Abstract
Recent technological advances that allow faster and cheaper DNA sequencing are now driving biological and medical research. In this review, we provide an overview of state-of-the-art next-generation sequencing (NGS) platforms and their applications, including in genome sequencing and resequencing, transcriptional profiling (RNA-Seq) and high-throughput survey of DNA-protein interactions (ChIP-Seq) and of the epigenome. Particularly, we focus on how new methods made possible by NGS can help unravel the biological and genetic mechanisms of aging, longevity and age-related diseases. In the same way, however, NGS platforms open discovery not available before, they also give rise to new challenges, in particular in processing, analyzing and interpreting the data. Bioinformatics and software issues plus statistical difficulties in genome-wide studies are discussed, as well as the use of targeted sequencing to decrease costs and facilitate statistical analyses. Lastly, we discuss a number of methods to gather biological insights from massive amounts of data, such as functional enrichment, transcriptional regulation and network analyses. Although in the fast-moving field of NGS new platforms will soon take center stage, the approaches made possible by NGS will be at the basis of molecular biology, genetics and systems biology for years to come, making them instrumental for research on aging.
Keywords: bioinformatics, epigenetics, functional genomics, senescence, systems biology
1. Introduction
Next-generation sequencing (NGS) technologies allow massive-scale DNA sequencing at a low cost and are now driving biomedical research (Church, 2006; Mardis, 2008). In a near future, large-scale projects in the life and medical sciences will depend on DNA sequencing as a readout, given that NGS platform s offer superior performance and specificity in many applications (Ansorge, 2009). The purpose of this review is to summarize the current state-of-the-art in sequencing technologies from an end-user point-of-view. Particularly, our goal is to provide an overview of how biogerontologists can employ these technologies and the methods derived thereof to advance our knowledge of aging, longevity and age-related diseases. Lastly, we discuss some of the potential problems inherent to such high-throughput approaches, in particular at the levels of bioinformatics, statistics and data interpretation, and suggest possible solutions.
2. State-of-the-art in sequencing technology
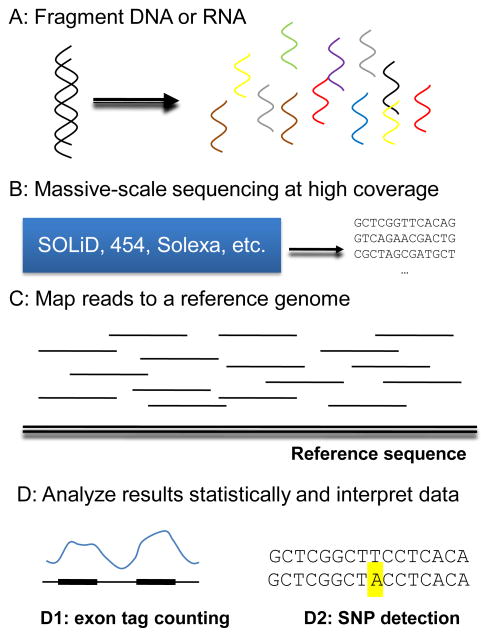
The current crop of NGS platforms, also called second-generation sequencing technologies, was driven by the initial sequencing of genomes using traditional Sanger sequencing and its variants. Genome assemblies provided a reference to which the shorter sequence reads generated by NGS methods could be mapped back to, allowing for cheaper and faster sequencing (Fig. 1). This has made second-generation platforms particularly adequate for studying humans and model organisms whose genomes had been sequenced already, as detailed below.
Figure 1.
Diagrammatic scheme of a NGS platform pipeline. Typically, DNA or RNA is fragmented into smaller pieces (A). Libraries are constructed from the fragments (or miRNAs) and sequenced at a high coverage (B). The sequenced reads are aligned to a reference genome (C) and the results are analyzed statistically and interpreted (D). Depending on the specific application, reads may be counted across genes (D1), SNPs detected (D2) or other analyses carried out.
Generally, the principle behind second-generation platforms is to randomly fragment DNA or RNA into smaller pieces and construct a DNA or cDNA library—given their smaller sizes, micro RNAs (miRNAs) can be used directly to make cDNA libraries. Libraries are sequenced at a high coverage and the sequenced reads are then mapped into the reference genome of the species (Fig. 1). For genome resequencing, genetic variants can then be identified in the sample genome and for transcriptional profiling the number of reads mapping to each exon or mRNA can be quantified. Though mapping reads to a reference genome is much more common now, for an increasing number of projects, reads can also be assembled de novo. All these applications are further detailed below.
Current second-generation platforms include the 454 system from Roche, which can yield 500 bp reads, plus the Illumina/Solexa platform and the SOLiD platform from Applied Biosystems, which both yield shorter reads while generating considerably more data per run and are thus more cost-effective in terms of price per bp sequenced (Table 1). For more details on the techniques employed on each platform, please see Ansorge (2009).
Table 1.
Comparison between the most common NGS platforms
| Platform | 454 | Illumina/Solexa | SOLiD |
|---|---|---|---|
| Sequence data per run | 400–600 Mb | >25,000 Mb | >20,000 Mb |
| Read length (bases) | 400–500 | 35–75 | 50 |
| Cost per MB | ~$60 | ~$2 | ~$2 |
Notes: Platform specifications for 454 (http://www.454.com), Illumina/Solexa (http://www.illumina.com) and SOLiD (http://solid.appliedbiosystems.com). Costs are estimates taken from Shendure and Ji (2008). Given the dynamic nature of the NGS field, the above values are meant as a snapshot of current NGS platforms and are likely to be outdated soon.
While the read lengths offered by NGS are still lower than what can be obtained with traditional Sanger sequencing (up to 1000 bp), these technologies are faster and much more cost-effective, meaning much more sequence data can be generated in a given experiment. Sanger sequencing costs approximately $500 per Mb at the time of writing, or roughly 100 times more than sequencing with the 454 system (Table 1). It should be noted, however, that not only are read lengths shorter when using NGS but raw accuracy is inferior when compared to Sanger sequencing, albeit both read lengths and accuracy are improving in NGS technologies and the lower costs of NGS platforms means a higher coverage can be used resulting in an improved accuracy of the consensus sequences (Shendure and Ji, 2008).
