A previously unrecognized pore membrane protein, Pom33, stabilizes the interface between the nuclear envelope and the NPC to facilitate NPC biogenesis and spatial organization.
Abstract
The biogenesis of nuclear pore complexes (NPCs) represents a paradigm for the assembly of high-complexity macromolecular structures. So far, only three integral pore membrane proteins are known to function redundantly in NPC anchoring within the nuclear envelope. Here, we describe the identification and functional characterization of Pom33, a novel transmembrane protein dynamically associated with budding yeast NPCs. Pom33 becomes critical for yeast viability in the absence of a functional Nup84 complex or Ndc1 interaction network, which are two core NPC subcomplexes, and associates with the reticulon Rtn1. Moreover, POM33 loss of function impairs NPC distribution, a readout for a subset of genes required for pore biogenesis, including members of the Nup84 complex and RTN1. Consistently, we show that Pom33 is required for normal NPC density in the daughter nucleus and for proper NPC biogenesis and/or stability in the absence of Nup170. We hypothesize that, by modifying or stabilizing the nuclear envelope–NPC interface, Pom33 may contribute to proper distribution and/or efficient assembly of nuclear pores.
Introduction
Building sophisticated and dynamic macromolecular machines is a hallmark of cellular life. In eukaryotic cells, one such highly complex multiprotein assembly is the nuclear pore complex (NPC). NPCs, which mediate bidirectional exchanges of molecules between the nucleus and the cytoplasm, are embedded within the nuclear envelope (NE) at fusion points between the inner and the outer nuclear membranes (for review see Tran and Wente, 2006). Proteomic and genetic analyses of budding yeast and mammalian NPCs have detailed their protein composition (Rout et al., 2000; Cronshaw et al., 2002; for review see D’Angelo and Hetzer, 2008). NPCs are composed of multiple copies of ∼30 proteins, called nucleoporins (Nups), most of which can be biochemically isolated as subcomplexes. Knowledge on the spatial organization of NPC components has recently been refined by the computational integration of biochemical data into a map of the yeast NPC (Alber et al., 2007a,b) and by the availability of 3D structures for an increasing number of Nups (for review see Brohawn et al., 2009).
Over the last several years, major insights have also been gained into the mechanisms of NPC assembly. In organisms that undergo an open mitosis in which the NE breaks down and NPCs dismantle, nuclear pore assembly can follow two pathways: de novo NPC biogenesis that allows the insertion of new NPCs into an intact NE during interphase, and postmitotic NPC reassembly that is tightly coordinated with NE reformation (for review see Fernandez-Martinez and Rout, 2009). The latter process has been extensively studied, and the chronological recruitment of different factors is now well established. At an early step, ELYS/MEL-28 binds to chromatin and recruits the Nup107-160 subcomplex, in a Ran-dependent manner, to form pre-pore structures to which membrane vesicles or sheets containing the pore membrane proteins Pom121 and Ndc1 subsequently associate. The sequential recruitment of the remaining soluble pore subcomplexes then allows completion of NPC assembly (for review see Antonin et al., 2008; Kutay and Hetzer, 2008).
Much less is known about the molecular mechanisms involved in de novo NPC biogenesis, the only process accounting for NPC assembly in yeasts that undergo a closed mitosis. A genetic screen for Saccharomyces cerevisiae NPC assembly mutants initially revealed the contribution of the Ran GTPase cycle to this process (Ryan et al., 2003, 2007). Using a cell-free assay, D’Angelo et al. (2006) demonstrated that de novo NPC biogenesis during NE expansion in interphase Xenopus laevis nuclei shares common players with postmitotic NPC assembly, including, in addition to Ran and karyopherin β, the Nup107-160 complex (D’Angelo et al., 2006). In addition, major vault protein-containing particles were recently reported to contribute to this process (Vollmar et al., 2009).
Because de novo pore insertion requires local fusion events between the inner and outer NE membranes, integral membrane proteins are anticipated to play a key role in this process. Among the three integral membrane Nups identified so far in yeast and vertebrates, only Ndc1 is universally conserved in eukaryotes (Stavru et al., 2006; Mansfeld et al., 2006). In S. cerevisiae, the three pore membrane proteins, Ndc1, Pom34, and Pom152, form a trimeric complex that binds to Nup53 or Nup59, which in turn interact with Nup157 and Nup170, generating an interaction network critical for NPC assembly (Madrid et al., 2006; Onischenko et al., 2009; Flemming et al., 2009; Makio et al., 2009). Besides these bona fide Nups, proper NPC assembly was also shown to involve the inner nuclear membrane–associated protein Esc1 (Lewis et al., 2007); Apq12 and Brr6, two NE/ER integral membrane proteins contributing to lipid homeostasis (Scarcelli et al., 2007; Hodge et al., 2010); and the RSC chromatin-remodeling complex (Titus et al., 2010). Finally, a recent study pointed to a role of members of the reticulon (Rtn) and Yop1/DP1 families in de novo NPC assembly. These integral membrane proteins were previously demonstrated to have membrane-bending properties and to be involved in both tubular ER maintenance and postmitotic NE formation (Kiseleva et al., 2007; Audhya et al., 2007; Anderson and Hetzer, 2008; Hu et al., 2008; for review see Yang and Strittmatter, 2007). Dawson et al. (2009) further showed that S. cerevisiae Rtn1 and Yop1 are partially localized at NPCs and that their codeletion leads to a NPC clustering phenotype. In addition, antibody-mediated inhibition of vertebrate Rtn4a inhibited de novo NPC formation in a cell-free assay based on X. laevis egg extracts, which suggests a conserved function of Rtns in pore assembly (Dawson et al., 2009).
In S. cerevisiae, deletion of genes encoding several members of the Nup84 complex (the counterpart of the vertebrate Nup107-160 complex) also leads to a constitutive NPC clustering phenotype (for review see Doye and Hurt, 1997). The Nup84 complex is also required for mRNA export, a function shared by the Nup107-160 complex (Doye and Hurt, 1997; Vasu et al., 2001), and for maintenance of appropriate sumoylation patterns (Palancade et al., 2007). To gain further insights into the function of this complex, we previously undertook a systematic synthetic lethal screening of the collection of nonessential gene deletions using as bait a strain lacking one of the Nup84 complex subunits, Nup133 (Loeillet et al., 2005). Among the candidate genes of unknown function identified in the screen, three encoded proteins reportedly localized at the NE or ER according to the systematic annotation of the localization of GFP-tagged yeast proteins (Huh et al., 2003). One of them was Apq12, which was demonstrated in the meantime to be a transmembrane NE/ER protein functionally linked to nuclear pores (Scarcelli et al., 2007), whereas the second one, renamed Pml39, is associated with NPCs and functions in mRNA quality control before export (Palancade et al., 2005).
In this study, we have characterized the third NE-associated protein identified in our genetic screen. We show that this protein, renamed Pom33 (pore membrane protein, ∼33 kD), is an evolutionarily conserved membrane protein that is functionally linked to NPC components and required for proper NPC assembly and/or stability in S. cerevisiae.
Results
POM33 displays genetic interaction with NUP133 and encodes a previously uncharacterized transmembrane protein localized at NPCs
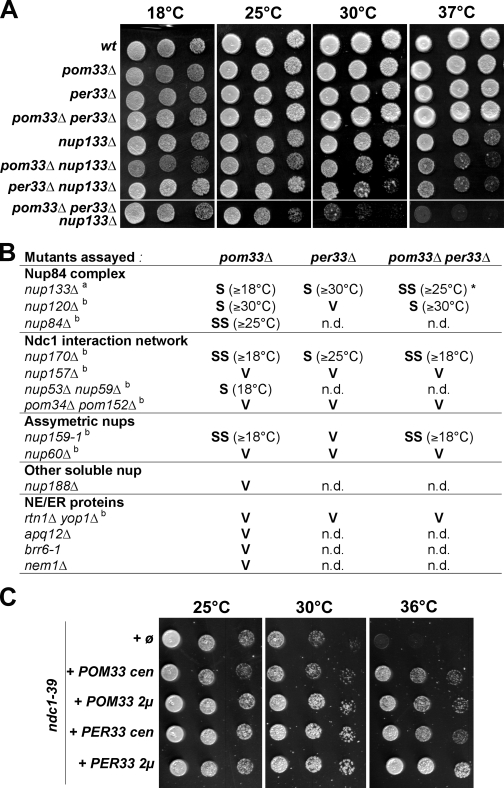
Systematic synthetic lethal screening of the collection of nonessential gene deletions using the nup133Δ mutant as bait identified the YLL023c open reading frame (ORF), which encodes a ∼33 kD protein with several putative transmembrane helixes (subsequently termed Pom33, see the following paragraph). Although deletion of YLL023c/POM33 did not impair growth at any temperature, its genetic interaction with NUP133 was confirmed by the enhanced growth defect of yll023cΔ nup133Δ mutant cells as compared with nup133Δ, which was recorded at all temperatures tested (see Fig. 4 A).
