Summary of recent advances
Existing drugs address a relatively narrow range of biological targets. As a result, libraries of drug-like molecules have proven ineffective against a variety of challenging targets, such as protein–protein interactions, nucleic acid complexes, and antibacterial modalities. In contrast, natural products are known to be effective at modulating such targets, and new libraries are being developed based on underrepresented scaffolds and regions of chemical space associated with natural products. This has led to several recent successes in identifying new chemical probes that address these challenging targets.
Introduction
Genome sequencing efforts and an increasingly molecular level understanding of biological processes have uncovered myriad new biological targets of both fundamental and potential therapeutic interest. Pharmacological studies of these targets with small molecule ligands provide a powerful means to dissect their functions and to validate their therapeutic potential. Increasing access to high-throughput screening technologies in academia has facilitated the discovery of such molecules [1]. However, several classes of targets have proven particularly challenging in these efforts, and some have even been labeled ‘undruggable’ as a result. Despite this moniker, the identification of small molecules to address these challenging targets is, at its core, a chemical problem in molecular recognition and, thus, one that begs a chemical solution.
In considering the limitations of chemical libraries in addressing challenging targets, it is important to recognize that the vast majority of these libraries are based on existing drugs. Leveraging these well-established structural classes has been viewed as a means to identify new small molecule ligands having desirable properties for subsequent drug development. However, a recent study by Overington and coworkers indicates that all current small molecule drugs address only 207 protein targets encoded in the human genome [2]. Moreover, 50% of all drugs are focused on only four protein classes: rhodopsin-like G-protein coupled receptors, nuclear receptors, and voltage- and ligand-gated ion channels. Additional factors further narrow the range of chemical structures found in drug-like libraries, including convenient synthetic accessibility, physicochemical properties thought to be desirable for various reasons, and, in the case of proprietary industrial libraries, each company’s intellectual property position. Accordingly, it has been estimated that only ≈10–14% of the proteins encoded in the human genome are ‘druggable’ using existing drug-like molecules [3].
How, then, might one develop small molecules to address the many challenging, but biologically and therapeutically interesting, targets that lie beyond this small subset? Chemical space [4], the complete set of all possible small molecules, has been variously calculated to comprise 1030–10200 structures, depending on the parameters used (for one example, see [5]). Only a tiny fraction of this total space can be tested in a typical large screening campaign involving on the order of 106 molecules – even at the lower limit of the estimates for chemical space, this fraction (10−24) is approximately equivalent to being able to test only one cell in one person out of the entire population of Earth! However, despite these seemingly daunting numbers, it seems likely that only a portion of total chemical space is relevant to biology, in that these molecules must be reasonably soluble and stable in aqueous media, and have appropriate structural features to bind to proteins and other biological targets with useful specificity; structural factors impacting cell permeability and pharmacokinetics impose further constraints upon molecules used in cellular and animal studies and beyond.
A recent study by Shoichet and coworkers provides important insights into how existing screening collections overcome this numerical problem and target biologically-relevant chemical space [6]. In their analysis of Reymond and coworker’s universe of synthetically accessible molecules with ≤11 heavy atoms (C, N, O, F) [7], commercially available compounds and libraries exhibited much higher similarity to metabolites and natural products than did the complete set of all 26.4 million possible molecules. The authors conclude that the reason existing libraries are effective at all in identifying new small molecule ligands is that they are based, albeit largely unintentionally, on structures in naturally occurring molecules, which have coevolved with proteins that bind them. Indeed, the tremendous historical impact of natural products upon drug discovery is well-established [8].
Thus, rather than sampling chemical space randomly to address challenging biological targets, there is considerable interest in developing new libraries based on other classes of molecules that are biologically validated, but remain underrepresented in current screening collections (for an example, see: [9]). Importantly, the Shoichet study indicates that 83% of natural product scaffolds and 20% of metabolite scaffolds (with ≤11 heavy atoms; the percentages are likely higher for larger molecules) are absent from commercially available collections [6]. Accordingly, libraries based on specific, underrepresented scaffolds may address challenging targets by providing new pharmacophores and binding geometries.