At present, the choice of NGS platform for a given project depends on the nature of the experimental design. Generally speaking, for tag counting (e.g., expression profiling), shorter reads are preferable as they are more cost-effective. For de novo sequencing, longer reads are more adequate since they facilitate assembly generation (Shendure and Ji, 2008). Nonetheless, the idiosyncrasies of each platform (e.g., the type and frequency of errors) must be considered when choosing the best system for a given experiment. Instrument costs are similar (~$500k) at the time of writing.
3. Emerging applications of NGS
Second-generation platforms are revolutionizing research in genomics. Below we offer an overview of the applications made possible by NGS platforms, having in mind specific foci in aging research and how these technologies can help biogerontologists.
3.1. Genome sequencing and resequencing
NGS platforms allow the resequencing of an organism’s genome at an affordable price. In fact, resequencing the human genome is now several orders of magnitude lower than the approximately $3 billion it initially cost. This ability to sequence an individual’s (or animal’s) genome means that NGS technologies are extremely powerful for studying genetic variation and help bridge the gap between genotype and phenotype in population studies in both humans and model organisms. Human genome resequencing and proof-of-principle of its power to discover DNA variation has been demonstrated using 454 (Wheeler et al., 2008), Illumina/Solexa (Bentley et al., 2008) and SOLiD (McKernan et al., 2009).
Succinctly, resequencing can be used for large-scale SNP and mutation discovery. Individual lifespans within humans and laboratory model species show modest heritability of 10–35% (Christensen et al., 2006; Finch and Tanzi, 1997; Vijg and Suh, 2005), which may be consistent with the growing evidence for modest allelic influences on chronic diseases of aging (Finch, 2007). Because of this considerable inter-individual variability in longevity in both humans and model organisms, genome resequencing is a powerful approach to identify genetic variants associated with longevity and/or age-related diseases (de Magalhaes, 2009). Already a number of studies have examined the association between gene variants and human longevity using a variety of experimental setups (Tan et al., 2006; Vijg and Suh, 2005). For example, two recent studies in different populations reported gene variants in FOXO3A associated with human longevity (Flachsbart et al., 2009; Willcox et al., 2008). As detailed below, NGS will make this type of studies more informative than ever.
In addition to studying longevity as a trait, there is great interest in determining genes contributing to age-related diseases. Considerable progress has been made already with the identification of genes associated with neurodegenerative diseases like Parkinson’s disease (Singleton et al., 2003), type 2 diabetes (Sladek et al., 2007) and age-related macular degeneration (Klein et al., 2005). Large-scale association studies focusing on multiple diseases have also revealed new associations between genes and age-related diseases (Wellcome Trust Case Control Consortium, 2007). Nonetheless, much remains to be discovered about the genetics of age-related diseases. NGS will be a powerful tool to confirm and extend previous associations between gene variants and age-related diseases. In fact, one recent study—employing the Illumina/Solexa system for genotyping—associated two loci with Alzheimer’s disease that had not been previously associated with the disease (Harold et al., 2009).
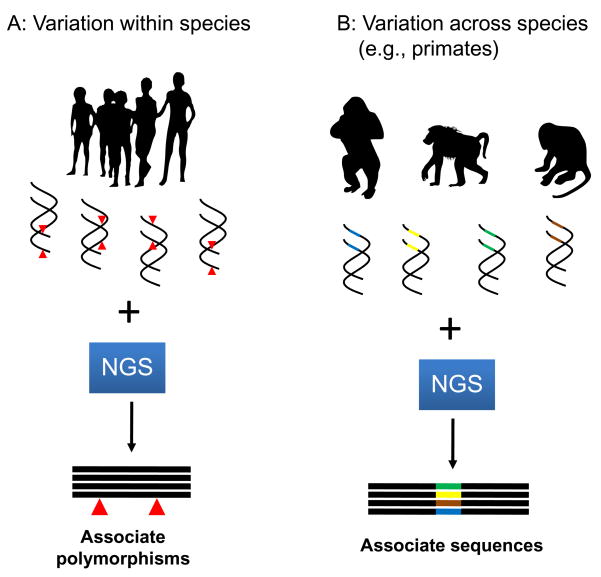
Compared to the typical genome-wide association study (GWAS) using SNPs, genome resequencing—if performed accurately—has the great advantage of allowing researchers to identify the specific nucleotide(s) associated with a given phenotype. For example, the specific frequency of all DNA variants can be compared between long-lived individuals and controls, offering a much more comprehensive picture of the genetics of longevity (but see further down for caveats of such studies). GWAS of longevity and age-related diseases using resequencing techniques will play a pivotal role to identify new alleles that determine longevity, susceptibility to age-related diseases and how environmental factors interact with genes to influence these phenotypes (Fig. 2).
Figure 2.
NGS as a tool to uncover the genetic variation within (A) and across species (B). Within the humans species there is great variability in longevity and in susceptibility to age-related diseases, which to a large degree has a genetic basis. The capacity to resequence the genome of multiple individuals affected by a given disease or with different longevities offers powerful opportunities to identify polymorphisms and mutations that contribute to longevity and to age-related diseases (A). A large variance in lifespans is observed among similar species (B). For example, among primates, humans can live over 100 years, gorillas over 50 and Old World monkeys up to 40 (de Magalhaes, 2009). With the decreasing costs of sequencing, it is now possible to explore the genetic basis of such differences and identify coding or regulatory sequences that contribute to a given organism having a shorter or longer lifespan (B). Animals were drawn using fonts by Alan Carr.
