Abstract
Mammalian cells possess a cell-autonomous molecular clock which controls the timing of many biochemical reactions and hence the cellular response to environmental stimuli including genotoxic stress. The clock consists of an autoregulatory transcription-translation feedback loop made up of four genes/proteins, BMal1, Clock, Cryptochrome, and Period. The circadian clock has an intrinsic period of about 24 hours, and it dictates the rates of many biochemical reactions as a function of the time of the day. Recently, it has become apparent that the circadian clock plays an important role in determining the strengths of cellular responses to DNA damage including repair, checkpoints, and apoptosis. These new insights are expected to guide development of novel mechanism-based chemotherapeutic regimens.
Keywords: Checkpoints, DNA repair, Apoptosis, XPA, Cryptochrome, Chemotherapy
Introduction
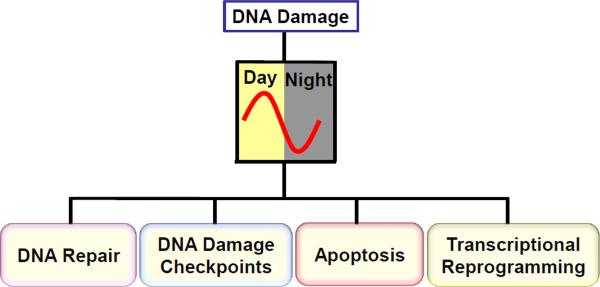
The circadian clock and the DNA damage response are two global regulatory mechanisms that control many aspects of cellular physiology and adaptation to the environment at the cellular and organismal level. The two regulatory systems can operate independently of one another. However, under physiological conditions, the two systems interface. As a consequence, a signal that primarily affects one system, to varying degrees, ultimately affects the other as well. In this review we will mainly focus on our current understanding of the control of the DNA damage response by the circadian clock. The DNA damage response includes DNA repair, DNA damage checkpoints, apoptosis, and transcriptional reprogramming, and current evidence indicates that all these responses are gated by the circadian clock (Figure 1). In the following, the basic molecular mechanism of the mammalian clock will be described first, followed by a survey of recent findings on the control of the DNA damage response by the clock, and how these findings may be used to develop new chemotherapy regimens. Finally, we discuss the possible evolutionary link between the circadian clock and the cellular response to DNA damage.
Figure 1.
Circadian gating of DNA damage responses. Major cellular response pathways to DNA damage including DNA repair, DNA damage checkpoints, apoptosis, and transcriptional reprogramming are gated by the circadian clock.
Molecular Circadian Clock
The circadian clock is the molecular system that confers daily rhythmicity to physiologic functions [1-4]. The clock is cell-autonomous and self-sustaining; that is, it keeps time in the absence of external input. However, in mammalian organisms the cell-autonomous clocks in the peripheral organs such as liver, heart, kidney, and skin are synchronized with one another by neural and humoral inputs from the “master clock” in the brain [1]. The master clock is located in the suprachiasmatic nucleus (SCN) which is in the anterior hypothalamus in two clusters of neurons above the optic chiasma. The basic molecular architecture of the SCN clock is the same as those of the peripheral clocks; however, the SCN has the capability of signaling to, and thus synchronizing, the peripheral clocks with itself and with one another.
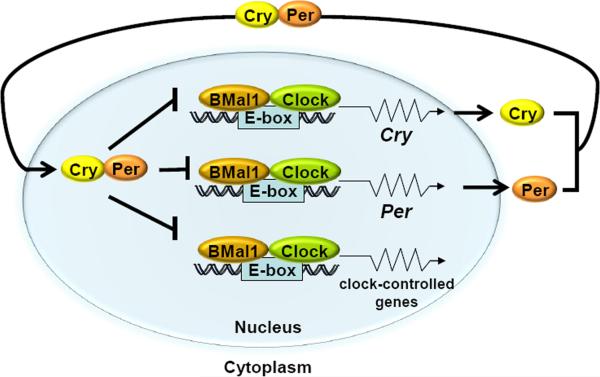
At the molecular level the clock is made up of 4 genes/proteins: Clock, BMal1, Crys (Cryptochrome 1 and 2), and Pers (Period 1 and 2) which operate in the following manner to generate a transcription-translation feedback loop (TTFL) (Figure 2): Clock and BMal1 are bHLH-PAS domain-containing transcriptional activators which make a heterodimer that binds to the E-boxes (CACGTG) in the promoters of the Cry and Per genes and activate their transcription. The Cry and Per proteins, in turn, make heterodimers or higher order multimeric complexes which, after a time lag, enter the nucleus and inhibit Clock-BMal1-activated transcription. Clock-BMal1 also controls the transcription of about 10% of the genes in a given cell, causing rhythmic expression of these so-called clock-controlled genes (CCG) [5]. In contrast to Crys and Pers, the proteins encoded by CCGs do not seem to directly feed back into the clock TTFL. However, the core clock circuitry is consolidated by additional transcriptional circuits as well as post-translational modifications that ensure high amplitude and a high-precision clock.
Figure 2.
Mammalian molecular clock. The bHLH-PAS domain-containing proteins Clock and BMal1 make a heterodimer which bind to the E-boxes (CACGTG) in the promoters of the Per and Cry genes, as well as in the promoters of the clock controlled genes, such as the excision repair gene Xpa and the checkpoint gene Wee1, activating their transcription. The Cry and Per proteins dimerize and, after a time lag, enter the nucleus and inhibit Clock-BMal1-activated transcription of their own genes as well as of those of clock-controlled genes, thus generating an oscillatory pattern of gene expression. Modified from [67].
Clock Control of DNA Repair
DNA repair is the ensemble of enzyme systems that eliminates DNA damage which might be defined as covalent change in the DNA structure [6]. There are several DNA repair mechanisms, including direct repair (photolyases and alkyl transferases), base excision repair (glycosylases and AP endonucleases), nucleotide excision repair, and double-strand break/crosslink repair. Currently available evidence indicates that of these repair mechanisms, only nucleotide excision repair is tightly controlled by the circadian clock. Hence, the mechanism of nucleotide excision repair and the control of this repair pathway by the circadian clock will be discussed in some detail.
