Abstract
Aedes aegypti utilizes blood for energy production, egg maturation and replenishment of maternal reserves. The principle midgut enzymes responsible for bloodmeal digestion are endoproteolytic serine-type proteases within the S1.A subfamily. While there are hundreds of serine protease-like genes in the A. aegypti genome, only five are known to be expressed in the midgut. We describe the cloning, sequencing and expression profiling of seven additional serine proteases and provide a genomic and phylogenetic assessment of these findings. Of the seven genes, four are constitutively expressed and three are transcriptionally induced upon blood feeding. The amount of transcriptional induction is strongly correlated among these genes. Alignments reveal that, in general, the conserved catalytic triad, active site and accessory catalytic residues are maintained in these genes and phylogenetic analysis shows that these genes fall within three distinct clades; trypsins, chymotrypsins and serine collagenases. Interestingly, a previously described trypsin consistently arose with other serine collagenases in phylogenetic analyses. These results suggest that multiple gene duplications have arisen within the S1.A subfamily of midgut serine proteases and/or that A. aegypti has evolved an array of proteases with a broad range of substrate specificities for rapid, efficient digestion of bloodmeals.
Keywords: Aedes aegypti, Midgut, Serine proteases
1. Introduction
Most mosquito species require blood from a vertebrate host to provide the necessary nutrients for survival and egg production. Adult female mosquitoes can consume more than their body weight in blood and therefore must be able to quickly digest the blood components and eliminate excess water and nitrogenous waste through diuresis. Digestion occurs in the mosquito midgut and is mediated by a robust release of exo- and endoproteolytic enzymes such as endoproteolytic serine proteases, carboxypeptidases and aminopeptidases (Noriega et al., 2002; Noriega and Wells, 1999; Sanders et al., 2003). Of particular interest are the midgut serine proteases that function as the principle enzymes in bloodmeal digestion.
Serine proteases are a diverse group of proteolytic enzymes identified by their nucleophilic Ser residue at the active site (Blow, 1997). They are found in all phylogenetic kingdoms including viruses and are involved in many physiological processes (Hedstrom, 2002). The serine protease clan of endoproteolytic enzymes can be further divided into families and subfamilies. The S1.A subfamily includes trypsins, chymotrypsins, elastases and some recently identified serine collagenases. These enzymes contain a His-Asp-Ser catalytic triad and the same basic three-dimensional structure consisting of two six-stranded β-barrels and containing catalytic, substrate recognition and zymogen activation domains (Blow, 1997). Despite the similarities among these enzymes, each has a unique set of accessory catalytic residues that are thought to be important in determining substrate specificity (Hedstrom, 2002).
The Aedes aegypti genome contains hundreds of serine protease-like genes, of which 66 are putative trypsins (Venancio et al., 2009). However, only five endoproteolytic serine proteases (three trypsins and two chymotrypsins) are known to be expressed in the midgut (Barillas-Mury and Wells, 1993; Bian et al., 2008; Jiang et al., 1997; Kalhok et al., 1993; Noriega et al., 1996). A sixth endoproteolytic enzyme reported here as AaSP VII, has recently been shown to be required for complete in vivo digestion and maximal fecundity (Isoe et al., 2009). Recent bioinformatics’ analysis of publicly available larval and adult expressed sequence tags (EST) revealed that there may be eight additional bloodmeal-induced midgut-associated putative trypsins (Venancio et al., 2009). However, these findings have not been confirmed by molecular or biochemical approaches.
The trypsins are thought to account for the majority of bloodmeal digestion and are expressed in a unique biphasic manner; 1–3 h post-bloodmeal (hpbm) and 8–36 hpbm (Felix et al., 1991). Trypsins are classified by their ability to C-terminally cleave Lys/Arg. The chymotrypsins, which C-terminally cleave Phe/Tyr/Trp, only moderately contribute to bloodmeal digestion and are continuously expressed (Bian et al., 2008; Jiang et al., 1997). This preference towards trypsin-mediated digestion may have arisen in response to greater amino acid frequencies of Lys/Arg over Phe/Tyr/Trp (White, 1992). The three midgut trypsins, early trypsin (AaET, GenBank accession no. X64362), late trypsin (AaLT, GenBank accession no. M77814) and Aa5G1 (GenBank accession no. X64363), were identified from deduced amino acid sequences and alignment data (Barillas-Mury and Wells, 1993; Kalhok et al., 1993; Noriega et al., 1996). While, trypsin specific enzymatic activity has not been demonstrated in vitro, RNA interference studies have shown that reduction of both AaET and Aa5G1 transcripts correlated with significant reductions in tryptic activity in vivo (Brackney et al., 2008; Isoe et al., 2009). Interestingly, when AaLT transcripts were reduced there was no loss in late phase tryptic activity (Brackney et al., 2008). Upon further analysis of the AaLT sequence, it was determined that AaLT lacks the critical Asp189 commonly associated with trypsins. Together these data suggest that AaLT lacks tryptic activity and may not be a trypsin. The chymotrypsins, AaCHYMO (GenBank accession no. U56423) and AaJA15 (GenBank accession no. AY957559), have been shown to have chymotryptic specific activity in vitrovia recombinant proteins and in vivo by RNA silencing, both of which corroborate the sequence alignments (Bian et al., 2008; Brackney et al., 2008; Jiang et al., 1997).
Here we present the cloning, sequencing and expression data of seven recently identified serine proteases from the midgut of A. aegypti. In addition, we provide extensive alignment and phylogenetic analyses for each of the genes along with genomic annotation. We found a strong correlation between the transcriptional induction of three of the genes following acquisition of a bloodmeal, while the other four are constitutively expressed pre- and post-blood feeding. Phylogenetic analyses reveal that there are three clades corresponding to the serine proteases, trypsins, chymotrypsins and serine collagenases, expressed in the midgut. Interestingly, AaLT, once thought to be a trypsin, consistently arose in the serine collagenase clade. Overall, these findings provide novel insights into the evolutionary complexity of mosquito bloodmeal digestion.
2. Materials and methods
2.1. Mosquito rearing
The Rockefeller strain of A. aegypti was maintained at a constant temperature 27 °C with a relative humidity of 80% and a 16:8 light:dark cycle. Adult mosquitoes were provided a 10% sucrose solution for maintenance. 3- to 4-day-old female mosquitoes were offered defibrinated porcine blood supplemented with 5 mM ATP.