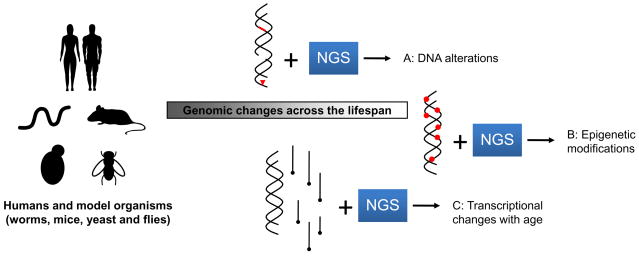
NGS can be used for discovery of structural rearrangements. These are likely to be important in GWAS and, for example, detecting copy number variation is made considerably easier since NGS platforms can employ paired-end sequencing (i.e., the sequencing from both ends of a DNA molecule), even if the analysis of such data can be complex (Korbel et al., 2007). Studies of structural variation might also offer new clues regarding genome changes during aging. It has even been suggested that it will be possible in a near future to resequence the genome of a large number of individual cells from humans (or another organism) across the lifespan to obtain a map of somatic nuclear and mitochondrial DNA mutations that accumulate in different tissues with age (Busuttil et al., 2007; Salipante and Horwitz, 2007). Tissue and cell type analysis of somatic mutations during aging can now be considered in longitudinal studies of individuals from various disease risk groups (Fig. 3).
Figure 3.
Employing NGS platforms to study age-related changes. During the course of an organism’s lifetime, a number of genomic changes occur. NGS allows these changes to be quantified at a whole-genome level. Changes to be DNA, from single nucleotide mutations to large chromosome rearrangements, can be detected (A). Likewise, genome-wide epigenetic changes across the lifespan (or between different lifestyles or diets) can be assayed. Lastly, transcriptional changes with age can be quantified with unprecedented accuracy using NGS (C). Mouse and human figures were drawn using fonts by Alan Carr.
Sequencing of the genome opens the door to a number of analytical techniques to be employed in the study of a given organism, such as transcriptional profiling (but see below) and advanced proteomics like mass spectrometry. With the lowering costs of sequencing, nontraditional model organisms are now becoming affordable targets for genome sequencing and consequently for modern molecular biology tools and high-throughput methods in particular. While Sanger sequencing is still required for de novo sequencing, at least for higher animals even if to a decreasing extent (i.e., Sanger sequencing might be employed at a low coverage followed by high coverage 454 sequencing), it is anticipated that de novo sequencing of virtually any species with NGS will soon be plausible. Sequencing of genomes at much more affordable prices will allow individual laboratories to sequence the genome of their organism(s) of choice, allowing non-traditional model organisms to join the post-genome era.
Affordable, fast sequencing of whole genomes will greatly advance the field of comparative genomics (Fig. 2). Considering the large variation in longevity and aging rates between similar species and even strains of the same species (e.g., in dogs or mice) the comparative biology of aging is bound to greatly benefit from NGS platforms (Austad, 2009; de Magalhaes, 2009). For example, studies of molecular evolution rates may allow researchers to identify genes with patterns of selection associated with the evolution of lifespan across species with unprecedented power and accuracy (de Magalhaes and Church, 2007).
One other area of potential interest not yet (to our knowledge) explored by biogerontologists is metagenomics. In metagenomics, en entire microbial community can be sequenced, for example to provide insights into microbial ecosystems in extreme environments. One emerging area is the survey of gut flora, which for example has been applied to the honey bee (Cox-Foster et al., 2007). Obesity may also be influenced by gut flora, as shown in microbial transfer experiments between lean and obese mice and in identical twins discordant for obesity (Turnbaugh and Gordon, 2009). Gut microbial flora might also play a role in longevity (including in humans), as suggested from caloric restriction studies in dogs (Wang et al., 2007), which metagenomics will be able to survey.
3.2. Transcriptional profiling: RNA-Seq
Several studies of aging gene expression profiles have been carried out already using microarrays. Briefly, such studies have identified age-related changes in key pathways and processes, such as inflammatory and mitochondrial processes (de Magalhaes et al., 2009b), provided new mechanistic insights into aging and its modulation by genes and interventions like caloric restriction (Lee et al., 2000), and have helped identify biomarkers of aging (de Magalhaes et al., 2009b; Lee et al., 2000).
Although the ability of microarrays to interrogate thousands of transcripts in a cost-effective manner has provided key insights into a number of processes, including aging, microarrays have important limitations. One intrinsic problem of microarrays is their lack of sensitivity to low abundance transcripts, potentially missing important information such as certain transcription factors that tend to have low expression levels. Moreover, microarrays have limitations for profiling the emerging mRNA complexity of different transcripts originating from a single gene and noncoding transcripts (Cloonan et al., 2008; Marioni et al., 2008). To understand transcriptional networks, it is also crucial to examine miRNAs. This new family of non-coding small 21–25-nucleotide RNAs has a major role in gene regulation during development and in adult maintenance (He and Hannon, 2004). Therefore, in order to unravel transcriptional networks of aging and of interventions that modulate aging, it is necessary to obtain a global view of the transcriptome.
One major application of NGS is in transcriptional studies (Fig. 3). The idea, based on previous technologies like serial analysis of gene expression (SAGE), involves generating libraries which are then fragmented and sequenced. By deeply sequencing the transcriptome and determining the frequency of each gene in the sequence sample by matching it to the genome sequence (also called RNA-Seq), one can obtain a digital measure of the presence and levels of known and unknown genes (Mortazavi et al., 2008). Succinctly, the relative expression of each gene’s mRNA—and, depending on coverage and experimental design, even of each exon—can be calculated by counting the number of sequenced tags mapping to it. Normalized gene signals can then be used to calculate the differential expression between samples and conditions, just like in a typical microarray experiment.
