A natural substrate for UGT1 is confirmed, revealing how the enzyme functions in the calnexin chaperone system as a quality control step in protein folding.
Abstract
An endoplasmic reticulum (ER) quality control system assists in efficient folding and disposal of misfolded proteins. N-linked glycans are critical in these events because their composition dictates interactions with molecular chaperones. UDP-glucose:glycoprotein glucosyltransferase 1 (UGT1) is a key quality control factor of the ER. It adds glucoses to N-linked glycans of nonglucosylated substrates that fail a quality control test, supporting additional rounds of chaperone binding and ER retention. How UGT1 functions in its native environment is poorly understood. The role of UGT1 in the maturation of glycoproteins at basal expression levels was analyzed. Prosaposin was identified as a prominent endogenous UGT1 substrate. A dramatic decrease in the secretion of prosaposin was observed in ugt1−/− cells with prosaposin localized to large juxtanuclear aggresome-like inclusions, which is indicative of its misfolding and the essential role that UGT1 plays in its proper maturation. A model is proposed that explains how UGT1 may aid in the folding of sequential domain–containing proteins such as prosaposin.
Introduction
Protein maturation in the mammalian secretory pathway largely occurs in the ER, where maturation intermediates and products are subjected to an evaluation or quality control process (Ellgaard and Helenius, 2003). The majority of the proteins that traverse the eukaryotic secretory pathway receive multiple N-linked glycans (Apweiler et al., 1999). These covalent modifications can act as maturation and quality control tags with their composition providing information about the fitness of the modified protein (Helenius and Aebi, 2004; Hebert and Molinari, 2007). The UDP-glucose:glycoprotein glucosyltransferase 1 (UGT1) is a key ER quality control folding sensor that modifies the composition of glycans based on the structural integrity of the modified protein (for review see Caramelo and Parodi, 2008). UGT1 reglucosylation is proposed to support folding assistance and ER retention of maturing proteins by directing their persistent binding by ER-resident carbohydrate-binding molecular chaperones (Van Leeuwen and Kearse, 1997; Wada et al., 1997; Molinari et al., 2005; Pearse et al., 2008).
The ER lectin chaperone system is comprised of the type I membrane protein calnexin and its soluble paralogue calreticulin. These two chaperones bind proteins possessing monoglucosylated glycans (Glc1Man9GlcNAc2; Hammond et al., 1994). The initial round of binding to the chaperones is triggered by the rapid, sequential, and frequently co-translational trimming of two glucoses from the glycans by glucosidase I followed by glucosidase II to generate the monoglucosylated substrate (Chen et al., 1995; Hebert et al., 1997). Chaperone binding also supports the recruitment of the chaperone-associated oxidoreductase ERp57 that can assist in disulfide bond formation or rearrangement (Oliver et al., 1997; Zapun et al., 1998; Soldà et al., 2006). Glucosidase II cleavage of the terminal glucose abrogates lectin chaperone association by generating unglucosylated proteins (Hebert et al., 1995). If the protein folds and assembles properly, it is free to be transported out of the ER to the Golgi. In contrast, proteins possessing nonnative structures are recognized by UGT1, reglucosylated, and targeted for chaperone rebinding and ER retention.
UGT1 is a large soluble ER protein (170 kD) that consists of an N-terminal folding sensor domain responsible for the selection of malformed substrates and a C-terminal catalytic domain, which transfers glucose residues (Arnold and Kaufman, 2003; Guerin and Parodi, 2003). In vitro studies have revealed that UGT1 recognizes near-native molten globule substrates via surface-exposed hydrophobic patches (Sousa and Parodi, 1995; Caramelo et al., 2003, 2004). Deletion of ugt1 is lethal for mice; however, ugt1−/− mouse embryonic fibroblast (MEF) cells have been generated (Molinari et al., 2005). Overexpression of the temperature-sensitive misfolded VSVG (vesicular stomatitis virus G protein) in ugt1−/− cells resulted in its accumulation in covalent-linked aggregates. A study involving the heterologous expression of influenza hemagglutinin demonstrated that the magnitude of substrate misfolding dictates the degree of reglucosylation (Pearse et al., 2008). Understanding the cellular role of UGT1 has been hindered by the inability to follow its activity in live cells and the dependence on overexpressed substrates, and in vitro studies have relied largely on the analysis of nonnatural or engineered substrates (Sousa et al., 1992; Caramelo et al., 2003, 2004; Taylor et al., 2003, 2004; Keith et al., 2005). The biological limitation of these studies underscores the importance of understanding the physiological role of UGT1 in the maturation and quality control of endogenous cellular proteins in their natural environment.
In this study, the role of UGT1 in endogenous protein maturation and quality control was investigated. Using a cellular assay to isolate endogenous substrates of the enzyme, prosaposin was identified as a prominent natural substrate for UGT1 reglucosylation. In the absence of UGT1, there was a significant decrease in monoglucosylated prosaposin and prosaposin secretion. Prosaposin formed nonnative conformations and accumulated in cytoplasmic perinuclear aggresome-like inclusions in ugt1−/− cells. Together, these results provide evidence for an integral involvement of UGT1 in glycoprotein maturation, as without this critical folding sensor, dramatic protein folding and trafficking defects occurred.
Results
Prosaposin is an endogenous UGT1 substrate
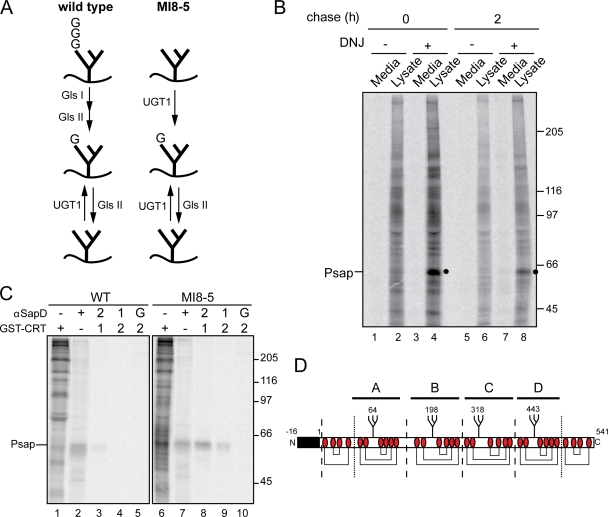
To understand the role of UGT1 in the maturation and quality control of glycoproteins within the cell, the identification of endogenous cellular substrates of UGT1 was explored. The oligosaccharyl transferase of MI8-5 CHO cells transfers truncated unglucosylated N-linked glycans (Man9GlcNAc2) as the result of a deficiency in the dolichol-P glucose–dependent glycosyltransferase (Alg6; Quellhorst et al., 1999). This permits the isolation of glycoproteins reglucosylated by UGT1 because in these cells, reglucosylation is the exclusive mechanism by which proteins can reach a monoglucosylated state (Fig. 1 A; Pearse et al., 2008). As CHO cells lack an endomannosidase activity (Karaivanova et al., 1998), glucosidase inhibition with N-butyl deoxynojirimycin (DNJ) traps reglucosylated glycans in their monoglucosylated state in MI8-5 CHO cells. Monoglucosylated proteins can then be isolated based on their affinity for GST-calreticulin (Pearse et al., 2008), a soluble carbohydrate-binding chaperone which binds monoglucosylated high mannose glycans (Peterson et al., 1995; Kapoor et al., 2003).
Figure 1.