Figure 4.
POM33 displays genetic interactions with core components of NPC. (A) Growth properties of wt, pom33Δ, per33Δ, nup133Δ, and of corresponding double and triple mutants. Equivalent amounts of cells were spotted as fivefold dilutions on YEPD plates, and were incubated for 5 d at 18°C or for 3 d at 25, 30, and 37°C. (B) Summary of the genetic interactions between pom33Δ, per33Δ, or pom33Δ per33Δ, and Nups or NE/ER mutants. Synthetic interactions were scored as: SS, strong synergistic interaction; S, synergistic interaction; V, viable; n.d., not determined. The temperature above which the synthetic phenotype was observed is indicated. Footnote a, see Fig. 4 A; footnote b, see Fig. S2; *, note that PER33 deletion partially rescues the pom33Δ nup133Δ synthetic phenotype at 18°C. (C) Growth assay of ndc1-39 mutant cells transformed with an empty pRS315 plasmid (ø), with centromeric (cen), or with multicopy (2µ) plasmids encompassing the POM33 or PER33 genes. Transformants were spotted as fivefold dilutions on selective medium, and plates were incubated for 3 d at the indicated temperatures.
Fluorescence microscopy analysis of Yll023c C-terminally tagged at its genomic locus with GFP revealed a punctuate labeling at the NE (Fig. 1 A). The genetic interaction of YLL023c with NUP133 prompted us to analyze whether Yll023c was associated with nuclear pores. For this purpose, Yll023c was assayed for colocalization with clustered NPCs in nup133Δ mutant cells. Unlike yll023cΔ, YLL023c-GFP did not enhance the growth defect of nup133Δ cells (unpublished data), which suggests that the Yll023c-GFP fusion protein is functional. In nup133Δ cells, all the NE-associated Yll023-GFP relocalized in foci coinciding with Nup49-mCherry clusters (Fig. 1 A), which indicates that Yll023c specifically associates with NPCs. Because this protein contains transmembrane helixes (Fig. S1 C and the following section), YLL023c was subsequently designated POM33 (pore membrane protein, ∼33 kD).
Figure 1.
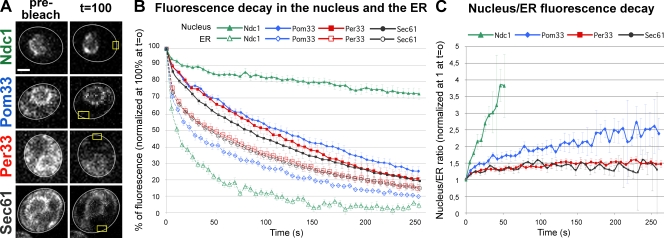
Pom33, and to a lower extent its paralogue Per33, are associated with NPCs. (A and B) Fluorescence microscopy analysis of Pom33-GFP (A) and Per33-GFP (B) in wt and nup133Δ cells expressing Nup49-mCherry. Spinning disk confocal images of single-channel fluorescence for GFP and mCherry are shown (left) as well as the merge (right). Bar, 5 µm. (C) Immunolocalization of GFP-tagged Pom33 at nuclear pores. Cryosections of Pom33-GFP cells were successively labeled with anti-GFP (detected using ProtA 10-nm gold particles) and with mAb414 (detected using ProtA 15-nm gold particles). (a) Typical patterns of Pom33-GFP localization at NPCs (detected based on NE structure and mAb414 labeling) as well as a section encompassing a nucleus are presented. Arrows point to 10-nm gold particles recognizing the GFP moiety of Pom33-GFP at nuclear pores. Arrowheads point to 10-nm gold particles localized outside of the NE. The cytoplasmic faces of the NE are oriented toward the top of each micrograph. (b) Statistical analysis of the distribution of anti-GFP–associated gold particles in control (expressing no GFP fusion protein), Pom33-GFP, Per33-GFP, and Ndc1-GFP cells. For each strain, the total number of anti-GFP–associated gold particles (n1) and of cryosectioned cells (n2) analyzed, as well as the percentage of GFP-associated gold particles localized at NPCs relative to the entire NE (NPC + NE), are indicated.
To further characterize the localization of Pom33, immunogold labeling on cryosections of Pom33-GFP cells was performed using polyclonal anti-GFP antibodies and the mAb414 antibody that labels yeast NPCs (Aris and Blobel, 1989). In control cells, anti-GFP–bound gold particles were rarely detected at the NE and never at NPCs (Fig. 1 C, b). In contrast, a significant fraction of anti-GFP–bound gold particles colocalized with mAb414 at NPCs in Pom33-GFP cells (Fig. 1 C). Quantitative analysis of the distribution of anti-GFP–bound gold particles in Pom33-GFP cells revealed that 60% of the NE-associated gold particles were localized at NPCs, a fraction comparable to the one obtained with similarly processed cells expressing Ndc1-GFP (Fig. 1 C, b). The increased cytoplasmic fraction of anti-GFP–bound particles detected in Pom33-GFP cells as compared with Ndc1-GFP cells probably represents an ER-associated pool of Pom33 (Fig. 1 A).
Because a minor fraction of Pom33-GFP also localizes within the ER, fluorescence loss in photobleaching (FLIP) was used to investigate its dynamics between these two compartments. As controls, the dynamics of GFP-tagged versions of Ndc1 and of Sec61, a subunit of the translocon that localizes to both the cortical and perinuclear ER, was similarly analyzed. As shown in Fig. 2, repetitive photobleaching of a small area of the cortical ER led to a decay in Pom33-GFP fluorescence within the NE that was significantly faster as compared with the one displayed by Ndc1-GFP (Fig. 2). Yet, as best revealed upon comparison of the variations of the nucleus/ER fluorescence ratio over time (Fig. 2 C), Pom33 exchanges between these two compartments in a slightly less dynamic fashion than Sec61. Together, these data therefore indicate that Pom33 behaves as a dynamic transmembrane Nup.
Figure 2.
Pom33 is a dynamic component of nuclear pores. (A) FLIP analysis in Ndc1-GFP–, Pom33-GFP–, Per33-GFP–, and Sec61-GFP–expressing cells. Shown are confocal microscopy images of representative cells before photobleaching and after ∼15–20 bleaching pulses (100 s). The cell shape is outlined in white and the bleached area within the cortical ER is outlined in yellow. Bar, 2 µm. (B) For each strain, the fluorescence decay in the nucleus (closed symbols) and in the ER (open symbols) was quantified as described in Materials and methods. Each curve represents the mean of a least 16 independent cells. SD is also indicated. (C) For each cell, the ratio of fluorescence in the nucleus over the ER, normalized to 1 at t = 0, was quantified at each time point. The median value and SD (error bars) are plotted over time.
