SUMMARY
Background
The membrane-bound mucins, MUC17 (human) and Muc3 (mouse), are highly expressed on the apical surface of intestinal epithelia and have cytoprotective properties. Their extracellular regions contain two EGF-like Cys-rich domains (CRD1 and CRD2) connected by an intervening linker segment with SEA module (L), and may function to stimulate intestinal cell restitution. The purpose of this study was to determine the effect of size, recombinant host source, and external tags on mucin CRD1-L-CRD2 protein activity.
Methods
Four recombinant Muc3 CRD proteins and three MUC17-CRD proteins were generated using E.coli or baculovirus-insect cell systems and tested in colonic cell cultures for activity related to cell migration and apoptosis.
Results
N-terminal glutathione-S-transferase (GST) or C-terminal His8 tags had no effect on either the cell migration or anti-apoptosis activity of Muc3-CRD1-L-CRD2. His-tagged Muc3-CRD1-L-CRD2 proteins with truncated linker regions, or the linker region alone, did not demonstrate biologic activity. The human recombinant MUC17-CRD1-L-CRD2-His8 was shown to have anti-apoptotic and pro-migratory activity, but did not stimulate cell proliferation. This protein showed similar in vitro biologic activity, whether produced in E.coli or a baculovirus-insect cell system.
Conclusions
Recombinant mucin proteins containing a bivalent display of Cys-rich domains accelerate colon cell migration and inhibit apoptosis, require a full length intervening linker segment for optimal biologic activity, and are functional when synthesized in either E. coli and insect cell systems.
Significance
These results indicate that an E. coli-derived full length His8-tagged human MUC17 CRD1-L-CRD2 recombinant protein is a biologically active candidate for further development as a therapeutic agent.
Keywords: mucin, cell migration, inflammatory bowel disease, MUC17, epidermal growth factor
INTRODUCTION
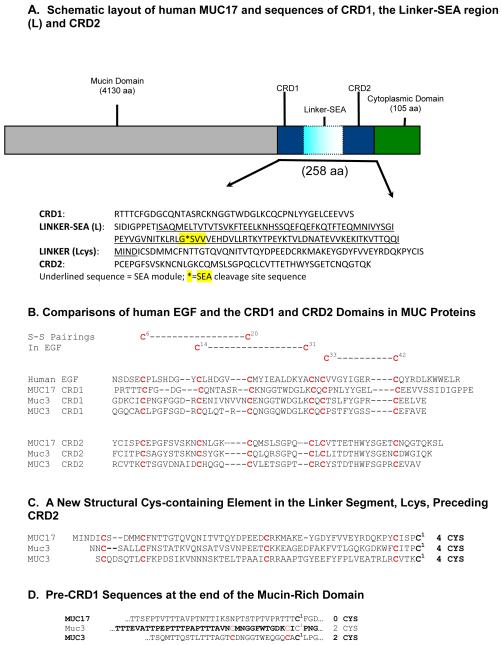
Mucin-type proteins, categorized as secretory or membrane bound (1), represent the major structural proteins of mucous gels that are integral to the epithelial defense of respiratory, digestive, ocular, and reproductive surfaces. The membrane-bound mucins are characterized by an extracellular region with a short amino terminal domain, followed by a large, heavily O-glycosylated tandem repeat domain which accounts in part for their protective function. This large (>4000 amino acids) structural domain is followed by a globular region just proximal to the membrane that is made up of two Cys-rich motifs (CRD1 and CRD2), each with similarity to epidermal growth factor (EGF), separated by a Linker-SEA (L) domain which contains an SEA (sea urchin sperm protein, enterokinase, and agrin) module and Linker residues before and after the module. SEA domains are found in several membrane associated mucins and other membrane proteins and are thought to be important in the noncovalent association of protein subunits, and may play a role in the release of membrane protein subunits at the cell surface [1, 2]. C-terminal to the CRD1-L-CRD2 unit is a transmembrane segment followed by a small cytoplasmic domain. An overview of this structure is shown schematically in Figure 1A, together with a breakdown of amino acid sequences in each part of the CRD1-L-CRD2 segment of human MUC17 [3]. The related membrane bound mucin genes MUC3A/B, MUC12, and MUC17 are clustered on chromosome 7q22, and are highly expressed in intestinal tissues at the apical surface of enterocytes. The mouse Muc3 gene[4] is most similar in sequence and chromosomal localization to the human MUC17 gene [3, 5-7]. The similarity of Cys spacings in mouse Muc3 and human MUC3 and MUC17 mucin CRD proteins and EGF are indicated in Figure 1B. The amino acid sequence of the remaining areas contain the 167 amino acid Linker-SEA domain, which contains a 106 amino acid SEA module, a 7 amino acid sequence preceding the SEA module, and a 54 amino acid region of unknown function that flanks the SEA module. The latter region of the Linker-SEA domain contains several cysteine residues (defined as Lcys). These regions and the pre-CRD1 sequences at the end of the tandem repeat or mucin type domain are indicated in Figures 1C and 1D.
Figure 1.
(A). Schematic representation of MUC17 and amino acid sequences of units within the 259-residue Cys-rich region, including CRD1, the Linker-SEA (L) segment including the L”cys.”segment, and CRD2 (see references [2, 3]). These sequences correspond to the core structure of MUC17-CRD1-L-CRD2 discussed in the text, and defined in Figure 2. (B). Comparative sequence analysis of the Cys-rich regions of human MUC17 and MUC3, and mouse Muc3 - comparisons of CRD units with themselves and with human EGF. (C.) Comparisons within the segment of structure preceding CRD2, herein defined as (D.) Comparisons within the segment preceding CRD1. Note that these residues are seen in MUC3 and Muc3, but not MUC17; however, whether this results in differences in structure or aggregation of these proteins is not determined.
We have previously shown that GST-tagged recombinant mouse Muc3-CRD1-L-CRD2 inhibits cellular apoptosis and accelerates cell migration over surfaces in vitro, and also promotes healing in mouse models of ulcerative colitis [8]. The Muc3-GST-CRD1-L-CRD2 protein does not directly activate EGF receptors, but the specific mechanisms responsible for the actions of the mucin-derived recombinant proteins have not been determined to date. These findings constitute the rationale for continuing evaluation of recombinant mucin proteins containing a bivalent display of these Cys-rich EGF-like CRD domains and to develop a human recombinant mucin protein for potential therapeutic use.