(1) Nucleotide Excision Repair
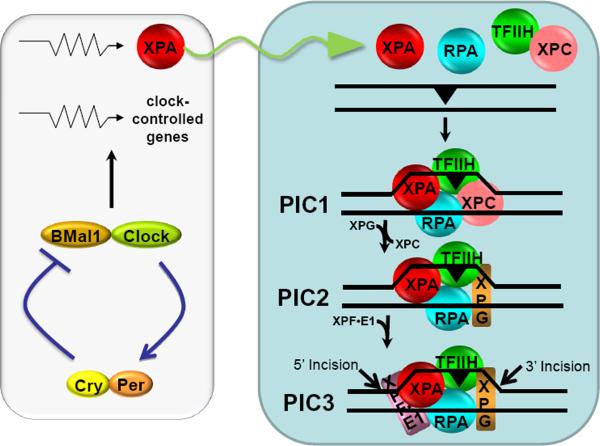
Nucleotide excision repair (excision repair) is a universal repair system that acts on virtually all types of base lesions. In many organisms, including humans and mice, it is the only repair system for bulky DNA adducts such as UV-induced cyclobutane pyrimidine dimers (Pyr<>Pyr) and cisplatin induced cisplatin-1,2-d(GpG) diadducts. In humans, excision repair is carried out by 6 factors [7-10]: RPA, XPA, XPC-HR23, TFIIH, XPG, and XPF-ERCC1, and proceeds as follows (Figure 3): RPA, XPA, and XPC, acting cooperatively, bind to the damage site and recruit TFIIH through its interactions with XPA and XPC. The two helicases within the TFIIH complex (XPB and XPD) unwind the DNA by about 20 bp around the damage to form a stable complex called pre-incision complex 1 (PIC1). Then, XPG enters the assembly as XPC-HR23 exits, to form PIC2. Finally, XPF-ERCC1 is recruited through a strong interaction with XPA to form PIC3. At each of these steps ATP is hydrolyzed, and the free energy of ATP hydrolysis is used to unwind the helix as well as to amplify the damage recognition specificity of the enzyme system by “kinetic proofreading” [9]. Within PIC3, XPG makes the 3′ incision 6±3 nucleotides 3′ to the lesion and XPF makes the 5′ incision 20±5 nucleotides 5′ to the damage [10]. The resulting 24-32 nucleotide-long oligomer carrying the lesion is released along with the repair factors, except RPA. The RPA-bound excision gap recruits RFC clamp loader and PCNA clamp, and eventually DNA polymerases δ/ε which fill the excision gap, and the repair patch is ligated by DNA ligase I.
Figure 3.
Model for nucleotide excision repair in humans and its regulation by the circadian clock. Excision of DNA damage is accomplished by sequential and partially overlapping functions of 6 core repair factors. Damage is recognized by cooperative activities of RPA, XPA, and XPC, followed by recruitment of TFIIH by XPC and XPA. The DNA around the damage site is unwound by the helicase activity of TFIIH to form a stable pre-incision complex 1 (PIC1) which recruits XPG, and XPC is displaced from the complex to form PIC2. Then XPF-ERCC1 is recruited to form PIC3. Within PIC3, XPG makes the 3′ incision 6±3 phosphodiester bonds 3′ and XPF makes the 5′ incision 20±5 phosphodiester bonds 5′ to the damage. The excised 24-32 nucleotide-long oligomer carrying the damage is released and the resulting gap is filled by DNA polymerases and ligated. The XPA protein which plays an essential role in damage recognition and is a rate-limiting factor is regulated by the clock, and as a consequence the daily oscillation of XPA (sinusoidal arrow) causes the entire excision repair activity to exhibit a daily rhythm, increasing during the day and decreasing during the night. Modified from [9, 66].
Mutations in excision repair proteins XPA, XPB, XPC, XPD, XPF, and XPG cause xeroderma pigmentosum. This disease is characterized by extreme sensitivity to sunlight, ~10,000-fold increase in the incidence of skin cancer, and, in some of the complementation groups, by mental and developmental abnormalities [7, 8]. In addition to the core excision repair factors essential for dual incision/excision, an accessory protein, DDB2, which is encoded by the XPE gene and is in a complex with a ubiquitin E3-ligase called DDB1 (UV-DDB=DDB1-DDB2=UV-damaged DNA binding protein), prevents UV carcinogenesis by an unknown mechanism [11]. Finally, in a form of XP called XP variant (XPV), a mutation in the XPV gene which encodes DNA polymerase η leads to high error-prone translesion synthesis by other polymerases across UV-photoproducts resulting in a high rate of mutation and cancer [12].
(2) Regulation of Nucleotide Excision Repair
Among the XP proteins, the regulation of XPE (DDB2), XPC-HR23, and XPA have been studied in some detail. The XPE gene is induced after DNA damage in a p53-dependent manner [13-15]. Then, the induced protein is ubiquitinated by the RING-type CUL4a-Roc1-DDB1 E3 ligase and degraded by the proteasome [16]. Similarly, XPC transcription is induced by DNA damage including UV-induced damage [14, 15]. However, in contrast to DDB2, this transcriptional induction does not result in elevated levels of XPC protein after DNA damage [17-19]. Like DDB2, XPC is ubiquitinated by CUL4a-Roc1-DDB1 following DNA damage, but unlike DDB2, ubiquitinated XPC is not targeted for proteolytic degradation by the proteasome [17, 18]. Currently, the physiological relevance of DDB2 ubiquitination and degradation, and of XPC ubiquitination, but not proteasomal degradation, are not known [20-22]. In contrast, XPA is ubiquitinated independently of DNA damage, and the ubiquitinated protein is targeted for proteolytic degradation [23, 24]. The regulation of mammalian excision repair by XPA ubiquitination and proteolysis is discussed in more detail below.