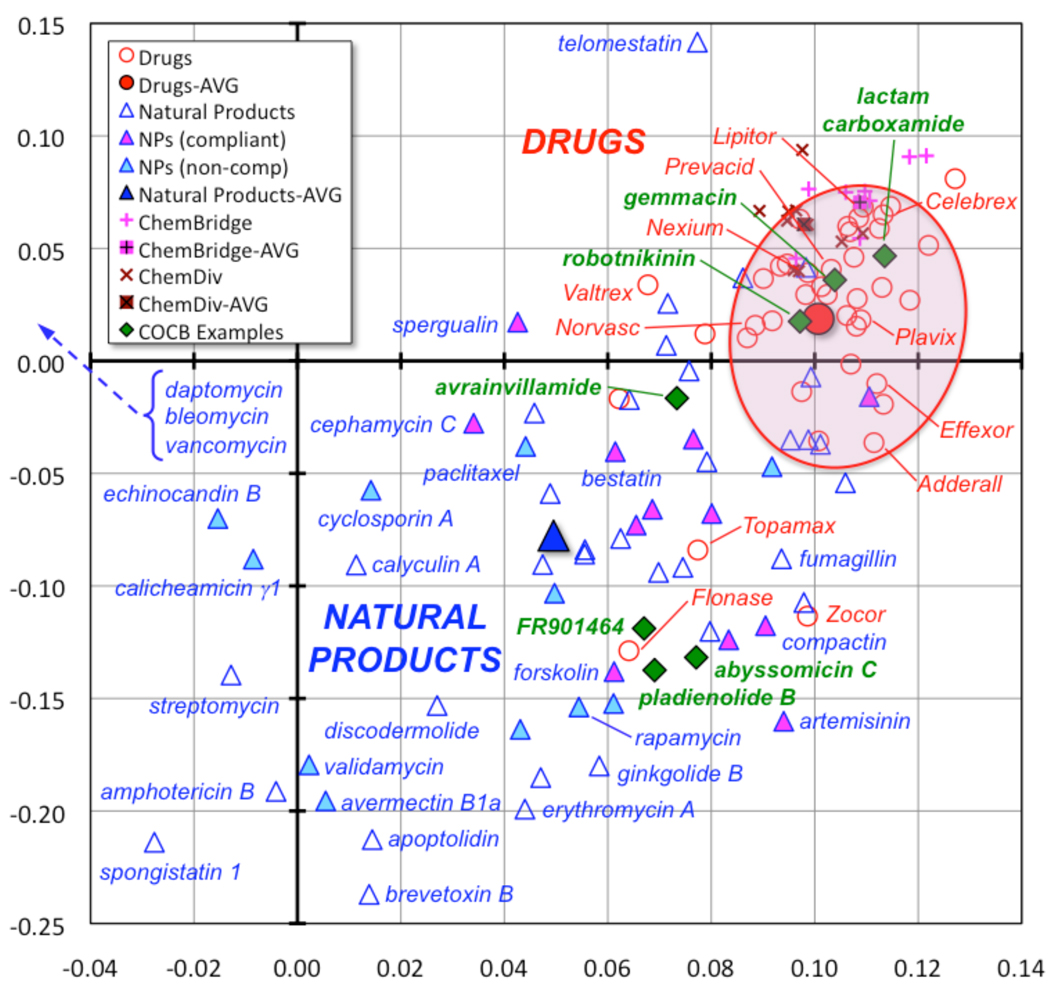
In addition to specific underrepresented scaffolds, more general differences between biologically active natural products and existing synthetic drugs, based on structural and physicochemical properties, have also been described [10,11]. While large datasets are often used in such analyses, we favor the use of smaller datasets and commonly available software programs to provide increased accessibility to synthetic chemists while retaining robustness of the results. We have now updated our own previous analysis [12] to include 40 top-selling small molecule drugs [13] (39 of which are orally bioavailable), a collection of 60 diverse natural products (including the 24 identified by Ganesan as having led to an approved drug from 1970–2006 [14]) and 20 drug-like compounds from ChemBridge and Chem Div (see Supplementary Information). Each compound was analyzed for 20 calculated structural and physicochemical parameters, then principal component analysis was used to replot the data in a two-dimensional format representing 73% of the information in the full 20-dimensional dataset (Figure 1). The two unitless, orthogonal axes represent linear combinations of the original 20 parameters.
Figure 1.
Principal component analysis of 20 structural and physicochemical characteristics of 40 top-selling drugs (red circles), 60 natural products (blue triangles), including Ganesan’s rule-of-five compliant (pink filled) and non-compliant (blue filled) subsets, and 20 compounds from commercial drug-like libraries (ChemBridge, pink plusses; Chem Div, maroon crosses) illustrates the narrow focus of existing drugs and drug-like libraries in chemical space in contrast to natural products (see Supplementary Information for full details). The two unitless, orthogonal axes represent 73% of the information in the full 20-dimensional dataset. Recent examples of natural products and library-derived probes that address challenging targets discussed herein are also shown (green diamonds).
Notably, the top-selling drugs cluster largely in one region of the plot, and the drug-like libraries overlap with this region. The few outlier drugs are natural products or derivatives, and these molecules, along with the 60 natural products, span a much broader range of chemical space. Analysis of component loadings indicates that, in general, the natural products in this analysis feature higher polarity/decreased hydrophobicity and higher molecular weights (to left on x-axis) and more stereochemical features and fewer aromatic rings (to bottom on y-axis) compared to synthetic drugs and drug-like libraries (see Supplementary Information). Interestingly, two subsets of rule-of-five [15] compliant and non-compliant natural products identified by Ganesan do cluster in distinct regions of our plot, although both subsets are equal in size (12 molecules) and have resulted in equal numbers of orally bioavailable drugs (7 each) [14]. Thus, libraries that explore underrepresented regions of biologically-relevant chemical space with respect to structural and physicochemical parameters may also address challenging targets by providing, for example, larger binding surfaces, polarity/charge states, and functional groups that are often excluded from drug-like libraries.
