Abstract
1,3-butadiene (BD)1 is an important industrial and environmental chemical classified as a human carcinogen based on epidemiological evidence for an increased incidence of leukemia in workers occupationally exposed to BD and its potent carcinogenicity in laboratory rats and mice. BD is metabolically activated to epoxide intermediates that can react with nucleophilic sites of cellular biomolecules. Among these, 1,2,3,4-diepoxybutane (DEB) is considered the ultimate carcinogenic species of BD due to its potent genotoxicity and mutagenicity attributed to the ability to form DNA-DNA cross-links and exocyclic nucleoside adducts. DEB mutagenesis studies suggest that adducts formed at adenine bases may be critically important, as DEB induces large numbers of A → T transversion mutations. We have recently identified two regioisomeric exocyclic DEB-dA adducts, 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2'-deoxyadenosine (1,N6-γ-HMHP-dA) and 1,N6-(1-hydroxymethyl-2-hydroxypropan-1,3-diyl)-2'-deoxyadenosine (1,N6-α-HMHP-dA) (Seneviratne et al. Chemical Research in Toxicology 23, 118-133, 2010), which were detected in DEB-treated calf thymus DNA and in tissues of BD-exposed laboratory animals. In the present work, we describe a column switching HPLC-ESI-MS/MS methodology for the quantitative analysis of 1,N6-HMHP-dA isomers in DNA of laboratory mice exposed to BD by inhalation. Based on their exocyclic structure, which prevents normal Watson-Crick base pairing, these adducts could be responsible for mutations at the A:T base pairs observed following exposure to DEB.
Introduction
1,2,3,4-Diepoxybutane (DEB) is the proposed ultimate carcinogenic form of 1,3-butadiene (BD), a high volume industrial chemical used commonly in rubber and plastic manufacturing (1), and a known human and animal carcinogen (2;3). BD is also an environmental pollutant present in automobile exhaust and in cigarette smoke (4). Although DEB is a relatively minor metabolite of BD, it is by far the most mutagenic, inducing large numbers of base substitutions, sister chromatid exchanges, and chromosomal aberrations (5-7). Because of the widespread human exposure to BD, and the key role of DEB, there is a great need for biomarkers of BD metabolism to DEB.
Our laboratory has previously developed quantitative HPLC-ESI+-MS/MS methods for two types of DEB-specific DNA-DNA cross-links, 1,4-bis-(guan-7-yl)-2,3-butanediol (bis-N7G-BD) and 1-(guan-7-yl)-4-(aden-1-yl)-2,3-butanediol (N7G-N1A-BD) (8-10). Both types of DEB-induced cross-linked adducts were observed in DNA extracted from tissues of laboratory mice that were exposed to BD by inhalation, with bis-N7G-BD present at a higher concentration than N7G-N1A-BD (3.95 ± 0.89 adducts/107 nts and 0.27 ± 0.08 adducts/107 nts, respectively) (8-10). While the mutagenicity of DEB-induced DNA-DNA cross-links is unknown, the mutational spectra of DEB in human lymphocytes show large numbers of point mutations at the A:T base pairs, specifically A → T transversions (11), suggesting that it forms strongly mispairing adducts at adenine nucleobases.
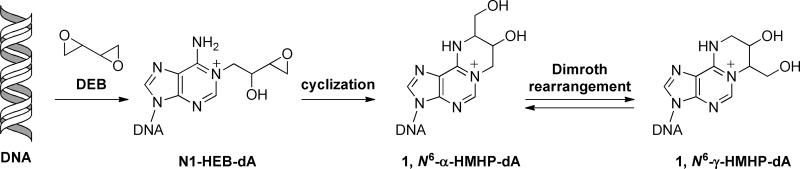
We have recently identified two novel DEB-dA adducts that are formed by sequential alkylation of the N-1 and the N6 positions of adenine by DEB: 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-dA (1,N6-γ-HMHP-dA) and 1,N6-(1-hydroxymethyl-2-hydroxypropan-1,3-diyl)-dA (1,N6-α-HMHP-dA) (12). We hypothesized that DEB initially alkylates the N1 position of adenine to form N1-(2-hydroxy-3,4-epoxybut-1-yl)-dA (N1-HEB-dA), which spontaneously cyclizes to 1,N6-α-HMHP-dA adducts (Scheme 1) (12). 1,N6-α-HMHP-dA undergoes a slow, reversible Dimroth-like rearrangement to 1,N6-γ-HMHP-dA (Scheme 1) (12). In the present work, we describe the development of quantitative column switching capillary HPLC-ESI+-MS/MS methods for 1,N6-HMHP-dA adducts of DEB in liver DNA of laboratory mice exposed to 1,3-butadiene by inhalation.