(3) Regulation of Nucleotide Excision Repair by the Circadian Clock
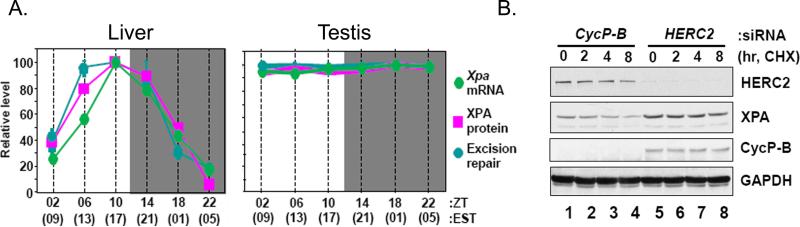
Analysis of nucleotide excision repair activity over the course of a day in various mouse tissues revealed that the repair activity has a robust circadian rhythm in brain [23] and liver but not in testis [24] (Figure 4A). Of the 6 core excision repair factors, only the XPA protein oscillates, and this oscillation is in phase with the excision repair activity and anti-phase with the Cry1 protein which functions as a transcriptional repressor in the core clock mechanism [23]. Further analysis revealed that Xpa is a clock-controlled gene that contains two canonical E-boxes in its promoter. Accordingly, in Cry1-/-Cry2-/- mice, Xpa transcription and XPA protein levels are constitutively high and no longer rhythmic [24]. However, robust transcriptional rhythm is not necessarily sufficient for rhythmic protein levels. If the rhythmically transcribed protein is stable, the transcriptional rhythm might confer protein level rhythmicity of only modest amplitude. However, if the rhythmically transcribed protein is targeted by the ubiquitin-proteasome system for ubiquitination and subsequent degradation by proteolysis, then high amplitude protein oscillation can be achieved. In fact, all core clock proteins that exhibit rhythmicity, Cry1 and Cry2 [25, 26], Per1 and Per2 [27, 28], and BMal1 [29], are targeted by specific E3 ubiquitin/SUMO ligases for ubiquitination or sumoylation and subsequent degradation by the proteasome. As a consequence, all of these proteins have half-lives of ~3 hrs. To find out if proteolytic degradation to achieve rhythmicity applied to a clock output protein, the half-life of XPA was measured and it was found that XPA also has a half-life of ~3 hrs [24].
Figure 4.
Circadian regulation of XPA and excision repair by the clock and the ubiquitin-proteasome system in the liver but not in testis. (A) Circadian rhythm of XPA transcript, protein, and nucleotide excision repair in mouse liver (left panel) but not testis (right panel) (ZT=0 is light on and ZT=12 is light off; EST=Eastern Standard Time) [23]. (B) HERC2 E3 ligase controls XPA stability. A549 cells were transfected with either cyclophilin-B siRNA (control) or HERC2 siRNA and then incubated with cycloheximide (CHX, a protein synthesis inhibitor) for the indicated times and the XPA protein levels were determined by immunoblotting [24].
Further analysis revealed that XPA is ubiquitinated by a HECT family E3 ligase called HERC2 [24]. Downregulation of HERC2 stabilizes XPA (Figure 4B) and increases cellular repair capacity moderately [24]. Similarly, in Cry1-/-Cry2-/- cells, XPA levels and excision repair activity are elevated. Interestingly, HERC2 itself is a clock-controlled protein that oscillates in-phase with XPA in liver, but it does not oscillate in the brain (T.H.K. and A.S., unpublished data), even though Xpa transcription and XPA protein levels oscillate in both organs. Thus, it appears that a protein such as XPA that is made in a rhythmic manner may exhibit an oscillatory pattern of accumulation whether it is degraded at a constant rate or in an oscillatory manner.
In addition to XPA and excision repair, it has been reported that O6-methylguanine-DNA methyl transferase [30] and alkylguanine DNA glycosylase [31] also exhibit circadian oscillation. However, the ratio of the nadir to the zenith for these enzymes is only about 1.2-fold (as opposed to 5- to 10-fold for XPA), and therefore the physiological significance of those oscillations is unclear at present.
Clock Control of DNA Damage Checkpoints
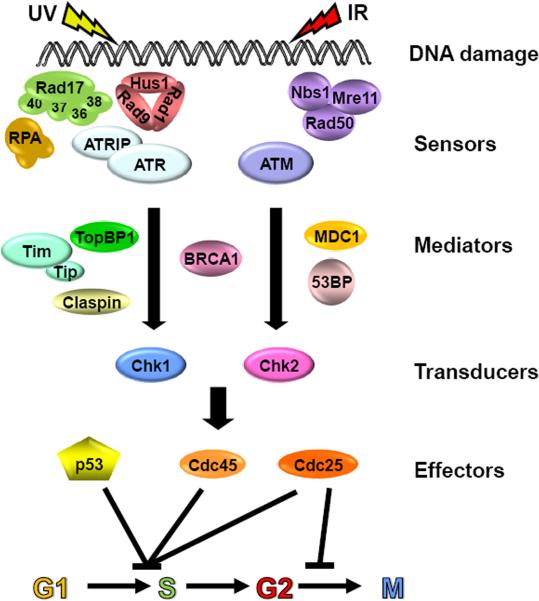
DNA damage checkpoints are cellular surveillance mechanisms that slow down or arrest cell cycle progression in response to DNA damage [6]. In mammalian cells, two checkpoint signaling pathways have been described: the ATR Chk1 pathway that is activated by UV, UV-mimetic agents, and chemical agents that inhibit replication fork progression, and the ATM→Chk2 pathway which is mainly activated by double strand breaks that are induced by ionizing radiation and radiomimetic agents (Figure 5). Both pathways encompass damage sensors, mediators, signal transducers, and effectors. The damage is detected by the ATR and ATM sensor kinases with the aid of accessory proteins, and is transmitted to signal transducing kinases, Chk1 and Chk2, with the aid of mediators. Signal transducing kinases then phosphorylate effector proteins including p53, Cdc25, and Cdc45. Phosphorylation of these proteins eventually leads to inhibition of two key kinases, Cdc2 and Cdk2, which cause cell cycle arrest at G2/M and G1/S, respectively. In addition to halting cell cycle progression, the DNA damage checkpoint response activates some DNA repair pathways that rescue stalled replication forks and repair double-strand breaks. The circadian clock affects the DNA damage checkpoint response in two ways: at the transcriptional level by controlling the transcription of the checkpoint proteins and by direct participation of the clock proteins in the checkpoint response. Accordingly, two mechanisms of circadian clock-DNA damage checkpoint connection have been described, serial and direct connections [32] (Figure 6).
Figure 5.