To meet the need for new libraries to address challenging targets, many academic labs are developing libraries that are based on underrepresented scaffolds and that probe underrepresented regions of chemical space. In so doing, it is often appropriate to ignore, or at least avoid strict adherence to, various ‘rules’ that have been developed for drug-like libraries. First, many of these rules have been established with a view toward developing drugs that are orally bioavailable; of course, this is not a primary concern in academic screening, where new probes are needed to investigate fundamental biological processes in biochemical and cellular systems and, in the limit, to validate new targets in animal models. Second, natural products are often cited as being exempt from such rules, due to the influence of carrier-mediated and active transport; interestingly, however, it has recently been suggested that such transport may be much more common across all drugs than previously assumed [16] and, thus, the exclusion of natural products from rule definitions becomes less straightforward. Third, and perhaps most importantly, these rules are based on retrospective analysis of existing drugs and do not provide a blueprint on how to escape from the narrow range of targets addressed by such drugs in the future.
Herein, we discuss three classes of biological targets that have proven challenging to address using existing drug-like libraries. While there is certainly some overlap between these categories, they provide a useful framework for discussion. For each, we provide recent examples of natural products and novel molecules derived from academic libraries that successfully engage these targets.
Protein–Protein Interactions
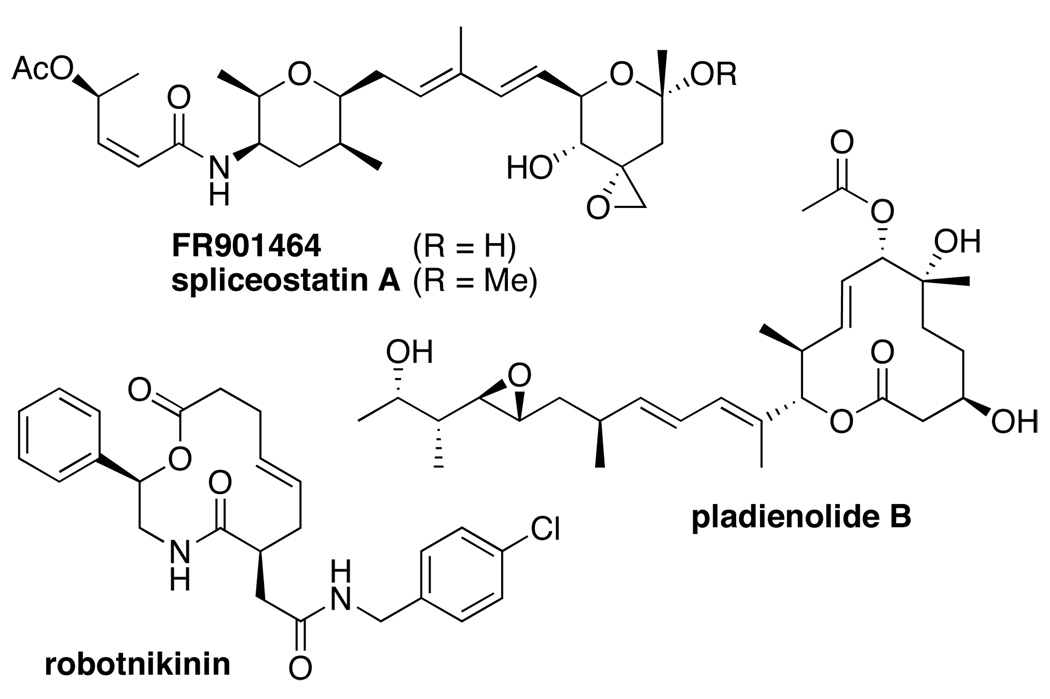
Protein–protein interactions are pervasive in biology, but are classically challenging targets because they often involve large, flat binding interfaces comprised of non-contiguous amino acid residues, and lack cognate small-molecule binding partners [17,18]. As such, they have frequently proven problematic in screens of drug-like libraries [19]. However, there is, a priori, no thermodynamic reason why such interactions should not be amenable to modulation with small molecules [20], and promising early results with Bcl-2 antagonists suggest that such molecules can, indeed, be translated to the clinic [21,22]. Three examples of recently identified small molecules that modulate protein–protein interaction targets are the natural products FR901464 and pladienolide B and the library-derived macrocycle robotnikinin (Figure 2).
Figure 2.
Small molecule inhibitors of protein–protein interaction targets.