Scheme 1.
Reactions of 1,2,3,4-diepoxybutane with DNA to yield exocyclic DEB-dA adducts.
Experimental
Materials
Authentic 1,N6-HMHP-dA and 15N4-1,N6-HMHP-dA (internal standards for quantitation) were prepared as previously reported (12). Phosphodiesterase I, phosphodiesterase II, and DNase I were obtained from Worthington Biochemical Corp. (Freehold, NJ). All other reagents were from Sigma (Milwaukee, WI) unless otherwise noted.
Methods
Animals and DNA extraction
Female B6C3F1 mice were acclimated for about 10 days before initiation of chemical exposures. Animals were randomly separated into air-control and exposure groups by weight and were housed individually in hanging wire stainless steel cages according to NIH guidelines (NIH Publication 86-23, 1985). Mice were exposed to 0, 62.5, 200, or 625 ppm 1,3-butadiene (99+%, Aldrich, Milwaukee, WI) inhalation for 2 weeks at the Lovelace Respiratory Research Institute (LRRI) (Albequerque, NM) as described previously (9;10). All animal procedures were approved by the LRRI Institutional Animal Care and Use Committee.
Briefly, experimental animals were exposed using multi-tiered whole-body exposure chambers (H-2000, Lab Products, Aberdeen, MD). Rodents in one chamber received filtered air only as a control group, and rodents in the other chambers received nominal 62.5, 200, or 625 ppm BD for 2 weeks (6 h/day, 5 days/week). Animals were housed within exposure chambers throughout the experiment, and had free access to food and water except for removal of food during the 6-h exposure periods. BD was metered through a mass flow controller to achieve the desired concentration. Chamber concentrations were monitored every hour using a Foxboro Wilkes 1A IR detector.
Following BD exposure, animals were sacrificed via cardiac puncture. Liver tissues were flash frozen and shipped to the University of Minnesota on dry ice. DNA was extracted using NucleoBond-AXG500 extraction kits (Macherey-Nagel, Bethlehem, PA) following the manufacturer's protocol. DNA amounts and purity were determined by UV spectrophotometry, and by dG quantitation.
DNA Hydrolysis and sample preparation
Mouse liver DNA (100 μg, from 200-400 mg of tissue) was subjected to neutral thermal hydrolysis (60 min at 70 °C), followed by ultrafiltration using YM-10 Centricon filters (Millipore Corp., Billercia, MA). The resulting partially depurinated DNA was spiked with 15N4-1,N6-HMHP-dA (27.5 fmol) and enzymatically digested with phosphodiesterase I (1.3 mU/μg DNA), phosphodiesterase II (1.4 mU/μg DNA), DNAse (0.67 U/μg DNA), and alkaline phosphatase (0.26 U/μg DNA) in 20 mM Tris-HCl/30 mM MgCl2 at 37 °C overnight. DNA digests were filtered through YM-10 Centricon filters, and 1,N6-HMHP-dA adducts were enriched by solid phase extraction on Extract Clean Carbo cartridges (150 mg, Grace Davidson Discovery Science, Deerfield, IL). Cartridges were prepared by equilibrating with methanol (2 × 3 mL) and water (2 × 3 mL). Samples (1 mL in water) were loaded by gravity, washed with water (3 mL) and 5 % methanol (3 mL), and eluted in 30 % methanol (3 mL). The SPE fractions containing 1,N6-HMHP-dA were dried under reduced pressure and dissolved in 0.05% acetic acid (25 μL) prior to HPLC-ESI+-MS/MS analysis.