A number of studies have demonstrated that RNA-Seq is considerably more sensitive than traditional microarrays, has a much greater dynamic range and can detect splice variants and non-coding RNAs that would otherwise go undetected (Berezikov et al., 2006; Marioni et al., 2008; Mortazavi et al., 2008; Sultan et al., 2008). The capacity of RNA-Seq to survey transcriptomes in a near-complete fashion has been recently demonstrated in mouse embryonic stem cells and shown to detect up to 50% more genes than microarrays (Cloonan et al., 2008). Because the dynamic range of RNA-Seq is higher, microarrays and RNA-Seq correlate relatively well for genes with medium expression levels but not for genes with high or low expression levels (Wang et al., 2009). Discrepancies between results obtained with microarrays and RNA-Seq have been tested by qRT-PCR and most errors were found to occur in microarrays (Marioni et al., 2008).
One large-scale gene expression study of aging is AGEMAP which stands for Atlas of Gene Expression in Mouse Aging Project (Zahn et al., 2007). This project aims to identify and catalogue genes differentially expressed with age in all possible tissues of mice. AGEMAP studies conducted to date employed traditional microarrays (Xu et al., 2007; Zahn et al., 2007), so while this approach has already proven useful we suggest that many more insights await when NGS is employed in this type of study.
NGS platforms will allow researchers to characterize the aging transcriptome with exceptional resolution and identify transcripts associated with age as well as with life-extension due to genetic or environmental interventions in order to provide new insights about aging and its underlying molecular and genetic mechanisms. As mentioned above, although 454 has been used for RNA-Seq, Illumina/Solexa and SOLiD are likely to be more cost-efficient in terms of coverage and depth (Table 1).
3.3. DNA-protein interactions: ChIP-Seq
Following from microarray (RNA-Chip) and RNA-Seq, a similar relationship exists between ChIP-Chip and ChIP-Seq. Chromatin immunoprecipitation (ChIP) is a method used to discover DNA-protein associations, revealing the DNA interactions of a protein of interest—often its transcriptional targets. In a typical experiment, cross-links of the DNA-protein complexes are induced, usually with formaldehyde. After chromatin fragmentation and immunoprecipitation of the DNA linked protein, unlinking of the DNA-protein association can reveal the DNA sequence the protein was associated with (Solomon et al., 1988). A readout of the data can be obtained by hybridizing the sequences to a microarray (ChIP-Chip).
Several studies have highlighted the importance of using ChIP-Chip, by itself or in combination with other approaches, in aging research. One of these discoveries was the finding that histones are modified at the telomeres in senescent human cells (Meier et al., 2007). With NGS technologies, however, it is now possible to sequence the immunoprecipitated DNA in a high-throughput manner (ChIP-Seq) which results in greater power, specificity and cost-effectiveness (Robertson et al., 2007; Valouev et al., 2008).
Despite their usefulness, ChIP-Chip studies have limitations, many of which are reminiscent of the microarray limitations mentioned above. It has been pointed out by Valouev et al. (2008) that in higher organisms ChIP-chip data prove to be noisy and have low resolution. In a study by Robertson et al. (2007), it was found that clear advantages existed of ChIP-Seq over ChIP-Chip. To begin, when performed on genomes of large size, ChIP-Seq was estimated at being around one order of magnitude less expensive than ChIP-Chip. Also, ChIP-Seq requires less input material than ChIP-Chip and can cover a larger fraction of the genome at a higher resolution. Furthermore, because ChIP-Seq is based on genome resequencing it could permit the detection of mutations within the DNA binding site’s sequence. Just like RNA-Seq has replaced microarrays as the state-of-the-art technology for gene expression profiling, ChIP-Seq has taken over ChIP-Chip.
3.4. Sequencing the epigenome
Another level of genome regulation is the epigenome. Epigenetics represents chemical alterations of the DNA and histones that impact on function. Such alterations have been suggested to play an important role in aging (Fraga and Esteller, 2007; Richardson, 2003). Second-generation sequencing platforms have also given rise to more powerful techniques with which to study the epigenome. Chip-Seq, in fact, can be used to identify histone methylation marks (Barski et al., 2007). Data from the epigenome may be employed in a complementary fashion to RNA-Seq to more fully understand transcriptional regulation.
Emerging techniques in epigenomics include methyl-DNA immunoprecipitation (MeDIP), which allows a genome-wide analysis of DNA methylation (Pomraning et al., 2009). Bisulphite sequencing has also been used to study patterns of cytosine methylation in a genome-wide fashion, for example in cancer (Korshunova et al., 2008). Whole-genome analysis of epigenetic marks at the finer resolution delivered by NGS platforms promise to be of great value to biogerontologists (Fig. 3). Epigenetic regulation of transcription is essential to understand how organisms respond to their environment and to diet in particular. For example, prenatal exposure to famine in the Dutch Hunger winter of 1944–1945 altered methylation of the IGF2 gene as observed 60 years later (Heijmans et al., 2008). Thus these new techniques offer new strategies for analysis of dietary manipulations of aging across the life course. Epigenetic signatures might also be useful to identify biomarkers of aging and age-related diseases, potentially leading to improved diagnosis and risk prediction of the latter.
4. Problems, pitfalls and possible solutions
NGS technologies open discovery not available before but also new challenges, in particular in processing, analyzing and interpreting the data. Specifically, and since gigabase-scale data are generated (Table 1), the statistical and bioinformatics analyses are among the most challenging aspects of any such projects (Pop and Salzberg, 2008). One of the consequences of the decreasing costs of sequencing is the way the bottleneck in biomedical research is shifting from data generation to data analysis. Biological, medical and health sciences are becoming increasingly more dependent on information technology.
A number of software packages have already been developed for processing data from NGS platforms, including software provided by the manufacturers for aligning reads, detecting variants and generating assemblies from second-generation sequencing platforms (Shendure and Ji, 2008; Trapnell and Salzberg, 2009). Examples include ELAND (from Illumina) and Maq (http://maq.sourceforge.net). These and other packages are more suitable than software previously used in genomics. The popular BLAST tool, for example, is not optimized for aligning short reads and tends to be time-consuming when used for processing data from NGS platforms. For a recent review of the fast-changing software available for mapping short reads please see Trapnell and Salzberg (2009).