FR901464 and Pladienolide B
The spliceosome is a macromolecular complex that orchestrates the process of mRNA splicing, in which non-coding intron sequences in pre-mRNA are excised to produce mature mRNA [23,24]. Aberrant splicing is associated with numerous genetic diseases, including cancer. Spliceosome function involves dynamic interactions of over a hundred proteins and five RNA components that form snRNP (small nuclear ribonucleoprotein) subunits. These snRNPs then undergo a dynamic process of assembly, rearrangement, and release in association with the pre-mRNA substrate, followed by two transesterification reactions that comprise the actual chemical splicing process. The microbial natural products FR901464 and pladienolide B, originally discovered as anticancer agents, were recently identified as spliceosome inhibitors by the Yoshida and Mizui groups [25,26]. In both cases, inhibition was attributed to binding to the SAP130 and/or SAP155 proteins in the U2 snRNP SF3b subcomplex, which coordinates pre-mRNA recognition and spliceosome assembly. While the individual functions of these proteins are still being elucidated, they are thought to be scaffolding proteins not directly associated with active site functions [27], suggesting that these compounds likely act by modulating protein–protein or other macromolecular interactions [24]. Spliceostatin A, a methylated derivative of FR901464, was also shown to bind reversibly, despite the presence of the potentially reactive epoxide moiety, and caused aberrant nuclear release and translation of pre-mRNA, suggesting possible mechanisms for the selective antitumor activity of these compounds [25]. Notably, E7107, an analog of pladienolide B, was also advanced to Phase I clinical trials [26].
Robotnikinin
The hedgehog signal transduction pathway is involved in embryonic development and cancer [28]. Signaling is initiated by binding of the extracellular protein Sonic hedgehog (Shh) to a 12-transmembrane cell surface receptor Patched (Ptch1). Previously identified small molecule agonists and antagonists of this pathway act downstream of the Shh–Ptch1 interaction [28]. Robotnikinin, a 12-membered macrocycle related to scaffolds found in many macrolide natural products, was recently developed as an inhibitor of this pathway, following screens of a 2070-membered macrocycle library by Peng, Schreiber, and coworkers [29,30]. This compound was shown to bind the active, 20 kDa N-terminal fragment of Shh and to disrupt the ability of Shh to relay its signal to Ptch1. While it is not yet known if robotnikinin acts by direct physical disruption of the Shh–Ptch1 interaction or by modulating interactions with other Shh-associated proteins, this work demonstrates that it is feasible to identify molecules that disrupt protein–protein interaction function using macrocycle libraries based on natural product scaffolds.
Nucleic Acid Complexes
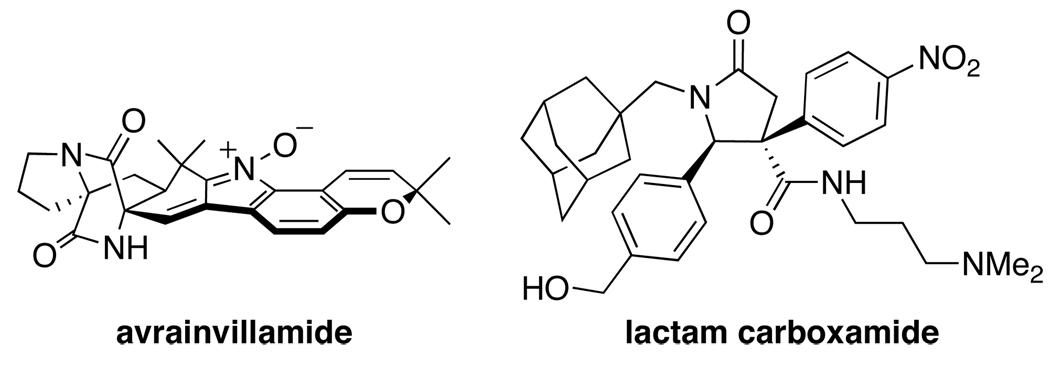
Nucleic acids are central to diverse biological processes, acting as substrates in the case of DNA and RNA, and as catalysts and regulators in the case of ribonucleoproteins and ribozymes. However, complexes involving nucleic acids are generally considered challenging targets for modulation with small molecules, due to the macromolecular nature of the interactions (protein–nucleic acid, nucleic acid–nucleic acid, and protein–protein), and the distinctive electrostatic features of nucleic acids [31,32]. However, many natural products do, in fact, modulate such targets, including a variety of clinically approved drugs [33,34]. Two examples of recently identified small molecules that target nucleic acid complexes are the natural product avrainvillamide and a library-derived lactam carboxamide (Figure 3).
Figure 3.
Small molecule inhibitors of nucleic acid complexes.
Avrainvillamide
Nucleophosmin is a multifunctional protein that is implicated in numerous processes relevant to cell growth, proliferation, and transformation [35]. Aberrant nucleophosmin function is associated with acute myeloid leukemia and anaplastic large-cell lymphomas [36]. Due to its complex role in many different cellular activities, involving interactions with DNA, RNA, and other proteins, small molecules that modulate individual functions of nucleophosmin would be powerful pharmacological tools to dissect those functions. The fungal natural product avrainvillamide was originally discovered based on its anticancer activity [37]. Recently, the antiproliferative effects of avrainvillamide have been attributed to its interaction with nucleophosmin by Myers and coworkers [36]. Site-directed mutagenesis studies indicated that avrainvillamide binds nucleophosmin covalently at Cys-275. This site has been implicated in nucleic acid binding [38], suggesting that this natural product acts by directly targeting this protein–nucleic acid interaction.