Pom33 and its paralogue, Per33, are evolutionarily conserved transmembrane proteins
Database analyses revealed that YLL023c/POM33 has a paralogue in S. cerevisiae, namely the nonessential YLR064w gene (subsequently termed PER33). Unlike Pom33-GFP, subcellular localization analysis of Ylr064w/Per33-GFP revealed a more typical ER localization, with no major enrichment at the nuclear periphery when compared with cortical ER staining (Fig. 1 B). Moreover, Per33-GFP was still present at the nuclear periphery in nup133Δ cells, although a minor fraction of this fusion protein colocalized with Nup49-mCherry aggregates (Fig. 1 B). Consistently, immunogold labeling of Per33-GFP cryosections revealed an increased proportion of gold particles outside the NE and a less significant enrichment of gold particles at NPCs over the entire NE as compared with Pom33-GFP or Ndc1-GFP cells (Fig. 1 C, b). Finally, FLIP experiments indicated that Per33-GFP dynamically exchanges between the ER and the NE, with kinetics similar to those displayed by Sec61-GFP (Fig. 2). Because Ylr064w can associate with NPCs, but unlike Pom33 is mainly associated with the ER/NE, YLR064w was renamed PER33 (pore and ER protein, 33 kD).
A phylogenic analysis revealed that Pom33 and Per33 belong to a protein family conserved during evolution (Fig. 3 A). Unlike in S. cerevisiae and more generally in the Saccharomycotina clade, only one orthologue was found in all other species. In vertebrates, this orthologue was previously termed TMEM33 (transmembrane No. 33). Fluorescence analysis of stable HeLa cell lines, mildly overexpressing GFP-HsTMEM33 as compared with endogenous HsTMEM33 (5–10-fold as established by RT-PCR analyses; unpublished data), revealed that this fusion protein localized to the NE and the ER, with a significant enrichment at the NE as compared with protein disulfide isomerase (PDI), a luminal ER protein, or to BAP31, an integral membrane protein of the ER (Fig. S1, A and B). Although the significant fraction of GFP-HsTMEM33 localized at the ER could be partly attributed to its mild overexpression, this localization may also reflect the fact that in vertebrates, TMEM33 fulfills the function of both S. cerevisiae Pom33 and Per33.
Figure 3.
S. cerevisiae Pom33 and Per33 and human TMEM33 are evolutionarily conserved NE/ER-associated proteins. (A) Phylogenic tree of S. cerevisiae Pom33 and Per33 homologues. The tree, based on ClustalW alignment of sequences identified by BLAST searches using ScPom33, ScPer33, and HsTMEM33 (highlighted in bold), was constructed with the Jalview software (www.jalview.org; Waterhouse et al., 2009) based on the percentage of identity between the proteins. (B) ClustalW alignment of ScPom33, ScPer33, and HsTMEM33. Identical residues are colored in black, and similar residues are colored in gray. Asterisks in the consensus line denote residues that are identical in all three proteins sequences, and periods indicate residues that are either identical or similar in at least two out of the three sequences. Approximate positions of the hydrophobic stretches are shown on top of the alignment (black line, see also Fig. S1 C).
Sequence analyses revealed that Pom33, Per33, and HsTMEM33 (as well as the various TMEM33 family members in other species) share three long hydrophobic regions of 38–46 amino acids, each potentially fitting two transmembrane helices (Figs. 3 B and S1 C; see Discussion). Moreover, a recent large-scale study using a topology reporter determined experimentally that the C termini of both Pom33 and Per33 are located in the cytosolic/pore face of the membrane (Kim et al., 2006). Various programs further predicted that Pom33, Per33, and HsTMEM33 are multipass transmembrane proteins, with the N and C termini both positioned on the cytosolic/pore face (Fig. S1 C). Consistently, topology analysis of HsTMEM33, based on the accessibility of the GFP tags within HsTMEM33-GFP and GFP-HsTMEM33 upon digitonin treatment of transfected HeLa cells, revealed that the N- and C-terminal domains of HsTMEM33 are both positioned on the cytoplasmic/pore face of the NE/ER (Fig. S1 B).
POM33 genetically interacts with a subset of Nups including members of the Nup84 complex and of the Ndc1 interaction network
Analysis of pom33Δ per33Δ cells did not reveal any growth defect as compared with wild-type (wt) cells, which indicates that these paralogous genes do not perform an essential redundant function (Fig. 4 A). Analysis of the per33Δ nup133Δ double mutant revealed a synergistic growth defect that was however only detected at 30°C and 37°C (Fig. 4 A). As a result of an additive effect, cell growth was further impaired at 30°C and 37°C in the pom33Δ per33Δ nup133Δ triple mutant (Fig. 4 A).
To gain insight into the function of Pom33 and Per33, we then investigated whether genetic interactions occurred between POM33 or PER33 and genes encoding other proteins physically or functionally associated with NPCs. This analysis revealed impaired growth of nup120Δ cells upon deletion of POM33, but not PER33 (Figs. 4 B and S2 A), which strengthens the evidence for a genetic connection between POM33 and members of the Nup84 complex. This study also uncovered a strong genetic interaction between pom33Δ (but not per33Δ) and nup159-1, which carries a mutation within an essential cytoplasmic Nup contributing to mRNA export and NPC distribution (Figs. 4 B and S2 C; Gorsch et al., 1995). Interestingly, a synergistic growth defect was also observed when pom33Δ or per33Δ were combined with deletion of NUP170, but not of its orthologue NUP157 (Figs. 4 B and S2, B and F). In contrast, deletion of POM33, PER33, or both POM33 and PER33 in pom34Δ pom152Δ cells did not lead to any synergistic growth defect at any temperature (Figs. 4 B and S2 E). Because of the strong growth defects caused by the ndc1 alleles available, it was difficult to assess their synergistic interactions with pom33Δ. However, we observed that mild overexpression of Pom33 or Per33 partially rescued the temperature-sensitive growth defect of the ndc1-39 mutant that affects NPC assembly, but not of the ndc1-1 mutant that was reported to mainly affect spindle pole body insertion (Fig. 4 C and not depicted; Chial et al., 1998; Lau et al., 2004).
The rtn1Δ yop1Δ mutant was recently demonstrated to genetically interact with mutants in genes encoding Poms and a subset of structural Nups (Dawson et al., 2009). However, the pom33Δ per33Δ rtn1Δ yop1Δ quadruple mutant displayed no growth defects at any temperature (Figs. 4 B and S2 H). Finally, no synergistic growth defect could be detected when pom33Δ was combined with nup188Δ, nup60Δ, or with mutants in genes encoding non-NPC ER/NE proteins (apq12Δ, nem1Δ, and brr6-1; Figs. 4 B and S2 G; and not depicted).
Collectively, these data thus reveal strong genetic connections between POM33 and a subset of Nups, notably with two structural NPC subcomplexes: the Nup84 complex and the Ndc1 interaction network. PER33 exhibits a more restricted pattern of genetic interactions, with the most prominent connection being to the Ndc1 interaction network, which indicates that although only a minor fraction of Per33 is localized at NPCs, it might fulfill partially overlapping functions with Pom33.