Column-switching HPLC-ESI+-MS/MS Method
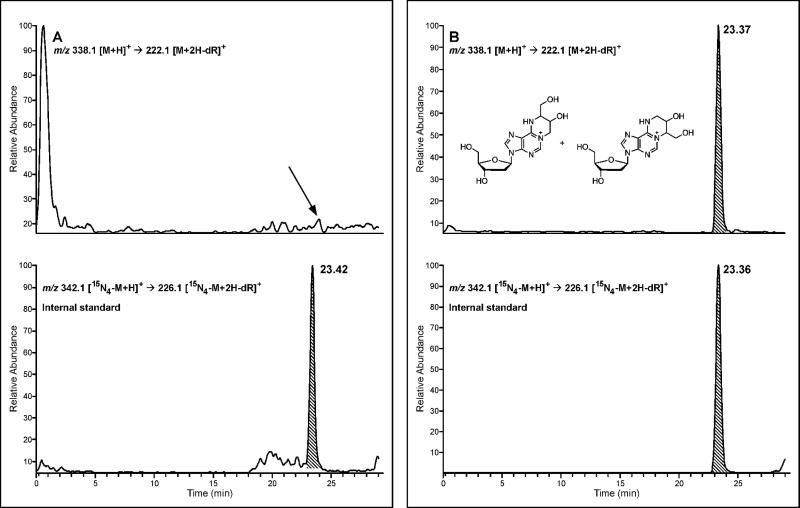
A Thermo-Finnigan TSQ Quantum Ultra mass spectrometer (Thermo Fisher Scientific Corp., Waltham, MA) interfaced to a Waters Acquity HPLC system (or a Thermo-Finnigan TSQ Discovery Max mass spectrometer interfaced to an Agilent 1100 HPLC system) was used in all analyses. Samples were loaded onto an SCX trap column (300 Å, 5 μm, nanoAcquity Waters Corp., Millford, MA) using an auxiliary Agilent 1100 pump delivering 0.5 % methanol in water isocratically at 15 μL/min for 10 min. During this time, the divert valve was in position A (Figure S-1). Following a 10 min loading period, the divert valve was switched to position B, and the adducts were released from the trap column and transferred to the analytical Synergi Hydro-RP (250 × 0.5 mm, Phenomenex) by backflushing for five minutes with a gradient starting at 0.5% MeOH (B) in 0.05% acetic acid (A). The solvent composition was maintained at 0.5% B for 1 minute, linearly increased to 5% B in 2 minutes, and maintained at 5% B for 4 minutes. The divert valve was returned to position A for equilibration of the trap column, and the analytical column gradient was further increased to 20% B over 9 minutes and then returned to 0.5% in 1 minute. Under these conditions, 1,N6-α-HMHP-dA and 1,N6-γ-HMHP-dA eluted together as a sharp peak at 23.5 min (Figure 1).
Figure 1.
Column switching HPLC-ESI+-MS/MS analysis of pure standards of 1,N6-HMHP-dA and [15N4]-1,N6-HMHP-dA (internal standard).
The mass spectrometer was operated in the positive ion mode, with nitrogen used as a sheath gas (5 L/min). Electrospray ionization was achieved at a spray voltage of 4.0 kV and a capillary temperature of 250 °C. Collision induced dissociation (CID) was achieved with Ar as a collision gas (1 mTorr) and a collision energy of 20 V. The mass spectrometer parameters were optimized for maximum response during the infusion of standard solutions.
The exocyclic DEB-dA lesions were quantified by isotope dilution with the corresponding 15N4-1,N6-HMHP-dA internal standards. The mass spectrometer was operated in the selected reaction monitoring mode by following the mass transition m/z 338.1 [M + H]+ → 222.1 [M +2H – dR]+, and the corresponding transition for the 15N4-1,N6-HMHP-dA internal standard (m/z 342.1 → 226.1). Quantitative analyses were based on the ratios of areas under the peak in the selected ion chromatogram corresponding to the analyte and the internal standard, respectively (relative response ratios). Standard curves were constructed by analyzing the solutions containing 1,N6-HMHP-dA (0 – 5 fmol) and 15N4-1,N6-HMHP-dA (2.5 fmol), followed by regression analysis of the relative response ratios calculated from HPLC-ESI+-MS/MS peak area ratios corresponding to 1,N6-HMHP-dA isomers and their internal standards. A good correlation was observed for standard calibration curves with a y-value of 1.05 and R2 = 0.997 (Figure S-2).
Validation of the quantitative column switching HPLC-MS/MS method
Control DNA was extracted from human cervical carcinoma (HeLa) cell cultures using NucleoBond AXG kits (Macherey-Nagel, Bethlehem, PA). DNA (0.1 mg) was spiked with 1,N6-HMHP-dA (0-5 fmol) and 15N -1,N46-HMHP-dA (5.5 fmol). DNA was enzymatically digested to deoxynucleosides and filtered by Centricon YM-10 size exclusion cartridges. Following SPE purification, 1,N6-HMHP-dA adducts were analyzed by column switching HPLC-ESI-MS/MS as described above. A good correlation between 1,N6-HMHP-dA spiked and measured was observed with an R2 value of 0.997 (Figure S-3). Three replicates spiked with 0.5 fmol 1,N6-HMHP-dA were also processed as above and analyzed on three separate days to determine the accuracy and the precision of the method (Table 1).