In order to define the optimal structure of a functional recombinant mucin CRD protein, we sought to determine: 1) the size constraints and result of altering the Linker-SEA regions of recombinant CRD1-L-CRD2 proteins on biologic activity, 2) whether a human (MUC17) derived recombinant CRD1-L-CRD2 mucin protein has biologic activity, and 3) compare the in vitro activity of recombinant human MUC17-CRD1-L-CRD2 produced in either bacterial E. coli or eukaryotic insect cell systems. Preliminary findings were previously presented [9]. The findings from these studies demonstrate for the first time that a recombinant CRD1-L-CRD2 unit from human MUC17 is a candidate for further development as a potential mucosal restitution agent for the treatment of inflammatory bowel disease and other mucosal disorders, and can be feasibly synthesized in a bacterial expression system.
MATERIALS AND METHODS
Recombinant proteins
Figure 2 depicts the structure and amino acid sequences of recombinant proteins synthesized for this study. His-tagged fusion proteins: The mouse Muc3CRD1-L-CRD2 DNA fragment was cloned into pET28a vector, sequenced, and transformed into BL21(DE3) cells (Figure 2.1). This sequence expresses the protein with a C-terminal His8 tag. Freshly transformed cells were inoculated and grown overnight in 5 ml of LB + 100 μg/ml kanamycin then induced with 1mM IPTG (isopropylthio-beta-D-galactoside (IPTG; Fisher, Pittsburgh, PA). Induced cells were collected by centrifugation and lysed by sonication in L buffer (50 mM Tris (pH 8.0), 100 mM NaCl, 5 mM EDTA, 0.5% (v:v) Triton-X-100, 0.1% (v:v) 2-ME, 100 uM PMSF). The lysate was centrifuged and the insoluble fraction (inclusion bodies) was washed in L buffer. The resultant insoluble protein was resuspended in PS buffer (50 mM Na2HPO4, pH 7.6, containing 100 mM NaCl) by sonication and then dissolved with the addition of urea to 6 M (PSU buffer). The solubilized protein was next purified by binding to PSU equilibrated Ni-NTA resin and eluted with PSU + 0.5M imidizole. The protein was refolded by dialyzing in a 5kDa MWCO membrane against 100 volumes of 50mM Tris (pH 8.2) and 137mM NaCl with three changes of buffer. Mouse Muc3CRD1-L-CRD2 coding region was amplified and subcloned with part of the L region deleted, specifically most of the SEA module including the cleavage site, and replaced by a coding region for 11 random amino acids (Figure 2.2), or for 20 random amino acids (Figure 2.3), or for the Linker-SEA domain only without the Lcys region (Figure 2.4). The PCR products were topo-cloned into pCR2.1 and sequenced. These Muc3-CRD1-(11 aa spacer-Lcys)-CRD2, Muc3 (CRD1-(20 aa spacer-Lcys)-CRD2, and Muc3(L without Lcys) coding sequences were subcloned into pET23a using NdeI and Hind III. These sequences express the proteins with a C-terminal His8 tag and were transformed into BL21(DE3) cells. Freshly transformed cells were inoculated and grown overnight in 5 ml of LB + 100 μg/ml ampicillin, induced with 1mM IPTG, and the cells were processed as described above. Similarly, the MUC17-CRD1-L-CRD2 coding region was amplified and the PCR product was topo-cloned into pCR2.1 and sequenced. The MUC17-CRD1-L-CRD2 coding sequence was subcloned into pET28a using NcoI and XhoI to create pET28 Muc17. This sequence expresses MUC17-CRD1-L-CRD2 with a C-terminal His8 tag. This plasmid was transformed into BL21(DE3) and the bacteria processed as described above (Figure 2.6).
Figure 2.
Amino acid sequences of mucin-derived proteins used in this study. Underlined residues indicate SEA (sea-urchin sperm protein, enterokinase and agrin) module and shaded regions indicate the SEA cleavage site. (1) Mouse Muc3 CRD1-L-CRD2-His8 (I-1 through G-266) containing a C-terminal 8-His tag. Residues from the pre-CRD1 domain (Fig. 1D) are in lower case. (2) Mouse Muc-3 CRD1-(11 amino acid spacer)-Lcys domain-CRD2 containing a C-terminal 8-His tag. Pre-CRD1 sequence is in lower case, and an 11-residue spacer region is defined in parentheses, followed by the Lcys domain of Muc 3 (Fig. 1C). The underlined region is a partial SEA sequence. (3) Mouse Muc-3 CRD1-(20 amino acid spacer)-Lcys-CRD2 containing a C-terminal 8-His tag. Protein is the same as in 2 above, except the linker (in parentheses) contains 20 amino acids. The underlined region is a partial SEA sequence. (4) Linker Domain-SEA module Muc 3. Protein contains the Muc3 SEA module (underlined) with shaded residues indicate SEA cleavage site. The Linker region pre-CRD2 Cys-containing region (Lcys) is lacking. (5) Human GST-MUC17CRD1-Linker-CRD2 (R-1 through S-260) containing an N-terminal GST tag.(6) Human MUC17CRD1-L-CRD2-His8 (R-1 through K-259) with a 5-residue extension shown in lower case, followed by a C-terminal 8-His tag. (7) Full-length sequence of MUC17 CRD1-L-CRD2 as expressed in baculovirus-insect cell system. Part of the expression vector is shown in brackets [ ]. The sequence in 2.7 begins with a portion of the pBAC vector remaining after removal of the gp64 signal peptide during secretion. This vector portion has a His6 tag, and sites for cleavage by thrombin and enterokinase, and this extra segment is present in the MUC17 derived from insect cells.