Pom33 physically interacts with Rtn1, a Rtn family member functionally linked to NPCs
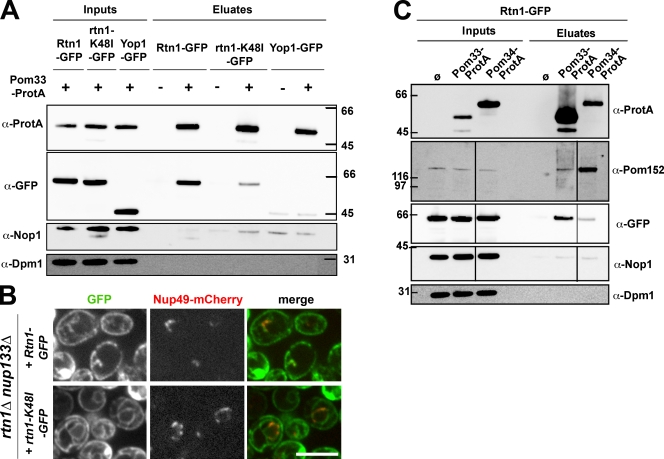
To identify proteins physically interacting with Pom33, the protein tagged with three IgG-binding domains of protein A (ProtA) was affinity-purified from yeast cells, and the copurifying proteins were analyzed by SDS gels followed by silver staining. This analysis revealed species migrating at ∼35–40 kD in the Pom33-ProtA fraction that were not detected in the control sample (unpublished data). Comparative mass spectrometry analysis of the proteins present within this area of the gel, as well as systematic mass-spectrometry analysis of all proteins copurifying with Pom33, led to the identification of the Rtn protein Rtn1 as one of the major species specifically present in the Pom33-ProtA fraction (17% coverage, see Materials and methods). Similar affinity purification from cells expressing either Rtn1-GFP or Yop1-GFP confirmed the interaction between Pom33 and Rtn1 but revealed no significant interaction between Pom33 and Yop1 (Fig. 5 A). Unexpectedly, Pom33-ProtA coprecipitated GFP-tagged Rtn1 more efficiently than rtn1-K48I, a mutant previously demonstrated to be enriched at the NE and to more efficiently complement the lethality of the pom34Δ nup59Δ mutant, as compared with wt Rtn1 (Fig. 5 A; Shibata et al., 2008; Dawson et al., 2009). However, fluorescence analysis performed in nup133Δ cells revealed that unlike Rtn1-GFP, rtn1-K48I-GFP is not specifically enriched at the clustered NPCs (Fig. 5 B). This result, along with the absence of a specific interaction between Pom33-ProtA and Dpm1, a transmembrane protein of the ER (Orlean et al., 1988), confirmed the specificity of the interaction between Pom33 and Rtn1, and further suggests that Pom33 may interact with the NPC-associated fraction of Rtn1. Finally, Western-blot analyses of proteins copurifying with Pom33-ProtA or Pom34-ProtA confirmed the strong interaction between Pom34 and Pom152 (Onischenko et al., 2009) and further revealed a modest interaction between Pom33-ProtA and Pom152 and between Pom34-ProtA and Rtn1-GFP (Fig. 5 C). Together, these data thus indicate that Pom33 physically interacts, albeit possibly indirectly, with Rtn1.
Figure 5.
Pom33 interacts with the Rtn protein Rtn1. (A) Affinity purification by IgG chromatography of Pom33-ProtA in Rtn1-GFP–, rtn1-K48I-GFP–, or Yop1-GFP–expressing strains. Total soluble extracts (inputs) and affinity-purified fractions (eluates, 2.5-fold equivalent for the anti–[α]-ProtA and 2,500-fold equivalent for the other antibodies) from strains expressing (+) or not expressing (−) Pom33-ProtA were analyzed by Western blotting using the indicated antibodies. Dpm1 and the nucleolar protein Nop1 were used as controls. (B) Spinning disk confocal images of rtn1Δ nup133Δ cells expressing Rtn1-GFP or rtn1K48I-GFP and Nup49-mCherry. Note that unlike wt Rtn1, the rtn1-K48I mutant is not enriched at the clustered pores labeled with Nup49-mCherry. Bar, 5 µm. (C) Affinity purification by IgG chromatography of Pom33-ProtA or Pom34-ProtA in Rtn1-GFP–expressing cells. Total soluble extracts (inputs) and affinity-purified fractions (eluates, fivefold equivalent for the anti-ProtA and 2,500-fold equivalent for the other antibodies) were analyzed by Western blotting using the indicated antibodies. Black lines indicate that intervening lanes have been spliced out. Size markers on the sides of the gel blots indicate kD.
Nucleocytoplasmic transport is not drastically impaired in the pom33Δ mutant
To functionally characterize Pom33, nucleocytoplasmic transport was analyzed in pom33Δ cells. In vivo analysis of the nuclear import of Mig1-GFP-LacZ (De Vit et al., 1997) and of the localization of the NLS–GFP2–nuclear export signal (NES) reporter (Stade et al., 1997) revealed no defect in the classical NLS- or NES-mediated transport pathways in pom33Δ cells (unpublished data). Similarly, no defect was detected in the kinetics of import of the Nab2-NLS-GFP reporter, recently reported to be impaired in the rtn1Δ yop1Δ NPCs clustering mutant (Fig. S3, A and B; Dawson et al., 2009). Finally, fluorescence in situ hybridization only revealed a very mild accumulation of poly(A)+ RNA in the nucleus of a fraction of pom33Δ cells (Fig. S3 C). Although we cannot exclude that Pom33 may be required for efficient import or export of other specific cargos, these data indicate that Pom33 is only modestly involved in nucleocytoplasmic transport.
NPC distribution is altered in pom33Δ but not in per33Δ cells
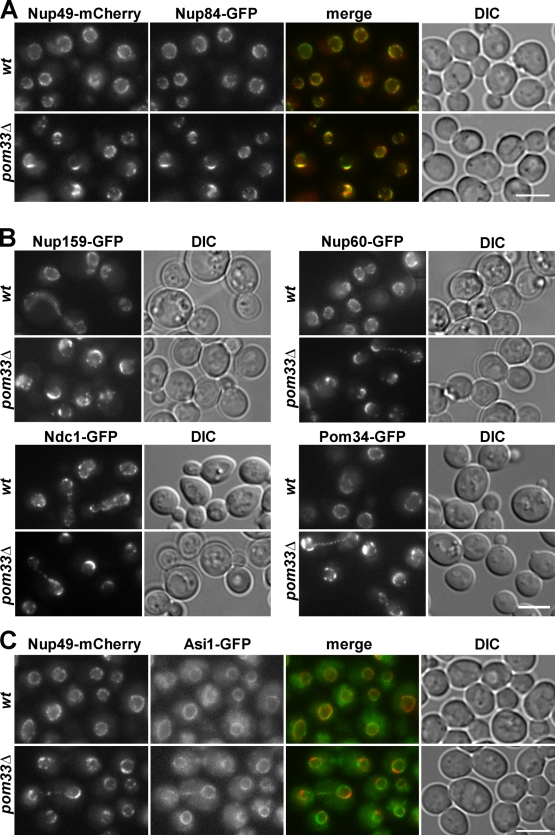
The genetic or physical interactions between Pom33 and proteins required for NPC assembly prompted us to analyze the localization of individual Nups in pom33Δ cells. Unlike in wt or per33Δ cells (see Fig. 8 A), live imaging of pom33Δ cells revealed clusters of Nup84-GFP colocalizing with Nup49-mCherry at the nuclear periphery (Fig. 6 A). A similar localization within NPC clusters was also recorded for all the GFP-tagged Nups analyzed, namely Nup159 (a cytoplasmic FG-Nup), Nup60, and Mlp2 that localize at the nuclear basket; Ndc1, Pom34, and Pom152 (the transmembrane Nups); and Nup157, a structural Nup of the inner ring (Fig. 6 B and not depicted). Unlike these various Nups, the inner nuclear membrane protein Asi1 tagged with GFP did not colocalize with clustered NPCs labeled by Nup49-mCherry, and its localization was not otherwise altered in pom33Δ cells (Fig. 6 C). Together, these data indicated that POM33 deletion alters NPC distribution without inducing profound alterations of the NE structure.
Figure 8.
Pom33 is required for normal pore density in the daughter nucleus in telophase. (A, a) Nup84-GFP fluorescence was analyzed by confocal microscopy in the indicated yeast strains. 3D reconstructions of total fluorescence from 13 planes covering the entire nuclei are shown. The cell shape of dividing cells is outlined in white. Bar, 5 µm. (b) The ratios of the total fluorescence intensity of Nup84-GFP in the mother versus the bud nuclei, quantified in telophase cells, are represented as box plots using KaleidaGraph (see Materials and methods). Mean values (m) and SD of the M/B ratios are indicated below the corresponding strains. *, 10−10 < P < 5 × 10−3 compared with wt using the Student’s t test; ***, P < 10−10. The total number of cells quantified is indicated (n; arising from at least two independent experiments). (B) M/B ratio distribution of Nup84-GFP fluorescence in wt and pom33Δ cells at early anaphase, telophase, and G1 stages of mitosis was plotted as in A. Confocal images of Nup84-GFP at the corresponding stages, revealed by the cell shape outlined in white, are shown on top. Bar, 2 µm.
Figure 6.