2.2. Genomic DNA library construction
Genomic DNA was isolated from A. aegypti using standard procedures (Sambrook et al., 1989). High molecular weight genomic DNA was partially digested with Sau3AI and size fractionated on a low melting point agarose gel. Fragments between 6 and 10 kb were excised and purified. The purified genomic DNA was ligated using a BamHI predigested λZapExpress vector and packaged using Gigapack III Gold Packaging Extract (Stratagene, Cedar Creek, TX).
2.3. Midgut cDNA library construction
Midguts were dissected from unfed and 24 hpbm mosquitoes. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacture’s instructions. Subsequently, 5 μg of mRNA were purified from total RNA using Oligotex mRNA Mini Kit (Qiagen, Valencia, CA). The cDNA were prepared using the ZAP Express cDNA Synthesis Kit and cloned into λZapExpress vector (Stratagene) according to the manufacture’s instruction.
2.4. Screening genomic DNA libraries
The A. aegypti genomic DNA and midgut cDNA libraries were probed with a full-length ORF of AaET or Aa5G1 genes labeled with digoxigenin-11-dUTP using the random priming method following manufacturer’s instructions (Roche Molecular Biochemicals, Basel, Switzerland). 2 × 105 and 1 × 105 plaques were screened for genomic DNA and cDNA libraries, respectively. The plaques were transferred to nylon membranes which were denatured, neutralized and DNA cross-linked using a Strata-linker (Stratagene). Hybridization was performed overnight at 55 °C. Positive plaques were detected colorimetrically using anti-digoxygenin conjugated with alkaline phosphatase, nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate as substrates. Phagemid DNAs for each putative trypsin gene were isolated by plasmid Miniprep (Promega, Madison, WI). All sequencing was performed at the University of Arizona Sequencing Facility using an automatic sequencer (Model 373, Applied Biosystems, Foster City, CA). Sequences have been deposited in GenBank (accession numbers in Table 1).
Table 1.
List of serine proteases used in the alignment and phylogenetic analyses.
| Species | Serine Protease | Accession # | Vector Base ID# |
|---|---|---|---|
| Aedes aegypti | AaET | X64362 | AAEL007818 |
| Aedes aegvpti | AaLT | M77814 | AAEL013284 |
| Aedes aegypti | Aa5G1 | X64363 | AAEL013712 |
| Aedes aegypti | AaCHYMO | U56423 | AAEL003060 |
| Aedes aegypti | AaJA15 | AY957559 | AAEL001703 |
| Aedes aegypti | AaSP I | GQ398043 | AAEL007432 |
| Aedes aegypti | AaSP II | GQ398044 | AAEL008093 |
| Aedes aegypti | AaSP III | GQ398045 | AAEL013623 |
| Aedes aegypti | AaSP IV | GQ398046 | AAEL013628 |
| Aedes aegypti | AaSP V | GQ398047 | AAEL008085 |
| Aedes aegypti | AaSP VI | GQ398048 | AAEL010196 |
| Aedes aegypti | AaSP VII | GQ398049 | AAEL010202 |
| Culex pipiens | PIPIENS ET | AY029276 | CPIJ019781 |
| Culex pipiens | PIPIENS LT | U65412 | CPIJ005132 |
| Aedes albopictus | ALBOPICTUS LT | AF268665 | NA |
| Anopheles gambiae | GAMBIAE TRYP1 | Z22930 | TRY1_ANOGA |
| Anopheles gambiae | GAMBIAE TRYP2 | Z18890 | TRY2_ANOGA |
| Anopheles gambiae | GAMBIAE CHYMO | Z18887 | CTR1_ANOGA |
| Simulium vittatum | VITTATUM TRYP | L08428 | NA |
| Phlebotomus papatasi | PHLEBOTOMUS TRYP | AY128110 | NA |
| Phlebotomus papatasi | PHLEBOTOMUS CHYMO | AY128106 | NA |
| Glossina moristans | GLOSSINA TRYP | AF252869 | NA |
| Glossina moristans | GLOSSINA CMYMO | AF252868 | NA |
| Drosophila melanogastor | DROSOPHILA TRYP | M96372 | NA |
| Manduca sexta | MANDUCATRYP | L16805 | NA |
| Hypoderma lineatum | HYPODERMA COLL | AB054066 | NA |
| Uca pugilator | CRAB COLL | U49931 | NA |
2.5. Quantitative reverse transcriptase polymerase chain reaction
The mRNA expression levels were measured by quantitative reverse transcriptase polymerase chain reaction (Q-RT-PCR). Midguts from adult female A. aegypti were dissected prior to blood feeding and 3, 6, 12, 24, 36, 48, 72, 96, and 120 hpbm. Total RNA was extracted using TRIzol Reagent and 2 μg of total RNA was used for cDNA synthesis with the Transcriptor First Strand cDNA Synthesis Kit according to the manufacturer’s protocol (Roche) with an oligo-dT primer. The synthesized cDNA was subsequently used as template for Q-RT-PCR using gene-specific primers and the FastStart Universal SYBR Green Master Mix (ROX) (Roche) (Table 2). Q-RT-PCR was performed in a 96-well plate on the ABI Prism 7300 Real-Time PCR System and data was analyzed with ABI Prism 7300 SDS Software (version 1.2.2; Applied Biosystems). The Q-RT-PCR parameters consisted of an initial denaturation step for 10 min at 95 °C followed by 30 cycles consisting of a 15 s denaturation step at 95 °C and a 1 min extension step at 60 °C. The expression profiles for AaLT, AaSP I, AaSP II-V (AaSP group I), AaSP VI, AaSP VII, AaVIII, AaSP IX and AaSP V were analyzed by this approach and relative levels of expression determined by comparison to ribosomal protein S7 RNA levels. The ribosomal protein S7 was used as the baseline control since its mRNA levels remain unchanged in unfed mosquitoes and during bloodmeal digestion (Dana et al., 2005). Because of the high sequence similarity between the genes in AaSP group I, each gene’s transcript levels could not be individually quantified, so the cumulative level of these four genes were obtained in these analyses.
Table 2.