Lactam carboxamide
The HOX family of transcription factors are master regulators of development, and aberrant expression is implicated in leukemia and other cancers [39]. Recently, a lactam carboxamide inhibitor of a HOX transcription factor was discovered from a 400-membered library of carboxy-γ-lactams and carboxy-2-quinolones having stereochemically and functionally diverse substituents by Stadler, Shaw, and coworkers [40]. Although the library was not explicitly inspired by natural products, a variety of highly substituted γ-lactam natural products are known, including lactacystin and salinosporamide (see also [41]). Screening of the library in 150 assays of protein–biopolymer interactions yielded the lactam carboxamide as an inhibitor of HOXA13 binding to DNA. The compound also inhibited transcriptional repression by HOXA13 in a luciferase reporter gene assay. This work demonstrates that cell-permeable inhibitors of transcription factor protein–DNA interactions can, indeed, be identified using novel chemical libraries.
Antibacterial Targets
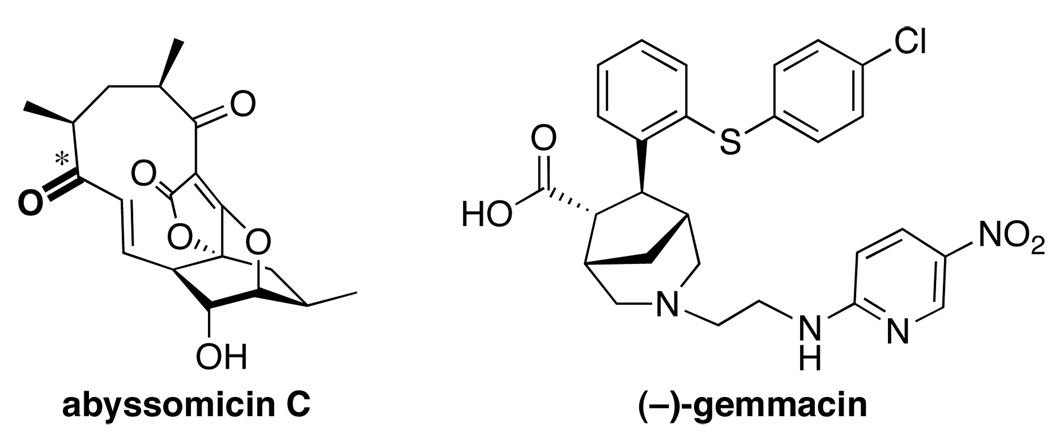
The increasing incidence of antibacterial resistance poses a major threat to human health [42]. Unfortunately, many pharmaceutical companies have abandoned their antibacterial discovery programs, and those that have continued them have found such targets to be difficult to address with existing libraries [43]. At least part of this problem can be attributed to the relatively narrow range of chemical structures present in industrial libraries, resulting from the corresponding focus on a narrow range of human protein targets. In contrast, known antibacterials address a distinct set of bacterial targets, including non-protein targets, and exhibit significantly different structural and physicochemical properties compared to other classes of drugs [44]. Accordingly, novel chemical libraries are needed to provide new starting points for antibacterial discovery. Two recent examples of new scaffolds that address antibacterial targets are the marine natural product abyssomycin C and the library-derived compound gemmacin (Figure 4).
Figure 4.
New scaffolds that address antibacterial targets. The atropisomeric position in abyssomicin C is indicated with a *.
Abyssomicin C
Tetrahydrofolate (THF) is a metabolite involved in myriad biological processes shared by humans and bacteria, including DNA synthesis and repair. While inhibitors of THF biosynthesis have been used heavily to combat bacterial infections, many target biosynthetic steps that are also found in the mammalian host. In contrast, para-aminobenzoic acid biosynthesis is specific to bacterial THF biosynthesis and is not used in the human pathway. The natural product abyssomicin C was discovered from a marine actinomycete Verrucosispora as an inhibitor of para-aminobenzoic acid biosynthesis by Süssmuth and coworkers [45]. This compound was proposed to act as a novel chorismate mimic, possibly involving covalent binding via a Michael acceptor moiety. Subsequently, total synthesis by Nicolaou and coworkers revealed that the isomer atrop-abyssomicin C was actually more active against methicillin-resistant Staphylococcus aureus [46], and reevaluation of the fermentation broth confirmed that this was, in fact, the major isomer produced. Further investigations by Süssmuth demonstrated that atrop-abyssomicin C binds the PabB subunit of 4-amino-4-deoxychorismate synthase covalently at Cys-263 [47]. Interestingly, the initial Michael addition to the enone may be followed transannular attack of the resulting enol on the butenolide moiety, and this may lead to additional conformational changes impacting activity. It is worth noting that, despite the role of such electrophilic functionalities in many biologically important natural products, they are often excluded from drug-like libraries a priori.