4.1. Sequencing just the right regions, the right amount of times
Resequencing an entire genome or the full transcriptome is still not only relatively expensive in terms of the reagents necessary for the sequencing itself, but time-consuming in regard to the statistical and bioinformatic analyses. Fortunately, a number of methods are now available that are capable of restricting regions for sequencing in order to increase sample size and/or coverage. In fact, in a number of experiments it is preferable to sequence a subset of the genome in a larger number of samples than to sequence the whole genome of a few samples. Interestingly, given the massive increases in sequencing capacity, accompanied by an equally impressive decrease in costs, in many experiments it is now adequate to sequence multiple samples in a single NGS instrument run. Some NGS platforms allow samples to be physically separated in the same run plus multiplexing can be achieved by means of barcoding methods that essentially add an index sequence into each DNA fragment allowing it to be associated with a given sample (Hamady et al., 2008; Kim et al., 2007). Pooled designs have also been proposed as an alternative to barcoding in resequencing (Prabhu and Pe’er, 2009).
A number of techniques have recently emerged that allow researchers to capture thousands of genomic regions which can later be sequenced (Shendure and Ji, 2008). For example, NimbleGen microarrays allow the capture by hybridization of thousands of predefined genomic regions, such as exons, which can then be used for targeted resequencing (Sugarbaker et al., 2008). One recent proof-of-concept study employed microarrays for targeted enrichment of protein-coding sequences, the subsequent sequencing of which provided candidate genes for Freeman–Sheldon syndrome, a rare Mendelian disorder (Ng et al., 2009). Multiplex PCR amplification can also be used for a candidate gene approach as these allow specific regions to be amplified and used to make libraries prior to sequencing.
In a given transcriptional study, a number of different strategies can be employed depending on its aims. Full length cDNA sequencing is not always necessary if quantifying the expression of genes is enough for a given analysis. As such, it is possible to sequence specific regions of transcripts, typically from the 5′-end, which decrease costs (Kim et al., 2007). RNA-Seq can be carried out at varying degrees of sequencing coverage. Transcriptional profiling with NGS at a low coverage is called digital gene expression profiling or DGE by some authors (though not all semantic issues in NGS have been resolved) and is suitable for inferring gene expression levels. Deeper, more expensive RNA-Seq, however, is capable of quantifying the expression levels of exons and of specific alleles as well as detect alternative transcripts and new splice junctions.
As described above, RNA-Seq is based on aligning sequences to a reference genome. With the improved read lengths and capacity of NGS platforms, however, it is now possible with RNA-Seq to do transcriptional studies in species without a reference genome and instead align the reads using the reference genome of a related species. Moreover, because “junk” DNA composes ~95% of the human genome, RNA-Seq may be used to sequence the regions of the genome that are known to be more important. Certainly, some transcripts may be missed, as even a complex cDNA library will not cover all transcripts and highly divergent or even novel genes may be difficult to assemble, but by and large genes can be unambiguously mapped to other genomes and annotated accordingly. For example, Vera et al. (2008) generated a de novo assembly of the transcriptome of a non-traditional eukaryote model organism (Melitaea cinxia, a butterfly), for which genomic data was not previously available. Succinctly, they created a cDNA library with equal levels across the cDNA population—since the large prevalence range of different mRNAs in cells would result in under-representation of low abundance transcripts. The normalized cDNA library was then sequenced using the 454 system (Vera et al., 2008). Of course, this approach depends on the availability of a closely related species, but with genomes from a large variety of taxa now available and with increased read lengths anticipated, RNA-Seq may be performed on a large number of species without a reference genome and can be used to sequence the regions of the genome that are more likely to be important.
4.2. Statistical challenges
Quality control issues are crucial when dealing with the massive amounts of data generated by NGS technologies, in particular because of the lower raw accuracy of these platforms when compared to Sanger sequencing. One concern is the fact that each NGS platform has its own systematic biases which need to be considered when designing and analyzing data, for example in association studies and SNP and mutation discovery experiments. A number of biases due to library preparation protocols also need to be considered plus each specific application has its own biases. Though these are too numerous to list herein, for example, in RNA-Seq it is often crucial to correct for gene length when calculating gene signals.
Resequencing individual genomes, while powerful, raises significant statistical problems. The future of human genomics lies in identifying and characterizing the natural variation found between individual genomes, in particular variants associated with diseases and major traits, such as longevity. Nonetheless, large-scale genome resequencing is expected to give rise to major mathematical and statistical challenges. The goal of GWAS is to identify correlations between a given allele or genomic region and a given trait, yet an increase in the amount of data generated requires stringent statistical thresholds to avoid false positives. Besides, many diseases and indeed longevity are affected by a large number of loci, each explaining only a small percentage of the phenotypic variance observed and often interacting with each other and with the environment in complex ways, making their detection difficult. It is thus not surprising that even GWAS employing cohorts with thousands of individuals can struggle to statistically demonstrate some of the associations found (Wellcome Trust Case Control Consortium, 2007). Longevity studies are particularly problematic because often the number of older individuals is limited and the penetrance of any longevity-conferring gene may be low since people with favorable alleles might perish due to accidents and other aging-unrelated causes. Genome resequencing efforts with NGS, like the 1000 Genomes project (http://www.1000genomes.org/), will eventually provide a comprehensive picture of normal human genetic variation to which genetic variation in specific phenotypes can be evaluated to predict the contributions of genes (Mardis, 2008). For the time being, however, rare alleles with no phenotypic consequences may be a source of false positives in GWAS using resequencing if sample size is low (Li and Leal, 2009). Conversely, given the lower raw accuracy of NGS platforms, identifying biologically-relevant rare alleles with confidence promises to be troublesome from a statistical perspective. These issues may be overcome by having suitable cohorts and experimental designs, but they are important to consider early in such projects.