Primers used for Q-RT-PCR. Name, sequence and position within the respective genes of the primers used for the expression profiles as determined by Q-RT-PCR.
| Primer name | 5′-sequence-3′ | Position |
|---|---|---|
| AaLT F | CTGTGCGCCAAGGGTGAACA | Nucleotides 580–599 |
| AaLT R | TCTGCTTGATCCAATCGACGAA | Nucleotides 739–760 |
| AaSP 1 F | CCGACGGCCGATGAGCAGTA | Nucleotides 433–452 |
| AaSP 1 R | CGTCCAGTTTCCGGTGTAAC | Nucleotides 583–602 |
| SP group II F | GATCATCGGCGGTTTTCC | Nucleotides 84–101 |
| SP group II R | ACACGACGCTTGCCCTC | Nucleotides 247–263 |
| AaSP VI F | ACTCTTGCCAGGGTGATTCT | Nucleotides 644–663 |
| AaSP VI R | TATTTTTAATCATTTCATTA | Nucleotides 831–850 |
| AaSP VII F | CCGCAGTACAACCCATCCAC | Nucleotides 337–356 |
| AaSP VII R | GATTCTGAAGGGTTTTGAGTATA | Nucleotides 490–512 |
| S7 F | ACCGCCGTCTACGATGCCA | Nucleotides 435–453 |
| S7 R | ATGGTGGTCTGCTGGTTCTT | Nucleotides 546–565 |
2.6. Statistical analyses
For the Q-RT-PCR data, raw relative levels of change compared to S7 ribosomal RNA were log10 transformed and compared with pairwise t-tests using pooled standard deviations and a Bonferroni adjustment. Pearson’s correlation coefficient was calculated among the log transformed means. All analyses were performed in R (version 2.9.1) and plotted in GraphPad Prism (version 5.02; GraphPad Software, Inc.).
2.7. Sequence alignments and phylogenetic analysis
The nucleic acid sequences of the seven recently identified midgut serine proteases were translated and the corresponding predicted amino acid sequences were used in all analyses. Similarly, the predicted amino acid sequences for the remaining 20 serine proteases were retrieved from the NCBI Entrez database (Table 1). Prior to alignment, SignalP 3.0 predicted the mature peptides used in the analysis (Bendtsen et al., 2004). Amino acid sequences were initially aligned using ClustalW and for both the multiple alignment and pairwise alignment parameters the GONNET protein-weight matrix was applied (Thompson et al., 1994). The alignments were visually verified and manually realigned as needed prior to phylogenetic analyses. Following alignment of the amino acids, the sequences were converted back to nucleic acids and the corresponding aligned codons were analyzed. Phylogenetic analyses were performed with PHYLIP version 3.67. Alignments were tested with multiple out-group comparisons using maximum parsimony (MP) and gaps were treated as a fifth character. Bootstrap support was evaluated based on 1000 pseudo-replicates.
Phylogenies were also derived using the Tamura–Nei distance (Tamura and Nei, 1993) and neighbor-joining analyses (Saitou and Nei, 1987) and bootstrap support was evaluated based on 1000 pseudo-replicates. Maximum likelihood (ML) phylogenies were also obtained using the HKY85 model, although bootstrapping was not applied.
2.8. Gene annotation
Nucleic acid sequences for all 12 A. aegypti serine protease genes were subjected to BLAST searches against the A. aegypti genome using VectorBase (http://aaegypti.vectorbase.org/index.php). Sequences were queried using the BLOSUM62 matrix and gapped alignment. Often a sequence aligned to several supercontigs. Using E-values, bit scores, alignment identities and sequence lengths as parameters, each of the genes was directionally assembled so that each gene was positioned within a single supercontig. The locations of the assembled genes were then annotated in VectorBase.
3. Results
3.1. Genomic and midgut cDNA library screens
Identification of novel serine proteases from A. aegypti was performed by screening genomic and midgut cDNA libraries with probes designed against AaET and Aa5G1. Approximately 300,000 clones were screened, of which 11 and 97 were positive from the genomic and midgut cDNA libraries, respectively. Aa5G1 was detected from both libraries, thus validating the sensitivity of these screens. Of the 11 positive clones from the genomic library, 6 were different from AaET and Aa5G1. Some of the positive clones shared ≤55% sequence similarity with either AaET or Aa5G1. However, when the coding sequences of the mature peptides were aligned a higher degree of conservation was observed (75% sequence similarity). The ability to identify these divergent serine proteases was most likely the result of using low stringency hybridization conditions. The DNA sequences of 24 clones among the 97 positives from the midgut cDNA library were determined, resulting in 6 unique cDNA clones. In total we detected 12 serine protease genes.
3.2. Serine protease expression profiles
Gene-specific primers were designed for each of the 12 serine proteases to determine relative amounts of expression in adult female midguts pre- and post-bloodmeal. We found that nine (AaSP I-IX) of these 12 clones were expressed in the midgut. Subsequently, expression profiles for these nine midgut serine proteases were determined. As a control for these experiments, AaLT expression levels were also measured. In accordance with previous studies, AaLT was up-regulated, 1400 fold following a bloodmeal and reached maximal expression levels 24–36 hpbm (Fig. 1A). AaSP VIII and AaSP IX were expressed in the midgut at extremely low levels in comparison to ribosomal protein S7 RNA and were not up-regulated by blood feeding (data not shown). Analysis of AaSP I showed expression at low levels in unfed midguts, but immediately following a bloodmeal, transcript levels increased 316-fold by 12 hpbm. This increase was followed by a slow decline in transcript levels through the remainder of bloodmeal digestion (Fig. 1B). Serine protease genes AaSP II thru V (AaSP group I) have an extremely high degree of sequence similarity and therefore we were unable to design gene-specific primers. The expression profile of the AaSP group I genes therefore represented transcripts from the entire gene family. Interestingly, these genes were expressed at constant levels throughout the experiment and were not significantly different from ribosomal protein S7 RNA levels (Fig. 1C). Conversely, both AaSP VI and AaSP VII had expression profiles similar to AaLT (Barillas-Mury et al., 1991). AaSP VI mRNA increased 407-fold and reached a maximum level by 24 hpbm (Fig. 1D), whereas AaSP VII mRNA increased 12,302-fold and peaked at 24 hpbm (Fig. 1E). The mRNA expression results for AaSPs II–VII strongly support the bioinformatics-based predicted expressions as previously described (Venancio et al., 2009). The induction and expression levels of AaLT, AaSP I, AaSP VI and AaSP VII transcripts were strongly correlated (Fig. 2) suggesting either co-transcription of these genes under a common promoter or co-regulation based on a common transcription factor.
Fig. 1.