GST-fusion proteins
The extracellular, globular region of mouse Muc3 including both EGF-like domains (Muc3CRD1-L-CRD2, previously labeled m3EGF1,2) was amplified from mouse intestinal cDNA, as described in an earlier publication [1]. The resulting fragments were cloned into the pGEX-2TK vector (Amersham, Piscataway, NJ), sequenced, and introduced into E. coli strain BL21 (Invitrogen, Carlsbad, CA). GST-fusion proteins were then expressed in E. coli by induction with 0.5mM IPTG and purified by affinity chromatography using glutathione agarose (Sigma Chemical Co, St. Louis, MO). Similarly, the Muc17-CRD1-L-CRD2 coding region was amplified from human intestinal cDNA, cloned into the pGEX-2TK vector (Amersham, Piscataway, NJ), sequenced, and introduced into E. coli strain BL21 (Invitrogen, Carlsbad, CA). GST-fusion proteins were then expressed in E. coli by induction with 0.5mM isopropylthio-beta-D-galactoside and purified by affinity chromatography using glutathione agarose (Sigma Chemical Co, St. Louis, MO) (Figure 2.5). The yield of proteins synthesized in E. coli ranged from 40 – 50 mg/liter and they were >90% pure based on coomassie blue staining following reducing SDS-PAGE, with an approximate molecular weight of 55-72 kDa (GST-tagged Muc3 and MUC17 CRD1-L-CRD2) and 30-35kDa (his-tagged Muc3 and MUC17 CRD1-L-CRD2) (data not shown).
Baculovirus-insect cell recombinant proteins
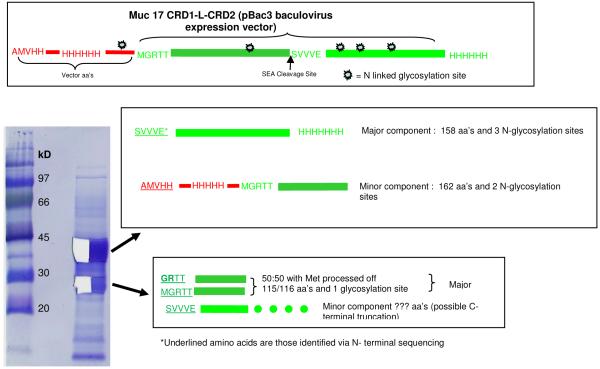
Proteins produced as inclusion bodies in E. coli require dissolution under denaturing conditions and refolding to yield the protein soluble in physiological buffers. There is always the question as to whether the soluble protein is properly refolded to an active species. In order to assure that the His-tagged MUC17-CRD1-L-CRD2 was in a native conformation, the protein was produced in insect cells. The sequence was cloned into the transfer vector pBac3 (Novagen) and co-transfected into Sf9 insect cells with baculovirus DNA. The MUC17-CRD1-L-CRD2 gene isolated from the E. coli vector was cloned as a HndIII/XhoI fragment. The gene already had a carboxy terminal His8 tag identical to what was expressed in E. coli. Transcription starts with the methionine of the gp64 signal peptide. During secretion, the gp64 signaling peptide is cleaved, leaving a portion of the pBAC vector in the final protein. Notable features encoded 5′ to the gene include 1.) an N-terminal His6 tag 2.) an N-terminal S-tag (both used for purification) 3.) a thrombin cleavage site and 4.) an enterokinase cleavage site. His-tagged protein was then purified by immobilized metal-ion affinity chromatography (IMAC) and dialyzed into PBS. The protein was secreted and purified from cell supernatant. The sequence expressed is given in Figure 2.7. The protein was ~95% pure based on SDS-PAGE analysis and N-terminal sequencing (Figure 7). In addition, recombinant proteins were endotoxin-purified using Detoxi-Gel Endotoxin Removal Gel (Pierce, Rockford, IL), which did not alter the activity of the proteins.
Figure 7.
N terminal sequence analysis of baculovirus-insect cell derived MUC17 CRD1-L-CRD2. Automated Edman degradation was performed with the aid of an Applied Biosystems Model 494 Protein Sequencer fitted with an HPLC for analysis of PTH amino acids. Protein samples (MW) were cut from PVDF-blots of the SDS-PAGE gels as indicated and subjected to N terminal amino acid sequence analysis. There were 2 major bands accounting for >95 % of the proteins present. Half of each stained band was cut out and separately subjected to Edman degradation. The upper band consists of a major component beginning at the SEA cleavage site with 3 glycosylation sites and a minor component beginning at the vector His tag. The lower band is a mixture of proteins with the N terminal with or without the Met and a minor component with the N-terminal end at the SEA cleavage site (see sequence data in Table 1).
Cell lines and cell culture
Several types of colonic cells lines were used for these experiments. LoVo cells are a human colon cancer cell line known to respond to Muc3 CRD protein as described previously [8], and express ErbB1 and low level ErbB2 receptors [10, 11]. Colon cell lines commonly used as models of “normal” colon cells were used, including the rat intestinal cell line IEC-6 (American Type Culture Collection (Manassas, VA) and the Young Adult Mouse Colon (YAMC) cell line [8]. YAMC are conditionally immortalized mouse colon cells grown in RPMI 1640 supplemented with 5% FCS + 50 U penicillin/ml and 0.05 ug streptomycin/ml, as described previously [12, 13]. YAMC cells have previously been shown to respond to Muc3 CRD protein and are known to express EGF-type receptors [8]. Cells were grown in 24-well plates for cell migration and proliferation experiments or T-25 flasks for immunoblotting experiments. Lovo and IEC-6 cells were grown using DMEM supplemented with 10% fetal calf serum + 50 U penicillin/ml and 0.05 ug streptomycin/ml (Invitrogen, Carlsbad, CA). Cells were cultured at 37°C, 5% CO2, 10% FCS until the desired confluence was reached. The monolayers were washed with PBS 24 h before experiments and switched to serum-free media for cell migration and immunoblotting experiments.
SDS-PAGE and N-terminal sequencing
After quantification by DC Protein Assay (Bio-Rad, Hercules, CA) equal amounts of total protein were resolved by 10% SDS-PAGE. Automated Edman degradation was performed with the aid of an Applied Biosystems Model 494 Protein Sequencer (Foster City, CA) fitted with an HPLC for analysis of PTH amino acids.
Cell Proliferation
Cells in triplicate were seeded into 24 well plates, exposed to media with and without serum and/or recombinant proteins. The cells were subsequently trypsinized at various time points and counted using a hemocytometer.
Cell Migration (Wound Healing) Assay
Cells were seeded onto 6 well plates, coated with poly-L-lysine, and cultured until confluent. When cells were 90% confluent, an area of cells was scraped with a razor, producing a sharp, clean scrape. Several scrapes were made per well. Cells surrounding the scrape were allowed to migrate over the scrape over 48 h in complete media. Cells were then fixed and stained with Dip Quick (Jorgensen Laboratories, Loveland, CO). The number of cells migrated across scrape was counted at 400x. For inhibition experiments, cells were pretreated with the ERK inhibitor, U0126 (Calbiochem, San Diego, CA), when indicated, for one hour in fresh media followed by treatment with recombinant proteins.