DNA damage checkpoint pathways. The pathways encompass damage sensors, mediators, signal transducers, and effectors. DNA damage in the form of double-strand breaks induced by ionizing radiation or radiomimetic agents activates the ATM→Chk2 pathway. DNA damage by UV and UV-mimetic agents activates the ATR→Chk1 pathway. Modified from [6].
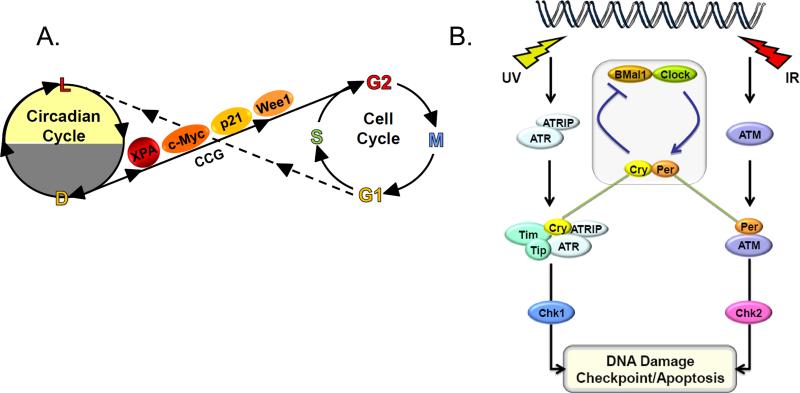
Figure 6.
Two models for coupling the circadian clock to the cell cycle/checkpoint response. (A) Serial coupling. The XPA repair factor, the p21 and Wee1 cell cycle proteins, and the c-Myc transcription factor, which are involved in DNA repair, the cell cycle, and cellular proliferation, respectively, are encoded by clock-controlled genes (CCG), and hence DNA repair, checkpoint activation, cell cycle regulation, and cellular proliferation are gated by the clock (solid line). Conversely, the cell cycle influences the circadian cycle by halting transcription during mitosis, thus causing a phase shift of the circadian rhythm (broken line). (B) Direct coupling. The core circadian clock protein Cry, in conjunction with Tim participates in the ATR-Chk1 signaling pathway in response to UV and UV-mimetic agents. Similarly, Per1 participates in the ATM-Chk2 signaling pathway in response to IR and radiomimetic agents.
(1) Serial connection of the circadian clock to the cell cycle checkpoints
In this model of coupling of the clock to the cell cycle checkpoints, the core clock system regulates genes that are involved in generating checkpoint signals, in sensing these signals, and in transducing the signals and executing the cell cycle arrest (Figure 6A). Examples for each of these are discussed below.
First, regulation of nucleotide excision repair by the clock [23, 24] results in generation of single-stranded DNA in the form of excision gaps in a rhythmic manner. Single-stranded DNA is a universal signal for ATR-mediated checkpoint signaling [6]. In non-dividing cells exposed to UV or UV-mimetic agents such as cisplatin, nucleotide excision repair is solely responsible for generating single-stranded DNA in the form of 24-32 nt-long excision gaps. Although unprocessed damage itself and RNA polymerases stalled at damage sites are capable of initiating checkpoint signaling [6, 33], it is most likely that in non-dividing cells at all times, and even in proliferating cells in G1 and G2 phases of the cell cycle, the excision gaps are the strongest signal for ATR activation. Because the XPA protein, and therefore excision repair activity, exhibits robust circadian rhythmicity, it is expected that UV-induced checkpoint signaling in such cells would manifest similar rhythmicity. However, this deductive prediction awaits experimental verification.
Second, the c-myc oncogene, which plays a key role in cellular proliferation as a bHLH transcription factor regulating a number of genes critical for G1/S transition, is a first-order clock controlled gene [34]. The mouse c-myc gene contains a canonical E-box in its promoter to which BMal1-NPas2(Clock) bind and inhibit c-myc transcription. In Per2 mutant mice, c-Myc is elevated because Per2 positively regulates BMal1 transcription; and in the absence of Per2, the BMal1-NPas2 level is reduced causing overexpression of c-Myc and promotion of cellular proliferation. Indeed, it has been reported that in Per2 mutant mice the overexpression of c-Myc is a major contributor to high-incidence of IR-induced lymphomas [34].
Third, the p21 gene is a second-order clock controlled gene which is negatively regulated by Rev-Erbα, which in turn is positively regulated by Clock-BMal1 [35]. Upon DNA damage the transcription of p21 is induced, and the elevated p21 protein level plays a critical role in the maintenance of G1/S checkpoint by binding to Cdk2 and inhibiting its activity. Furthermore, p21 binds to PCNA, inhibits DNA replication, and thus participates in intra-S checkpoint. Because of these important functions, it appears that in BMal1 mutant mice, reduced Rev-Erbα activity and hence elevation of p21 causes the mutant cells to exhibit a delay in the G1/S transition [35].
Finally, the Wee1 gene, which plays a key role in mitosis, is a first-order clock-controlled gene [36]. The Wee1 kinase phosphorylates the mitotic cyclin-dependent kinase, Cdc2, causing its inactivation and preventing the G2/M transition. In Cry mutant mice, Wee1 is elevated in the liver, and as a consequence, after partial hepatectomy, even though the wild-type and mutant hepatocytes reportedly progress through the G1 and S phases of the cell cycle similarly, the Cry mutants are delayed in entering mitosis resulting in slower regeneration of the liver [36]. However, despite this effect on normal hepatocyte progression through the cell cycle, it appears that elevated Wee1 does not affect the G2/M checkpoint significantly: Cry1-/-Cry2-/- mouse fibroblasts which overexpress Wee1 exhibit growth properties and G2/M checkpoint response to IR or UV indistinguishable from wild-type animals; they do not have prolonged arrest at the G2/M boundary and recover from the damage induced G2/M arrest essentially at the same rate as wild-type cells [37]. It is thus likely that the consequences of elevated Wee1 levels could be context-dependent and modulated by cellular physiology, cell-type, and the cellular environment.
(2) Direct connection of the circadian clock to the DNA damage checkpoints
In this mode of clock control of the checkpoint response, one or more proteins in the core clock circuitry also participate in DNA damage checkpoints. Currently, two examples for this mode of coupling are known (Figure 6B).