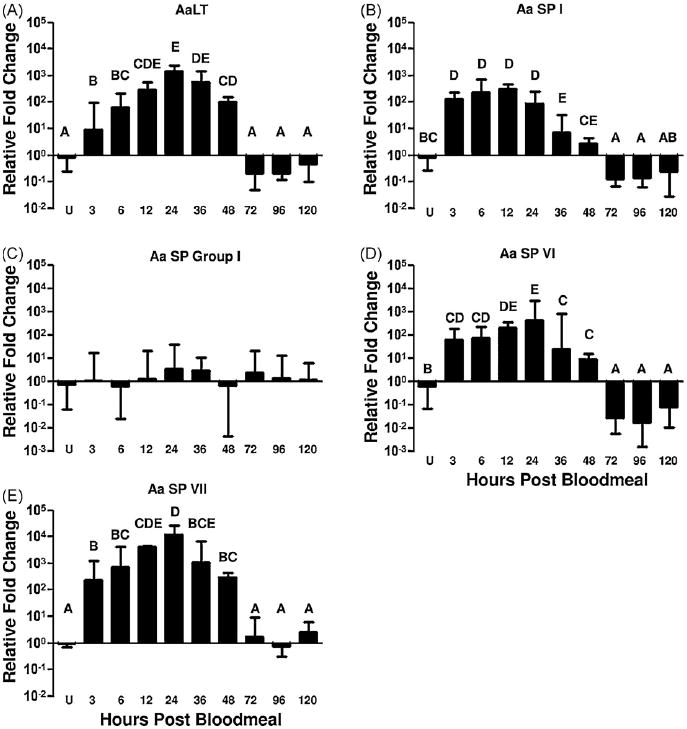
mRNA expression profiles of the midgut serine proteases. Relative levels of gene transcripts are presented as the ratio of gene transcript abundance relative to ribosomal protein S7 RNA abundance. Relative mRNA levels were measured in unfed (U) midguts and at 3, 6, 12, 24, 36, 48, 72, 96 and 120 h post-bloodmeal. Data are included for AaLT (A), AaSP I (B), AaSP group I (C), AaSP VI (D) and AaSP VII (E). Statistically homogeneous (p > 0.05) groupings appear under a common letter and were identified with pairwise t-tests among the different time points. Values in (C) were not significantly different.
Fig. 2.
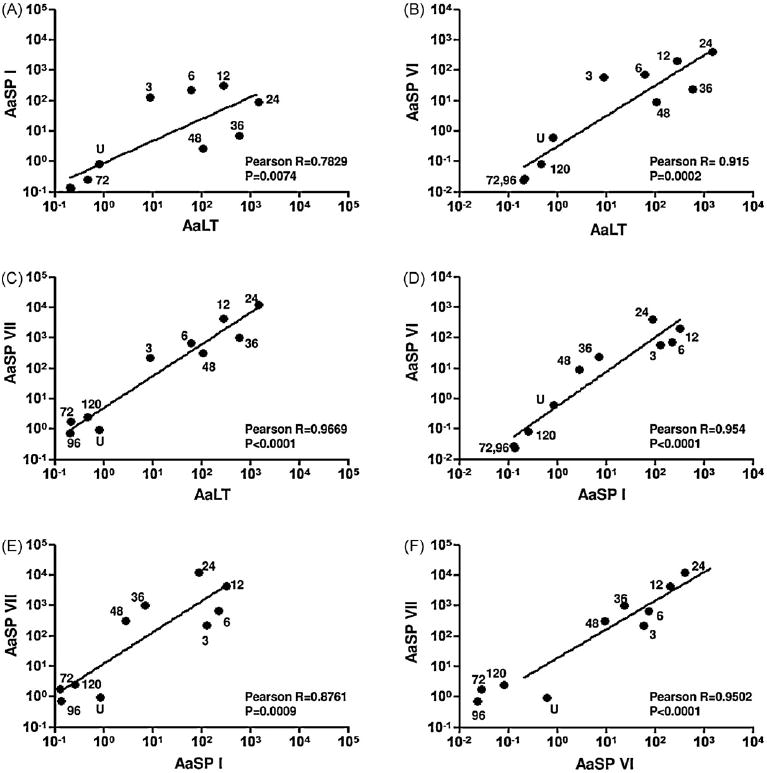
Transcriptional control of midgut serine proteases is correlated. The average transcription levels of AaLT, AaSP I, AaSP VI and AaSP VII plotted against one another. Each point is labeled as to whether it was measured in unfed (U) midguts or at 3, 6, 12, 24, 36, 48, 72, 96, and 120 h post-bloodmeal. The line on each graph represents the linear regression model obtained and Pearson’s correlation coefficients and associated probability values are listed on each graph. Numbers within figures define the timepoints post-bloodmeal. The values for timepoints 96 and 120 are not labeled within (A) because they overlap.
3.3. Amino acid alignments
A total of 26 insect serine proteases were aligned with a crab (Uca pugilator) collagenase. The analyses included 25 dipteran and 1 lepidopteran amino acid sequences from the following families; 18 Culicidae, 2 Psychodidae, 2 Glossinidae, 1 Simuliidae, 1 Drosophilidae, 1 Hypodermatidae, and 1 Sphingidae (Lepidoptera). References to specific locations within the amino acid alignments are based on the numbering of the prototypical serine protease, bovine chymotrypsinogen.
All of the sequences contained six highly conserved cysteines that correspond to the three disulfide bonds characteristic of arthropod serine proteases (Davis et al., 1985). Furthermore, all of the serine proteases contained either an Ile or Val at the first position of the mature peptide. Aside from the Glossina moristans CHYMO which has a Glu194, all of the other proteins contained an Asp at this site which buries the hydrophobic residue at the first position during folding of the mature peptide (Fig. 3) (Hedstrom, 2002). Interestingly, AaSP I and Aa SP V did not contain the archetypal His57-Asp102-Ser195 catalytic triad. In the case of AaSP I, His57 has been replaced with Gln57 and Ser195 changed to a Val195. On the other hand, AaSP V has Leu57 in place of His57 (data not shown).
Fig. 3.

Protein alignment of serine proteases. Protein alignment of all serine proteases used in this analysis from positions ~170–220. Numbering is based on bovine α-chymotrypsinogen. Residues of importance are represented as follows; (*) Ser195 catalytic triad residue, (◆) accessory catalytic residues, (▲) the third and final disulfide bridge and (Δ) Asp194 where position 1 (Ile/Val) becomes buried following activation of the mature peptide.
The S1.A subfamily of serine proteases can be further differentiated into trypsins, chymotrypsins and collagenases which, to some degree, can be identified by accessory catalytic residues. Typically, trypsins can be identified by the presence of Asp189. This was found in all of the trypsins included in these analyses with two exceptions, AaLT and Aedes albopictus LT, both of which have a Ser at this position. In addition, six of the seven novel A. aegypti serine proteases (AaSP II thru AaSP VII) contain this accessory catalytic residue (Fig. 3). Chymotrypsins usually possess a Ser189, but of the known chymotrypsins used in these analyses, only AaCHYMO conformed to this rule. The Glossina, Anopheles gambiae and AaJA15 chymotrypsins all have Gly189 while the Phlebotomus papatasi chymotrypsin encodes Ala189 (Fig. 3).