Isolation and identification of an endogenous substrate of UGT1. (A) Schematic of the glycoforms generated in wild-type and MI8-5 cells. Wild-type cells transfer triglucosylated N-glycans to nascent secretory proteins, which are trimmed by glucosidases I/II (Gls I/II) to generate the chaperone-binding monoglucosylated state. This last glucose is trimmed by a second digestion by glucosidase II, resulting in an unglucosylated N-glycan. UGT1 transfers a single glucose back to N-glycans to reinitiate chaperone interactions. In MI8-5 cells, unglucosylated N-glycans are transferred and monoglucosylated form is generated exclusively through UGT1 activity. (B) MI8-5 CHO cells were pulse labeled for 2 h and chased for the indicated times. 0.5 mM DNJ was included in the pulse and chase media where indicated. Radiolabeled monoglucosylated proteins were isolated by GST-calreticulin pull-down from either cell lysates or media and subjected to 7.5% SDS-PAGE. The ∼60-kD substrate was identified as prosaposin (Psap) and is marked with a closed circle. (C) Wild-type (WT) and MI8-5 cells were pulse labeled for 1 h with DNJ. Lysates were immunoprecipitated with saposin D antisera (αSapD; lane 2, 4, 7, and 9) or pulled down with GST-calreticulin (CRT; lanes 1, 3, 5, 6, 8, and 10). G indicates the initial binding to protein G to rule out nonspecific binding. Lanes 3–5 and 8–10 were sequentially precipitated as indicated. (B and C) Molecular mass is indicated in kilodaltons. (D) Schematic of mouse prosaposin (available from Protein Knowledgebase [UniProtKB] under accession number Q61207). The signal sequence is depicted as a black box. N-glycans are marked by branched structures with the modified Asn residue identified. Cys residues are labeled as red ovals, and disulfide bonds are drawn as connecting lines. Dashed lines represent the position of cathepsin D cleavage. Dotted lines represent the position of cleavage by unidentified lysosomal proteases.
To isolate endogenous substrates of UGT1, MI8-5 cells were pulse labeled in the presence of [35S]Met/Cys for 2 h, and total monoglucosylated glycoproteins were isolated from the cell lysates and media using GST-calreticulin. Several distinct bands were visible in the cell lysate (Fig. 1 B, lane 2), whereas the cell media was devoid of monoglucosylated proteins (Fig. 1 B, odd numbered lanes). The addition of DNJ prevented the removal of the UGT1-transferred glucose by glucosidase II, resulting in an accumulation of monoglucosylated substrates (Fig. 1 B, lane 4). After a 2-h chase period, several potential substrates were cleared from the cell lysates, whereas others continued to be recognized by GST-calreticulin in the presence of DNJ. An ∼60-kD protein was readily distinguishable by its abundance, and it remained bound to GST-calreticulin after a 2-h chase, which is suggestive of it being a prominent UGT1 substrate (Fig. 1 B, lane 8; and Fig. S1 A). Treatment with jack bean α-exomannosidase confirmed the presence of monoglucosylated glycans on the 60-kD protein because only a slight shift was observed upon jack bean α-exomannosidase digestion, which is indicative of the protection of A-branch mannose residues by glucosylation (Fig. S1 B; Hammond et al., 1994; Hebert et al., 1995).
Numerous biochemical characterizations were performed to gain insight to the identity of the 60-kD protein. First, alkaline fractionation revealed that it was a soluble protein (Fig. S3 A), likely bearing three to four N-linked glycans as predicted by the size of the mobility shift observed after endoglycosidase digestion (Fig. S1 B). In addition, two-dimensional isoelectric focusing gel electrophoresis yielded a pI of ∼5 for the putative UGT1 substrate (Fig. S2, A and B). The 60-kD protein associated with both calnexin and calreticulin, as determined by coimmunoprecipitation experiments using antibodies directed against the two lectin chaperones (Fig. S3, A and B).
After a bioinformatics search using these various experimentally determined parameters, potential matches were screened with specific antisera during pulse-chase analysis. The 60-kD protein immunoprecipitated with antisera recognizing prosaposin. Prosaposin migrated at the same position as the 60-kD protein (Fig. 1 C, compare lane 6 with lane 7). Sequential pull-down of monoglucosylated substrates with GST-calreticulin followed by saposin D antisera revealed prosaposin as the 60-kD UGT1 substrate. This was confirmed by a reciprocal sequential pull-down of prosaposin followed by GST-calreticulin, demonstrating that prosaposin contained monoglucosylated glycans in MI8-5 cells (Fig. 1 C, lanes 8 and 9).
Prosaposin is a soluble secretory protein that contains an N-terminal signal sequence. It is comprised of four sequential homologous domains, each containing three disulfide bonds and a single N-linked glycan (Fig. 1 D; Kishimoto et al., 1992). After passage through the ER, prosaposin has dual fates: it can traffic to lysosomes where it is processed by cathepsin D into four separate proteins, or it can be secreted as a full-length protein lacking its signal sequence (Lefrancois et al., 2003; O’Brien et al., 1994).
Prosaposin reglucosylation supports persistent lectin chaperone binding
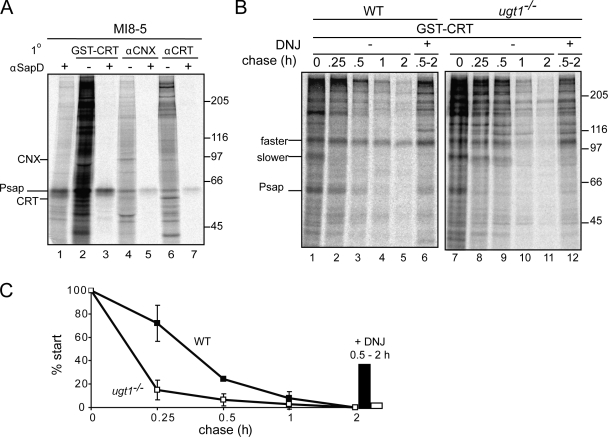
Reglucosylation by UGT1 drives the lectin chaperone binding cycle or chaperone rebinding (Hebert et al., 1995; for reviews see Helenius, 1994; Caramelo and Parodi, 2008). This is believed to increase the fidelity of the glycoprotein folding process and support the ER retention of immature or aberrant proteins (Rajagopalan et al., 1994; Hebert et al., 1996; Vassilakos et al., 1996). The topology of a substrate and the location of its glycans typically dictate whether it interacts with the membrane-bound calnexin, the soluble calreticulin, or both chaperones (Wada et al., 1995; Hebert et al., 1997; Danilczyk et al., 2000). To determine whether reglucosylated prosaposin was redirected for calnexin and/or calreticulin binding, radiolabeled substrates of the lectin chaperones were isolated with calnexin and calreticulin antisera followed sequentially by a pull-down with prosaposin antisera. Reglucosylated prosaposin was bound by both calnexin and calreticulin in MI8-5 cells (Fig. 2 A, lanes 5 and 7).
Figure 2.
Reglucosylation supports persistent lectin chaperone association. (A) MI8-5 cells were pulse labeled for 1 h in the presence of 0.5 mM DNJ. Lysates were subjected to GST-calreticulin (CRT) pull-down (lanes 2 and 3) or immunoprecipitated with calnexin (CNX; lanes 4 and 5) or calreticulin (lanes 6 and 7) antisera. Some samples (+) were sequentially precipitated with saposin D antisera (αSapD). (B) Wild-type (WT) and ugt1−/− cells were pulse labeled for 15 min and chased for the indicated times. Monoglucosylated prosaposin (Psap) was isolated by GST-calreticulin pull-down. DNJ was added at the 0.5-h time point until the completion of the assay for the indicated lanes. Samples were resolved via 7.5% SDS-PAGE. (A and B) Molecular mass is indicated in kilodaltons. (C) Quantifications of B using n = 3 and mean ± SD (error bars).
Prosaposin is reglucosylated by UGT1 and targeted to the lectin chaperones. To determine the extent to which prosaposin is reglucosylated by UGT1, ugt1−/− MEF cells were used (Molinari et al., 2005; Soldà et al., 2007). Wild-type and ugt1−/− MEF cells were pulse labeled and chased for the designated times (Fig. 2 B). Total monoglucosylated glycoproteins were isolated at each time point with GST-calreticulin. In wild-type cells, 50% of monoglucosylated prosaposin remained after ∼22 min. However, in ugt1−/− cells, there was a significant reduction in monoglucosylated prosaposin, exhibiting a monoglucosylated half-time of 8 min. A portion of reglucosylated prosaposin could be observed in wild-type cells when DNJ was added at the 30-min chase time point (39%). The DNJ-trapped prosaposin fraction was largely absent from ugt1−/− (7%; Fig. 2, B and C). These results indicate that UGT1 plays a significant role in regenerating monoglucosylated endogenous prosaposin. In addition to prosaposin, several other potential endogenous substrates displayed significant alterations in their monoglucosylated state in ugt1−/− cells. An unknown protein of ∼100 kD exhibited a dramatic decrease in GST-calreticulin binding from ugt1−/− cells, a similar phenotype as observed for prosaposin, which is indicative of reglucosylation. Interestingly, an additional protein of ∼80 kD displayed an increase in GST-calreticulin binding in the absence of UGT1 (Fig. 2 B). This phenotype has been observed previously in an analysis of endogenous calnexin substrates (Soldà et al., 2007). However, the mechanism by which glucose trimming is slowed is not understood. In summary, prosaposin was efficiently reglucosylated by UGT1 during its maturation.