The pom33Δ mutant displays an NPC clustering phenotype. (A) The localization of Nup84-GFP was analyzed in wt and pom33Δ cells carrying the Nup49-mCherry plasmid. Wide-field images of single-channel fluorescence for mCherry and GFP; merge and DIC images are also shown. (B) The localization of the indicated GFP-tagged Nups was analyzed by direct fluorescence microscopy in wt and pom33Δ cells. The bright Ndc1-GFP dots in wt and pom33Δ cells correspond to the spindle pole body. (C) Fluorescence microscopy analysis of the localization of the inner nuclear membrane protein Asi1-GFP in wt and pom33Δ cells expressing Nup49-mCherry. Bars, 5 µm.
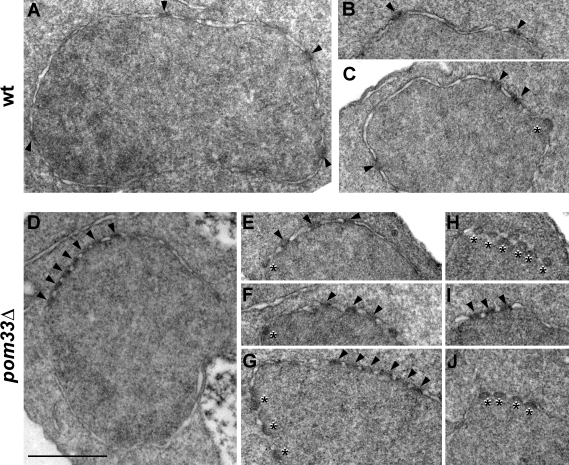
Consistent with this observation, thin-section EM analyses of pom33Δ cells revealed the presence of NPC clusters that were not associated with major distortions of the NE structure (Fig. 7, D–J). In most cases, these NPCs appeared, as in wt cells, as electron-dense structures spanning the two membranes of the NE (Fig. 7, arrowheads). Additional electron-dense structures that appeared to be associated with only one side of the NE were also observed in pom33Δ cells (Fig. 7, E–H and J, asterisks) and, albeit more rarely, in wt cells (Fig. 7 C, asterisk). These structures were somehow similar to those described in rtn1Δ yop1Δ cells and could, as proposed by Dawson et al. (2009), represent intermediates of NPC assembly. However, as similar observations were also made in other NPC-clustering mutants such as the nup82Δ108 mutant, which affects a peripheral (cytoplasmic) Nup (Belgareh et al., 1998), these apparently distorted structures may also correspond to tilted views of NPCs that would be more frequently detected in NPC clustering mutants because of the nearby distribution of multiple pores.
Figure 7.
Thin-section EM analysis of the NE and NPCs in wt and pom33Δ cells. wt cells (A–C) and pom33Δ cells (D–J) were grown to early log phase at 30°C and processed for thin-section EM (as described in Materials and methods). Arrowheads point to typical NPCs and asterisks denote distorted NPC-like structures. Bar, 500 nm.
Pom33 is required for normal pore density in the daughter nucleus during telophase
Shcheprova et al. (2008) have previously demonstrated that from anaphase on, a barrier restricts the diffusion of outer-membrane proteins and large structures such as NPCs through the bud neck. Consequently, de novo NPC insertion is likely to significantly contribute to the number of NPCs in the bud nuclei in telophase cells. These authors further showed that in the pore assembly mutants nup133Δ and to a lower extent nic96-1, the relative intensity of GFP-tagged Nup49 between the mother and the bud nucleus (subsequently called the mother/bud [M/B] ratio; see Materials and methods) is increased as compared with wt cells.
Using the structural Nup Nup84 as a reporter and in agreement with Shcheprova et al. (2008), we observed a slightly asymmetric distribution of Nup84-GFP between the mother and the bud nuclei of wt telophase cells that was significantly increased in nup133Δ telophase cells (Fig. 8 A). A similar analysis revealed an altered incorporation of Nup84-GFP in the daughter nuclei of pom33Δ cells, comparable to the one observed in nup133Δ cells (Fig. 8 A). In contrast, PER33 deletion only induced a slight increase of the Nup84-GFP M/B ratio and did not enhance the phenotype of pom33Δ cells (Fig. 8 A). This bias toward the mother in Nup84-GFP fluorescence intensity did not merely reflect a defect in the nuclear or NE growth of pom33Δ daughter nuclei, as probed using a soluble nuclear reporter (TetR-mCherry, see Fig. S4 A). An increased M/B ratio was also observed in pom33Δ cells upon fluorescence intensity quantification of two other NPC markers: the cytoplasmic Nup159 and the symmetrically localized Nup49 (Fig. S4 B). In the latter case, however, the phenotype was more pronounced, likely reflecting the fact that Nup49-GFP is not fully functional (Fig. S4 B; Makio et al., 2009).
In nup133Δ and pom33Δ cells, the decreased density of pores in the daughter cell could have two possible origins: (1) a defect in de novo NPC biogenesis or stabilization in the bud nuclei, or (2) an altered transit of clustered and/or defective NPCs from the mother to the bud. However, the mean M/B ratio was not significantly different between wt and pom33Δ cells in early anaphase cells (whose nuclei are just entering the bud; Fig. 8 B). This indicates that despite the clustering phenotype of pom33Δ cells, a similar fraction of NPCs passes through the bud in pom33Δ and wt cells during early anaphase, before the establishment of a tight diffusion barrier. In addition, disrupting BUD6 in the pom33Δ mutant partially decreased the M/B ratio of telophase cells as compared with the parental pom33Δ strain (Fig. S4 C), which indicates that alteration of the diffusion barrier allows clustered NPCs present in pom33Δ mother cells to reach the daughter nuclei. Finally, the altered M/B ratio observed in nup133Δ cells was partially rescued by a plasmid encoding the ΔN-nup133 mutant allele, in which NPCs are still clustered (Fig. 8 A; Doye et al., 1994). Together, these data indicate that the altered M/B ratio observed in nup133Δ or pom33Δ cells is unlikely to solely result from a primary defect in NPC distribution, which suggests a contribution of Pom33 to efficient NPC biogenesis or stabilization in the daughter nucleus. However, the M/B ratio is no longer significantly distinct in G1 between wt and pom33Δ cells expressing Nup84-GFP (Fig. 8 B). This implies that NPCs finally reach a proper density in pom33Δ daughter cells, although less efficiently, a result consistent with the lack of defects in growth and transport of these mutant cells.
Loss of Pom33 impairs NPC assembly upon depletion of Nup170
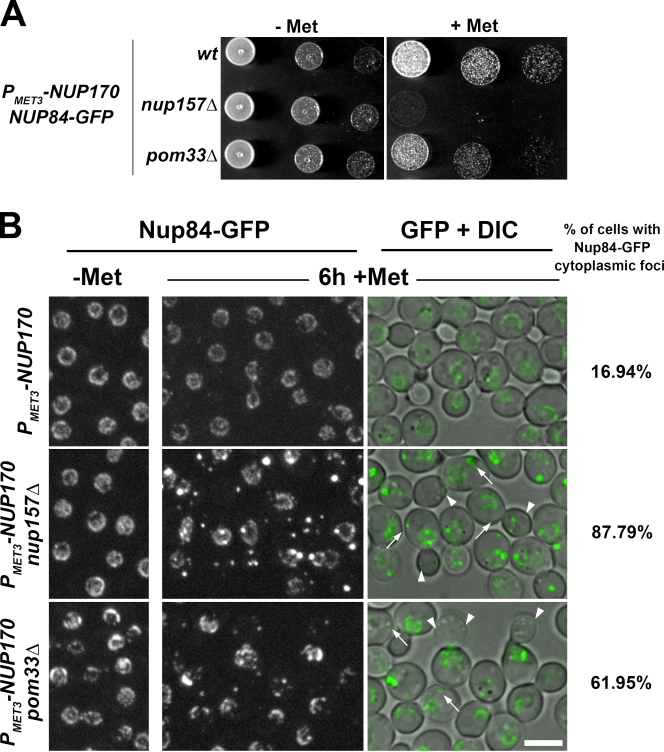
Recently, Makio et al. (2009) reported that repression of NUP170 in the absence of NUP157 causes a defect in NPC assembly leading to the accumulation of newly synthesized Nups in cytoplasmic foci and inner NE-associated structures. Our data suggesting a contribution of Pom33 to NPC biogenesis or stabilization (Fig. 8), along with the genetic interaction between POM33 and NUP170 (Figs. 4 B and S2 B), prompted us to investigate NPC assembly upon repression of NUP170 in the absence of Pom33. As shown in Fig. 9 A, depletion of Nup170 recapitulated the synergistic growth defect previously uncovered upon deletion of NUP170 in pom33Δ strains (Fig. S2 B). Analysis of Nup84-GFP localization upon repression of NUP170 in pom33Δ cells revealed a clear accumulation of this Nup into cytoplasmic foci (Fig. 9 B). The less-pronounced NPC defects induced upon Nup170 depletion in pom33Δ cells as compared with nup157Δ cells (Fig. 9 B; Makio et al., 2009) suggest that NPC assembly is delayed in nup170Δ pom33Δ cells, whereas it is completely inhibited in nup170Δ nup157Δ cells. Together, these data thus confirmed the requirement of Pom33 for efficient NPC assembly or stabilization.