Gemmacin
Selective membrane disruption is one mechanism by which several known peptides exert their antimicrobial properties [48]. Gemmacin was identified as a new antibacterial agent from a 242-membered library based on 18 natural product scaffolds by Spring and coworkers [49]. This compound has broad-spectrum activity against Gram-positive bacteria, including methicillin-resistant S. aureus, and exhibits low toxicity against fungal and mammalian cells. To elucidate the mechanism of growth inhibition, gemmacin was assayed for common modes of antibacterial activity, including dihydrofolate reductase inhibition, protein synthesis, and ATP synthesis uncoupling. Detection of reactive oxygen species in a Spodoptera frugiperda cell line suggested membrane disruption as a likely candidate. Although this mode of action is shared by other molecules, the previous examples were peptidic and of considerably higher molecular weight. Notably, gemmacin is zwiterionic, a feature seldom found in drug-like libraries. This work emphasizes the utility of libraries with new scaffolds in identifying compounds to address drug-resistant bacteria.
Structural Analysis of New Probes
Two facets of natural products discussed earlier are their specific scaffolds and their general structural and physicochemical properties. Along these lines, PubChem substructure searches for all three library-derived probes described above revealed that these scaffolds are, indeed, poorly represented in the NIH Molecular Libraries Small Molecule Repository (see Supplementary Information). Of the ≈342,000 compounds currently listed in the MLSMR (as of January 2010), the 12-membered ring core scaffold of robotnikinin was found only in the 249 members of the same library that was used to discover it, and only two 12-membered macrolactones were found. Substituted γ-lactams based on the lactam carboxamide scaffold were also underrepresented, with only 45 compounds having β,β-disubstitution and none having the β-carbonyl-β-aryl substituent pattern. Finally, the [3.2.1]bicyclic amine scaffold of gemmacin was found in only 30 compounds, almost all of which were N-substituted variants of a single cyclic imide scaffold, with no compounds having carbon substitution patterns similar to gemmacin.
In our PCA analysis, we were interested to find that all three of these new probes overlap with existing top-selling drugs (Figure 1). In contrast, the four natural products discussed above fall well outside of this region, overlapping with other natural products. This suggests that our current analysis of structural and physicochemical properties, while effective at identifying differences between synthetic drugs and natural products, is insufficient to discriminate between specific scaffolds that are common or underrepresented. This is, perhaps, not surprising, as the three-dimensional pharmacophores and binding geometries provided by these new scaffolds are difficult to incorporate into such parameter-based vector analyses. Thus, alternative computational approaches must be used to identify such scaffolds [6,50]. Nonetheless, it is important to note that two out of the three library-derived probes violate ‘rules’ that have been developed previously for orally bioavailable drugs and are often used to filter drug-like libraries (lactam carboxamide MW=589, XlogP=5.4, rotatable bonds=11; gemmacin MW=539, XlogP=5.3). This emphasizes the need to develop novel libraries that do not adhere strictly to these parameters in the search for molecules that can address challenging targets.
Conclusions
Efforts to identify small molecules that address challenging biological targets have met with increasing success using novel libraries based on natural products. Continued advances in chemical synthesis to enable efficient production of complex molecules, coupled with accessible computational tools to facilitate library design, should provide increased efficiency in this discovery process in the future. The ultimate goal of these ventures into underexploited regions of chemical space is to expand the range of ‘druggable’ targets, such that the identification of new ligands for currently challenge targets ultimately becomes routine. Success in this endeavor would have major positive impacts in chemical biology and drug discovery.
Supplementary Material
Acknowledgments
We thank Prof. Ganesan (University of Southampton) for stimulating discussions. D.S.T. is an Alfred P. Sloan Research Fellow. Financial support for our laboratory from the NIH (P41 GM076267), Starr Cancer Consortium, Tri-Institutional Stem Cell Initiative, William H. Goodwin and Alice Goodwin and the Commonwealth Foundation for Cancer Research, and MSKCC Experimental Therapeutics Center is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1. Lazo JS, Brady LS, Dingledine R. Building a pharmacological lexicon: small molecule discovery in academia. Mol Pharmacol. 2007;72:1–7. doi: 10.1124/mol.107.035113. • An excellent overview of academic screening in the context of the NIH Molecular Libraries Screening Center Network (MLSCN).
- 2. Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. •• A comprehensive effort to identify and classify the biological targets of all existing drugs. One key finding is that small molecule drugs modulate only 207 targets encoded in the human genome.
- 3.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 4. Dobson CM. Chemical space and biology. Nature. 2004;432:824–828. doi: 10.1038/nature03192. • An excellent introduction to the concept of chemical space and its interactions with biology.