Table 1.
Accuracy and precision of column switching HPLC-ESI-MS/MS analysis of 1,N6-HMHP-dA (0.5 fmol) spiked into control human DNA (0.1 mg).
| 1,N6-HMHP-dA (fmol) | ||
|---|---|---|
|
Day 1
|
mean | 0.4708 |
| RSD (%) | 14.3 | |
| accuracy (%) | 94.2 | |
| n |
3 |
|
|
Day 2
|
mean | 0.3716 |
| RSD (%) | 11.8 | |
| accuracy (%) | 74.3 | |
| n |
3 |
|
|
Day 3
|
mean | 0.4621 |
| RSD (%) | 9.45 | |
| accuracy (%) | 94.2 | |
| n |
3 |
|
| Interday | mean | 0.4348 |
| RSD (%) | 15.2 | |
| accuracy (%) | 87.0 | |
| n | 9 |
Results and Discussion
Column switching HPLC-ESI+-MS/MS method development
While it is known that DEB is the most genotoxic metabolite of BD (5), the specific DNA adducts responsible for its mutagenicity remain to be identified. In particular, large numbers of A → T transversions are observed (6;13;14), suggesting that DEB forms strongly mispairing deoxyadenosine adducts. We have recently identified regioisomeric exocyclic DEB-dA adducts, 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2'-deoxyadenosine (1,N6-γ-HMHP-dA) and 1,N6-(1-hydroxymethyl-2-hydroxypropan-1,3-diyl)-2'-deoxyadenosine (1,N6-α-HMHP-dA) (12). Both lesions were observed in vitro in DEB-treated DNA and in liver DNA of 1,3-butadiene treated mice (12). While our previous method separated 1,N6-HMHP-dA stereoisomers, it was not sensitive enough to detect the adducts at BD exposures lower than 625 ppm (12). Therefore, the objective of the present work was to develop more sensitive quantitative HPLC-ESI-MS/MS methods for the analysis of the novel exocyclic DEB-dA adducts in vivo.
Our initial experiments employed conventional reversed phase HPLC columns. However, since the 1,N6-HMHP-dA adducts have a positive charge at the N1 of adenine under physiological conditions (Scheme 1), they are not well retained on reversed phase HPLC columns, eluting with the solvent front and suffering from signal suppression (results not shown). Although somewhat better retention was obtained on a Synergi Hydro-RP C18 column, this column could not resolve 1,N6-HMHP-dA from dC, which is naturally present in enzymatic digests of DNA, leading to analyte signal suppression. To overcome this problem, we have developed a column switching method that takes advantage of the positive charge present on 1,N6-HMHP-dA under neutral pH conditions (Scheme 1). In our approach, the exocyclic adducts and their internal standards are captured on a strong cation exchange (SCX) trap column, and the bulk of natural nucleosides present in DNA digests are washed away. The analytes are then eluted with 0.05% acetic acid to enter an analytical Synergi Hydro-RP C18 column for capillary HPLC-ESI+-MS/MS analysis.
The mass spectrometer is operated in the selected reaction monitoring (SRM) mode by following a loss of 2'-deoxyribose from protonated molecules of the adduct, [M + H]+ → [M + H – dR]+ (m/z 338.1 → 222.1), and the corresponding transitions of the [15N4]- labeled internal standard (m/z 342.1 → 226.1) (Figure 1). Note that our current methods intentionally do not separate α and γ 1,N6-HMHP-dA, since both regioisomers elute as a single sharp peak at ~23.5 min, maximizing method sensitivity (Figure 1). We determined that the HPLC-ESI+-MS/MS responses for 1,N6-HMHP-dA using isotope dilution with 15N4-internal standards were linear between 0.3 and 5 fmol, with R2 values > 0.99 (Figure S-2). The method detection limit for 1,N6-HMHP-dA was calculated as 0.125 fmol for pure standards, and 0.3 fmol for standards spiked into 0.1 mg of blank human DNA (i.e. 0.1 1,N6-HMHP-dA adducts/108 nts) (S/N > 3).
