HR, Davey E, Schuster J, Dahl N. Posttranscriptional down-regulation of small ribosomal subunit proteins correlates with reduction of 18S rRNA in RPS19 deficiency. FEBS Lett. 2009 doi: 10.1016/j.febslet.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. Rna. 2008;14:1918–29. doi: 10.1261/rna.1132008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ivanov AV, Malygin AA, Karpova GG. Human ribosomal protein S26 suppresses the splicing of its pre-mRNA. Biochim Biophys Acta. 2005;1727:134–40. doi: 10.1016/j.bbaexp.2004.12.011. [DOI] [PubMed] [Google Scholar]
- [16].Malygin AA, Parakhnevitch NM, Ivanov AV, Eperon IC, Karpova GG. Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic Acids Res. 2007;35:6414–23. doi: 10.1093/nar/gkm701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Macias S, Bragulat M, Tardiff DF, Vilardell J. L30 binds the nascent RPL30 transcript to repress U2 snRNP recruitment. Mol Cell. 2008;30:732–42. doi: 10.1016/j.molcel.2008.05.002. [DOI] [PubMed] [Google Scholar]
- [18].Malygin A, Baranovskaya O, Ivanov A, Karpova G. Expression and purification of human ribosomal proteins S3, S5, S10, S19, and S26. Protein Expr Purif. 2003;28:57–62. doi: 10.1016/s1046-5928(02)00652-6. [DOI] [PubMed] [Google Scholar]
- [19].Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–30. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- [21].Miyake K, Utsugisawa T, Flygare J, Kiefer T, Hamaguchi I, Richter J, Karlsson S. Ribosomal protein S19 deficiency leads to reduced proliferation and increased apoptosis but does not affect terminal erythroid differentiation in a cell line model of Diamond-Blackfan anemia. Stem Cells. 2008;26:323–9. doi: 10.1634/stemcells.2007-0569. [DOI] [PubMed] [Google Scholar]
- [22].Idol RA, Robledo S, Du HY, Crimmins DL, Wilson DB, Ladenson JH, Bessler M, Mason PJ. Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol Dis. 2007 doi: 10.1016/j.bcmd.2007.02.001. [DOI] [PubMed] [Google Scholar]
- [23].Doherty L, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Clinton C, Schneider HE, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Glader B, Arceci RJ, Farrar JE, Atsidaftos E, Lipton JM, Gleizes PE, Gazda HT. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am J Hum Genet. 86:222–8. doi: 10.1016/j.ajhg.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Farrar JE, Nater M, Caywood E, McDevitt MA, Kowalski J, Takemoto CM, Talbot CC, Jr., Meltzer P, Esposito D, Beggs AH, Schneider HE, Grabowska A, Ball SE, Niewiadomska E, Sieff CA, Vlachos A, Atsidaftos E, Ellis SR, Lipton JM, Gazda HT, Arceci RJ. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112:1582–92. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gazda HT, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Zaucha JM, Glader B, Niemeyer C, Meerpohl JJ, Atsidaftos E, Lipton JM, Gleizes PE, Beggs AH. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–80. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gazda HT, Grabowska A, Merida-Long LB, Latawiec E, Schneider HE, Lipton JM, Vlachos A, Atsidaftos E, Ball SE, Orfali KA, Niewiadomska E, Da Costa L, Tchernia G, Niemeyer C, Meerpohl JJ, Stahl J, Schratt G, Glader B, Backer K, Wong C, Nathan DG, Beggs AH, Sieff CA. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79:1110–8. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007 doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- [28].Liu JM, Ellis SR. Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood. 2006;107:4583–8. doi: 10.1182/blood-2005-12-4831. [DOI] [PubMed] [Google Scholar]
- [29].Ellis SR, Massey AT. Diamond Blackfan anemia: A paradigm for a ribosome-based disease. Med Hypotheses. 2006;66:643–8. doi: 10.1016/j.mehy.2005.09.010. [DOI] [PubMed] [Google Scholar]
- [30].Perry RP. Balanced production of ribosomal proteins. Gene. 2007 doi: 10.1016/j.gene.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Granneman S, Tollervey D. Building ribosomes: even more expensive than expected? Curr Biol. 2007;17:R415–7. doi: 10.1016/j.cub.2007.04.011. [DOI] [PubMed] [Google Scholar]
- [32].Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, Avruch J, Meyuhas O. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol Cell Biol. 2001;21:8671–83. doi: 10.1128/MCB.21.24.8671-8683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]