The positioning and importance of accessory catalytic residues in serine collagenases are not well defined as evident from the botfly (Hypoderma lineatum) and crab collagenases. The botfly collagenase contains Ser189, Gly216 and Gly226 whereas the crab collagenase contains Gly189, Gly216 and Asp226 (Fig. 3). Interestingly, AaSP I which has a Pro189, Lys216 and Arg226 arrangement shares little sequence similarity at these accessory catalytic residues (Fig. 3).
3.4. Phylogenetic analyses
Once amino acids were aligned, the nucleic acid alignment was manually adjusted to insure correct codon alignment. Phylogenetic trees based upon codon sequences were then derived by maximum parsimony (MP). Support for individual clades was tested with 1000 bootstrap replicates. There were 909 total characters analyzed by this approach of which 791 were parsimony informative. Analysis of the bootstrap tree revealed three major clades; clade A (56% bootstrap support), clade B (<50% bootstrap support) and clade C (62% bootstrap support) (Fig. 4). Clade A contained all of the known trypsin genes with the exceptions of AaLT and A. albopictus LT both of which arise in clade C.2 (91% bootstrap support). Two of the novel A. aegypti serine proteases, AaSP VI and VII, arose within Clade A and on a common branch with Aa5G1 (Fig. 4). Clade B contains AaSP group I, A. gambiae CHYMO and AaJA15. Clade C contains the known chymotrypsins and serine collagenases (62% bootstrap support). This clade can be further subdivided into clade C.1 (the chymotrypsins, 92%) and clade C.2 (the serine collagenases, 91%). In addition to A. albopictus LT and AaLT, AaSP I arises within the serine collagenases in clade C.2 (95% bootstrap support) (Fig. 4). Overall the topologies and distance resolved by the MP analyses were also supported by the distance-neighbor-joining and ML analyses (data not shown).
Fig. 4.
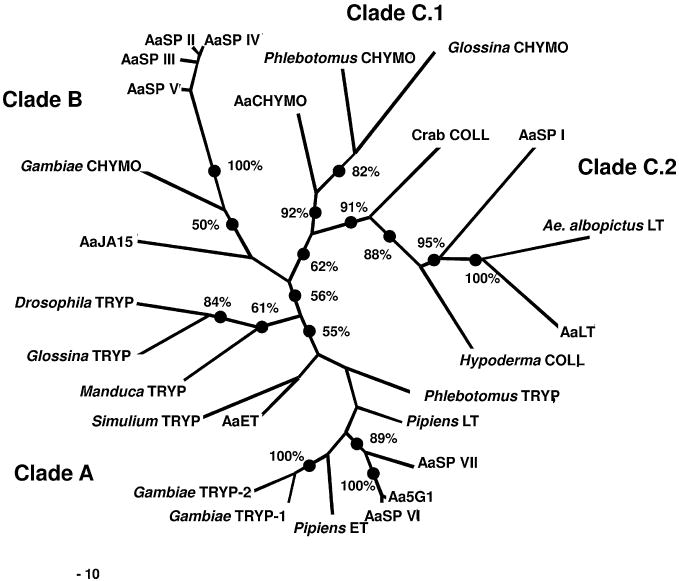
Maximum Parsimony analysis of serine protease codons. Aligned codons were used for this analysis. Clade A corresponds with the known trypsins, clade B represents a unique group of chymotrypsin-like serine proteases including A. gambiae CHYMO, AaJA15 and AaSP group I, clade C.1 contains the known chymotrypsins and clade C.2 the serine collagenases. Bootstrap analysis was performed with 1000 replicates.
3.5. Genomic annotation
Analysis of the genomic orientation and location of the serine proteases revealed that the majority of these genes occur on supercontigs that contain additional predicted serine proteases. Exceptions to this include AaCHYMO, AaET and AaSP I which are individually located on supercontigs 76, 284 and 255, respectively. AaLT is encoded by a single exon on supercontig 817. It is ~120 kb upstream of two predicted serine proteases also encoded by a single exon (Fig. 5). Similarly, Aa5G1 on supercontig 901 is encoded by a single exon and is in close proximity to five predicted serine protease ORFs (Fig. 5). Within the AaSP group I genes, AaSP II and AaSP V each have two introns and are located on supercontig 300 whereas AaSP IV and AaSP III are located in supercontig 887 with each containing three and four exons, respectively. AaJA15 has a single intron and is located on supercontig 39. It is positioned between two predicted serine protease ORFs that are in opposite orientation (Fig. 5). Finally, AaSP VI and AaSP VII are both located in supercontig 460 as single exons. Two additional predicted serine proteases are located in supercontig 460 in the same orientation; however, they contain multiple introns (Fig. 5).
Fig. 5.
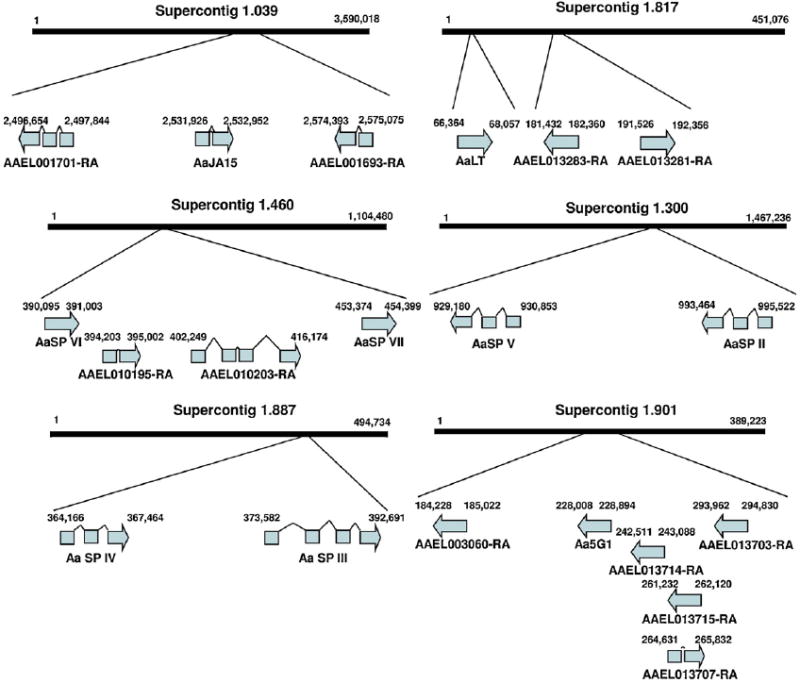
Genome annotation of A. aegypti midgut serine proteases. Each of the serine proteases were mapped to their respective supercontigs and oriented. The supercontigs were further analyzed for the presence of putative serine protease sequences and their positional relation to the characterized serine proteases schematically represented. Not represented are AaET, AaCHYMO and AaSP I which are found on supercontigs 284, 76 and 255, respectively.