Figure 9.
POM33 deletion impairs Nup84-GFP incorporation into NPCs upon Nup170 depletion. (A) Growth properties of wt, nup157Δ, and pom33Δ cells expressing NUP170 under the control of the PMET3 repressible promoter in a NUP84-GFP background. Serial dilutions were spotted on selective media lacking methionine (−Met) or on YEPD (+Met). (B) The localization of Nup84-GFP was analyzed in the indicated strains by spinning disk confocal microscopy before (−Met) or after a 6-h repression of NUP170 expression (+Met). Cytoplasmic Nup84-GFP foci are observed upon repression of NUP170 in nup157Δ and to a lower extent in pom33Δ cells (arrows, foci in mother cells; arrowheads, foci in buds). The percentage of cells with Nup84-GFP cytoplasmic foci is indicated. Bar, 5 µm.
Discussion
Pom33 is a novel integral membrane protein dynamically associated with nuclear pores
In this study, we identified the nonessential YLL023c/POM33 gene based on its functional interaction with nup133Δ. Immunofluorescence and EM studies further revealed that Pom33-GFP is highly enriched at the level of nuclear pores. In contrast, only a fraction of its paralogue, Ylr064w/Per33-GFP, associated with NPCs.
The identification of a fourth integral membrane protein of the yeast NPCs was surprising in view of the multiple high-throughput genetic, functional, and biochemical approaches that might have been anticipated to cover the entire NPC proteome. However, purification of NPC fractions followed by mass spectrometry (Rout et al., 2000; Alber et al., 2007b) may have missed Pom33 either because of technical limitations inherent to the small size of this highly hydrophobic protein, or because of its dynamic association with NPCs, a property revealed by our photobleaching experiments. In this respect, Pom33 somehow resembles gp210, a vertebrate transmembrane Nup that dynamically interacts with NPCs (Rabut et al., 2004).
Genetic and physical connections between Pom33, Rtns, and the Nup84 complex define a novel NPC-associated network required for NPC distribution
The most striking phenotype resulting from POM33 deletion was the clustering of the nuclear pores within a restricted region of the NE, which was not associated with any significant alteration in nucleocytoplasmic transport pathways. This clustering phenotype was observed in pom33Δ cells grown at all temperatures tested, and thus differed from the reversible phenotype described in mutants affecting peripheral Nups such as nup159-1, nup82Δ108, or gle2-1 (Gorsch et al., 1995; Murphy et al., 1996; Belgareh et al., 1998). EM images further revealed that pom33Δ cells do not exhibit major alterations of the NE, such as herniations documented previously for nup116Δ, gle2-1, gle2Δ, nup188Δ, and brr6–1 (Wente and Blobel, 1993; Murphy et al., 1996; Zabel et al., 1996; Bailer et al., 1998; de Bruyn Kops and Guthrie, 2001; Hodge et al., 2010) or other NE abnormalities reported for mutants in genes encoding NE/ER-associated proteins including Acc1, Spo7, Nem1, Npl4, or Apq12 (DeHoratius and Silver, 1996; Schneiter et al., 1996; Siniossoglou et al., 1998; Scarcelli et al., 2007). In addition, analysis of a representative panel of GFP-tagged Nups indicated that the clustered NPCs in pom33Δ cells apparently encompassed the entire Nup content. These NPC clusters are thus very distinct from the inner nuclear membrane–associated NPC-like structures or the punctuated cytoplasmic foci described in mutants of the Ndc1 interaction network or in apq12Δ cells, which only comprised subsets of Nups (Scarcelli et al., 2007; Flemming et al., 2009; Makio et al., 2009; Onischenko et al., 2009). Rather, the phenotype observed in the pom33Δ mutant strikingly resembles the one recently reported for rtn1Δ yop1Δ cells (Dawson et al., 2009) and for Nup84 complex mutants, although NE alterations leading to grapelike NPC clusters were also observed for some of the latter ones, notably at restrictive temperature (for review see Doye and Hurt, 1997).
Besides their closely related NPC clustering phenotype, both pom33Δ and rtn1Δ yop1Δ are synthetically lethal when combined with mutants of the Nup84 complex, although with slightly distinct patterns of genetic interactions (this paper; Dawson et al., 2009). It is noteworthy that a minor pool of Rtn1 reproducibly copurified with Pom33. This may reflect the fact that the interaction between Pom33 and Rtn1 is dynamic, or restricted to the NPC-associated fraction of Rtn1. Although this interaction might be indirect, it is remarkable that it specifically connects NPC-associated proteins whose depletion leads to very similar phenotypes.
Pom33 contributes to NPC biogenesis and/or stabilization
In addition to NPC clustering, measurements of the intensities of distinct GFP-tagged Nups in pom33Δ cells uncovered an altered NPC density in the daughter nucleus during telophase. A similarly altered M/B ratio was also observed for nup133Δ (this paper; Shcheprova et al., 2008) and for rtn1Δ yop1Δ (Nup84-GFP M/B ratio for rtn1Δ yop1Δ = 1.88 ± 0.67 compared with wt = 1.53 ± 0.46; P < 10−10; unpublished data). It cannot be formally excluded that the NPC clustering phenotype of these mutant cells may contribute to the transiently altered NPC density observed in the daughter nucleus. However, the involvement of Pom33 in efficient NPC biogenesis and/or stabilization was corroborated by the accumulation of Nup84-GFP in cytoplasmic foci upon depletion of Nup170 in pom33Δ cells, a previously reported readout for NPC assembly defects (Makio et al., 2009).
The implications of the Nup84 complex and of Rtn1/Yop1 in NPC assembly in yeast were inferred from in vitro data demonstrating the implications of their vertebrate orthologues, namely the Nup107-160 complex and Rtn4a, in de novo NPC biogenesis in interphase (D’Angelo et al., 2006; Dawson et al., 2009). So far, preliminary experiments based on siRNA-mediated depletion of HsTMEM33 in HeLa cells revealed no significant defects in NPC assembly (unpublished data). This might reflect the specific contribution of HsTMEM33 to efficient de novo but not postmitotic NPC assembly. However, as previously discussed (Madrid et al., 2006; Mansfeld et al., 2006; Stavru et al., 2006), and in view of the nonessential function of Pom33 in S. cerevisiae, the lack of a detectable phenotype upon HsTMEM33 depletion may also highlight redundancy among the transmembrane Nups, or a yet undefined function of TMEM33, possibly shared by S. cerevisiae Per33.
Pom33 association with membranes: a key to its function in NPC integrity?
As an integral membrane protein, Pom33, along with the other NPC-associated transmembrane proteins, may contribute to nuclear pore insertion in the lipid bilayer of the NE. This first hypothesis is supported by the genetic interactions occurring between POM33 and the Ndc1 interaction network, notably the complementation of the ndc1-39 phenotype upon overexpression of Pom33 or its paralogue Per33, a property possibly reflecting redundancy among pore membrane Nups (see also Miao et al., 2006; Dawson et al., 2009). This rescue further underscores the shared predicted topology of yeast Pom33, Per33, and Ndc1; i.e., three pairs of transmembrane segments flanked by N- and C-terminal domains exposed to the cytosolic/pore face of the NE (Fig. S1 B; Stavru et al., 2006; Lau et al., 2006; Kim et al., 2006).