Apoptosis
Apoptosis was induced by incubating cells with either interferon gamma-100 ng/ml for 24 h followed by removal of the interferon and the addition of anti-Fas antibody at 500 ng/ml for 48 h (R&D Systems, Minneapolis, MN) [14], or 125 μM etoposide (Calbiochem, San Diego, CA). Lovo cells were previously shown to be sensitive to anti-Fas treatment [8]. Cells were pretreated with or without recombinant proteins 1 hour prior to addition of anti-Fas. Cells were fixed in 4% paraformaldehyde in PBS pH 7.4 for 5 minutes, then washed twice in PBS. The cells were stained with the nuclear dye, Hoechst 33258 (Polysciences Inc., Warrington, PA), at a concentration of 5ug/ml in PBS for 30 min, rinsed, cover-slipped with Slowfade Antifade (Molecular Probes, Eugene, OR), and then immediately imaged using an ultraviolet microscope. Apoptotic nuclei were identified by morphology [15]. The total number of normal and apoptotic nuclei were counted in three 400x lens fields per dish (representing >200 nuclei per dish). Three or more dishes were used for each experimental condition. Data is also represented as normalized to control with anti-Fas in order to compare experiments done at different times with variations in the maximal apoptosis observed with control anti-Fas (range 14-28%).
Statistical Analysis
Data was tested for normality using the two-sample Kolmogorov-Smirnov tests. The data was found to be normally distributed, satisfying the assumption as needed for parametric testing. Mean +/− SEM was calculated for variables in each experimental group, then analyzed by ANOVA and Student’s t-test (two-tailed) or Fishers exact test, as appropriate. A p-value of <0.05 was considered significant.
RESULTS
Expression and Characterization of Recombinant Muc3 CRD proteins
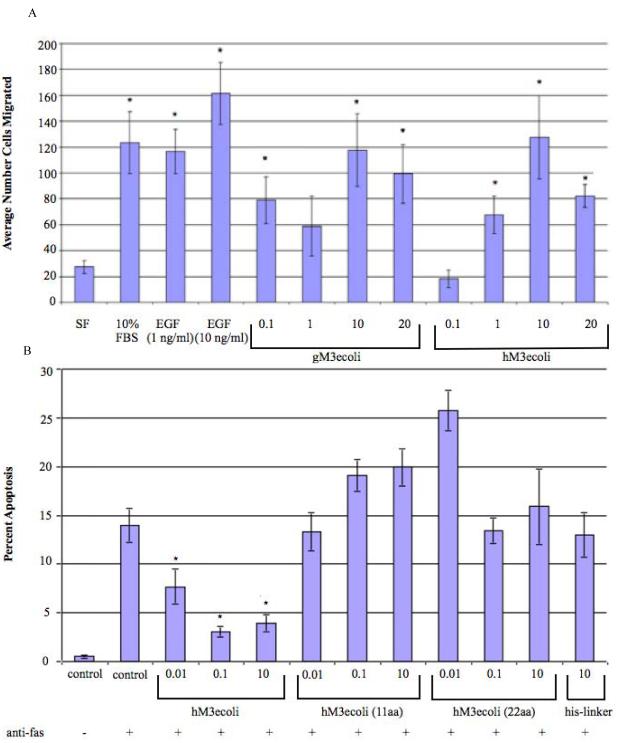
The structures of the mucin CRD proteins used in this study are indicated in Figure 1 and Figure 2. We have previously reported that a GST-tagged recombinant mouse Muc3 cysteine-rich domain protein (GST-Muc3-CRD1-L-CRD2) demonstrated anti-apoptotic and pro-migratory activity in colonic cell lines [8]. In order to determine if replacement of the large GST purification tag with a smaller His8-tag altered the biologic activity of the CRD recombinant proteins, we expressed a full length His-tagged Muc3-CRD1-L-CRD2 (Figure 2.1) in E.coli. This protein was shown to have an ability to stimulate cell migration in a colon epithelial cell line similar to GST-Muc3-CRD1-L-CRD2 (Figure 3A). Next, in order to determine the effect of the spacing between the CRD domains on biologic activity of the Muc3-CRD1-L-CRD2 protein, two recombinant proteins were made with a truncated Linker-SEA (L) domain lacking most of the SEA module, including the cleavage site, but in which the last 56 residues in the Linker-SEA region were retained; this region we define as the L“cys” domain. In these two proteins the L“cys” domains were preceded by 11 (Figure 2.2) or 20 (Figure 2.3) amino acid stretches designed to be of random structure. These recombinant proteins with truncated linker region lacking most of the SEA module demonstrated no anti-apoptotic activity in a colonic cell line (Figure 3B). In addition, a recombinant linker (L) domain protein containing the SEA module sequence and lacking the L“cys” domain and the CRD domains (Figure 2.4) had no anti-apoptotic activity (Figure 3B). We have previously shown that individual mouse Muc3 GST-tagged CRD1 and CRD2 proteins alone or in combination did not demonstrate anti-apoptosis cell migration properties [8]. Taken together, these data indicate that 1) smaller His8-tagged recombinant CRD proteins retain biological activity compared to the previously described GST-tagged protein, and 2) anti-apoptotic activity requires a “full length” CRD1-L-CRD2 molecule with an intact SEA module. Proteins with truncated L domains lacking the SEA module, but containing L“cys”; proteins containing the SEA module alone, lacking CRD domains and L“cys” and proteins with only individual CRD domains (shown previously) are inactive.
Figure 3.
Cell migration induced by recombinant mucin CRD proteins: (A) Induction of cell migration: comparison of GST-tagged Muc3-CRD1-L-CRD2 (gM3ecoli) and His-tagged Muc3-CRD1-L-CRD2 (Figure 2.1; hM3ecoli) (numbers indicate concentration in ug/ml) expressed in E. coli, and purified EGF (ng/ml) in Lovo cell migration. Results indicate similar stimulation of cell migration in Lovo colon cells ( *p<0.027 vs SF, n=5-6). (B) Evaluation of Muc3-CRD1-L-CRD2His8 (hM3ecoli; Figure 2.1) in inhibition of apoptosis in response to anti-Fas, as compared to derivatives having truncation in the Linker-SEA region, and to the Linker-SEA region itself (missing Lcys). The Linker-SEA region truncates are referred to as hM3ecoli(11aa spacer) + Lcys (Figure 2.2), hM3ecoli(20aa spacer) + Lcys (Figure 2.3), and the His-Linker-SEA (L) region alone minus LcysFigure 2.4) (numbers indicate concentration in ug/ml). C = Control (no protein). Lovo colon cells treated with full length hM3ecoli showed an anti-apoptotic effect when compared to cells without treatment, p<0.05, whereas cells treated with hM3ecoli having Linker-SEA truncates, or the Linker-SEA region alone, showed no anti-apoptotic effect.