Method validation
Aliquots of control DNA from human cervical carcinoma (HeLa) cells (0.1 mg) were spiked with 0-5 fmol 1,N6-HMHP-dA and 27.5 fmol of the corresponding 15N4-labelled internal standard, followed by sample processing and column switching HPLC-MS/MS analysis by usual methods. We found that the method's lower limit of quantitation (S/N ratio of 10 or better) was 0.5 fmol 1,N6-HMHP-dA in 0.1 mg DNA (0.15 1,N6-HMHP-dA adducts per 108 nts). A good correlation was observed between the expected and the measured levels of 1,N6-HMHP-dA spiked into human DNA, with an R2 value of 0.997. The method accuracy and precision were determined for three replicates of 0.5 fmol 1,N6-HMHP-dA spiked into 0.1 mg DNA. The samples were analyzed on three separate days, and the accuracy and precision were determined to be 87.0 ± 15.2 % (Table 1).
Analysis of 1,N6-HMHP-dA in mouse liver DNA
The newly validated quantitative HPLC-MS/MS method was employed to analyze the formation of 1,N6-HMHP-dA adducts in liver DNA of B6C3F1 mice that were exposed to 0 - 625 ppm BD by inhalation for 2 weeks. We found that DNA extracted from tissues of mice treated with 625 ppm contained 0.44 ± 0.08 1,N6-HMHP-dA adducts per 108 normal nucleotides. This value is much lower than the concentrations of DEB-induced DNA-DNA cross-links in the same samples (Table 2) (8;9). DNA of control animals did not contain detectable amounts of 1,N6-HMHP-dA lesions (N = 5) (limit of detection, 0.15 1,N6-HMHP-dA per 108 normal nucleotides).
Table 2.
Comparison of bifunctional DEB-DNA adduct amounts in liver DNA of B6C3F1 mice exposed to 625 ppm BD for 2 weeks (N=4 - 5).
| # adducts/108 nts | |
|---|---|
| bis-N7G-BD | 39.5 ± 8.9 |
| N7G-N1A-BD | 2.7 ± 0.7 |
| 1,N6-HMHP-dA | 0.44 ± 0.08 |
Representative extracted ion chromatograms from HPLC-ESI+-MS/MS analysis of 1,N6-HMHP-dA in liver DNA samples from control and BD-exposed mice are shown in Figure 2. Although the concentrations of 1,N6-HMHP-dA in animals exposed to 625 ppm BD are quite low, they are 3-fold higher than the method's limit of quantitation, ensuring accurate measurement. The column switching method is very selective for 1,N6-HMHP-dA, leading to excellent signal to noise ratios for the in vivo samples (Figure 2B).
Figure 2.
Representative column switching HPLC-ESI+-MS/MS extracted ion chromatograms of 1,N6-HMHP-dA in liver DNA from control B6C3F1 mouse (A) and a mouse exposed to 625 ppm BD for 2 weeks (B).
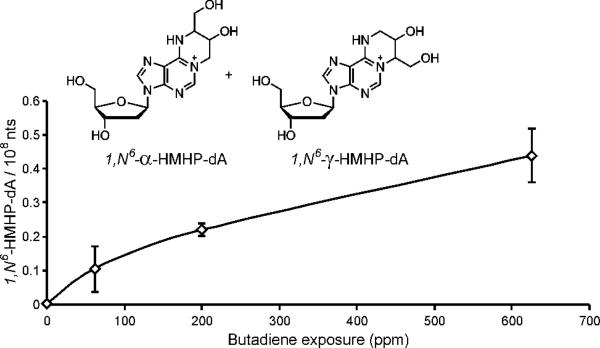
We next quantified 1,N6-HMHP-dA adducts in liver DNA of mice exposed to 62.5 and 200 ppm BD to obtain dose response relationships (Figure 3). We observed a curvilinear dose response curve similar to those previously obtained for other bifunctional DEB-DNA adducts, bis-N7G-BD and N7G-N1A-BD, in the same animals (10), suggesting that the metabolic activation of BD to DEB in the mouse does not reach saturation at the exposures studied here, but that 1,N6 -HMHP-dA adduct formation is more efficient at lower BD concentrations. The 62.5 ppm BD exposure was the lowest examined, however, 1,N6 -HMHP-dA levels at this exposure (0.1 adducts/108 nts) were at the LOD.
Figure 3.
Dose response curve of 1,N6-HMHP-dA in liver DNA isolated from female B6C3F1 mice exposed to 0, 62.5, 200, and 625 ppm BD for 2 weeks (N=5).
Even though 1,N6-HMHP-dA are the least abundant of the three bifunctional DEB-DNA adducts quantified (Table 2), we hypothesize that they are highly mutagenic adducts based on the mutagenicity of other known exocyclic dA lesions (15;16). Exocyclic DNA adducts are highly mutagenic because of their effects on the molecular shape and hydrogen bonding properties of the parent nucleobase (17-20). For example, 1,N6-ethenoadenine and 1,N6-ethanoadenine are highly blocking during DNA polymerization, and induce A → T transversions in human cells (16;21). Interestingly, low levels of 1,N6-ethenoadenine adducts (0.12 – 0.45 εA/107 nucleotides) have been detected in control mice with no carcinogen exposure as a result of inflammation-associated lipid peroxidation (22-24).