UGT1 is critical for the proper maturation of prosaposin
Prosaposin is reglucosylated by UGT1, and this reglucosylation occurs readily during the maturation of prosaposin. A portion of prosaposin is secreted from cells as full-length protein (Griswold et al., 1986; Collard et al., 1988). To investigate the importance of UGT1 in prosaposin secretion, wild-type and ugt1−/− cells were pulse labeled with [35S]Met/Cys, and prosaposin was isolated from cell lysates and media. Secreted prosaposin was detectable after 0.5 h of chase and reached 72% secretion after 1 h of chase (Fig. 3, A and B). In ugt1−/− cells, secretion was greatly reduced, reaching a maximum of only 24% after 1 h of chase (Fig. 3 A, compare lane 6 with lane 12). Only a minimal level of prosaposin was processed to saposins in either cell type after a 1-h chase (Fig. 3 A and Fig. S5 A, low molecular bands in cell lysates labeled SAPs).
Figure 3.
UGT1 is critical for prosaposin secretion. (A) Wild-type (WT) and ugt1−/− cells were pulse labeled for 15 min and chased for the indicated time periods. Radiolabeled prosaposin (Psap) was immunoprecipitated with saposin C antisera from the lysate (lys) and media fractions. SAPs, saposins. (B) Quantifications of A using n = 4 and mean ± SD (error bars). (C) ugt1−/− cells were transfected with either empty vector or pUGT1-C (wild type or catalytic mutant [D1360A]) and radiolabeled for 15 min with or without a 1-h chase period. Radiolabeled prosaposin was immunoprecipitated from lysate and media fractions with saposin C antisera. Transfection of pUGT1-C was confirmed by immunoblotting in E. GAPDH (glyceraldehyde 3-phosphate dehydrogenase) served as a loading control. (A, C, and E) Molecular mass is indicated in kilodaltons. (D) Quantifications of C with n = 3 and mean ± SD (error bars).
To ensure that the decreased secretion of prosaposin observed in ugt1−/− cells was caused by the absence of UGT1, human UGT1 tagged at its C terminus with a protein C epitope (pUGT1-C) was transfected into ugt1−/− cells (Fig. 3 E). Cells transfected with pUGT1-C or empty vector were subjected to pulse-chase analysis. As observed previously (Fig. 3, A and B), only ∼20% of total prosaposin was secreted from ugt1−/− cells (Fig. 3, C and D). However, expression of pUGT1-C restored prosaposin secretion to a level similar to that observed in wild-type cells (Fig. 3, C and D).
To confirm that the deficient secretion of endogenous prosaposin in ugt1−/− cells was the result of the lack of UGT1 reglucosylation activity, a catalytic mutant of UGT1 (D1360A) was expressed, and the trafficking of prosaposin was analyzed. The mutation of the DxD motif in the C-terminal domain of UGT1 has been shown to prevent the catalytic transfer of glucose in vitro (Tessier et al., 2000). Protein C epitope–tagged UGT1D1360A was transfected into ugt1−/− cells. Expression levels were similar to those observed for wild-type UGT1 (Fig. 3 E). Whereas overexpression of pUGT1-C was able to rescue the secretion defect of endogenous prosaposin, the expression of the corresponding catalytic mutant was unable to increase the levels of prosaposin found in the media (pUGT1D1360A-C; Fig. 3, C and D). Together, these results implicate UGT1 in playing a critical role in prosaposin maturation, as the absence of UGT1 resulted in a significant decrease in the secretion of prosaposin, and its secretion can be efficiently rescued by overexpression of the folding sensor. This defect arises as the result of the absence of the catalytic reglucosylation activity of UGT1 because expression of a catalytic mutant of UGT1 was incapable of improving secretion levels of endogenous prosaposin in ugt1−/− cells.
Prosaposin is located in juxtanuclear aggresome-like inclusions in ugt1−/− cells
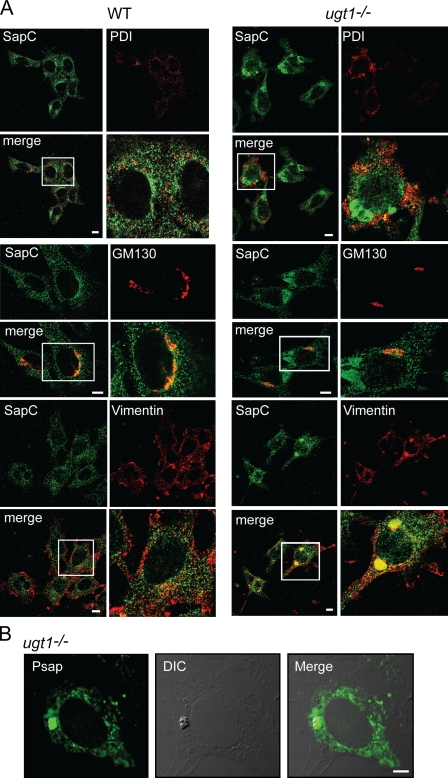
Because there was a striking decrease in the level of secreted prosaposin in ugt1−/− cells, the subcellular localization of prosaposin was investigated by immunofluorescence microscopy using a saposin antibody. In wild-type cells, prosaposin was found in punctate structures dispersed throughout the cell, with a portion of prosaposin colocalizing with the ER marker protein disulfide isomerase (PDI; Fig. 4 A). Only minimal levels of saposin were localized to lysosomes in wild-type or ugt1−/− cells (Fig. S4 A). Surprisingly, in approximately one fifth of the ugt1−/− cells, prosaposin was located in an additional region that corresponded to large dense juxtanuclear structures. These structures did not colocalize with the ER markers PDI or calnexin (Fig. 4 A and Fig. S4 B).
Figure 4.
Prosaposin is localized to aggresome-like inclusions in ugt1−/− cells. Prosaposin localization in wild-type (WT) and ugt1−/− cells was determined by confocal immunofluorescence microscopy. Cells were fixed in 4% paraformaldehyde/PBS and permeabilized in 0.1% Triton X-100. (A) Samples were double labeled with saposin C (SapC) antisera and PDI (ER), GM130 (Golgi), or vimentin (aggresome) antisera. The white boxed regions denote the area magnified in the panels to the right. (B) Samples labeled with full-length prosaposin (Psap) antisera were compared with DIC images. Bars, 10 µm.
Because the trafficking of properly folded prosaposin involves passage through the Golgi, colocalization of prosaposin with the Golgi marker GM130 was analyzed. In wild-type cells, a portion of prosaposin was found localized with GM130. The large intracellular prosaposin structures observed in ugt1−/− cells did not colocalize with GM130 (Fig. 4 A). In addition, neither the ER nor the Golgi appeared to be morphologically disturbed in the ugt1−/− cells.
There are several studies that have revealed intracellular inclusions to sequester malformed proteins from the cellular milieu in mammalian cells. One such inclusion is the aggresome, a perinuclear inclusion generated by misfolded proteins delivered by microtubule motor proteins and localized to the microtubule-organizing center (Johnston et al., 1998, 2002). Unlike Russell bodies (Valetti et al., 1991) or the ER-derived quality control compartment (Kamhi-Nesher et al., 2001), aggresomes are not surrounded by a membrane coat. Instead, aggresomes are encapsulated by the reorganized intermediate filament protein vimentin (Johnston et al., 1998). Because the prosaposin inclusions observed in ugt1−/− cells were juxtanuclear and did not colocalize with PDI, calnexin, or GM130 (Fig. 4 A and Fig. S4 B), their colocalization with vimentin was investigated (Fig. 4 A). In wild-type cells, colocalization with prosaposin and vimentin was not observed as vimentin was mainly localized to the cell periphery. However, in ugt1−/− cells, vimentin localized just outside the nucleus in large punctate structures, which colocalized with the prosaposin inclusions.