- 5.Bohacek RS, McMartin C, Guida WC. The art and practice of structure-based drug design: A molecular modeling perspective. Med Res Rev. 1996;16:3–50. doi: 10.1002/(SICI)1098-1128(199601)16:1<3::AID-MED1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 6. Hert J, Irwin JJ, Laggner C, Keiser MJ, Shoichet BK. Quantifying biogenic bias in screening. Nat Chem Biol. 2009;5:479–483. doi: 10.1038/nchembio.180. •• A seminal analysis of the structural similarity between existing libraries and naturally occurring molecules, leading to the proposal that new libraries should be developed based on other naturally occuring scaffolds that are also biologically validated but remain underrepresented in screening collections.
- 7.Fink T, Reymond J-L. Virtual exploration of the chemical universe up to 11 atoms of C, N, O, F: Assembly of 26.4 million structures (110.9 million stereoisomers) and analysis for new ring systems, stereochemistry, physicochemical properties, compound classes, and drug discovery. Journal of Chemical Information and Modeling. 2007;47:342–353. doi: 10.1021/ci600423u. [DOI] [PubMed] [Google Scholar]
- 8.Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981–2002. J Nat Prod. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 9. Driggers EM, Hale SP, Lee J, Terrett NK. The exploration of macrocycles for drug discovery - An underexploited structural class. Nat Rev Drug Discov. 2008;7:608–624. doi: 10.1038/nrd2590. • An excellent discussion of the functional capabilities of macrocycles and their underutilization in drug discovery.
- 10. Feher M, Schmidt JM. Property distributions: Differences between drugs, natural products, and molecules from combinatorial chemistry. J Chem Inf Comput Sci. 2003;43:218–227. doi: 10.1021/ci0200467. • An important large-scale analysis of structural differences between approved drugs, natural products, and commercial combinatorial libraries.
- 11. Shelat AA, Guy RK. The interdependence between screening methods and screening. Curr Opin Chem Biol. 2007;11:244–251. doi: 10.1016/j.cbpa.2007.05.003. • A recent analysis of drugs, natural products, and diversity libraries using principal component analysis and radar plots of molecular descriptors.
- 12.Tan DS. Diversity-oriented synthesis: Exploring the intersections between chemistry and biology. Nat Chem Biol. 2005;1:74–84. doi: 10.1038/nchembio0705-74. [DOI] [PubMed] [Google Scholar]
- 13.Top 200 Brand-Name Drugs by Retail Dollars in 2006 on World Wide Web URL. http://drugtopics.modernmedicine.com/
- 14. Ganesan The impact of natural products upon modern drug discovery. Curr Opin Chem Biol. 2008;12:306–317. doi: 10.1016/j.cbpa.2008.03.016. •• Identification of 24 natural products that have led directly to approved drugs since 1970 and discussion of their structural properties.
- 15.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 16.Dobson PD, Kell DB. Carrier-mediated cellular uptake of pharmaceutical drugs: An exception or the rule? Nat Rev Drug Discov. 2008;7:205–220. doi: 10.1038/nrd2438. [DOI] [PubMed] [Google Scholar]
- 17. Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. • An excellent review of recent successes in developing small molecule that disrupt protein-protein interactions.
- 18.Whitty A, Kumaravel G. Between a rock and a hard place? Nat Chem Biol. 2006;2:112–118. doi: 10.1038/nchembio0306-112. [DOI] [PubMed] [Google Scholar]
- 19.Spencer RW. High-throughput screening of historic collections: Observations on file size, biological targets, and file diversity. Biotechnol Bioeng. 1998;61:61–67. [PubMed] [Google Scholar]
- 20.Cochran AG. Antagonists of protein-protein interactions. Chem Biol. 2000;7:R85–R94. doi: 10.1016/s1074-5521(00)00106-x. [DOI] [PubMed] [Google Scholar]
- 21.Chonghaile TN, Letai A. Mimicking the BH3 domain to kill cancer cells. Oncogene. 2009;27:S149–S157. doi: 10.1038/onc.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang MH, Reynolds CP. Bcl-2 inhibitors: Targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res. 2009;15:1126–1132. doi: 10.1158/1078-0432.CCR-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wahl MC, Will CL, Luehrmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Rymond B. Targeting the spliceosome. Nat Chem Biol. 2007;3:533–535. doi: 10.1038/nchembio0907-533. [DOI] [PubMed] [Google Scholar]
- 25.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 26.Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 27.Golas MM, Sander B, Will CL, Luehrmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980–984. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 28.Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 29.Stanton BZ, Peng LF, Maloof N, Nakai K, Wang X, Duffner JL, Taveras KM, Hyman JM, Lee SW, Koehler AN, et al. A small molecule that binds Hedgehog and blocks its signaling in human cells. Nat Chem Biol. 2009;5:154–156. doi: 10.1038/nchembio.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng LF, Stanton BZ, Maloof N, Wang X, Schreiber SL. Syntheses of aminoalcohol-derived macrocycles leading to a small-molecule binder to and inhibitor of Sonic Hedgehog. Bioorg Med Chem Lett. 2009;19:6319–6325. doi: 10.1016/j.bmcl.2009.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berg T. Inhibition of transcription factors with small organic molecules. Curr Opin Chem Biol. 2008;12:464–471. doi: 10.1016/j.cbpa.2008.07.023. • A review focused on recent successes in developing small molecules that inhibit dimeric transcription factors.