Expression and Characterization of Recombinant MUC17 CRD proteins
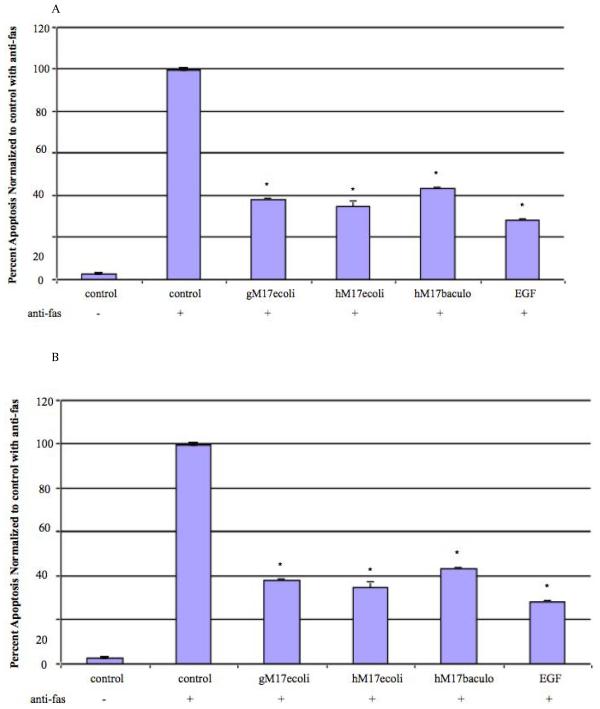
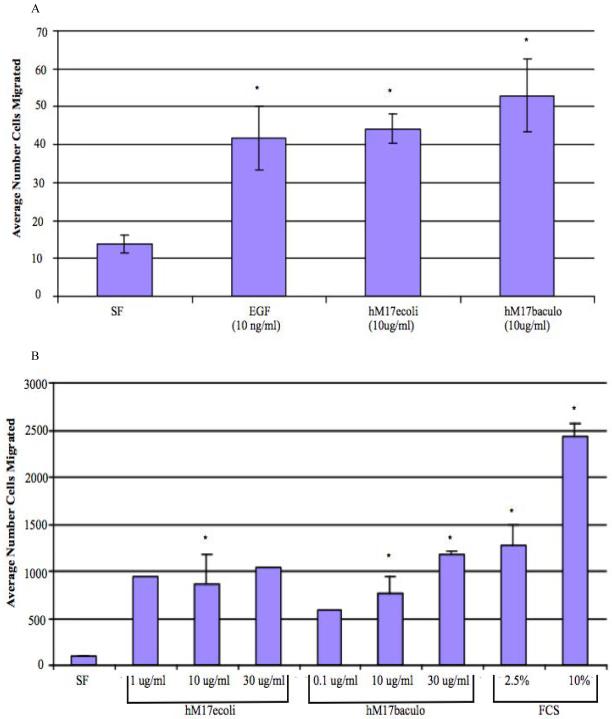
In order to determine if the human homolog of the mouse Muc3 CRD protein was biologically active, we generated a variety of GST- and His-tagged MUC17-CRD1-L-CRD2 proteins in both E.coli and baculovirus-insect cell systems (Figure 2.5-2.7). As shown in Figure 4A, incubation of Lovo colonic cells with 10 ug/ml of each of these proteins significantly inhibited apoptosis induced by anti-Fas, which was equivalent to inhibition induced by 10 ng/ml EGF. Treatment of colon cells with increasing doses of His-tagged MUC17-CRD1-L-CRD2 demonstrated a dose response with increased levels of inhibition of apoptosis with increased doses of the recombinant protein (Figure 4B). Both E .coli and baculovirus derived His-tagged-MUC17-CRD1-L-CRD2 protein induced cell migration in Lovo cells to an equivalent degree when compared with 10 ng/ml EGF (Figure 5A). In order to test whether this response occurred in other intestinal cell lines, we tested migration in IEC-6 intestinal cells, and again found a similar stimulation of cell migration by proteins derived from E. coli and insect cells (Figure 5B).
Figure 4.
Inhibition of apoptosis by MUC17-CRD1-L-CRD2 in response to anti-Fas. (A) Percent apoptotic Lovo colon cells with and without pretreatment with E. coli-derived GST-tagged MUC17-CRD1-L-CRD2 (gM17ecoli; Figure 2.5), His-tagged MUC17-CRD1-L-CRD2 (hM17ecoli; Figure 2.6), insect cell derived His-tagged MUC17-CRD1-L-CRD2 (hM17baculo; Figure 2.7), (all 10 ug/ml) and purified EGF (10 ng/ml). (Data is represented as normalized to maximum apoptosis observed with control with anti-Fas from experiments performed at different times). (B). Inhibition of apoptosis induced by anti-fas in Lovo cells indicating a dose response of E.coli-derived MUC17-CRD1-L-CRD2His8(hM17ecoli), p<0.01 vs no treatment.
Figure 5.
Cell migration studies. The number of cells migrating across a razor scrape were measured after 24-48 h at 37°C. (A) Migration of Lovo cells in response to no treatment (serum free medium or SF) and treatment with MUC17-CRD1-L-CRD2His8 (hM17ecoli) from E. coli (10 ug/ml), his-MUC17 CRD1-L-CRD2 (hM17baculo) from baculovirus-insect cells (10 ug/ml), and purified EGF (10ng/ml). *p< 0.01 vs SF. (B) Migration of IEC6 cells in response to no treatment (serum free medium or sf), and treatment with increasing doses of hMUC17ecoli, hMUC17baculo, and fetal calf serum (FCS). *p<0.05 SF vs other conditions.