To further examine the aggresome-like inclusions visualized by immunofluorescence microscopy, ugt1−/− cells were examined by differential interference contrast (DIC) microscopy and compared with the localization of prosaposin using prosaposin antisera. A large juxtanuclear punctate structure was found in ugt1−/− cells, which colocalized with endogenous prosaposin (Fig. 4 B). Altogether, endogenous prosaposin at steady-state in ugt1−/− cells existed in dense perinuclear inclusions that were distinct from the ER and Golgi. These inclusions shared the characteristic vimentin colocalization with aggresomes, intracellular sequestrations that function to compartmentalize malformed protein substrates from the active folding/maturation events occurring in the cell.
Prosaposin exhibits nonnative conformations in ugt1−/− cells
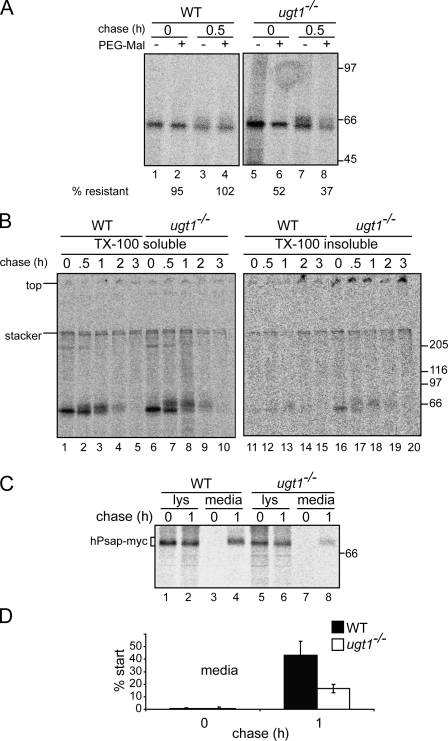
The localization of proteins to intracellular inclusions is usually the result of a folding defect. Because prosaposin failed to be secreted in significant quantities and was found in aggresome-like inclusions in ugt1−/− cells, the folding status of endogenous prosaposin was analyzed. To investigate the extent of the nonnative conformations found in prosaposin derived from ugt1−/− cells, the accessibility of free sulfhydryls was probed by polyethylene glycol (PEG)–maleimide modification.
Prosaposin contains several rapidly forming disulfide bonds (Fig. 1 D; Ruiz-Canada et al., 2009). Wild-type and ugt1−/− cells were radiolabeled, lysed, and subjected to PEG-maleimide treatment. After quenching with DTT, radiolabeled prosaposin was immunoprecipitated. In wild-type cells, prosaposin was nearly completely resistant to PEG-maleimide modification with or without a chase period, indicating that there were no exposed free sulfhydryls on Cys residues (Fig. 5 A, lanes 2 and 4). Interestingly, prosaposin was highly sensitive to PEG-maleimide addition in ugt1−/− cells (52% resistant), and this sensitivity increased with chase time, with only 37% resistant to modification after 0.5 h of chase (Fig. 5 A, lanes 6 and 8).
Figure 5.
Prosaposin displays nonnative characteristics. (A) Wild-type (WT) and ugt1−/− cells were pulse labeled for 15 min, chased for 30 min where indicated, lysed, and treated with 5 mM PEG-maleimide (PEG-Mal) for 30 min on ice. PEG-maleimide was quenched in DTT, and prosaposin was immunoprecipitated with saposin C antisera. Samples were resolved via 7.5% SDS-PAGE. (B) Wild-type and ugt1−/− cells were pulse labeled for 15 min and chased for the indicated times. After cell lysis, the Triton X-100 (TX-100)–insoluble pellet was solubilized in 1% SDS and quenched with excess Triton X-100. Samples were then subjected to immunoprecipitation with saposin C antisera. Samples were resolved via 7.5% nonreducing SDS-PAGE. (C) Myc/His-tagged human prosaposin (hPsap) was transiently overexpressed in wild-type and ugt1−/− cells, pulse labeled for 15 min, and chased for the indicated times. Human prosaposin was isolated from cell lysates (lys) and media with antisera recognizing the myc epitope and analyzed via 7.5% SDS-PAGE. (A–C) Molecular mass is indicated in kilodaltons. (D) Quantifications of C using n = 3 and mean ± SD (error bars).
To further evaluate the status of prosaposin in ugt1−/− cells, the detergent solubility of prosaposin was analyzed. In a previous study, proteins localized to aggresomes have been found to display detergent insolubility (Johnston et al., 1998). Wild-type and ugt1−/− cells were pulsed with [35S]Met/Cys for 15 min, and endogenous prosaposin was isolated from Triton X-100–soluble and –insoluble fractions under nonreducing conditions after various times of chase. The detergent solubility levels of prosaposin were similar in wild-type and ugt1−/− cells under nonreducing conditions (Fig. 5 B, lanes 1–10). This is in agreement with the analysis of intracellular prosaposin levels under reducing conditions (Fig. 3, A and B). However, in ugt1−/− cells, there was a visible amount of prosaposin found in the detergent-insoluble fractions, as some of the prosaposin resided in high molecular weight disulfide-associated aggregates that did not enter the resolving gel regardless of the duration of the chase period (Fig. 5 B, lanes 16–20). Therefore, endogenous prosaposin in ugt1−/− cells possessed exposed reactive Cys residues, which resulted in a portion of prosaposin forming large disulfide-linked detergent-insoluble aggregates.
To verify that the prosaposin maturation defect and the resultant deficiency in secretion was not the result of a pleiotropic effect of the ugt1−/− cells, affinity-tagged human prosaposin was transfected in wild-type and ugt1−/− cells. Cells were pulse labeled with [35S]Met/Cys, and the overexpressed human prosaposin was isolated from cell lysates and media using antisera directed against their C-terminal myc tag. Nearly 50% of the overexpressed prosaposin was secreted from wild-type cells after 1 h of chase (Fig. 5, C and D). In contrast, there was a significant decrease in the secretion efficiency of prosaposin in ugt1−/− cells (∼17%; Fig. 5, C and D). In conclusion, the secretion defect that was observed with endogenous prosaposin in ugt1−/− cells (Fig. 3, A and B) was also observed for overexpressed human prosaposin, indicating that the folding defect that arises in the absence of UGT1 is robust and not caused by a defect in the prosaposin gene in ugt1−/− cells.
Discussion
A complete picture of the role of UGT1 in glycoprotein maturation within the mammalian cell has remained elusive since its discovery more than 20 yr ago in pioneering studies by Parodi et al. (1984), Trombetta et al. (1989), and Sousa et al. (1992). This can be attributed in part to technical difficulties in following its activity within the mammalian ER. In this study, we have identified an endogenous substrate of UGT1 reglucosylation using a mutant mammalian cell line that allows the monitoring of the reglucosylation reaction and the isolation of UGT1 substrates. Prosaposin was efficiently reglucosylated by UGT1, and in its absence, prosaposin was unstable. Endogenous prosaposin secretion was drastically reduced without UGT1 as it existed in nonnative conformations sequestered in large intracellular perinuclear aggregates. Together, these results demonstrate the importance of UGT1 in glycoprotein maturation.
Identifying cellular substrates of UGT1 has proven difficult because monoglucosylated glycoproteins that have been generated via glucosidase I/II trimming versus those generated by reglucosylation are indistinguishable. To circumvent this issue, we have used the alg6-deficient mammalian cell line MI8-5, which allows for the isolation of products of UGT1 reglucosylation (Quellhorst et al., 1999; Pearse et al., 2008). Substrates of reglucosylation were enriched by the inhibition of glucosidase II to prevent glucose removal and isolated with purified GST-calreticulin. Furthermore, cellular maturation studies commonly rely on the use of overexpressed proteins (Wada et al., 1997; Johnston et al., 1998; Danilczyk et al., 2000; Kamhi-Nesher et al., 2001; Kawaguchi et al., 2003; Molinari et al., 2005; Soldà et al., 2007; Pearse et al., 2008). The use of overexpressed proteins can be problematic because the ER homeostasis is tightly controlled by the unfolded protein response (Schröder and Kaufman, 2005). The unfolded protein response pathway can be activated by protein overexpression, supporting a change in the balance of the ER-resident proteins, which are frequently the proteins under study (Schröder and Kaufman, 2005; Farhan et al., 2008). The ugt1−/− cells have previously been shown to possess an adaptive stress response (Rutkowski et al., 2006). Furthermore, aggregation is a concentration-dependent reaction, which can be nonphysiologically favored by protein overexpression, and quality control/sorting steps can be saturated, resulting in the aberrant trafficking of overexpressed proteins. Therefore, it is of important significance to study the reglucosylation of endogenous proteins by this critical quality control factor in the intact ER.