However, only the first hydrophobic stretch within the primary sequence of Pom33 and Per33 is predicted with very high reliability to encompass two membrane-spanning segments (Fig. S1 B). As demonstrated for the vertebrate counterparts of Rtn1 and Yop1 (Rtn4c/NogoC and DP1, respectively), the two other hydrophobic domains of Pom33, Per33, and TMEM33 may adopt a hairpin-like structure in which hydrophobic segments do not fully span the membrane (Fig. S1 B; Voeltz et al., 2006). The resulting interaction of Pom33 with membranes could, as reported for the Rtns and Yop1/DP1 family members (Hu et al., 2008), induce and/or stabilize positive membrane curvatures required along with negative curvature for nuclear pore formation (discussed in Antonin, 2009). Unlike Rtns, Pom33 mainly localizes at the NPC and is thus properly positioned to act at the site of pore formation or stabilization. Interestingly, members of the Nup84 complex that are required for both NPC biogenesis and distribution also exhibit biochemical features, which suggests a role in stabilization or sensing of membrane curvatures (Devos et al., 2006; Drin et al., 2007; Kampmann and Blobel, 2009). Although affecting the Ndc1 interaction network may block NPC assembly at an early step (for review see Rexach, 2009), lack of Pom33, Rtns, or constituents of the Nup84 complex may impair stabilization of the pore–membrane interface at various stages of pore formation, thus leading to a delay in proper NPC assembly. In these mutants, defective curvature stabilization may ultimately allow assembly of complete NPCs, which could be further stabilized through clustering.
Further investigations of the biochemical properties of Pom33 with relation to membranes and its additional partners at NPCs will now be required, along with studies in other organisms, to decipher the specific function of this protein that contributes to proper distribution and to efficient assembly or stabilization of NPCs. After the first identification of NPC clustering yeast mutants in the 1990s, such studies may provide clues on the molecular mechanisms connecting NPC distribution and biogenesis in yeast.
Materials and methods
Yeasts strains and plasmids
The strains used are listed in Table S1. Deletion strains were obtained from the EUROSCARF deletion collection (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/), and GFP C-terminally tagged strains were obtained from Invitrogen (http://clones.invitrogen.com/cloneinfo.php?clone=yeastgfp). Unless otherwise indicated, all strains were obtained by successive crosses and are isogenic to S288C. Yeast cells were grown in standard yeast extract peptone dextrose (YEPD) or synthetic complete (SC) media lacking appropriate amino acids. Transformation, mating, and sporulation were performed as described previously (Palancade et al., 2005). Construction of plasmids used here (Table S2) was performed using standard molecular cloning techniques.
Purification of Pom33/34-ProtA-associated proteins and mass spectrometry analysis
Pom33 or Pom34 tagged with IgG-binding domains of the Staphylococcus aureus ProtA were affinity-purified from whole cell lysates using IgG-conjugated magnetic beads essentially as described in Alber et al. (2007a). In brief, 2.4 g of frozen yeast grindate was homogenized in 10 ml of extraction buffer (20 mM Hepes, pH 7.5, 110 mM KOAc, 2 mM MgCl2, 0.1% Tween 20, and 1% Triton X-100) supplemented with 1 mM DTT, 0.5 mM PMSF, 2 µg/µl Leupeptine, and 1× protease inhibitor cocktail (complete EDTA-free; Roche). The soluble fraction, isolated by centrifugation at 2,000 gmax, at 4°C for 15 min, was incubated for 30 min at 4°C with IgG-conjugated magnetic beads. The beads were then washed three times with extraction buffer, three additional times with extraction buffer supplemented with 150 mM NaCl and 1 mg/ml heparin, and once with 0.1 M NH4OAc, 0.1 mM MgCl2, and 0.02% Tween 20. The proteins were eluted with 0.5 M NH4OH and 0.5 mM EDTA. The eluates were lyophilized and resuspended in SDS-PAGE sample buffer. The proteins were then separated on 4–12% SDS-PAGE gels and either silver stained or transferred to nitrocellulose filters. The resulting blots were probed with the following antibodies: 1:5,000 rabbit IgG-HRP polyclonal antibody (PAP; code Z0113; Dako), 1:500 anti-GFP (monoclonal, clones 7.1/13.1; Roche), 1:100 anti-Pom152 (mAb118C3; gift from M. Rout, The Rockefeller University, New York, NY; Strambio-de-Castillia et al., 1995), 1:200 anti-Nop1 (mAbA66; Tollervey et al., 1991), or 1:1,000 anti-Dpm1 (monoclonal 5C5; Invitrogen). The blots were saturated with TBS, 0.1% Tween, 5% dried milk, and, except for ProtA detection, 10% human serum. Antibodies were detected with anti–mouse-HRP (Jackson ImmunoResearch Laboratories, Inc.).
For mass spectrometry analyses, gel slices were reduced, alkylated, and subjected to digestion with trypsin (Roche) as described previously (Fevrier et al., 2004). The extracted peptides were dried and resolubilized in solvent A (95:5 water/acetonitrile in 0.1% [wt/vol] formic acid). The total digestion product of a gel slice was used for liquid chromatography-tandem MS (LC-MS/MS) analysis. The extracted peptides were concentrated and separated on an HPLC system (LC Packings; Dionex) coupled to the nano-electrospray II ionization interface of a mass spectrometer (QSTAR Pulsar i; Applied Biosystems) using a PicoTip emitter (10 µm in diameter; New Objectives). HPLC mobile phases contained solvent A and solvent B (20:80 water/acetonitrile in 0.085% [wt/vol] formic acid). Bound peptides were eluted with a gradient of 5–50% of solvent B. Information-dependent acquisition was used to acquire MS/MS data, with the experiments designed such that the two most abundant peptides were subjected to collision-induced dissociation, using nitrogen as collision gas. Data from the information-dependent acquisition experiments were searched twice using MASCOT (Matrix Science) and PHENYX (Geneva Bioinformatics) software on the Saccharomyces Genome Database (SGD ORF 062007). All data were manually validated using myProMS (Poullet et al., 2007).
Fluorescence microscopy of yeast and HeLa cells
Yeast live cell imaging was performed using exponentially growing cultures. Nuclear import assays were performed according to Shulga et al. (1996) after metabolic poisoning of Nab2-NLS-GFP–expressing cells in the presence of deoxyglucose and sodium azide at 4°C. FISH to detect poly(A)+ RNAs was performed essentially as described previously (Doye et al., 1994), except that the (dT)50 probe used was directly coupled to Cy3, and that the methanol/acetone fixation step was replaced by a treatment with 70% ethanol.
HeLa cells were cultured as described previously (Loïodice et al., 2004). Plasmid transfections were performed using Lipofectamine 2000 according to manufacturer’s instructions (Invitrogen). To establish stable cell lines, individual clones were isolated after transfection by applying G418 (0.5 mg/ml) selection. Immunofluorescence was performed on paraformaldehyde-fixed HeLa cells as described previously (Loïodice et al., 2004) using the following primary antibodies: anti-BAP31 (mouse monoclonal antibody No. A1/182; Enzo Life Sciences, Inc.), anti-RanGAP1 (goat polyclonal antibody, clone N-19; Santa Cruz Biotechnology, Inc.), anti-PDI (mouse monoclonal antibody, clone No. 1D3; Enzo Life Sciences) and anti-GFP (rabbit polyclonal antibody; Invitrogen). For digitonin treatment, the cells were fixed in 3% paraformaldehyde for 20 min and permeabilized with 0.004% digitonin for 5 min.
Wide-field fluorescence images were acquired using a microscope (DM6000B; Leica) with a 100×, NA 1.4 (HCX Plan-Apo) oil immersion objective and a charge-coupled device camera (CoolSNAP HQ; Photometrics). Confocal GFP and mCherry fluorescence images were acquired on spinning disk microscopes (Ultraview CSU10; Leica [described in Katsani et al., 2008]; or Eclipse TE2000-U; Nikon [described in Almonacid et al., 2009]) using a 491-nm laser and a 561-nm laser, respectively, and a triple-band filter (515–535, 590–615, and 690–730). The corresponding differential interference contrast (DIC) images were also acquired. Images were scaled equivalently using MetaMorph 5 (Universal Imaging) and processed with Photoshop CS2 9.0 software (Adobe).