In one case, the Timeless (Tim) protein couples the clock to the ATR→Chk1 signaling pathway [32]. Tim is an accessory clock protein [1-3] that consolidates the core clock circuitry in mammalian organisms: Through its direct interaction with cryptochrome it participates in the molecular clock. Similarly, Tim participates in ATR→Chk1 signaling through its direct interaction with both ATR and Chk1 [32]. As a consequence, down-regulation of Tim disrupts both the circadian clock and the ATR→Chk1 DNA damage signaling pathway. Downregulating Tim in SCN slices drastically alters the levels of Crys and Pers and abolishes the oscillatory pattern of the SCN electrochemical output indicating that Tim is a bona fide clock protein in mammals [38]. However, Tim knockout in mice causes embryonic lethality [1], and therefore, it has not been possible to investigate the effect of Tim mutation on behavioral rhythmicity because of its essential function. Essential functions for Tim likely include its role in sister chromatid cohesion and its role in both intra-S and replication checkpoints [32].
In the second example of direct coupling of the clock to the DNA damage checkpoints, Per1 participates both in the core clock machinery and in the ATM→Chk2 DNA damage signaling pathway [39]. The role of Per1 in the circadian clock as a co-repressor is well-understood [1-3]. It participates in the negative arm of the autoregulatory transcription-translation loop. Per1-/- mice have long periods and are prone to become arrhythmic in constant darkness. In its role in the DNA damage checkpoints, Per1 participates in ATM→Chk2 signaling by directly interacting with both ATM and Chk2. Downregulation of Per1 interferes with phosphorylation of Chk2 by ATM after IR treatment and reduces DNA damage-induced apoptosis. Conversely, overexpression of Per1 inhibits proliferation of a number of cancer cell lines by promoting apoptosis [39]. In agreement with the cell biological data, it has been reported that Per1 is downregulated in a number of cancers, including lung and breast cancers [39].
Clock Control of Apoptosis
Apoptosis is programmed cell death that has important physiologic and pathologic consequences [40]. From a physiologic perspective, apoptosis plays an essential role during development in sculpting organs during embryogenesis. From a pathologic standpoint, it is an important contributor to cell death in several pathologic conditions such as tissue damage in cardiovascular and cerebrovascular ischemia by initiating extensive cell death in response to reactive oxygen species (ROS) generated by transient ischemia [40]. Finally, one of the most significant advances in cancer biology in recent years has been the realization of the importance of apoptosis in preventing oncogenically transformed cells from proliferation and hence from producing overt cancer. It appears that, as a rule, oncogenically transformed cells are more prone to apoptosis than normal cells [41].
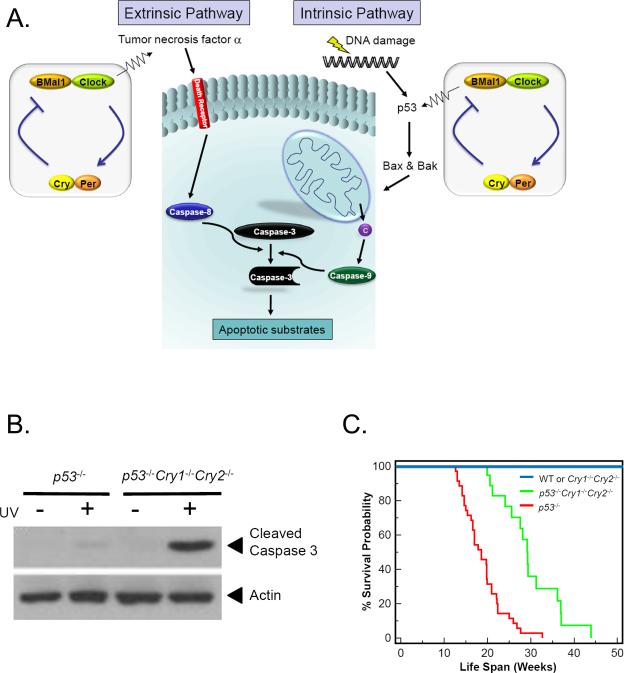
Formally, two biochemical pathways of apoptosis have been defined [40], extrinsic and intrinsic pathways (Figure 7A). In the mitochondria-independent, extrinsic (death receptor) pathway, a member of the tumor necrosis factor family, such as TNFα, binds to its cognate death receptor in the plasma membrane causing clustering of the receptor and the cytoplasmic adaptor molecules, and as a consequence, dimerization of pro-caspase 8. The latter is activated by autoproteolysis in trans to generate initiator caspase 8, which in turn converts pro-caspase 3 to the executioner caspase 3 that dismantles cellular architecture, destroys some key cellular enzymes, and activates caspase activated DNase (CAD) which attacks chromosomes, generating the apoptosis signature feature, the “nucleosome ladder” [42]. Recent work indicates that the core circadian clock participates in the extrinsic pathway by regulating the synthesis of TNFα [43]. Moreover, as TNFα is known to be involved in the inflammatory response, Cry1-/-Cry2-/- mice which overexpress TNFα are uniquely sensitive to inflammatory stimuli and exhibit a rheumatoid arthritis-like syndrome caused by such stimuli [44].
Figure 7.
Regulation of apoptosis by the circadian clock. (A) The extrinsic and intrinsic apoptosis pathways. Circadian regulation of the extrinsic pathway occurs by regulating the synthesis of TNFα which binds to death receptors, which through death adaptors transduce the signal to caspase 8 which in turn activates executioner caspases including caspase 3. In the intrinsic pathway, DNA damage activates p53 (in a circadian regulated manner) which activates transcription of Bax and Bak pro-apoptotic members of Bcl2 family. Bax and Bak cause release of cytochrome c (c) from mitochondrial intermembrane space. Cytochrome c helps assemble Apaf1 into an apoptosome which leads to cleavage and activation of transducer caspase 9 which in turn cleaves and activates executioner caspase 3. (B) Elimination of Crys in immortal p53-deficient cells makes cells more sensitive to killing by apoptosis induced by genotoxic agents including UV [46]. (C) Cry mutations increase the life-span of p53-/- mice [46]. The Kaplan-Meier survival analysis of mice of the indicated genotype is shown. Significant differences were observed between p53–/– (n=35) and p53–/–Cry1–/–Cry2–/– (n=19) mice (P < 0.0001).