A prominent endogenous cellular UGT1 substrate identified was prosaposin. Prosaposin is a soluble glycoprotein comprised of four domains that each contain three overlapping disulfide bonds and a single N-linked glycan (Fig. 1 D; Kishimoto et al., 1992). After passage through the ER, prosaposin has multiple fates. It can be secreted as a full-length protein, whose function appears to be wide ranging, with proposed involvement in neurological development, stress and antiapoptotic signaling, reproductive development, and cancer (O’Brien et al., 1994; Hiraiwa et al., 1997; Morales et al., 2000; Misasi et al., 2001). Alternatively, prosaposin can traffic to lysosomes via a mannose 6-phosphate receptor–independent mechanism involving the Golgi receptor sortilin (Lefrancois et al., 2003). Cathepsin D, a lysosomal protease, cleaves prosaposin into four individual proteins called saposin A–D (Fig. 1 D). In the lysosome, the individual saposins act as cofactors for different lipid hydrolases (Hiraiwa et al., 1997; Gopalakrishnan et al., 2004).
Each of the saposins exists as a stable protein, displaying resistance to elevated temperatures, low pH, and several proteases (Kondoh et al., 1991; Hiraiwa et al., 1993; Vaccaro et al., 1995). In wild-type MEF cells, prosaposin was largely secreted with only a minor fraction being diverted to the lysosomes (Figs. 3 and 4 and Figs. S4 A and S5 A). Prosaposin secretion was significantly reduced in ugt1−/− MEF cells (Fig. 3, A and B). This is likely caused by the persistent conformational instability of prosaposin found in the absence of UGT1, as disulfide bonding was found to be incomplete and aberrant, and prosaposin displayed an increase in detergent insolubility (Fig. 5). A recent study in mice, in which the fifth Cys of saposin C and D was mutated, revealed that the combined C/D mutation resulted in increased ER retention of prosaposin and decreased processing to the individual saposin domains (Sun et al., 2007). Surprisingly, the loss the fifth or fourth Cys of the saposin D and B domains, respectively, supported trafficking to the lysosome and normal processing of prosaposin with the exception that the saposin containing the mutation was unstable and absent from the lysosome (Sun et al., 2007, 2008). The remaining three saposins were present at normal levels. Therefore, it appeared that the folding defect in prosaposin that was evident in ugt1−/− cells was more severe than the loss of a single prosaposin Cys residue. Evidently, only one of the two disulfides that connect the two saposin helices are needed to pass the ER quality control test; however, both disulfides appear to be required to survive the harsh conditions encountered in the lysosome.
Like the decreased secretion of endogenous prosaposin in ugt1−/− cells, the overexpressed human protein also failed to traffic out of cells in significant quantities. This indicated that the folding defect that occurs in ugt1−/− cells was evident even at elevated levels of the substrate protein, illustrating the effectiveness of the ER quality control process and the necessity of the folding sensor of UGT1 for the proper folding of prosaposin.
In the secretory pathway, nonnative proteins are targeted for ER-associated degradation by quality control machinery, resulting in their subsequent degradation by the cytoplasmic proteasome (Hebert and Molinari, 2007). The aberrant prosaposin created in ugt1−/− cells was not efficiently turned over by the proteasome but rather was sequestered in large intracellular juxtanuclear inclusions. These inclusions colocalized with a known aggresomal marker protein, vimentin. Aggresomes have been defined as a cellular response to protein misfolding (Johnston et al., 1998). A driving force for aggresome formation appears to be evasion of proteasomal degradation. Aggresomes are generally observed after cells overexpressing a mutant protein are treated with proteasome inhibitors (Johnston et al., 1998; Kawaguchi et al., 2003). In the present study, aggresome-like inclusions were found to accumulate in ugt1−/− cells in the absence of proteasome inhibition when proteins were expressed at their endogenous levels. The rapid self-association of prosaposin is likely caused by structural defects that develop in the absence of UGT1, which precludes efficient degradation by the proteasome. The large number of hydrophobic residues within prosaposin, which are packed into folded domains and are also involved in lipid binding in the lysosome, may contribute to the inability of the proteasome to degrade the aberrant protein (Rossmann et al., 2008). Because the aggresome-like structures were sufficiently large to be viewed by DIC microscopy, they likely contain additional secretory cargo that is defective in the ugt1−/− cells.
The loss of secreted prosaposin in ugt1−/− cells implicates a role for the calnexin/calreticulin cycle in the normal maturation of prosaposin. Because calnexin and calreticulin binding can slow the folding of a protein in a domain-specific manner, it has been proposed that glycans can help to direct the molecular choreography of the maturation process (Hebert et al., 1997; Daniels et al., 2003). Carbohydrate trimming and reglucosylation determine which glycans will be bound by the lectin chaperones and the timing and duration of the interaction. Glycans reglucosylated by UGT1 support persistent binding to a given domain, which in turn can delay the folding of the reglucosylated domain. We have previously analyzed the reglucosylation of a maturing model glycoprotein, influenza hemagglutinin, in the intact mammalian ER of MI8-5 CHO cells (Pearse et al., 2008). UGT1 preferentially targets slow-folding domains of a glycoprotein to specifically direct chaperone binding to immature regions. In the case of hemagglutinin, this involved the reglucosylation of the N-terminal portion of a nonsequential domain comprised of two distal regions, thereby providing protection to the N-terminal domain until its C-terminal portion was translated.
UGT1 appears to recognize and reglucosylate near-native structures, as demonstrated using purified enzyme (Caramelo et al., 2003, 2004; Taylor et al., 2004; Keith et al., 2005). Studies in Schizosaccharomyces pombe and Trypanosoma cruzi demonstrated that UGT in these organisms recognizes substrates with near-native disulfide bonds and not fully reduced proteins (Fernández et al., 1998; Labriola et al., 1999). That UGT1 does not appear to recognize grossly misfolded substrates is in agreement with our results that incorporation of AZC (l-azetidine-2-carboxylic acid) into glycoprotein in MI8-5 cells does not increase the level of reglucosylation observed (Fig. S5 B).
UGT1 selectivity appears to be driven by exposed hydrophobic residues that are hallmarks of nonnative or immature proteins (Sousa and Parodi, 1995; Caramelo et al., 2003; Taylor et al., 2003). Purified UGT1 was found to preferentially reglucosylate glycopeptides possessing dual hydrophobic patches situated C-terminal to the glycan (Taylor et al., 2003). Analysis of the hydrophobicity of the regions C-terminal to the glycans of prosaposin found that the two C-terminal glycans on saposin C and D possess the signature hydrophobic patches associated with UGT1 recognition (Fig. 6). Protein folding for secretory proteins is initiated co-translationally and co-translocationally, supporting a vectorial folding reaction whereby N-terminal domains can fold first, helping to taper the number of available folding conformations or the width of the associated protein folding funnel (Clark, 2004). As prosaposin is comprised of four sequential homologous folding domains, a single round of calnexin/calreticulin binding to the N-terminal domains directed by glucose trimming of the triglucosylated modification may minimize their time encumbered by chaperone binding and support their rapid folding. It is of interest to note that saposin A in humans contains an additional N-linked glycan that increases the overall hydrophilicity of this domain, which would presumably further disfavor the reglucosylation of its glycans by UGT1 and ensure transient binding to the lectin chaperones (Wang et al., 2008). Persistent chaperone binding to the C-terminal domains as dictated by UGT1 reglucosylation could aid in this vectorial reaction by extending the time permitted for the N-terminal domains to fold before being hindered by exposure to the immature C-terminal domains.
Figure 6.