While there is considerable debate regarding the adequate statistical treatment in genome-wide studies and new methods continue to be proposed, traditional Bonferroni corrections tend to be too strict for genome-wide projects, often resulting in a large number of false negatives. Calculating a false discovery rate (FDR) by random permutations of the data and then using the FDR to set a statistical cutoff for determining statistical significance has proven a powerful approach in genome-wide studies (Storey and Tibshirani, 2003), including in aging (de Magalhaes et al., 2009b). Further developments are warranted, however, in the specific statistical tests used in each application. For example, to determine differentially expressed genes between experimental conditions in RNA-Seq experiments an empirical Bayes method has been shown to moderate standard errors (Cloonan et al., 2008). Other authors have reported that using a Poisson model allowed them to detect 30% more statistically significant genes than using standard analysis (Marioni et al., 2008). Further work is required to better define the appropriate statistical methods for RNA-Seq. Moreover, in the context of biogerontology, gene expression profiles of aging have idiosyncrasies that make them difficult to analyze and interpret (de Magalhaes et al., 2009b). Specific methods are also being developed to analyze ChIP-Seq, such as the statistical framework developed by Valouev et al. (2008) to determine positions where protein complexes contact DNA, and a Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis (Down et al., 2008).
4.3. Moving from gene lists to pathways and mechanisms
It is crucial in large-scale studies, in particular transcriptional studies that tend to generate large numbers of statistically significant genes, that the data be clustered and pathways analyzed (Slonim, 2002). Despite the attractive discoveries of single gene mutations that increase the lifespan of laboratory model organisms, it is unlikely that aging is controlled by single genes. Only by studying pathways and networks as an integrated system can complex processes of aging be understood in a quantitative and experimentally amenable framework—the emerging “systems biology of aging paradigm”. Therefore, while ranked lists of genes obtained from high-throughput methods may be useful to prioritize candidates for follow-up, lists of genes and results need to be analyzed as a whole to help interpret data biologically and mechanistically. As further described below, results can also be integrated with other sources of genomic data to more faithfully understand the underlying biological processes (Giallourakis et al., 2005).
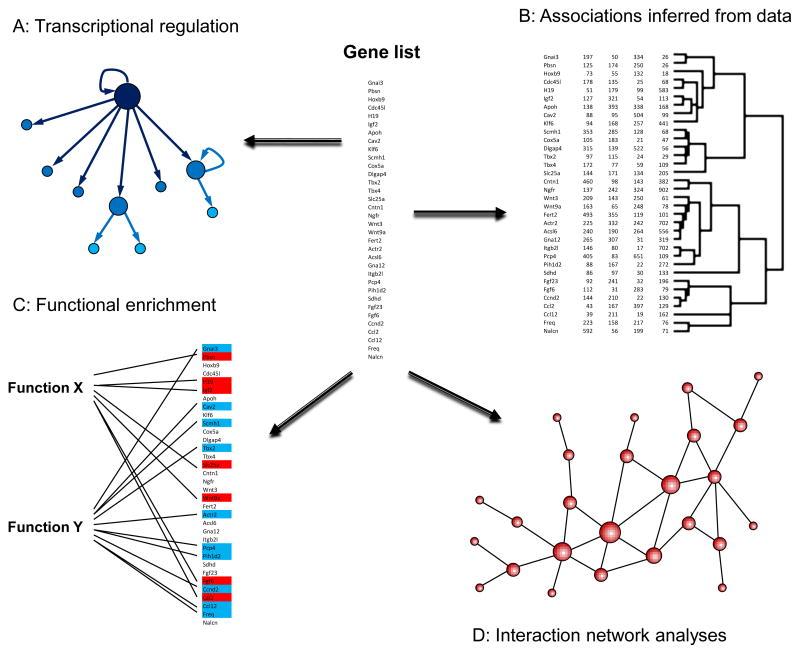
A variety of bioinformatics methods and tools are now available to analyze high-throughput data (Fig. 4). One powerful approach involves incorporating gene annotation to identify enriched functions and processes. In other words, calculate for a given list of genes which functions the genes tend to be more associated than expected by change. Functional enrichment analysis often provides insights beyond what can be obtained from lists of genes, as we recently demonstrated by analyzing age-related profiles in multiple mammalian tissues to reveal a large number of enriched functions and processes whose expression is associated with age (de Magalhaes et al., 2009b). An almost limitless number of software packages and online tools are available for functional enrichment and discover common features in small or large lists of genes, such as the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) and the Gene Set Enrichment Analysis (GSEA) (http://www.broad.mit.edu/gsea/).
Figure 4.
Moving from gene lists to biological insights. A number of data analysis methods are available to decrease the complexity of high-throughput data and gather biological information. In certain applications, transcriptional regulation information, either by incorporating existing knowledge or constructing transcriptional networks de novo, can be used to infer the causal structure of the data and infer regulatory nodes (A). Data can also be analyzed to identify new functional associations (B). For example, gene expression data can be used to cluster genes with similar profiles and identify putative functional interactions. Gene annotation information can also be integrated with experimental data to identify enriched functions and processes in large datasets (C). Lastly, data can be overlaid on protein-protein interaction networks to identify highly-connected hubs which tend to exert a regulating influence (D).
Other techniques used to analyze gene expression profiles involve clustering genes together using the readout data to identify co-expressed genes that may be under similar regulation. Gene co-expression analysis can be used to identify groups within the data and thus simplify the analysis and interpretation of the results, often coupled with visualization tools (Slonim, 2002). Another method involves overlaying results over known pathways for comparing disease vs. non-disease states, for example using software from companies like Ingenuity Systems (http://www.ingenuity.com/) and Ariadne Genomics (http://www.ariadnegenomics.com/). Most current regulatory network models are far from complete, however, in part because the technologies used thus far, such as microarrays, fail to provide a complete assessment of transcriptional complexity (Cloonan et al., 2008). As such, transcriptional networks can be constructed de novo. A variety of methods are available, such as the weighted gene-expression network analysis method (Zhang and Horvath, 2005). Briefly, in this method, co-expression concordance is determined using a Pearson correlation to identify modules of co-expressed genes. Rather than merely identify clusters of related genes, however, the algorithm derives a gene co-expression network which can be further analyzed, as discussed below.