In the intrinsic (or mitochondrial) pathway, cellular damage, including DNA damage and stress caused by unfolded proteins, leads to increased expression of pro-apoptotic members (Bax and Bak) of the Bcl-2 family. The p53 tumor suppressor plays an important role in DNA damage-initiated intrinsic apoptosis pathway [41]. Indeed, it appears that p53 acts a tumor suppressor to a large extent by inducing apoptosis in oncogenically transformed cells by upregulating the expression of pro-apoptotic Bax and Bak in response to genotoxic stress (Figure 7A). With respect to clock-intrinsic apoptosis pathway, there appears to be a direct connection between the core clock and p53: BMal1 upregulates [45] and Cry downregulates [46] p53 expression, even though p53 does not have a robust circadian rhythm because of multiple other inputs into the regulation of this multipurpose tumor suppressor. In addition, in a cell-based assay, knockdown of p53 reduces the amplitude of the circadian rhythm [47], but p53 mutant mice have an apparently normal clock. Clearly, the clock-p53-apoptosis connection is rather complex. Of special significance, p53-/-Cry1-/-Cry2-/- fibroblasts are more sensitive to genotoxicant-induced apoptosis than p53-/- cells (Figure 7B) [46], suggesting that Crys negatively regulate a p53-independent apoptotic pathway that becomes more relevant in a p53-/- background.
Clock, Cancer, and Chemotherapy
The interfacing of the clock at multiple points with cellular responses to DNA damage leads to the inevitable prediction that circadian clock disruption may contribute to carcinogenesis and that the clock would play significant roles in cancers induced by genotoxicants as well as in treating cancers by DNA damaging agents. Indeed, such connections do exist and these topics have been the subject of several recent reviews [48-56] and will be discussed here briefly.
(1) Circadian Clock and Cancer
Based largely on epidemiologic data, it was widely believed that circadian clock disruption predisposes humans to cancer [48, 49]. A study with Per2 mutant mice seemed to support this view [34]. It was reported that after IR treatment these mice developed lymphomas at about 10-fold higher incidence than wild-type mice. The higher incidence of lymphoma was ascribed to upregulation of c-myc oncogene and downregulation of p53 tumor suppressor, and hence reduced apoptosis of transformed cells in the mutant mice [34, 48]. However, studies with Cry mutant mice revealed a more complex pattern of interactions among the clock, apoptosis, and oncogenic transformation. First, Cry1-/-Cry2-/- mice, which lack a circadian clock, are indistinguishable from wild-type mice with respect to the incidence of both spontaneous and IR-induced cancers [37]. Second, when the Cry mutation was combined with the p53 mutation, with the expectation that clock disruption would increase cancer incidence in mice already predisposed to cancer because of the p53 mutation, a paradoxical result was obtained [46]: The Cry mutation protected p53 mutant mice from early onset of cancer and extended their median lifespan by ~50% from 19 weeks to 28 weeks (Figure 7C).
To explain this unexpected finding, immortal cell lines of p53-/- and p53-/-Cry1-/-Cry2-/- genotypes were analyzed for the integrity of the DNA damage response pathways that contribute to cancer prevention, including DNA repair, DNA damage checkpoints, and apoptosis. The two cell lines behaved similarly with respect to repair of UV-induced DNA damage and UV-induced activation of ATR→Chk1 and ATM→Chk2 signaling pathways [46]. In contrast, as noted above, it was found that elimination of Cry in immortal p53-deficient cells made the cells more sensitive to killing by apoptosis by genotoxic agents including UV (Figure 7B). Therefore, it was proposed that the increased sensitivity to apoptosis by endo-toxicants might be the mechanism by which the Cry mutation in p53 mutant mice prevented oncogenically transformed cells from proliferating to overt cancers. The mechanism by which the Cry mutation activates the p53-independent apoptosis pathway remains to be elucidated.
Finally, studies with Clock or BMal1 mutant mice have reinforced the notion that clock disruption in itself does not predispose animals to cancer. Thus, even though Clock mutants are more sensitive than wild-type to acute lethal effects of cyclophosphamide, presumably because of increased toxicity to B lymphocytes [57], the mutants were indistinguishable from wild-type with respect to the incidence of spontaneous and IR-induced cancers [58]. Similarly, BMal1 mutants do not exhibit increased rates of cancers; however, these mice exhibit a premature aging phenotype, presumably because of increased rate of ROS production [59] and chronic oxidative stress.
To summarize, the hypothesis that clock disruption predisposes animals to cancer, as a general rule, has not been supported by experimental tests. A more conservative view that clock disruption by certain mechanisms (such as Per mutations) or by unorthodox lifestyles may promote cancer is plausible and it deserves further critical investigations.
(2) Circadian Clock and Chemotherapy
Chronochemotherapy is the administration of anticancer drugs at specific times of the day so as to achieve optimal outcomes with tolerable side effects [51-56]. The effect of genotoxic chemotherapeutic drugs such as cisplatin on cancer cells is dictated, in addition to pharmacokinetic and pharmacodynamic factors, by cellular response to DNA damage including DNA repair, DNA damage checkpoints, and apoptosis. Because these responses are, to varying degrees, controlled by the clock it is to be expected that the time of drug delivery would affect its efficacy. Indeed, some small scale clinical trials have reported dramatic effects of chronochemotherapy [51]. However, as a rule, the effects of chronotherapy in large scale trials have been modest [56], and as a consequence chronotherapy is not currently practiced by oncologists. This is, in part, due to the empirical nature of the past clinical trials. The recent advances in understanding the mechanistic links between the clock and the DNA damage response summarized here are expected to lead to mechanism-based chronotherapy regimens in the near future. In fact, the high amplitude circadian oscillation of excision repair has led to the proposal of the following specific cisplatin chronotherapy schedule in a mouse model.