Our current column switching isotope dilution HPLC-ESI-MS/MS method is among the most sensitive MS-based methods reported for quantitative analyses of DNA adducts. The limit of quantitation of the new method (0.15 1,N6-HMHP-dA/108 nts) is similar or better than those of other recently published column switching methods for DNA adducts, which report LODs of 0.03 – 7.2 adducts/108 nts (25-31). The accuracy of our column switching HPLC-ESI-MS/MS method (9.4 – 14% RSD at the LOQ) is comparable to previously published column switching methods for DNA adducts (26-29). The excellent sensitivity observed with our new methodology can be attributed to highly selective trapping of the positively charged 1,N6-HMHP-dA adducts on an SCX trap column, facilitating the removal of interferences and improving adduct detection by HPLC-ESI-MS/MS.
Our future efforts will focus on enhancing HPLC-ESI-MS/MS method sensitivity and optimizing HPLC separation to enable separate detection and quantitation of 1,N6-α-HMHP-dA and 1,N6-γ-HMHP-dA adducts in vivo. Recent studies revealed that the two regioisomers can be resolved with a Hypercarb Hypersil HPLC column (Thermo Fisher Scientific Hypersil-Keystone). It is possible that the column switching methodology can be modified by incorporating an analytical Hypercarb Hypersil HPLC column to separate 1,N6-HMHP-dA regioisomers.
Conclusions
In summary, an isotope-dilution column switching LC-MS/MS method has been developed to quantify DEB-induced 1,N6-HMHP-dA adducts in DNA extracted from tissues of BD-exposed mice. This method is highly sensitive, accurate, and selective for 1,N6-HMHP-dA, allowing detection of the low abundance BD lesion in 100 μg DNA at exposures as low as 62.5 ppm BD. The online clean-up method based on column switching saves time and materials, and prevents sample loss. It also provides separation of the exocyclic DEB-dA lesions from the large excess of unmodified nucleosides, which was not possible by conventional sample cleanup methods. This new column switching method can be further employed to investigate 1,N6-HMHP-dA adduct formation in repair deficient animals to gain insight into their repair mechanisms, and to facilitate butadiene risk assessment in humans.
Supplementary Material
Acknowledgements
We thank Bob Carlson (University of Minnesota Cancer Center) for preparing figures for this manuscript, and Brock Matter (University of Minnesota Cancer Center) for helpful suggestions on HPLC and MS method development. This work is supported by grants from the National Cancer Institute (C.A. 100670), the National Institute of Environmental Health Sciences (ES 12689), the Health Effects Institute (Agreement 05-12), and the American Chemistry Council (OLF-163.0).
Footnotes
List of Abbreviations: BD, 1,3-butadiene; bis-N7G-BD, 1,4-bis-(guan-7-yl)-2,3-butanediol; DEB, 1,2,3,4-diepoxybutane; HPLC-ESI+-MS/MS, liquid chromatography-electrospray ionization tandem mass spectrometry; N7G-N1A-BD, 1-(guan-7-yl)-4-(aden-1-yl)-2,3-butanediol; 1,N6-γ-HMHP-dA, 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-dA; 1,N6-α-HMHP-dA, 1,N6-(1-hydroxymethyl-2-hydroxypropan-1,3-diyl)-2'-deoxyadenosine; N1-HEB-dA, N1-(2-hydroxy-3,4-epoxybut-1-yl)-dA; SRM, selected reaction monitoring
References
- 1.White WC. Butadiene production process overview. Chem. Biol. Interact. 2007;166:10–14. doi: 10.1016/j.cbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Melnick RL, Huff JE. 1,3-Butadiene induces cancer in experimental animals at all concentrations from 6.25 to 8000 parts per million. IARC Sci. Publ. 1993:309–322. [PubMed] [Google Scholar]
- 3.Melnick RL, Kohn MC. Mechanistic data indicate that 1,3-butadiene is a human carcinogen. Carcinogenesis. 1995;16:157–163. doi: 10.1093/carcin/16.2.157. [DOI] [PubMed] [Google Scholar]