One crucial aspect of high-throughput experiments, such as RNA-Seq, is the difficulty in determining cause and effect. Thus, it is often not easy to identify for two co-expressed genes the causal hierarchy over a biologically relevant time span like aging. The ultimate goal, of course, is to reduce the complexity of the data by identifying transcription factors that regulate numerous downstream targets in the shifts of gene expression in disease or during aging. The studies on early development in many species have identified sequences of transcriptional programs with core modules that recur in many different cell-type lineages. We anticipate that many aspects of aging will prove to involve core modules in damage responses that will be in continuity with repair processes necessary for successful development in humans and other species with prolonged pre- and postnatal development times.
In addition to ChIP-Seq described above, which provides direct evidence for transcriptional interaction between two genes, there are also in silico methods (Kim et al., 2009). For example, it is possible to search conserved functional motifs in related gene sets, such as genes clustered based on data (e.g., gene expression changes) obtained under different conditions or from functional annotation (enriched functional categories can be seen as modules of coordinately regulated genes). A number of databases and tools, such as PReMod (http://genomequebec.mcgill.ca/PReMod/), may be employed to identify regulatory candidates and reconstruct transcriptional networks. Analyses of regulatory networks can provide important insights by allowing key regulatory nodes likely to play a mechanistic role in these processes to be identified. One previous analysis of cis-regulatory motifs from gene expression profiles identified an NF-kB motif as strongly associated with aging gene expression changes (Adler et al., 2007). Not only can key regulatory nodes (e.g., transcription factors) be identified but those genes with central positions in networks—called hubs, which are attractive candidates for further studies—can be identified, for example using visualization tools like VisANT (http://visant.bu.edu).
Similar methods to those used to analyze regulatory networks and identify hubs can be employed by overlaying the experimental data (e.g., genes differentially expressed in a given condition or genes that interact with a given protein) into protein-protein interaction maps, also known as the “interactome”. Although still far from complete, such maps can be obtained from several websites, like the Human Protein Reference Database (http://www.hprd.org/) and the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (http://string.embl.de/). A number of software platforms, like Cytoscape (http://www.cytoscape.org/), can be used to visualize and analyze networks constructed based not only on protein-protein interactions but also gene co-expression, transcriptional regulation and signaling pathways. In addition to identifying central hubs, network analyses can be useful in identifying interactions modified between two or more experimental conditions.
A number of genes have already been shown to regulate aging and longevity in model organisms and many others have been associated with longevity and age-related diseases in model organisms and humans alike (Finch, 2007). Such genes are available for search and download in the GenAge database and Gene Aging Nexus that we developed (de Magalhaes et al., 2009a; Pan et al., 2007). Databases of genes associated with specific age-related diseases are also available online, such as the AlzGene database which focuses on Alzheimer’s disease (http://www.alzgene.org/). Integrating data from high-throughput studies with knowledge of genes with known associations with aging, longevity and/or age-related diseases can help identify promising candidates for follow-up and help gather additional insights on the causal structure of the data.
Overall, the goal of the integrative approaches described above is to simplify complex datasets into information that is easier to interpret biologically, for example, by detecting regulatory modules, common pathways and central hubs. These may in turn help understand age-related changes in vivo or in vitro, alterations caused by caloric restriction or mutations that confer life-extension, the genetics of longevity, age-related diseases and other processes of interest to biogerontologists.
Finally, we think it is crucial that large-scale data generated from projects employing NGS platforms, just like for microarrays and other high-throughput methods, be properly annotated and made available to the scientific community as it is often be beneficial to other researchers through re- or meta-analysis. The NCBI Short Read Archive provides such a system and researchers are encouraged to submit their data to it.
5. Concluding remarks
In this work, we provided a snapshot of NGS technologies, their applications and problems, in particular in the context of research on aging. We are fully aware that, as companies put forward new advances, the specific features of each of the platforms described above will be outdated soon and new machines are continually being developed. Another platform recently released was the Polonator (http://www.polonator.org/), developed by George Church’s laboratory at Harvard, which is considerably cheaper than its competitors, albeit reads are also shorter (13 bp or 26 bp per run for paired-end sequencing). Moreover, third-generation sequencing technologies, possibly single-molecule sequencing such as the HeliScope developed by Helicos Biosciences (http://www.helicosbio.com/), may provide even greater advances by making sequencing faster, easier and cheaper in a near future (Gupta, 2008). Human genome resequencing has already been demonstrated with Helicos (Pushkarev et al., 2009). As we approach the $1,000 cost of resequencing a human genome set by the NIH in 2004, we are confident that the approaches made possible by second-generation sequencing will be at the basis of molecular biology and genetics for years to come as genomics takes centre stage of biological and medical research.