In all organs tested in mouse, with the exception of testis [24], excision repair of cisplatin adducts is at its zenith at ~5 pm and its nadir at ~5 am [24]. Therefore, a cisplatin treatment regimen that would maximize cisplatin-DNA adduct formation at ~5 am should be most effective against mouse cancers responsive to cisplatin. Moreover, it has been found that after intra-peritoneal injection of a single cisplatin dose, the cisplatin-DNA adduct level reaches its maximum in about 1 hr in mouse liver and likely other major organs except the brain [60]. Thus, ignoring other factors that might influence the cytotoxicity of the drug, it would appear that ~5 am would be the best time to treat cancer with cisplatin in mice. Because circadian gene expression and circadian physiology of humans are out of phase with those of nocturnal mice by 12 hr [52], administering cisplatin at ~5 pm might be optimal or treatment of human cancers except testicular cancer. Indeed, there are reports of higher success rate in treating ovarian cancer with cisplatin when the drug is delivered at 6 pm compared to 6 am [61, 62]. However, these reports have not been confirmed in subsequent studies [52], underscoring the need for mechanism-based approaches. Clearly, chemotherapeutic regiments that take into consideration the circadian phases of both the cancer and the normal tissue are needed to further refine chronotherapy and make it a standard part in the cancer treatment arsenal.
Concluding Remarks
According to the “Escape from Light” hypothesis for the evolution of the circadian clock [63], an ancient aquatic organism employed a blue-light sensor (because blue light penetrates the deepest in water) to synchronize its movements to and from the surface of the ocean so as to optimize nutrient availability and minimize exposure to the harmful effect of the UV component of sunlight. It is conceivable that the last common ancestor of the present day photolyase/cryptochrome family was the blue light sensor directing the diel vertical movement as well as using the blue light as an energy source to repair the UV-induced DNA damage that occurred under those conditions [64, 65]. This primitive flavoprotein with blue-light absorption maxima then diverged to give rise to the present day photolyases which repair DNA and to cryptochromes that control the circadian clock by light-dependent and light-independent mechanisms [64, 65].
Thus, even though the possible common evolutionary origin of the circadian clock and the mechanisms to maintain genomic integrity were recognized early-on in the circadian research fields [63], only recently have the multiple links between the two systems been described. It should be noted, however, that even though photolyase no longer has a photosensory function and cryptochrome no longer has a direct DNA repair function, cryptochrome still participates in the maintenance of genomic integrity against DNA damage induced by UV and UV-mimetic agents [24, 66]. Cryptochrome contributes to DNA repair/genome maintenance by regulating nucleotide excision repair which is the sole repair mechanism for repairing UV-induced photodimers in placental mammals which lack photolyase [65], and by coordinating the circadian clock with DNA damage checkpoints which also aids in cell survival and hence in “escape from light” [63, 65].
Abbreviations
- TTFL
transcription-translation feedback loop
- CCG
clock-controlled genes
- XP
xeroderma pigmentosum
- UV
ultraviolet
- IR
ionizing radiation
- SCN
suprachiasmatic nucleus
- Cry
Cryptochrome
- Per
Period
- Tim
Timeless
- PIC
pre-incision complex
- ROS
reactive oxygen species
- CAD
caspase activated DNase
- EST
Eastern Standard Time
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–61. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–75. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sancar A. Regulation of the mammalian circadian clock by cryptochrome. J Biol Chem. 2004;279:34079–82. doi: 10.1074/jbc.R400016200. [DOI] [PubMed] [Google Scholar]
- 5.Hughes M, Deharo L, Pulivarthy SR, Gu J, Hayes K, Panda S, Hogenesch JB. High-resolution time course analysis of gene expression from pituitary. Cold Spring Harb Symp Quant Biol. 2007;72:381–6. doi: 10.1101/sqb.2007.72.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 7.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 8.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272:23465–8. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 9.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 10.Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5' and the 6th phosphodiester bond 3' to the photodimer. Proc Natl Acad Sci U S A. 1992;89:3664–8. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols AF, Itoh T, Graham JA, Liu W, Yamaizumi M, Linn S. Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J Biol Chem. 2000;275:21422–8. doi: 10.1074/jbc.M000960200. [DOI] [PubMed] [Google Scholar]
- 12.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–83. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 13.Vaisman A, Chaney SG. Induction of UV-damage recognition protein by cisplatin treatment. Biochemistry. 1995;34:105–14. doi: 10.1021/bi00001a013. [DOI] [PubMed] [Google Scholar]
- 14.Amundson SA, Do KT, Shahab S, Bittner M, Meltzer P, Trent J, Fornace AJ., Jr. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 2000;154:342–6. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Rieger KE, Chu G. Portrait of transcriptional responses to ultraviolet and ionizing radiation in human cells. Nucleic Acids Res. 2004;32:4786–803. doi: 10.1093/nar/gkh783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Zhang Y, Douglas L, Zhou P. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J Biol Chem. 2001;276:48175–82. doi: 10.1074/jbc.M106808200. [DOI] [PubMed] [Google Scholar]
- 17.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Hanaoka F. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 18.Wang QE, Zhu Q, Wani G, El-Mahdy MA, Li J, Wani AA. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005;33:4023–34. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang QE, Praetorius-Ibba M, Zhu Q, El-Mahdy MA, Wani G, Zhao Q, Qin S, Patnaik S, Wani AA. Ubiquitylation-independent degradation of Xeroderma pigmentosum group C protein is required for efficient nucleotide excision repair. Nucleic Acids Res. 2007;35:5338–50. doi: 10.1093/nar/gkm550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–67. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 21.Nag A, Bondar T, Shiv S, Raychaudhuri P. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol Cell Biol. 2001;21:6738–47. doi: 10.1128/MCB.21.20.6738-6747.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–7. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 23.Kang TH, Reardon JT, Kemp M, Sancar A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc Natl Acad Sci U S A. 2009;106:2864–7. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci U S A. 2010;107:4890–5. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–4. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 26.Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–23. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–72. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 28.Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, Herzel H, Kramer A. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms. 2007;22:375–86. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- 29.Cardone L, Hirayama J, Giordano F, Tamaru T, Palvimo JJ, Sassone-Corsi P. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309:1390–4. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 30.Marchenay C, Cellarier E, Levi F, Rolhion C, Kwiatkowski F, Claustrat B, Madelmont JC, Chollet P. Circadian variation in O6-alkylguanine-DNA alkyltransferase activity in circulating blood mononuclear cells of healthy human subjects. Int J Cancer. 2001;91:60–6. doi: 10.1002/1097-0215(20010101)91:1<60::aid-ijc1010>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Matsunaga N, Koyanagi S, Ohdo S. Clock gene mutation modulates the cellular sensitivity to genotoxic stress through altering the expression of N-methylpurine DNA glycosylase gene. Biochem Pharmacol. 2009;78:1075–82. doi: 10.1016/j.bcp.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–16. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindsey-Boltz LA, Sancar A. RNA polymerase: the most specific damage recognition protein in cellular responses to DNA damage? Proc Natl Acad Sci U S A. 2007;104:13213–4. doi: 10.1073/pnas.0706316104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 35.Grechez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283:4535–42. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- 36.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–9. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 37.Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65:6828–34. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- 38.Barnes JW, Tischkau SA, Barnes JA, Mitchell JW, Burgoon PW, Hickok JR, Gillette MU. Requirement of mammalian Timeless for circadian rhythmicity. Science. 2003;302:439–42. doi: 10.1126/science.1086593. [DOI] [PubMed] [Google Scholar]
- 39.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–82. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 40.Hotchkiss RS, Strasser A, McDunn JE, Swanson PE. Cell death. N Engl J Med. 2009;361:1570–83. doi: 10.1056/NEJMra0901217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Junttila MR, Evan GI. p53--a Jack of all trades but master of none. Nat Rev Cancer. 2009;9:821–9. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- 42.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 43.Hashiramoto A, Yamane T, Tsumiyama K, Yoshida K, Komai K, Yamada H, Yamazaki F, Doi M, Okamura H, Shiozawa S. Mammalian Clock Gene Cryptochrome Regulates Arthritis via Proinflammatory Cytokine TNF-{alpha}. J Immunol. 2010;184:1560–5. doi: 10.4049/jimmunol.0903284. [DOI] [PubMed] [Google Scholar]
- 44.Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD, Kramer A, Maier B. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci U S A. 2009;106:21407–12. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullenders J, Fabius AW, Madiredjo M, Bernards R, Beijersbergen RL. A large scale shRNA barcode screen identifies the circadian clock component ARNTL as putative regulator of the p53 tumor suppressor pathway. PLoS One. 2009;4:e4798. doi: 10.1371/journal.pone.0004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc Natl Acad Sci U S A. 2009;106:2841–6. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, 3rd, Janes J, Su AI, Hogenesch JB, Kay SA. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–61. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 49.Stevens RG. Circadian disruption and breast cancer: from melatonin to clock genes. Epidemiology. 2005;16:254–8. doi: 10.1097/01.ede.0000152525.21924.54. [DOI] [PubMed] [Google Scholar]
- 50.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–96. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 51.Kobayashi M, Wood PA, Hrushesky WJ. Circadian chemotherapy for gynecological and genitourinary cancers. Chronobiol Int. 2002;19:237–51. doi: 10.1081/cbi-120002600. [DOI] [PubMed] [Google Scholar]
- 52.Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 53.Hogenesch JB. It's all in a day's work: Regulation of DNA excision repair by the circadian clock. Proc Natl Acad Sci U S A. 2009;106:2481–2. doi: 10.1073/pnas.0813323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hede K. Cancer and the circadian clock: has the time finally come? J Natl Cancer Inst. 2009;101:550–3. doi: 10.1093/jnci/djp085. [DOI] [PubMed] [Google Scholar]
- 55.Antoch MP, Kondratov RV. Circadian proteins and genotoxic stress response. Circ Res. 2010;106:68–78. doi: 10.1161/CIRCRESAHA.109.207076. [DOI] [PubMed] [Google Scholar]
- 56.Levi F, Okyar A, Dulong S, Innominato PF, Clairambault J. Circadian timing in cancer treatments. Annu Rev Pharmacol Toxicol. 2010;50:377–421. doi: 10.1146/annurev.pharmtox.48.113006.094626. [DOI] [PubMed] [Google Scholar]
- 57.Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, Antoch MP. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci U S A. 2005;102:3407–12. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antoch MP, Gorbacheva VY, Vykhovanets O, Toshkov IA, Kondratov RV, Kondratova AA, Lee C, Nikitin AY. Disruption of the circadian clock due to the Clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008;7:1197–204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–73. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnsson A, Olsson C, Nygren O, Nilsson M, Seiving B, Cavallin-Stahl E. Pharmacokinetics and tissue distribution of cisplatin in nude mice: platinum levels and cisplatin-DNA adducts. Cancer Chemother Pharmacol. 1995;37:23–31. doi: 10.1007/BF00685625. [DOI] [PubMed] [Google Scholar]
- 61.Hrushesky WJ. Circadian timing of cancer chemotherapy. Science. 1985;228:73–5. doi: 10.1126/science.3883493. [DOI] [PubMed] [Google Scholar]
- 62.Levi F, Benavides M, Chevelle C, Le Saunier F, Bailleul F, Misset JL, Regensberg C, Vannetzel JM, Reinberg A, Mathe G. Chemotherapy of advanced ovarian cancer with 4'-O-tetrahydropyranyl doxorubicin and cisplatin: a randomized phase II trial with an evaluation of circadian timing and dose-intensity. J Clin Oncol. 1990;8:705–14. doi: 10.1200/JCO.1990.8.4.705. [DOI] [PubMed] [Google Scholar]
- 63.Pittendrigh CS. Temporal organization: reflections of a Darwinian clockwatcher. Annu Rev Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 64.Gehring W, Rosbash M. The coevolution of blue-light photoreception and circadian rhythms. J Mol Evol. 2003;57(Suppl 1):S286–9. doi: 10.1007/s00239-003-0038-8. [DOI] [PubMed] [Google Scholar]
- 65.Sancar A. Cryptochrome: the second photoactive pigment in the eye and its role in circadian photoreception. Annu Rev Biochem. 2000;69:31–67. doi: 10.1146/annurev.biochem.69.1.31. [DOI] [PubMed] [Google Scholar]
- 66.Kang TH, Sancar A. Circadian regulation of DNA excision repair: implications for chrono-chemotherapy. Cell Cycle. 2009;8:1665–7. doi: 10.4161/cc.8.11.8707. [DOI] [PubMed] [Google Scholar]
- 67.Sancar A. The intelligent clock and the Rube Goldberg clock. Nat Struct Mol Biol. 2008;15:23–4. doi: 10.1038/nsmb0108-23. [DOI] [PubMed] [Google Scholar]