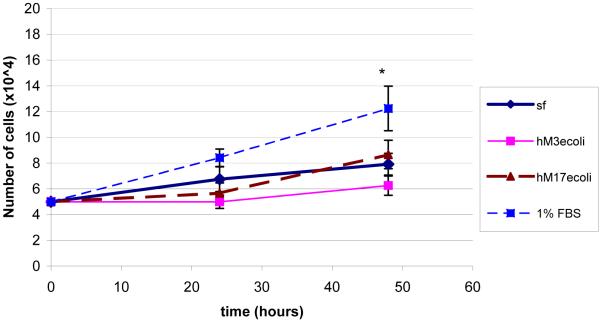
Proliferation of His-tagged Muc3 and MUC17 CRD1-L-CRD2 were compared over 48 h in the index Lovo colon cell line, using a protein concentration that was shown to maximally stimulate cell migration and inhibit apoptosis. As indicated in Figure 6, neither of these proteins stimulated cell growth over 48 h in comparison to cells grown in serum free medium, in contrast to a significant increase in cell growth induced by 1% FBS. We have previously shown that GST-tagged Muc3CRD1-L-CRD2 did not stimulate cell proliferation [8].
Figure 6.
Lovo cell proliferation was measured by cell counts at 24 and 48 h in serum free (sf) medium, with E. coli-derived HisMuc3 CRD1-L-CRD2 (hM3ecoli; 10 ug/ml), with E. coli-derived HisMUC17-CRD1-L-CRD2 (hM17ecoli; 10 ug/ml), and the positive control, 1% fetal bovine serum (FBS). His-tagged Muc3- and MUC17-CRD1-L-CRD2 proteins do not stimulate cell proliferation. N=6 samples each, SF vs FBS p<0.05.
N-terminal sequencing of purified MUC17 CRD1-L-CRD2
Membrane-bound mucins contain a SEA (sea urchin sperm protein, enterokinase and agrin) module that contains a proteolytic cleavage site and sequences that are responsible for noncovalent association of protein subunits [1]. In order to determine if the proteolytic cleavage site RLG/SVVVE within the SEA sequence in recombinant MUC17CRD1-L-CRD2 is in fact cleaved, we tested this protein made using the baculovirus-insect cell system by sequencing the subunits separated on SDS-PAGE. As indicated in Figure 7, N-terminal sequence analysis of baculovirus-insect cell derived his-tagged MUC17 CRD1-L-CRD2 was performed. This recombinant protein consists of two bands at 35 and 30kD when resolved on a reducing SDS-PAGE gel. A portion of each band was excised and the eluted protein analyzed separately, and the results are indicated in Table 1. N-terminal amino acid sequence analysis demonstrated that the upper band consists of a major component beginning at the SEA cleavage site (SVVVE…) with 3 glycosylation sites and a minor component beginning at the N-terminus of this construct (AMVHH…), which relates to the secretion sequence used for expression, the vector his tag. There is little, if any, full length MUC17 protein present; processing at the SEA sequence is nearly 100%. The lower band has 3 proteins, 2 of which were from the actual N-terminus of MUC17 with or without the Met, probably due to proteolytic release of N-terminal Met (GRTT… and MGRTT....). A minor component is also present starting at the SEA site with probable early C-terminal truncation to account for its smaller size. We also performed N-terminal sequencing of protein eluted from a gel of recombinant MUC17-CRD1-L-CRD2 produced in E. coli, and observed similar evidence of cleavage at the SEA site in a minor component (data not shown). These data indicate that the MUC17 recombinant protein is cleaved, as expected, at the SEA cleavage site, and the components co-purify with the final his-tagged protein.
Table 1. PTH amino acids and yields (picomol) at each cycle of Edman degradation.
| Cycle | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Upper Band [Fig. 7] |
Ser (102) Ala (34) |
Val (167) Met (30) |
Val (192)a Val (192) a |
Val (167) His (12) a |
Glu (119) His (16) a |
| Lower Band [Fig. 7] |
Gly (24) Met (120) Ser (9) |
Arg (9) Gly (63) Val (15) |
Thr (31) Arg (30) Val (14) a |
Thr (80) a Thr (80) a Val (14) a |
Thr (91) a Thr (91) a Glu (9) |
( ) = picomol.
Repetitive appearances of Val, Thr, and His in individual peptides and in mixtures lead to increasing apparent yields during the analysis, while yields would be expected to diminish as one proceeds to the next cycle. The Table indicates one major C-terminal polypeptide, the major component of the upper band, and 3 others coming from the N-terminus the sum of which is equivalent to the C-terminal protein.
DISCUSSION
Cell migration and apoptosis are critical for normal intestinal homeostasis and for mucosal healing in response to injury. The normal intestinal barrier is maintained by the continuous migration of cells from the proliferative compartment in the lower crypts to the intestinal villi or colon surface [16]. Following intestinal injury the ability of cells to migrate and close a wound allows for restitution of the epithelial barrier more rapidly than by enhanced proliferation. Increased cell migration occurs in response to experimental intestinal injury and peptic ulcer disease [17, 18], and inflammatory diseases of the bowel [19, 20]. Increased cell migration and anti-apoptosis are often associated and share some common pathways [21]. Thus, agents that enhance intestinal cell migration and reduce apoptosis are potentially therapeutic for conditions of epithelial injury.
We have previously shown that a recombinant mouse GST-Muc3 cysteine-rich domain (CRD) protein with two cysteine-rich EGF-like domains was able to inhibit apoptosis and stimulate cell migration in colonic epithelial cells, and was able to enhance healing of experimental mouse models of colitis [8]. In the current study we have shown that the activity of the mucin cysteine-rich domain proteins requires a full length linker segment between the two CRD units, including the full SEA module, and that a smaller sized recombinant protein can be effectively achieved by substituting a His8-tag for the GST purification tag. We have also extended these studies to include the demonstration that the human ortholog of the mouse Muc3 cysteine-rich protein, MUC17CRD1-L-CRD2, has similar biologic activity in vitro, regardless of whether the protein is synthesized in E. coli or in insect cells. We chose baculovirus-infected insect cells as a eukaryotic system that would afford properly folded material for production of MUC17CRD1-L-CRD2. This is important in providing evidence that protein refolded from bacterial inclusion bodies is equivalent to properly folded protein made in a eukaryotic host. In addition, it should be noted that glycosylation of proteins can occur when synthesized in eukaryotic cells, whereas proteins made in bacteria do not undergo glycosylation. Given the fact that the molecular weights of fragments observed upon analysis of the MUC17-CRD1-L-CRD2 produced in insect cells were considerably higher than expected on the basis of amino acid composition (Figure 7), this protein was most likely glycosylated at its various N-glycosylation sites. Testing of MUC17-CRD1-L-CRD2 proteins derived from bacteria and insect cells indicated that the function did not appear altered despite the fact that the insect cell protein was N-glycosylated. We did not use mammalian cells for protein synthesis, which would have resulted in proteins with the most “native” glycosylation pattern. This was because the insect cell system generally succeeds quickly in providing recombinant proteins that are properly folded and active in amounts needed for biological studies. Future studies would include developing a transformed mammalian cell line, such as CHO, which is a generally accepted host for making protein therapeutics that are both properly folded and glycosylated.