Hydropathic profiles of mouse prosaposin glycan regions. The hydropathic profiles of the amino acids C-terminal to the N-glycans of prosaposin are depicted. Kyte-Doolittle values were used in calculating the mean hydrophobicity with a window size of 3 aa. The Asn residue for each N-glycan is positioned at 0 on the plots. Note that the C and D glycans of prosaposin possess the oscillating hydrophobicity profiles associated with efficient reglucosylation by UGT1 (Taylor et al., 2003). Positive values indicate hydrophobicity.
The identification of the prosaposin as a prominent substrate of UGT1 implicates UGT1 and the calnexin-binding cycle assisting in the proper folding of sequential domain–containing proteins. The discovery of additional UGT1 substrates and their sites of reglucosylation will provide us with a more comprehensive knowledge of the magnitude of the role of UGT1 in cell homeostasis. UGT1 is not only a critical quality control factor that evaluates the maturation process for the ER retention and proper sorting of secretory cargo, but it also appears to be a critical protein folding factor that determines the timing and duration of chaperone binding to help optimize the fidelity of the folding process for complex multidomain proteins.
Materials and methods
Reagents
Wild-type and MI8-5 CHO cells were a gift from S. Krag (Johns Hopkins University, Baltimore, MD; Quellhorst et al., 1999). Wild-type and ugt1−/− MEFs and the protein C–tagged UGT1 expression plasmid were described previously (Molinari et al., 2005; Soldà et al., 2007). The pGEX-3X GST-calreticulin plasmid was a gift from M. Michalak (University of Alberta, Edmonton, Alberta, Canada). The following antibodies were obtained as indicated: saposin D antisera (K. Sandhoff, Universität Bonn, Bonn, Germany), saposin C antisera (Santa Cruz Biotechnology, Inc.), calreticulin (Thermo Fisher Scientific), PDI (Thermo Fisher Scientific), GM130 (BD), protein C epitope (Roche), and vimentin and myc (Sigma-Aldrich). Additional prosaposin and saposin antibodies used were described previously (Sun et al., 2007). Cell culture materials and Lipofectamine 2000 were purchased from Invitrogen. [35S]Met/Cys and reduced glutathione Sepharose 4B beads were acquired from PerkinElmer and GE Healthcare, respectively. All other reagents were purchased from Sigma-Aldrich.
Transfection, metabolic labeling, and immunoprecipitation
Cells were pulse labeled with [35S]Met/Cys and chased for the indicated times as described previously (Pearse et al., 2008). For the overexpression of pUGT1–protein C, nearly confluent cells were transfected with Lipofectamine 2000 according to manufacturer’s instructions. GST-calreticulin pull-down was performed as previously described (Pearse et al., 2008). In brief, bacterially expressed GST-calreticulin was purified according to a previous study with reduced glutathione Sepharose beads (Baksh and Michalak, 1991). After cell lysis in MNT (20 mM 2-[N-morpholino] ethane sulfate, 30 mM Tris-Cl pH 7.5, and 0.5% Triton X-100) containing 20 µM leupeptin, 1.5 µM aprotinin, 1 µM pepstatin A, 100 µM PMSF, and 50 µM LLnL, postnuclear supernatants were incubated with reduced glutathione Sepharose beads, which were prebound by 8 µg GST-calreticulin. Samples were rotated overnight at 4°C, subsequently centrifuged at 1,000 g for 5 min, and washed twice. Immunoprecipitations were performed as previously described (Pearse et al., 2008). In the case of sequential immunoprecipitation/pull-down, samples were eluted from the initial isolation by incubation in 1% SDS at 100°C for 10 min and quenched in excess buffer containing 2% Triton X-100. Radiolabeled samples were resolved via SDS-PAGE and visualized by phosphorimaging (FLA-500; Fujifilm). Quantifications were determined using Multigauge version 2.02 (Fujifilm).
Confocal immunofluorescence microscopy
Cells were split onto coverslips and were fixed by 4% paraformaldehyde in PBS for 10 min at room temperature and then permeabilized with 0.1% Triton X-100 for 1 min at 2°C in the immunostaining buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 1 mM EGTA, and 2% bovine serum albumin). Cells were incubated with primary antibody in immunostaining buffer and then stained with goat anti–rabbit or mouse IgG conjugated with Alexa Fluor 488 or 594 (Invitrogen). After a rinse with distilled water, coverslips were mounted face down on slide glasses with VectaShield (Vector Laboratories) and sealed with nail polish. Images were taken with a confocal microscope (LM510; Carl Zeiss, Inc.) and processed with Photoshop software (Adobe).
PEG-maleimide and detergent insolubility
The sensitivity of prosaposin to PEG-maleimide was determined as previously described (Ruiz-Canada et al., 2009). In brief, after metabolic labeling, cells were lysed in MNT containing the aforementioned protease inhibitors with or without 5 mM PEG-maleimide 5K (Sigma-Aldrich) and incubated on ice for 30 min. Samples were then diluted in MNT containing 20 mM DTT and immunoprecipitated as described in the section Transfection, metabolic labeling, and immunoprecipitation. For detergent solubility assays, cells were lysed in MNT containing protease inhibitors after metabolic labeling. After complete lysis, samples were centrifuged at 18,000 g for 15 min at 4°C. Detergent-soluble supernatants were immunoprecipitated with prosaposin antisera. Detergent-insoluble pellets were solubilized in 1% SDS in 100 mM Tris, pH 8.0, for 10 min at 100°C. The samples were then diluted in MNT and subjected to immunoprecipitation with prosaposin antisera.
Online supplemental material
Figs. S1 and S2 show the analysis of the prominent UGT1 substrate termed PUGST1 and its identification as prosaposin. Fig. S3 demonstrates that PUGST1 binds to both calnexin and calreticulin in MI8-5 cells treated with DNJ. Fig. S4 displays the intracellular localization of prosaposin in wild-type and ugt1−/− cells. Fig. S5 shows partial prosaposin processing to saposin and that AZC does not increase the reglucosylation of glycoproteins. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912105/DC1.
Acknowledgments
We are grateful to Dr. R. Gilmore (University of Massachusetts, Worcester, MA) and members of his laboratory including D. Kelleher and Dr. C. Ruiz-Canada for their generous help in identifying prosaposin. We would like to acknowledge the following members of the Hebert laboratory for fruitful discussions and critical reading of this manuscript: J. Cormier, K. Moody Giorda, and N. Woodward.
This work was supported by United States Public Health grants CA79864 and GM086874 (to D.N. Hebert), NS36681 (to G.A.Grabowski), and DK42394, HL52173, and PO1 HL057346 (to R.J. Kaufman). R.J. Kaufman is an investigator of the Howard Hughes Medical Institute. B.R. Pearse was partially supported by a National Institutes of Health Chemistry–Biology Interface training grant (T32GM00815) and a predoctoral University fellowship from the University of Massachusetts. This study was also supported by the Uehara Memorial Foundation (grant to T. Tamura). The Central Microscopy Facility at the University of Massachusetts, Amherst, is supported by a grant from the National Science Foundation (NSF BBS 8714235).