- 4.Hecht SS. Tobacco smoke carcinogens and lung cancer. J. Natl. Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 5.Cochrane JE, Skopek TR. Mutagenicity of butadiene and its epoxide metabolites: I. Mutagenic potential of 1,2-epoxybutene, 1,2,3,4-diepoxybutane and 3,4-epoxy-1,2- butanediol in cultured human lymphoblasts. Carcinogenesis. 1994;15:713–717. doi: 10.1093/carcin/15.4.713. [DOI] [PubMed] [Google Scholar]
- 6.Steen AM, Meyer KG, Recio L. Analysis of hprt mutations occurring in human TK6 lymphoblastoid cells following exposure to 1,2,3,4-diepoxybutane. Mutagenesis. 1997;12:61–67. doi: 10.1093/mutage/12.2.61. [DOI] [PubMed] [Google Scholar]
- 7.Meng Q, Henderson RF, Walker DM, Bauer MJ, Reilly AA, Walker VE. Mutagenicity of the racemic mixtures of butadiene monoepoxide and butadiene diepoxide at the Hprt locus of T-lymphocytes following inhalation exposures of female mice and rats. Mutat. Res. 1999;429:127–140. doi: 10.1016/s0027-5107(99)00105-0. [DOI] [PubMed] [Google Scholar]
- 8.Goggin M, Loeber R, Park S, Walker V, Wickliffe J, Tretyakova N. HPLC-ESI+-MS/MS analysis of N7-guanine-N7-guanine DNA cross-links in tissues of mice exposed to 1,3-butadiene. Chem. Res. Toxicol. 2007;20:839–847. doi: 10.1021/tx700020q. [DOI] [PubMed] [Google Scholar]
- 9.Goggin M, Anderson C, Park S, Swenberg J, Walker V, Tretyakova N. Quantitative high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry analysis of the adenine-guanine cross-links of 1,2,3,4-diepoxybutane in tissues of butadiene-exposed B6C3F1 mice. Chem. Res. Toxicol. 2008;21:1163–1170. doi: 10.1021/tx800051y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goggin M, Swenberg JA, Walker VE, Tretyakova N. Molecular dosimetry of 1,2,3,4-diepoxybutane-induced DNA-DNA cross-links in B6C3F1 mice and F344 rats exposed to 1,3-butadiene by inhalation. Cancer Res. 2009;69:2479–2486. doi: 10.1158/0008-5472.CAN-08-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Recio L, Steen AM, Pluta LJ, Meyer KG, Saranko CJ. Mutational spectrum of 1,3-butadiene and metabolites 1,2-epoxybutene and 1,2,3,4-diepoxybutane to assess mutagenic mechanisms. Chem. Biol. Interact. 2001;135-136:325–341. doi: 10.1016/s0009-2797(01)00220-4. [DOI] [PubMed] [Google Scholar]
- 12.Seneviratne U, Antsypovich S, Goggin M, Dorr DQ, Guza R, Moser A, Thompson C, York DM, Tretyakova N. Exocyclic deoxyadenosine adducts of 1,2,3,4-diepoxybutane: synthesis, structural elucidation, and mechanistic studies. Chem. Res. Toxicol. 2010;23:118–133. doi: 10.1021/tx900312e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cochrane JE, Skopek TR. Mutagenicity of butadiene and its epoxide metabolites: II. Mutational spectra of butadiene, 1,2-epoxybutene and diepoxybutane at the hprt locus in splenic T cells from exposed B6C3F1 mice. Carcinogenesis. 1994;15:719–723. doi: 10.1093/carcin/15.4.719. [DOI] [PubMed] [Google Scholar]
- 14.Meng Q, Redetzke DL, Hackfeld LC, Hodge RP, W. D. M, Walker VE. Mutagenicity of stereochemical configurations of 1,2-epoxybutene and 1,2:3,4-diepoxybutane in human lymphblastoid cells. Chem. Biol. Interact. 2007;166:207–218. doi: 10.1016/j.cbi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Hang B. Repair of exocyclic DNA adducts: rings of complexity. Bioessays. 2004;26:1195–1208. doi: 10.1002/bies.20130. [DOI] [PubMed] [Google Scholar]
- 16.Levine RL, Yang IY, Hossain M, Pandya GA, Grollman AP, Moriya M. Mutagenesis induced by a single 1,N6-ethenodeoxyadenosine adduct in human cells. Cancer Res. 2000;60:4098–4104. [PubMed] [Google Scholar]
- 17.Basu AK, McNulty JM, McGregor WG. Solution conformation and mutagenic specificity of 1,N6 -ethenoadenine. IARC Sci. Publ. 1999:325–333. [PubMed] [Google Scholar]