4. Discussion
The A. aegypti genome contains 369 serine protease-like genes of which 66 are putative trypsins (Venancio et al., 2009). However, to date, only five have been characterized from the adult female midgut. Several factors may contribute to this large discrepancy. While serine proteases are known to be important for digestion, these enzymes are also involved in many other critical processes, such as immunity, reproduction, development, signal transduction and wound healing (Barros et al., 1996; Coughlin, 2000; Hedstrom, 2002; Joseph et al., 2001; LeMosy et al., 1999; Neurath, 1984). In fact, many of the 369 serine protease-like genes encode for serpins, zinc metalloproteases and kinases which are clearly not involved in bloodmeal digestion. The broad physiological utility of serine proteases may explain their diversity. Further, mosquitoes have a fairly complex lifecycle with multiple life-stages. It would be logical to assume that many of these serine proteases are critical to other life-stages, tissues and physiological states. Nevertheless it remains possible that other uncharacterized serine proteases are present in the adult female midgut. In fact, recent bioinformatics analysis of A. aegypti ESTs suggests that there may be eight additional midgut-associated bloodmeal-induced trypsin-like serine proteases (Venancio et al., 2009). Therefore, we sought to more completely describe the serine protease profile of the adult midgut and provide a genomic and phylogenetic assessment of these findings.
A. aegypti bloodmeal digestion can be divided into two phases; early phase 1–3 hpbm and late phase 8–36 hpbm (Felix et al., 1991). AaET and AaCHYMO transcripts that accumulate after emergence and upon blood feeding become translated during the early phase of digestion (Bian et al., 2008; Jiang et al., 1997; Noriega et al., 1996). During the early phase of digestion the late phase enzymes, AaLT and Aa5G1, become transcriptionally induced and by 6–8 hpbm begin to be translated and reach maximal concentrations 24–36 hpbm (Barillas-Mury et al., 1991; Kalhok et al., 1993). AaSP I exists in unfed midguts at a minimal, yet detectable level and upon blood feeding becomes transcriptionally induced (Fig. 1B). Interestingly, this expression pattern is similar to the late phase serine proteases, yet transcription levels reach maximal levels much earlier (Fig. 1A, D and E). This suggests that AaSP I spans both the early and late phases of digestion. Analysis of the AaSP I deduced amino acid sequence revealed that AaSP I does not conform to the traditional His57-Asp102-Ser195 catalytic triad of S1.A serine proteases (Blow, 1997). In fact, the nucleophilic Ser195 residue has been replaced with a Val and the basic His57 replaced with a neutral Gln. Furthermore, it lacks the traditional accessory catalytic residues (Fig. 3). Despite its unique characteristics, phylogenetic analyses revealed that AaSP I was closely related to the botfly and crab serine collagenases with 91% bootstrap support (Fig. 4). These results were not surprising considering our incomplete understanding of serine collagenase architecture. It remains to be determined whether AaSP I is a serine collagenase or instead encodes for a non-catalytic protein.
As with AaSP I, both AaLT and A. albopictus LT arose within clade C.2 (Fig. 4). Collagen is not a normal component of blood, therefore the significance of a serine collagenase in hematophagous arthropod digestion is not known. However, C1q, the initiator of the classical complement cascade, has a collagen-like sequence of 81 residues (Kishore and Reid, 2000). It may be that AaLT is an inducible proteolytic defense mechanism required for degradation of C1q. In theory, this degradation would block downstream effects of the complement pathway and protect the midgut epithelial cells from complement-induced damage or prevent native bacterial flora already in the midgut from being targeted by ingested human complement. Further investigations into the function and substrate specificity of AaLT in bloodmeal digestion are needed. However, insights into its possible enzymatic activity can be revealed from studies with the botfly and crab serine collagenases.
The substrate specificity of serine collagenases is poorly understood. While both the botfly and crab collagenases can cleave collagen, only the crab collagenase has tryptic, chymotryptic and elastolytic activity (Grant and Eisen, 1980; Lecroisey and Keil, 1985). Crab collagenase has a unique rearrangement of the S1 binding pocket in which the trypsin Asp189 and Gly226 instead contain residues Gly and Asp, respectively (Grant et al., 1980). This rearrangement may, in part, be responsible for its broad substrate specificity, although studies have demonstrated that introducing these changes into trypsin did not affect the enzymes strict affinity for Lys/Arg (Perona et al., 1993). This suggests that such reorganization alone cannot account for the expansion of substrate specificity in crab collagenases. Alternatively, the broad substrate specificity may arise because of a two amino acid insertion after Gly216 (Tsu et al., 1997). In trypsins, positioning of the substrate scissile bond by Gly216 is thought to be dependent on the ionic interactions between Asp189 and substrate Lys/Arg. From crystal structure studies, it is predicted that this two amino acid insertion allows Gly216 positioning of the substrate scissile bond to act independently (Tsu et al., 1997). While these conclusions are well founded, they are based on the comparison of an arthropod collagenase and a mammalian trypsin. Therefore, it is difficult to conclude whether these two additional amino acids are the basis for the broad specificity or a result of evolutionary divergence. In fact, our alignments reveal that this two amino acid insertion is conserved among the many arthropod serine proteases analyzed in this manuscript (Fig. 3). If the insertion were the only determinant of broad substrate specificity then all of the enzymes analyzed should have broad substrate specificity, but this is not the case. For instance, the botfly collagenase specifically cleaves collagen and lacks tryptic and chymotryptic activity (Lecroisey and Keil, 1985).
While the botfly collagenase and AaLT maintain a fairly traditional S1 binding pocket organization, Ser189-Val216-Val226 and Ser189-Gly216-Gly216, respectively, they possess a hydrophobic Pro190 adjacent to the S1 binding pocket (Fig. 3). This differs from the nucleophilic amino acid located at this position in trypsins, chymotrypsins and the crab collagenase which is thought to stabilize binding to polar amino groups (Perona and Craik, 1995). This alteration near the S1 binding pocket may account for the discrepancy between the botfly and crab serine collagenases. However, it seems that multiple factors influence serine protease substrate specificity and individual factors alone cannot explain these differences. Furthermore, these results, in accordance with earlier RNAi suppression studies, suggest that AaLT is not a trypsin and may be a serine collagenase with a substrate specificity profile similar to botfly collagenase (Brackney et al., 2008).