- 32. Arndt H-D. Small molecule modulators of transcription. Angew Chem, Int Ed. 2006;45:4552–4560. doi: 10.1002/anie.200600285. • An excellent review of small molecules that inhibit transcription factors and interactions with coregulatory proteins.
- 33.Pommier Y, Marchand C. Interfacial inhibitors of protein-nucleic acid interactions. Current Medicinal Chemistry: Anti-Cancer Agents. 2005;5:421–429. doi: 10.2174/1568011054222337. [DOI] [PubMed] [Google Scholar]
- 34.Moore PB, Steitz TA. The structural basis of large ribosomal subunit function. Annu Rev Biochem. 2003;72:813–850. doi: 10.1146/annurev.biochem.72.110601.135450. [DOI] [PubMed] [Google Scholar]
- 35.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 36.Wulff JE, Siegrist R, Myers AG. The natural product avrainvillamide binds to the oncoprotein nucleophosmin. J Am Chem Soc. 2007;129:14444–14451. doi: 10.1021/ja075327f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fenical W, Jensen PR, Cheng XC. Avrainvillamide, a cytotoxic marine natural product, and the derivatives thereof. 6066635. US Patent. 2000
- 38.Grummitt CG, Townsley FM, Johnson CM, Warren AJ, Bycroft M. Structural consequences of nucleophosmin mutations in acute myeloid leukemia. J Biol Chem. 2008;283:23326–23332. doi: 10.1074/jbc.M801706200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grier DG, Thompson A, Kwasniewska A, McGonigle GJ, Halliday HL, Lappin TR. The pathophysiology of HOX genes and their role in cancer. J Pathol. 2005;205:154–171. doi: 10.1002/path.1710. [DOI] [PubMed] [Google Scholar]
- 40.Ng PY, Tang Y, Knosp WM, Stadler HS, Shaw JT. Synthesis of diverse lactam carboxamides leading to the discovery of a new transcription-factor inhibitor. Angew Chem, Int Ed. 2007;46:5352–5355. doi: 10.1002/anie.200700762. [DOI] [PubMed] [Google Scholar]
- 41.Nay B, Riache N, Evanno L. Chemistry and biology of non-tetramic γ-hydroxy-γ-lactams and γ-alkylidene-γ-lactams from natural sources. Nat Prod Rep. 2009;26:1044–1062. doi: 10.1039/b903905h. [DOI] [PubMed] [Google Scholar]
- 42.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. •• An important report on GlaxoSmithKline's antibacterial screening campaigns and reasons for the disappointing results, including the need for increased chemical diversity in screening collections.
- 44. O'Shea R, Moser HE. Physicochemical properties of antibacterial compounds: Implications for drug discovery. J Med Chem. 2008;51:2871–2878. doi: 10.1021/jm700967e. • A key illustration of the structural and physicochemical differences between antibacterials and other classes of drugs.
- 45.Bister B, Bischoff D, Stroebele M, Riedlinger J, Reicke A, Wolter F, Bull AT, Zaehner H, Fiedler H-P, Suessmuth RD. Abyssomicin C - A polycyclic antibiotic from a marine Verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway. Angew Chem, Int Ed. 2004;43:2574–2576. doi: 10.1002/anie.200353160. [DOI] [PubMed] [Google Scholar]
- 46.Nicolaou KC, Harrison ST. Total synthesis of abyssomicin C and atrop-abyssomicin C. Angew Chem, Int Ed. 2006;45:3256–3260. doi: 10.1002/anie.200601116. [DOI] [PubMed] [Google Scholar]
- 47.Keller S, Schadt HS, Ortel I, Suessmuth RD. Action of atrop-abyssomicin C as an inhibitor of 4-amino-4-deoxychorismate synthase PabB. Angew Chem, Int Ed. 2007;46:8284–8286. doi: 10.1002/anie.200701836. [DOI] [PubMed] [Google Scholar]
- 48.Walsh C. Antibiotics: Actions, Origins, Resistance. ASM Press; 2003. [Google Scholar]
- 49.Thomas GL, Spandl RJ, Glansdorp FG, Welch M, Bender A, Cockfield J, Lindsay JA, Bryant C, Brown DFJ, Loiseleur O, et al. Anti-MRSA agent discovery using diversity-oriented synthesis. Angew Chem, Int Ed. 2008;47:2808–2812. doi: 10.1002/anie.200705415. [DOI] [PubMed] [Google Scholar]
- 50. Shelat AA, Guy RK. Scaffold composition and biological relevance of screening libraries. Nat Chem Biol. 2007;3:442–446. doi: 10.1038/nchembio0807-442. • A network graph approach for assessing and visualizing scaffold similarity.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.