- 18.Marnett LJ, Burcham PC. Endogenous DNA adducts: potential and paradox. Chem. Res. Toxicol. 1993;6:771–785. doi: 10.1021/tx00036a005. [DOI] [PubMed] [Google Scholar]
- 19.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase iota. Nat. Struct. Mol. Biol. 2006;13:619–625. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- 20.VanderVeen LA, Hashim MF, Nechev LV, Harris TM, Harris CM, Marnett LJ. Evaluation of the mutagenic potential of the principal DNA adduct of acrolein. J. Biol. Chem. 2001;276:9066–9070. doi: 10.1074/jbc.M008900200. [DOI] [PubMed] [Google Scholar]
- 21.Hang B, Chenna A, Guliaev AB, Singer B. Miscoding properties of 1,N6-ethanoadenine, a DNA adduct derived from reaction with the antitumor agent 1,3-bis(2-chloroethyl)-1-nitrosourea. Mutat. Res. 2003;531:191–203. doi: 10.1016/j.mrfmmm.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Beland FA, Benson RW, Mellick PW, Kovatch RM, Roberts DW, Fang JL, Doerge DR. Effect of ethanol on the tumorigenicity of urethane (ethyl carbamate) in B6C3F1 mice. Food Chem. Toxicol. 2005;43:1–19. doi: 10.1016/j.fct.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 23.Churchwell MI, Beland FA, Doerge DR. Quantification of multiple DNA adducts formed through oxidative stress using liquid chromatography and electrospray tandem mass spectrometry. Chem. Res. Toxicol. 2002;15:1295–1301. doi: 10.1021/tx0101595. [DOI] [PubMed] [Google Scholar]
- 24.Pang B, Zhou X, Yu H, Dong M, Taghizadeh K, Wishnok JS, Tannenbaum SR, Dedon PC. Lipid peroxidation dominates the chemistry of DNA adduct formation in a mouse model of inflammation. Carcinogenesis. 2007;28:1807–1813. doi: 10.1093/carcin/bgm037. [DOI] [PubMed] [Google Scholar]
- 25.Chao MR, Wang CJ, Chang LW, Hu CW. Quantitative determination of urinary N7-ethylguanine in smokers and non-smokers using an isotope dilution liquid chromatography/tandem mass spectrometry with on-line analyte enrichment. Carcinogenesis. 2006;27:146–151. doi: 10.1093/carcin/bgi177. [DOI] [PubMed] [Google Scholar]
- 26.Chen YL, Wang CJ, Wu KY. Analysis of N7-(benzo[a]pyrene-6-yl)guanine in urine using two-step solid-phase extraction and isotope dilution with liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2005;19:893–898. doi: 10.1002/rcm.1868. [DOI] [PubMed] [Google Scholar]
- 27.Churchwell MI, Beland FA, Doerge DR. Quantification of O6-methyl and O6-ethyl deoxyguanosine adducts in C57BL/6N/Tk+/- mice using LC/MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2006;844:60–66. doi: 10.1016/j.jchromb.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 28.Gamboa da CG, Marques MM, Beland FA, Freeman JP, Churchwell MI, Doerge DR. Quantification of tamoxifen DNA adducts using on-line sample preparation and HPLC-electrospray ionization tandem mass spectrometry. Chem. Res. Toxicol. 2003;16:357–366. doi: 10.1021/tx020090g. [DOI] [PubMed] [Google Scholar]
- 29.Hillestrom PR, Weimann A, Poulsen HE. Quantification of urinary etheno-DNA adducts by column-switching LC/APCI-MS/MS. J. Am. Soc. Mass Spectrom. 2006;17:605–610. doi: 10.1016/j.jasms.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 30.Huang CC, Shih WC, Wu CF, Chen MF, Chen YL, Lin YH, Wu KY. Rapid and sensitive on-line liquid chromatographic/tandem mass spectrometric determination of an ethylene oxide-DNA adduct, N7-(2-hydroxyethyl)guanine, in urine of nonsmokers. Rapid Commun. Mass Spectrom. 2008;22:706–710. doi: 10.1002/rcm.3414. [DOI] [PubMed] [Google Scholar]
- 31.Loureiro AP, Marques SA, Garcia CC, Di MP, Medeiros MH. Development of an on-line liquid chromatography-electrospray tandem mass spectrometry assay to quantitatively determine 1,N2-etheno-2'-deoxyguanosine in DNA. Chem. Res. Toxicol. 2002;15:1302–1308. doi: 10.1021/tx025554p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.