The oxidation state of Cys residues in CRD1-L-CRD2 have not been determined. The Cys-rich region of mucins can be divided into various regions with respect to the EGF-like CRD units. This is done in Figure 1B, where we see that CRD1 and CRD2 have an arrangement of Cys residues suggestive of a relationship with EGF. This accounts for 12 Cys residues in CRD1; but there are additional Cys residues in our recombinant protein. The existence of 4 highly conserved Cys residues in the region of the Linker region just preceding CRD2 (Figure 1C), suggests that these residues are also paired in disulfide linkage; and they serve to define a Cys-containing structural element in the linker region that we call the L”cys” domain. L“cys” may be separate from the CRD units, and may have some other function in the mucin molecule. Our studies to date have not delineated a function for Thus far, we have considered 16 Cys residues in our recombinant protein that are best referred to as potential disulfide bonded Cys residues. Further delineation of the role of individual Cys residues in these recombinant proteins may be accomplished by site-directed mutagenesis studies, which are planned for the future. In addition, a comparison of the segment preceding CRD1 is indicated in Figure 1D. Note that Cys residues are seen in MUC3 and Muc3, but not MUC17; however, whether this results in differences in structure or aggregation of these proteins has not been determined.
Within the Linker-SEA region is the SEA module, which is highly conserved among membrane-bound mucins and contains a autocatalytic cleavage site and amino acid sequences required for the noncovalent association of cleaved subunits [1, 2]. Macao, et al, have previously shown that a recombinant MUC1 mucin SEA protein produced in E. coli undergoes autocatalytic cleavage at the GSSV sequence, indicating that the SEA cleavage process may occur in proteins produced in E. coli as well as eukaryotic cells [2]. We demonstrated that the purified his-tagged MUC17CRD1-L-CRD2 recombinant protein contains mixtures of proteins with evidence of cleavage at the predicted SEA cleavage site. This cleavage may be important for the functioning of the linker region of the recombinant protein, since smaller linker regions without the SEA module have no activity. We have previously shown that individual mouse Muc3 GST-tagged CRD1 and CRD2 proteins alone or in combination were not biologically active [8]. In addition, the recombinant Muc3 proteins in Figure 2.2 and 2.3 included a modified Linker-SEA domain in which a portion of the SEA domain was removed, including the SEA cleavage site, which resulted in a loss of biological activity. This strongly suggests that the presence of the SEA module is required for the observed activity of the recombinant protein. The protein in Figure 2.4 contained only the SEA module, without the remainder of the Linker region or the CRD domains, and this protein also demonstrated no biological activity. These data suggest that the full length CRD1-L-CRD2 protein with an intact SEA domain are required for the protein to be fully functional. He, et al. have shown that within the SEA module several N-glycosylation sites flanking the GSVVV cleavage site may control the extent of cleavage that occurs at this cleavage site, indicating the functional importance of the full SEA module [22]. Further experiments with site directed mutagenesis of the SEA cleavage site would be indicated to determine more specifically if this plays a role in recombinant mucin function, and also what part of the SEA module is important for functionality.
The mechanisms whereby membrane-bound mucins and their cysteine-rich domains influence cell migration and anti-apoptosis are largely unknown and will require further studies. The morphologic assay used to measure apoptosis was a rapid overall assay of total apoptosis, and does not distinguish between early and late apoptosis. Also, the specific mechanism for inhibition of apoptosis was not determined, and we speculate may involve induction of anti-apoptotic proteins via a PI3-kinase pathway or by targeting the caspace-9 enzyme, as has been shown for trefoil factor (TFF) proteins [23, 24]. We have previously demonstrated that the recombinant mouse GST-Muc3 cysteine-rich domain (CRD) protein did not stimulate erbB or EGF-type receptors [8]. Studies are currently underway to determine potential cell signaling pathways involved. However, we note that EGF-type receptors remain likely candidate interacting proteins, and further experiments using more sensitive techniques to determine whether mucin CRD proteins cause clustering or internalization of erbB receptors are required.
In conclusion, these results indicate that full length human MUC17 mucin-derived CRD protein produced in E. coli or in a eukaryotic expression system is a feasible and promising candidate for further development as a therapeutic for conditions associated with epithelial cell injury, such as inflammatory bowel disease or mucositis.