Footnotes
Abbreviations used in this paper:
- DIC
- differential interference contrast
- DNJ
- N-butyl deoxynojirimycin
- MEF
- mouse embryonic fibroblast
- PDI
- protein disulfide isomerase
- PEG
- polyethylene glycol
References
- Apweiler R., Hermjakob H., Sharon N. 1999. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta. 1473:4–8 [DOI] [PubMed] [Google Scholar]
- Arnold S.M., Kaufman R.J. 2003. The noncatalytic portion of human UDP-glucose: glycoprotein glucosyltransferase I confers UDP-glucose binding and transferase function to the catalytic domain. J. Biol. Chem. 278:43320–43328 10.1074/jbc.M305800200 [DOI] [PubMed] [Google Scholar]
- Baksh S., Michalak M. 1991. Expression of calreticulin in Escherichia coli and identification of its Ca2+ binding domains. J. Biol. Chem. 266:21458–21465 [PubMed] [Google Scholar]
- Caramelo J.J., Parodi A.J. 2008. Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 283:10221–10225 10.1074/jbc.R700048200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo J.J., Castro O.A., Alonso L.G., De Prat-Gay G., Parodi A.J. 2003. UDP-Glc:glycoprotein glucosyltransferase recognizes structured and solvent accessible hydrophobic patches in molten globule-like folding intermediates. Proc. Natl. Acad. Sci. USA. 100:86–91 10.1073/pnas.262661199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo J.J., Castro O.A., de Prat-Gay G., Parodi A.J. 2004. The endoplasmic reticulum glucosyltransferase recognizes nearly native glycoprotein folding intermediates. J. Biol. Chem. 279:46280–46285 10.1074/jbc.M408404200 [DOI] [PubMed] [Google Scholar]
- Chen W., Helenius J., Braakman I., Helenius A. 1995. Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc. Natl. Acad. Sci. USA. 92:6229–6233 10.1073/pnas.92.14.6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark P.L. 2004. Protein folding in the cell: reshaping the folding funnel. Trends Biochem. Sci. 29:527–534 10.1016/j.tibs.2004.08.008 [DOI] [PubMed] [Google Scholar]
- Collard M.W., Sylvester S.R., Tsuruta J.K., Griswold M.D. 1988. Biosynthesis and molecular cloning of sulfated glycoprotein 1 secreted by rat Sertoli cells: sequence similarity with the 70-kilodalton precursor to sulfatide/GM1 activator. Biochemistry. 27:4557–4564 10.1021/bi00412a050 [DOI] [PubMed] [Google Scholar]
- Daniels R., Kurowski B., Johnson A.E., Hebert D.N. 2003. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol. Cell. 11:79–90 10.1016/S1097-2765(02)00821-3 [DOI] [PubMed] [Google Scholar]
- Danilczyk U.G., Cohen-Doyle M.F., Williams D.B. 2000. Functional relationship between calreticulin, calnexin, and the endoplasmic reticulum luminal domain of calnexin. J. Biol. Chem. 275:13089–13097 10.1074/jbc.275.17.13089 [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181–191 10.1038/nrm1052 [DOI] [PubMed] [Google Scholar]
- Farhan H., Weiss M., Tani K., Kaufman R.J., Hauri H.P. 2008. Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J. 27:2043–2054 10.1038/emboj.2008.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández F., D’Alessio C., Fanchiotti S., Parodi A.J. 1998. A misfolded protein conformation is not a sufficient condition for in vivo glucosylation by the UDP-Glc:glycoprotein glucosyltransferase. EMBO J. 17:5877–5886 10.1093/emboj/17.20.5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan M.M., Grosch H.W., Locatelli-Hoops S., Werth N., Smolenová E., Nettersheim M., Sandhoff K., Hasilik A. 2004. Purified recombinant human prosaposin forms oligomers that bind procathepsin D and affect its autoactivation. Biochem. J. 383:507–515 10.1042/BJ20040175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold M.D., Roberts K., Bishop P. 1986. Purification and characterization of a sulfated glycoprotein secreted by Sertoli cells. Biochemistry. 25:7265–7270 10.1021/bi00371a003 [DOI] [PubMed] [Google Scholar]
- Guerin M., Parodi A.J. 2003. The UDP-glucose:glycoprotein glucosyltransferase is organized in at least two tightly bound domains from yeast to mammals. J. Biol. Chem. 278:20540–20546 10.1074/jbc.M300891200 [DOI] [PubMed] [Google Scholar]
- Hammond C., Braakman I., Helenius A. 1994. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. USA. 91:913–917 10.1073/pnas.91.3.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert D.N., Molinari M. 2007. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 87:1377–1408 10.1152/physrev.00050.2006 [DOI] [PubMed] [Google Scholar]
- Hebert D.N., Foellmer B., Helenius A. 1995. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 81:425–433 10.1016/0092-8674(95)90395-X [DOI] [PubMed] [Google Scholar]
- Hebert D.N., Foellmer B., Helenius A. 1996. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO J. 15:2961–2968 [PMC free article] [PubMed] [Google Scholar]
- Hebert D.N., Zhang J.-X., Chen W., Foellmer B., Helenius A. 1997. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J. Cell Biol. 139:613–623 10.1083/jcb.139.3.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A. 1994. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol. Biol. Cell. 5:253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A., Aebi M. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73:1019–1049 10.1146/annurev.biochem.73.011303.073752 [DOI] [PubMed] [Google Scholar]
- Hiraiwa M., Soeda S., Martin B.M., Fluharty A.L., Hirabayashi Y., O’Brien J.S., Kishimoto Y. 1993. The effect of carbohydrate removal on stability and activity of saposin B. Arch. Biochem. Biophys. 303:326–331 10.1006/abbi.1993.1291 [DOI] [PubMed] [Google Scholar]
- Hiraiwa M., Taylor E.M., Campana W.M., Darin S.J., O’Brien J.S. 1997. Cell death prevention, mitogen-activated protein kinase stimulation, and increased sulfatide concentrations in Schwann cells and oligodendrocytes by prosaposin and prosaptides. Proc. Natl. Acad. Sci. USA. 94:4778–4781 10.1073/pnas.94.9.4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.A., Ward C.L., Kopito R.R. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883–1898 10.1083/jcb.143.7.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.A., Illing M.E., Kopito R.R. 2002. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil. Cytoskeleton. 53:26–38 10.1002/cm.10057 [DOI] [PubMed] [Google Scholar]
- Kamhi-Nesher S., Shenkman M., Tolchinsky S., Fromm S.V., Ehrlich R., Lederkremer G.Z. 2001. A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell. 12:1711–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M., Srinivas H., Kandiah E., Gemma E., Ellgaard L., Oscarson S., Helenius A., Surolia A. 2003. Interactions of substrate with calreticulin, an endoplasmic reticulum chaperone. J. Biol. Chem. 278:6194–6200 10.1074/jbc.M209132200 [DOI] [PubMed] [Google Scholar]
- Karaivanova V.K., Luan P., Spiro R.G. 1998. Processing of viral envelope glycoprotein by the endomannosidase pathway: evaluation of host cell specificity. Glycobiology. 8:725–730 10.1093/glycob/8.7.725 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Kovacs J.J., McLaurin A., Vance J.M., Ito A., Yao T.P. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 115:727–738 10.1016/S0092-8674(03)00939-5 [DOI] [PubMed] [Google Scholar]
- Keith N., Parodi A.J., Caramelo J.J. 2005. Glycoprotein tertiary and quaternary structures are monitored by the same quality control mechanism. J. Biol. Chem. 280:18138–18141 10.1074/jbc.M501710200 [DOI] [PubMed] [Google Scholar]
- Kishimoto Y., Hiraiwa M., O’Brien J.S. 1992. Saposins: structure, function, distribution, and molecular genetics. J. Lipid Res. 33:1255–1267 [PubMed] [Google Scholar]
- Kondoh K., Hineno T., Sano A., Kakimoto Y. 1991. Isolation and characterization of prosaposin from human milk. Biochem. Biophys. Res. Commun. 181:286–292 10.1016/S0006-291X(05)81415-9 [DOI] [PubMed] [Google Scholar]
- Labriola C., Cazzulo J.J., Parodi A.J. 1999. Trypanosoma cruzi calreticulin is a lectin that binds monoglucosylated oligosaccharides but not protein moieties of glycoproteins. Mol. Biol. Cell. 10:1381–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois S., Zeng J., Hassan A.J., Canuel M., Morales C.R. 2003. The lysosomal trafficking of sphingolipid activator proteins (SAPs) is mediated by sortilin. EMBO J. 22:6430–6437 10.1093/emboj/cdg629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misasi R., Sorice M., Di Marzio L., Campana W.M., Molinari S., Cifone M.G., Pavan A., Pontieri G.M., O’Brien J.S. 2001. Prosaposin treatment induces PC12 entry in the S phase of the cell cycle and prevents apoptosis: activation of ERKs and sphingosine kinase. FASEB J. 15:467–474 10.1096/fj.00-0217com [DOI] [PubMed] [Google Scholar]