Acknowledgments
The authors wish to thank George Church, Andy Cossins and Alistair Darby for critical reading of the manuscript as well as the staff of the Centre for Genomic Research at the University of Liverpool for useful discussions. J. P. de Magalhães thanks the BBSRC (BB/G024774/1) for supporting work in his lab. C.E. Finch is grateful for NIA support (R21AG031723).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler AS, Sinha S, Kawahara TL, Zhang JY, Segal E, Chang HY. Motif module map reveals enforcement of aging by continual NF-kappaB activity. Genes Dev. 2007;21:3244–3257. doi: 10.1101/gad.1588507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Austad SN. Comparative biology of aging. J Gerontol A Biol Sci Med Sci. 2009;64:199–201. doi: 10.1093/gerona/gln060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Busuttil RA, Garcia AM, Reddick RL, Dolle ME, Calder RB, Nelson JF, Vijg J. Intra-organ variation in age-related mutation accumulation in the mouse. PLoS One. 2007;2:e876. doi: 10.1371/journal.pone.0000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7:436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM. Genomes for all. Sci Am. 2006;294:46–54. doi: 10.1038/scientificamerican0106-46. [DOI] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA, Quan PL, Briese T, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP. Aging Research in the Post-Genome Era: New Technologies for an Old Problem. In: Foyer CH, Faragher R, Thornalley PJ, editors. Redox Metabolism and Longevity Relationships in Animals and Plants. Taylor and Francis; New York and Abingdon: 2009. pp. 99–115. [Google Scholar]
- de Magalhaes JP, Budovsky A, Lehmann G, Costa J, Li Y, Fraifeld V, Church GM. The Human Ageing Genomic Resources: online databases and tools for biogerontologists. Aging Cell. 2009a;8:65–72. doi: 10.1111/j.1474-9726.2008.00442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Church GM. Analyses of human-chimpanzee orthologous gene pairs to explore evolutionary hypotheses of aging. Mech Ageing Dev. 2007;128:355–364. doi: 10.1016/j.mad.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009b;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE. The Biology of Human Longevity: Inflammation, Nutrition, and Aging in the Evolution of Lifespans. Academic Press; Burlington, MA: 2007. [Google Scholar]
- Finch CE, Tanzi RE. Genetics of aging. Science. 1997;278:407–411. doi: 10.1126/science.278.5337.407. [DOI] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Giallourakis C, Henson C, Reich M, Xie X, Mootha VK. Disease gene discovery through integrative genomics. Annu Rev Genomics Hum Genet. 2005;6:381–406. doi: 10.1146/annurev.genom.6.080604.162234. [DOI] [PubMed] [Google Scholar]
- Gupta PK. Single-molecule DNA sequencing technologies for future genomics research. Trends Biotechnol. 2008;26:602–611. doi: 10.1016/j.tibtech.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Shay T, O’Shea EK, Regev A. Transcriptional regulatory circuits: predicting numbers from alphabets. Science. 2009;325:429–432. doi: 10.1126/science.1171347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Porreca GJ, Song L, Greenway SC, Gorham JM, Church GM, Seidman CE, Seidman JG. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316:1481–1484. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova Y, Maloney RK, Lakey N, Citek RW, Bacher B, Budiman A, Ordway JM, McCombie WR, et al. Massively parallel bisulphite pyrosequencing reveals the molecular complexity of breast cancer-associated cytosine-methylation patterns obtained from tissue and serum DNA. Genome Res. 2008;18:19–29. doi: 10.1101/gr.6883307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Li B, Leal SM. Discovery of rare variants via sequencing: implications for the design of complex trait association studies. PLoS Genet. 2009;5:e1000481. doi: 10.1371/journal.pgen.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A, Fiegler H, Munoz P, Ellis P, Rigler D, Langford C, Blasco MA, Carter N, et al. Spreading of mammalian DNA-damage response factors studied by ChIP-chip at damaged telomeres. EMBO J. 2007;26:2707–2718. doi: 10.1038/sj.emboj.7601719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Chiu CH, Pulapura S, Mehan MR, Nunez-Iglesias J, Zhang K, Kamath K, Waterman MS, et al. Gene Aging Nexus: a web database and data mining platform for microarray data on aging. Nucleic Acids Res. 2007;35:D756–759. doi: 10.1093/nar/gkl798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47:142–150. doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Pop M, Salzberg SL. Bioinformatics challenges of new sequencing technology. Trends Genet. 2008;24:142–149. doi: 10.1016/j.tig.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S, Pe’er I. Overlapping pools for high-throughput targeted resequencing. Genome Res. 2009;19:1254–1261. doi: 10.1101/gr.088559.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nat Biotechnol. 2009;27:847–852. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B. Impact of aging on DNA methylation. Ageing Res Rev. 2003;2:245–261. doi: 10.1016/s1568-1637(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Salipante SJ, Horwitz MS. A phylogenetic approach to mapping cell fate. Curr Top Dev Biol. 2007;79:157–184. doi: 10.1016/S0070-2153(06)79006-8. [DOI] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Slonim DK. From patterns to pathways: gene expression data analysis comes of age. Nat Genet. 2002;32(Suppl):502–508. doi: 10.1038/ng1033. [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugarbaker DJ, Richards WG, Gordon GJ, Dong L, De Rienzo A, Maulik G, Glickman JN, Chirieac LR, et al. Transcriptome sequencing of malignant pleural mesothelioma tumors. Proc Natl Acad Sci U S A. 2008;105:3521–3526. doi: 10.1073/pnas.0712399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Tan Q, Kruse TA, Christensen K. Design and analysis in genetic studies of human ageing and longevity. Ageing Res Rev. 2006;5:371–387. doi: 10.1016/j.arr.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Salzberg SL. How to map billions of short reads onto genomes. Nat Biotechnol. 2009;27:455–457. doi: 10.1038/nbt0509-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol. 2008;17:1636–1647. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- Vijg J, Suh Y. Genetics of longevity and aging. Annu Rev Med. 2005;56:193–212. doi: 10.1146/annurev.med.56.082103.104617. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lawler D, Larson B, Ramadan Z, Kochhar S, Holmes E, Nicholson JK. Metabonomic investigations of aging and caloric restriction in a life-long dog study. J Proteome Res. 2007;6:1846–1854. doi: 10.1021/pr060685n. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zhan M, Duan W, Prabhu V, Brenneman R, Wood W, Firman J, Li H, et al. Gene expression atlas of the mouse central nervous system: impact and interactions of age, energy intake and gender. Genome Biol. 2007;8:R234. doi: 10.1186/gb-2007-8-11-r234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, Weeraratna AT, Taub DD, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]