Acknowledgements
This study was supported by a VA Merit Review grant (SBH), NIH STTR grant G1 R43 DK072629-01, NIH center grant (DK080506), and the Research Service of the Department of Veterans Affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Endnote: Mucin references MUC3 EGF paper converted
REFERENCES
- [1].Palmai-Pallag T, Khodabukus N, Kinarsky L, Leir SH, Sherman S, Hollingsworth MA, Harris A. The role of the SEA (sea urchin sperm protein, enterokinase and agrin) module in cleavage of membrane-tethered mucins. Febs J. 2005;272:2901–11. doi: 10.1111/j.1742-4658.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- [2].Macao B, Johansson DG, Hansson GC, Hard T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat Struct Mol Biol. 2006;13:71–6. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- [3].Moniaux N, Junker WM, Singh AP, Jones AM, Batra SK. Characterization of human mucin MUC17. Complete coding sequence and organization. J Biol Chem. 2006;281:23676–85. doi: 10.1074/jbc.M600302200. [DOI] [PubMed] [Google Scholar]
- [4].Shekels LL, Hunninghake DA, Tisdale AS, Gipson IK, Kieliszewsji M, Kozak CA, Ho SB. Cloning and characterization of mouse intestinal Muc3 mucin: 3′ sequence contains epidermal-growth-factor-like domains. Biochem J. 1998;330:1301–1308. doi: 10.1042/bj3301301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Crawley SC, Gum JR, Jr., Hicks JW, Pratt WS, Aubert JP, Swallow DM, Kim YS. Genomic organization and structure of the 3′ region of human MUC3: alternative splicing predicts membrane-bound and soluble forms of the mucin. Biochem Biophys Res Commun. 1999;263:728–36. doi: 10.1006/bbrc.1999.1466. [DOI] [PubMed] [Google Scholar]
- [6].Williams SJ, Munster DJ, Quin RJ, Gotley DC, McGuckin MA. The MUC3 gene encodes a transmembrane mucin and is alternatively spliced. Biochem Biophys Res Commun. 1999;261:83–89. doi: 10.1006/bbrc.1999.1001. [DOI] [PubMed] [Google Scholar]
- [7].Gum JR, Jr., Ho JJ, Pratt WS, Hicks JW, Hill AS, Vinall LE, Roberton AM, Swallow DM, Kim YS. MUC3 human intestinal mucin. Analysis of gene structure, the carboxyl terminus, and a novel upstream repetitive region. J Biol Chem. 1997;272:26678–86. doi: 10.1074/jbc.272.42.26678. [DOI] [PubMed] [Google Scholar]
- [8].Ho SB, Dvorak LA, Moor RE, Jacobson AC, Frey MR, Corredor J, Polk DB, Shekels LL. Cysteine-rich domains of muc3 intestinal mucin promote cell migration, inhibit apoptosis, and accelerate wound healing. Gastroenterology. 2006;131:1501–17. doi: 10.1053/j.gastro.2006.09.006. [DOI] [PubMed] [Google Scholar]
- [9].Shekels LL, Moor R, Phillips J, Baskerville CM, Evans DB, Heinrikson RL, Ho SB. Biological activity of the human MUC17 membrane-bound mucin cysteine-rich domain. Gastroenterology. 2007;132:A569. [Google Scholar]
- [10].Magne N, Fischel JL, Dubreuil A, Formento P, Poupon MF, Laurent-Puig P, Milano G. Influence of epidermal growth factor receptor (EGFR), p53 and intrinsic MAP kinase pathway status of tumour cells on the antiproliferative effect of ZD1839 (“Iressa”) Br J Cancer. 2002;86:1518–23. doi: 10.1038/sj.bjc.6600299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nyati MK, Maheshwari D, Hanasoge S, Sreekumar A, Rynkiewicz SD, Chinnaiyan AM, Leopold WR, Ethier SP, Lawrence TS. Radiosensitization by pan ErbB inhibitor CI-1033 in vitro and in vivo. Clin Cancer Res. 2004;10:691–700. doi: 10.1158/1078-0432.ccr-1041-03. [DOI] [PubMed] [Google Scholar]
- [12].Kaiser GC, Polk DB. Tumor necrosis factor alpha regulates proliferation in a mouse intestinal cell line. Gastroenterology. 1997;112:1231–40. doi: 10.1016/s0016-5085(97)70135-5. [DOI] [PubMed] [Google Scholar]
- [13].Frey MR, Golovin A, Polk DB. Epidermal growth factor-stimulated intestinal epithelial cell migration requires Src family kinase-dependent p38 MAPK signaling. J Biol Chem. 2004;279:44513–21. doi: 10.1074/jbc.M406253200. [DOI] [PubMed] [Google Scholar]
- [14].Quirk SM, Porter DA, Huber SC, Cowan RG. Potentiation of fas-mediated apoptosis of murine granulosa cells by interferon-gamma, tumor necrosis factor-alfa and cycloheximide. Endocrinology. 1998;139:4860–4869. doi: 10.1210/endo.139.12.6353. [DOI] [PubMed] [Google Scholar]
- [15].Fan G, Ma X, Kren BT, Steer CJ. The retinoblastoma gene product inhibits TGF-beta1 induced apoptosis in primary rat hepatocytes and human HuH-7 hepatoma cells. Oncogene. 1996;12:1909–19. [PubMed] [Google Scholar]
- [16].Cheng H, LeBlond CP. Origin, differentiation and renewal of the four main epithelial cell types of the mouse small intestine. V. Unitarian theory of the origin of the four epithelial cell types. Am J Anat. 1974;141:537–562. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- [17].Feil W, Wenzl E, Vattay P, Starlinger M, Sogukoglu T, Schiessel R. Repair of rabbit duodenal mucosa after acid injury in vivo and in vitro. Gastroenterology. 1987;92:1973–86. doi: 10.1016/0016-5085(87)90632-9. [DOI] [PubMed] [Google Scholar]
- [18].Argenzio RA. Comparative pathophysiology of nonglandular ulcer disease: a review of experimental studies. Equine Vet J Suppl. 1999:19–23. doi: 10.1111/j.2042-3306.1999.tb05163.x. [DOI] [PubMed] [Google Scholar]
- [19].Karayiannakis AJ, Syrigos KN, Efstathiou J, Valizadeh A, Noda M, Playford RJ, Kmiot W, Pignatelli M. Expression of catenins and E-cadherin during epithelial restitution in inflammatory bowel disease. J Pathol. 1998;185:413–8. doi: 10.1002/(SICI)1096-9896(199808)185:4<413::AID-PATH125>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- [20].Hermiston ML, Gordon JI. Inflammatory bowel disease and adenomas in mice expressing a dominant negative N-cadherin. Science. 1995;270:1203–1207. doi: 10.1126/science.270.5239.1203. [DOI] [PubMed] [Google Scholar]
- [21].Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- [22].He Y, Li Y, Peng Z, Yu H, Zhang X, Chen L, Ji Q, Chen W, Wang R. Role of N-glycosylation of the SEA module of rodent Muc3 in posttranslational processing of its carboxy-terminal domain. Glycobiology. 2009;19:1094–102. doi: 10.1093/glycob/cwp095. [DOI] [PubMed] [Google Scholar]
- [23].Taupin DR, Kinoshita K, Podolsky DK. Intestinal trefoil factor confers colonic epithelial resistance to apoptosis. Proc Natl Acad Sci U S A. 2000;97:799–804. doi: 10.1073/pnas.97.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bossenmeyer-Pourie C, Kannan R, Ribieras S, Wendling C, Stoll I, Thim L, Tomasetto C, Rio MC. The trefoil factor 1 participates in gastrointestinal cell differentiation by delaying G1-S phase transition and reducing apoptosis. J Cell Biol. 2002;157:761–70. doi: 10.1083/jcb200108056. [DOI] [PMC free article] [PubMed] [Google Scholar]