The AaSP group I genes (AaSP II–V) are constitutively expressed in mosquito midguts prior to and after bloodmeal acquisition (Fig. 1C). However, the degree to which these four serine proteases are individually expressed in the midgut, if in fact all four are expressed in the midgut, could not be determined by Q-RT-PCR because of their high sequence similarity and the fact that they were isolated from genomic DNA libraries. This high sequence similarity is evident in the alignments and phylogenetic analyses (Figs. 3 and 4). Despite the high degree of similarity AaSP V does not contain the conserved catalytic triad and has a neutral Leu57 in place of the basic His (data not shown). At this time the significance of this mutation is difficult to assess. Nevertheless, all four genes arise within the same clade (100% bootstrap support) (Fig. 4). Interestingly, both AaJA15 and the A. gambiae chymotrypsin do not occur in the same clade with the other three confirmed chymotrypsins. Typically, chymotrypsins, including AaCHYMO, cleave Phe/Tyr/Trp, but the A. gambiae chymotrypsin has also been shown to have specific activity against Leu (Vizioli et al., 2001). This broader substrate specificity may account for its phylogenetic divergence from the other known chymotrypsins and suggest that the AaSP group I and AaJA15 might have similar enzymatic activity.
The two serine proteases, AaSP VI and AaSP VII, each have similar expression profiles to the previously reported late phase serine proteases, AaLT and Aa5G1 (Fig. 1D/E, 2B, C and F). The strong induction of these genes suggests their concerted involvement in digestion. Both of these genes contain the traditional serine protease features as well as the characteristic Asp189 commonly associated with trypsins (Fig. 3). In fact, they fall within the same clade as the known trypsins (specifically Aa5G1) with 89% bootstrap support (Fig. 4). These results are consistent with those previously published and suggest that AaSP VI and AaSP VII are both trypsins (Venancio et al., 2009).
The approaches used in this study were not exhaustive. Gene annotations suggest that there may be many other serine proteases expressed in the midgut (Fig. 5). To this end, microarrays may eventually prove to be useful for the discovery of novel midgut-associated serine proteases. However, to date, this has not been the case. Two previous studies analyzed bloodmeal-induced midgut gene expression utilizing microarrays with a subset of expressed sequence tags (EST) from midguts and malphigian tubules in fourth instar larvae and adult female unfed A. aegypti (Sanders et al., 2005, 2003). Both studies failed to identify novel serine proteases. In fact, AaET whose mRNA is present in unfed midguts was not reported to have been influenced by blood feeding. This suggests that microarrays may not provide the sensitivity needed for elucidation of novel midgut serine proteases. However, had the EST libraries been derived from unfed and post-bloodfed midguts the outcome may have been more fruitful. Further, deep-sequencing approaches may provide the required depth and sensitivity needed to fully elucidate the midgut serine protease transcriptome.
We have isolated seven midgut serine proteases and provided phylogenetic insight into their relationship. It is apparent that the A. aegypti midgut expresses numerous early and late phase serine proteases which act in concert to digest the bloodmeal. Our data strongly suggest that many of the midgut serine proteases share a common mechanism controlling transcriptional induction. Further, based on the gene annotations it seems likely that other serine proteases may be expressed in the adult female midgut. The reasons for this diversity are unclear, but may be the result of multiple gene duplications and/or A. aegypti may have evolved a complex array of proteases, each with a slightly different enzymatic activity profile, allowing for a broad range of substrate affinities. Overall, our findings provide new insights into mosquito midgut physiology and highlight the complexity of mosquito digestion. Understanding the basic physiology of vectors is critical to deciphering pathogen/vector interactions and can lead to novel control strategies.
Acknowledgments
We would like to thank Dr. Fernando Torres-Perez and Dr. Gregory Ebel for their insightful discussions concerning the amino acid alignments and phylogenetic analysis. Furthermore, we thank Mary Hernandez for rearing mosquitoes. These studies were funded by the National Institute of Health (AI-25489) to KEO, the Fellowship Training Program (T01/CCT822307) to KEO provided by the Centers for Disease Control and Prevention and by the National Institute of Health (AI31951) to RLM.
References
- Barillas-Mury C, Graf R, Hagedorn HH, Wells MA. cDNA and deduced amino acid sequence of a blood meal-induced trypsin from the mosquito Aedes aegypti. Insect Biochemistry. 1991;21:825–831. [Google Scholar]
- Barillas-Mury C, Wells MA. Cloning and sequencing of the blood meal-induced late trypsin gene from the mosquito Aedes aegypti and characterization of the upstream regulatory region. Insect Molecular Biology. 1993;2:7–12. doi: 10.1111/j.1365-2583.1993.tb00119.x. [DOI] [PubMed] [Google Scholar]
- Barros C, Crosby JA, Moreno RD. Early steps of sperm–egg interactions during mammalian fertilization. Cell Biology International. 1996;20:33–39. doi: 10.1006/cbir.1996.0006. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bian GW, Ralkhel AS, Zhu JS. Characterization of a juvenile hormone-regulated chymotrypsin-like serine protease gene in Aedes aegypti mosquito. Insect Biochemistry and Molecular Biology. 2008;38:190–200. doi: 10.1016/j.ibmb.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow DM. The tortuous story of Asp.His.Ser: structural analysis of [alpha]-chymotrypsin. Trends in Biochemical Sciences. 1997;22:405–408. doi: 10.1016/s0968-0004(97)01115-8. [DOI] [PubMed] [Google Scholar]
- Brackney DE, Foy BD, Olson KE. The effects of midgut serine proteases on dengue virus type 2 infectivity of Aedes aegypti. American Journal of Tropical Medicine and Hygiene. 2008;79:267–274. [PMC free article] [PubMed] [Google Scholar]
- Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- Dana A, Hong Y, Kern M, Hillenmeyer M, Harker B, Lobo N, Hogan J, Romans P, Collins F. Gene expression patterns associated with blood-feeding in the malaria mosquito Anopheles gambiae. BMC Genomics 6, 5. 2005 doi: 10.1186/1471-2164-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Riddell DC, Higgins MJ, Holden JJA, White BN. A gene family in Drosophila melanogaster coding for trypsin-like enzymes. Nucleic Acids Research. 1985;13:6605–6619. doi: 10.1093/nar/13.18.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix CR, Betschart B, Billingsley PF, Freyvogel TA. Post-feeding induction of trypsin in the midgut of Aedes aegypti L. (Diptera, Culicidae) is separable into 2 cellular phases. Insect Biochemistry. 1991;21:197–203. [Google Scholar]
- Grant GA, Eisen AZ. Substrate specificity of the collagenolytic serine protease from Uca pugilator—studies with noncollagenous substrates. Biochemistry. 1980;19:6089–6095. doi: 10.1021/bi00567a022. [DOI] [PubMed] [Google Scholar]
- Grant GA, Henderson KO, Eisen AZ, Bradshaw RA. Amino acid sequence of a collagenolytic protease from the hepatopancreas of the fiddler crab, Uca pugilator. Biochemistry. 1980;19:4653–4659. doi: 10.1021/bi00561a018. [DOI] [PubMed] [Google Scholar]
- Hedstrom L. Serine protease mechanism and specificity. Chemical Reviews. 2002;102:4501–4523. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- Isoe J, Rascon AA, Kunz S, Miesfeld RL. Molecular genetic analysis of midgut serine proteases in Aedes aegypti mosquitoes. Insect Biochemistry and Molecular Biology. 2009;39:903–912. doi: 10.1016/j.ibmb.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang QJ, Hall M, Noriega FG, Wells M. cDNA cloning and pattern of expression of an adult, female specific chymotrypsin from Aedes aegypti midgut. Insect Biochemistry and Molecular Biology. 1997;27:283–289. doi: 10.1016/s0965-1748(97)00001-5. [DOI] [PubMed] [Google Scholar]
- Joseph K, Ghebrehiwet B, Kaplan AP. Activation of the kinin-forming cascade on the surface of endothelial cells. Biological Chemistry. 2001;382:71–75. doi: 10.1515/BC.2001.012. [DOI] [PubMed] [Google Scholar]
- Kalhok SE, Tabak LM, Prosser DE, Brook W, Downe AE, White BN. Isolation, sequencing and characterization of two cDNA clones coding for trypsin-like enzymes from the midgut of Aedes aegypti. Insect Molecular Biology. 1993;2:71–79. doi: 10.1111/j.1365-2583.1993.tb00127.x. [DOI] [PubMed] [Google Scholar]
- Kishore U, Reid KBM. C1q: Structure, function, and receptors. Immuno-pharmacology. 2000;49:159–170. doi: 10.1016/s0162-3109(00)80301-x. [DOI] [PubMed] [Google Scholar]
- Lecroisey A, Keil B. Specificity of the collagenase from the insect Hypoderma lineatum. European Journal of Biochemistry. 1985;152:123–130. doi: 10.1111/j.1432-1033.1985.tb09171.x. [DOI] [PubMed] [Google Scholar]
- LeMosy EK, Hong CC, Hashimoto C. Signal transduction by a protease cascade. Trends in Cell Biology. 1999;9:102–107. doi: 10.1016/s0962-8924(98)01494-9. [DOI] [PubMed] [Google Scholar]
- Neurath H. Evolution of proteolytic enzymes. Science. 1984;224:350–357. doi: 10.1126/science.6369538. [DOI] [PubMed] [Google Scholar]
- Noriega FG, Edgar KA, Bechet R, Wells MA. Midgut exopeptidase activities in Aedes aegypti are induced by blood feeding. Journal of Insect Physiology. 2002;48:205–212. doi: 10.1016/s0022-1910(01)00165-2. [DOI] [PubMed] [Google Scholar]
- Noriega FG, Pennington JE, Barillas-Mury C, Wang XY, Wells MA. Aedes aegypti midgut early trypsin is post-transcriptionally regulated by blood feeding. Insect Molecular Biology. 1996;5:25–29. doi: 10.1111/j.1365-2583.1996.tb00037.x. [DOI] [PubMed] [Google Scholar]
- Noriega FG, Wells MA. A molecular view of trypsin synthesis in the midgut of Aedes aegypti. Journal of Insect Physiology. 1999;45:613–620. doi: 10.1016/s0022-1910(99)00052-9. [DOI] [PubMed] [Google Scholar]
- Perona JJ, Craik CS. Structural basis of substrate specificity in the serine proteases. Protein Science. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona JJ, Tsu CA, McGrath ME, Craik CS, Fletterick RJ. Relocating a negative charge in the binding pocket of trypsin. Journal of Molecular Biology. 1993;230:934–949. doi: 10.1006/jmbi.1993.1211. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sanders H, Foy B, Evans A, Ross L, Beaty B, Olson K, Gill S. Sindbis virus induces transport processes and alters expression of innate immunity pathway genes in the midgut of the disease vector, Aedes aegypti. Insect Biochemistry and Molecular Biology. 2005;35:1293–1307. doi: 10.1016/j.ibmb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Sanders HR, Evans AM, Ross LS, Gill SS. Blood meal induces global changes in midgut gene expression in the disease vector, Aedes aegypti. Insect Biochemistry and Molecular Biology. 2003;33:1105–1122. doi: 10.1016/s0965-1748(03)00124-3. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsu CA, Perona JJ, Fletterick RJ, Craik CS. Structural basis for the broad substrate specificity of fiddler crab collagenolytic serine protease 1. Biochemistry. 1997;36:5393–5401. doi: 10.1021/bi961753u. [DOI] [PubMed] [Google Scholar]
- Venancio TM, Cristofoletti PT, Ferreira C, Verjovski-Almeida S, Terra WR. The Aedes aegypti larval transcriptome: a comparative perspective with emphasis on trypsins and the domain structure of peritrophins. Insect Molecular Biology. 2009;18:33–44. doi: 10.1111/j.1365-2583.2008.00845.x. [DOI] [PubMed] [Google Scholar]
- Vizioli J, Catteruccia F, della Torre A, Reckmann I, Muller HM. Blood digestion in the malaria mosquito Anopheles gambiae—molecular cloning and biochemical characterization of two inducible chymotrypsins. European Journal of Biochemistry. 2001;268:4027–4035. doi: 10.1046/j.1432-1327.2001.02315.x. [DOI] [PubMed] [Google Scholar]
- White SH. Amino-acid preferences of small proteins—implications for protein stability and evolution. Journal of Molecular Biology. 1992;227:991–995. doi: 10.1016/0022-2836(92)90515-l. [DOI] [PubMed] [Google Scholar]