- Molinari M., Galli C., Vanoni O., Arnold S.M., Kaufman R.J. 2005. Persistent glycoprotein misfolding activates the glucosidase II/UGT1-driven calnexin cycle to delay aggregation and loss of folding competence. Mol. Cell. 20:503–512 10.1016/j.molcel.2005.09.027 [DOI] [PubMed] [Google Scholar]
- Morales C.R., Zhao Q., El-Alfy M., Suzuki K. 2000. Targeted disruption of the mouse prosaposin gene affects the development of the prostate gland and other male reproductive organs. J. Androl. 21:765–775 [PubMed] [Google Scholar]
- O’Brien J.S., Carson G.S., Seo H.C., Hiraiwa M., Kishimoto Y. 1994. Identification of prosaposin as a neurotrophic factor. Proc. Natl. Acad. Sci. USA. 91:9593–9596 10.1073/pnas.91.20.9593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J.D., van der Wal F.J., Bulleid N.J., High S. 1997. Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science. 275:86–88 10.1126/science.275.5296.86 [DOI] [PubMed] [Google Scholar]
- Parodi A.J., Mendelzon D.H., Lederkremer G.Z., Martin-Barrientos J. 1984. Evidence that transient glucosylation of protein-linked Man9GlcNAc2, Man8GlcNAc2, and Man7GlcNAc2 occurs in rat liver and Phaseolus vulgaris cells. J. Biol. Chem. 259:6351–6357 [PubMed] [Google Scholar]
- Pearse B.R., Gabriel L., Wang N., Hebert D.N. 2008. A cell-based reglucosylation assay demonstrates the role of GT1 in the quality control of a maturing glycoprotein. J. Cell Biol. 181:309–320 10.1083/jcb.200712068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J.R., Ora A., Van P.N., Helenius A. 1995. Transient, lectin-like association of calreticulin with folding intermediates of cellular and viral glycoproteins. Mol. Biol. Cell. 6:1173–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quellhorst G.J., Jr., O’Rear J.L., Cacan R., Verbert A., Krag S.S. 1999. Nonglucosylated oligosaccharides are transferred to protein in MI8-5 Chinese hamster ovary cells. Glycobiology. 9:65–72 10.1093/glycob/9.1.65 [DOI] [PubMed] [Google Scholar]
- Rajagopalan S., Xu Y., Brenner M.B. 1994. Retention of unassembled components of integral membrane proteins by calnexin. Science. 263:387–390 10.1126/science.8278814 [DOI] [PubMed] [Google Scholar]
- Rossmann M., Schultz-Heienbrok R., Behlke J., Remmel N., Alings C., Sandhoff K., Saenger W., Maier T. 2008. Crystal structures of human saposins C andD: implications for lipid recognition and membrane interactions. Structure. 16:809–817 10.1016/j.str.2008.02.016 [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C., Kelleher D.J., Gilmore R. 2009. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell. 136:272–283 10.1016/j.cell.2008.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D.T., Arnold S.M., Miller C.N., Wu J., Li J., Gunnison K.M., Mori K., Sadighi Akha A.A., Raden D., Kaufman R.J. 2006. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 4:e374 10.1371/journal.pbio.0040374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder M., Kaufman R.J. 2005. The mammalian unfolded protein response. Annu. Rev. Biochem. 74:739–789 10.1146/annurev.biochem.73.011303.074134 [DOI] [PubMed] [Google Scholar]
- Soldà T., Garbi N., Hämmerling G.J., Molinari M. 2006. Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J. Biol. Chem. 281:6219–6226 10.1074/jbc.M513595200 [DOI] [PubMed] [Google Scholar]
- Soldà T., Galli C., Kaufman R.J., Molinari M. 2007. Substrate-specific requirements for UGT1-dependent release from calnexin. Mol. Cell. 27:238–249 10.1016/j.molcel.2007.05.032 [DOI] [PubMed] [Google Scholar]
- Sousa M., Parodi A.J. 1995. The molecular basis for the recognition of misfolded glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. EMBO J. 14:4196–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa M.C., Ferrero-Garcia M.A., Parodi A.J. 1992. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 31:97–105 10.1021/bi00116a015 [DOI] [PubMed] [Google Scholar]
- Sun Y., Witte D.P., Zamzow M., Ran H., Quinn B., Matsuda J., Grabowski G.A. 2007. Combined saposin C and D deficiencies in mice lead to a neuronopathic phenotype, glucosylceramide and alpha-hydroxy ceramide accumulation, and altered prosaposin trafficking. Hum. Mol. Genet. 16:957–971 10.1093/hmg/ddm040 [DOI] [PubMed] [Google Scholar]
- Sun Y., Witte D.P., Ran H., Zamzow M., Barnes S., Cheng H., Han X., Williams M.T., Skelton M.R., Vorhees C.V., Grabowski G.A. 2008. Neurological deficits and glycosphingolipid accumulation in saposin B deficient mice. Hum. Mol. Genet. 17:2345–2356 10.1093/hmg/ddn135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.C., Thibault P., Tessier D.C., Bergeron J.J., Thomas D.Y. 2003. Glycopeptide specificity of the secretory protein folding sensor UDP-glucose glycoprotein:glucosyltransferase. EMBO Rep. 4:405–411 10.1038/sj.embor.embor797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.C., Ferguson A.D., Bergeron J.J., Thomas D.Y. 2004. The ER protein folding sensor UDP-glucose glycoprotein-glucosyltransferase modifies substrates distant to local changes in glycoprotein conformation. Nat. Struct. Mol. Biol. 11:128–134 10.1038/nsmb715 [DOI] [PubMed] [Google Scholar]
- Tessier D.C., Dignard D., Zapun A., Radominska-Pandya A., Parodi A.J., Bergeron J.J., Thomas D.Y. 2000. Cloning and characterization of mammalian UDP-glucose glycoprotein: glucosyltransferase and the development of a specific substrate for this enzyme. Glycobiology. 10:403–412 10.1093/glycob/10.4.403 [DOI] [PubMed] [Google Scholar]
- Trombetta S.E., Bosch M., Parodi A.J. 1989. Glucosylation of glycoproteins by mammalian, plant, fungal, and trypanosomatid protozoa microsomal membranes. Biochemistry. 28:8108–8116 10.1021/bi00446a022 [DOI] [PubMed] [Google Scholar]
- Vaccaro A.M., Salvioli R., Barca A., Tatti M., Ciaffoni F., Maras B., Siciliano R., Zappacosta F., Amoresano A., Pucci P. 1995. Structural analysis of saposin C and B. Complete localization of disulfide bridges. J. Biol. Chem. 270:9953–9960 10.1074/jbc.270.17.9953 [DOI] [PubMed] [Google Scholar]
- Valetti C., Grossi C.E., Milstein C., Sitia R. 1991. Russell bodies: a general response of secretory cells to synthesis of a mutant immunoglobulin which can neither exit from, nor be degraded in, the endoplasmic reticulum. J. Cell Biol. 115:983–994 10.1083/jcb.115.4.983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leeuwen J.E.M., Kearse K.P. 1997. Reglucosylation of N-linked glycans is critical for calnexin assembly with T cell receptor (TCR) alpha proteins but not TCRbeta proteins. J. Biol. Chem. 272:4179–4186 10.1074/jbc.272.7.4179 [DOI] [PubMed] [Google Scholar]
- Vassilakos A., Cohen-Doyle M.F., Peterson P.A., Jackson M.R., Williams D.B. 1996. The molecular chaperone calnexin facilitates folding and assembly of class I histocompatibility molecules. EMBO J. 15:1495–1506 [PMC free article] [PubMed] [Google Scholar]
- Wada I., Imai S.-i., Kai M., Sakane F., Kanoh H. 1995. Chaperone function of calreticulin when expressed in the endoplasmic reticulum as the membrane-anchored and soluble forms. J. Biol. Chem. 270:20298–20304 10.1074/jbc.270.50.29889 [DOI] [PubMed] [Google Scholar]
- Wada I., Kai M., Imai S., Sakane F., Kanoh H. 1997. Promotion of transferrin folding by cyclic interactions with calnexin and calreticulin. EMBO J. 16:5420–5432 10.1093/emboj/16.17.5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Glidden E.J., Murphy S.R., Pearse B.R., Hebert D.N. 2008. The cotranslational maturation program for the type II membrane glycoprotein influenza neuraminidase. J. Biol. Chem. 283:33826–33837 10.1074/jbc.M806897200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A., Darby N.J., Tessier D.C., Michalak M., Bergeron J.J., Thomas D.Y. 1998. Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J. Biol. Chem. 273:6009–6012 10.1074/jbc.273.11.6009 [DOI] [PubMed] [Google Scholar]