FLIP experiments
Yeast cells grown on YEPD plates were resuspended in SC medium and immobilized on an agar pad containing SC medium and 1.6% agar. The cells were imaged on a confocal microscope (LSM510; Carl Zeiss, Inc.) using a Plan-Apochromat 63× objective, typically using 2% of the laser intensity of an argon laser (488 nm) at 45% output. Photobleaching was applied on selected areas within the cortical ER as indicated on Fig. 2 A. Bleaching pulses were iterated at 30% of the laser intensity at 45% output.
Quantification of fluorescence decay in the ER and in the nucleus over time was performed using MetaMorph (MDS Analytical Technologies) software. At each time point (i.e., after each pulse), the total fluorescence intensities within the entire cell and within the nucleus were measured, corrected for background signal, and normalized over the value of an unbleached neighboring cell (to compensate for loss of fluorescence due to image acquisition). ER fluorescence was calculated as total cell minus nuclear signal. ER and nuclear values were normalized to 100% at time 0 and graphs were generated using Excel (Microsoft).
Quantification of M/B ratio
To assess NPC assembly in the daughter nuclei of telophase yeast cells, freshly thawed or transformed cells were systematically used. Quantifications of spinning disk confocal images were performed using the MetaMorph software on additive projections of 13 z stacks spaced by 0.8 µm, sampling the entire nuclei. Total fluorescence intensities within circular regions of 30 pixels placed on the mother or daughter/bud nuclei or in the intercellular background were measured, and M/B ratios were calculated as M/B = (Mother − background)/(Bud − background). Box plots were generated using KaleidaGraph (Synergy Software): each box encloses 50% of the M/B ratios obtained, centered on the median value. The bars extending from the top and bottom of each box mark the minimum and maximum values within the dataset falling within an acceptable range. Values falling outside of this range are displayed as an individual point. Unpaired student’s t tests were used to evaluate differences in M/B ratios between wt and mutant cells.
Thin-section EM
For structural studies, parental (BY4742) and pom33Δ yeast strains were grown in YEPD at 30°C to early log phase and processed for EM as described previously (Doye et al., 1994). Ultrathin sections (60–70 nm) of the embedded samples were cut on an ultramicrotome (UC6; Leica) and collected on copper grids (200 mesh). The samples were contrasted by staining with uranyl acetate and citrate lead, and examined on a transmission electron microscope (Tecnai 12 Bio Twin; FEI) equipped with a charge-coupled device camera (Keenview; Olympus).
Immunogold labeling of yeast cryosections was performed by the Tokuyasu method (Slot and Geuze, 2007), adapted for yeast (Griffith et al., 2008) with some modifications. In brief, pellets from yeast strains grown to log phase were resuspended in fixative (2% [wt/vol] PFA in 0.1 M phosphate buffer) and incubated for 1 h at room temperature on a roller. Fixation was continued overnight at 4°C. After three washes with PBS, the pellets were resuspended in 1 ml of freshly prepared 1% periodic acid in PBS and incubated at room temperature for 1 h on a roller. Cells were washed again three times in PBS and embedded in 12% gelatin. Gelatin blocks were infused with 2.3 M sucrose at 4°C and frozen in liquid nitrogen. Ultrathin cryosections were double-labeled with polyclonal rabbit anti-GFP (1:200; Abcam), mAb414 (1/400; Covance) antibodies, and ProtA coupled to 10- or 15-nm gold particles (obtained from the Cell Microscopy Center, Utrecht Medical School), and examined as described in the previous paragraph. For statistical analyses, sections of yeast cells passing through the nuclei were randomly acquired. For each strain, a similar number of NPCs per nuclear section (three to four on average) were identified based on mAb414 labeling and/or a typical NE structure. Anti-GFP–bound gold particles were quantified on the same sections and scored as being (1) out of the NE, (2) at the NE but not colocalizing with NPCs, or (3) at NPCs. Note that a significant fraction of the gold particles localized out of the NE reflect nonspecific binding of the anti-GFP antibodies, as revealed by the analysis of control cells expressing no GFP fusion. The fraction of gold particles scored as being at NPCs might be underestimated due to the absence of mAb414 labeling or to the nonoptimal preservation of the NE structure.
Online supplemental material
Fig. S1 shows experimental determination of HsTMEM33 topology in differentially permeabilized HeLa cells stably expressing HsTMEM33-GFP or GFP-HsTMEM33, and in silico predictions of ScPom33, ScPer33, HsTMEM33, and HsRtn4c topologies. Fig. S2 shows the growth properties of the pom33Δ and/or per33Δ in combination with various other mutants. Fig. S3 shows nucleocytoplasmic transport assays in the pom33Δ mutant; i.e., nuclear import kinetics of a Nab2-NLS-GFP reporter and FISH visualization of mRNA export. Fig. S4 provides control experiments for M/B ratio measurements in pom33Δ cells. Table S1 details all the yeast strains used. Table S2 details all the plasmids used. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200910043/DC1.
Acknowledgments
We are very grateful to E. Doumazane, S. Loeillet, and A. Nicolas for valuable input during the first stages of this project; to the staffs of the Institut Curie and the Institut Jacques Monod Imaging Platforms; to S. Dokudovskaya and W. Faigle for their efficient help with imaging and biochemical approaches; to C. Cole, D. Goldfarb, C. Guthrie, P. Hentges, U. Kutay, S. Leon, S. Osmani, A. Paoletti, A. Schmidt, M. Rout, A. Taddei, R. Tsapis, R. Tsien, S. Wente, M. Winey, and R. Wozniak for sharing reagents and/or discussion; and to A-L Haenni for careful reading of the manuscript.
This research was supported by the Centre National de la Recherche Scientifique, the Institut Curie, the Ligue Nationale contre le Cancer (to V. Doye, Equipe Labelisée 2006 and 2009), the Agence Nationale de la Recherche (ANR 07-BLAN-0063-01 to V. Doye), and the Eidgenössische Technische Hochschule Zurich (to Y. Barral and B. Hess). A. Chadrin was the recipient of PhD fellowships from Ministère de l’Enseignement Supérieur et de la recherche and from the Association pour la Recherche contre le Cancer.
Footnotes
Abbreviations used in this paper:
- DIC
- differential interference contrast
- FLIP
- fluorescence loss in photobleaching
- M/B
- mother/bud
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- Nup
- nucleoporin
- ORF
- open reading frame
- PDI
- protein disulfide isomerase
- ProtA
- protein A
- Rtn
- reticulon
- SC
- synthetic complete
- YEPD
- yeast extract peptone dextrose
References
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007a. Determining the architectures of macromolecular assemblies. Nature. 450:683–694 10.1038/nature06404 [DOI] [PubMed] [Google Scholar]
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007b. The molecular architecture of the nuclear pore complex. Nature. 450:695–701 10.1038/nature06405 [DOI] [PubMed] [Google Scholar]
- Almonacid M., Moseley J.B., Janvore J., Mayeux A., Fraisier V., Nurse P., Paoletti A. 2009. Spatial control of cytokinesis by Cdr2 kinase and Mid1/anillin nuclear export. Curr. Biol. 19:961–966 10.1016/j.cub.2009.04.024 [DOI] [PubMed] [Google Scholar]
- Anderson D.J., Hetzer M.W. 2008. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J. Cell Biol. 182:911–924 10.1083/jcb.200805140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W. 2009. Nuclear envelope: membrane bending for pore formation? Curr. Biol. 19:R410–R412 10.1016/j.cub.2009.03.053 [DOI] [PubMed] [Google Scholar]
- Antonin W., Ellenberg J., Dultz E. 2008. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 582:2004–2016 10.1016/j.febslet.2008.02.067 [DOI] [PubMed] [Google Scholar]
- Aris J.P., Blobel G. 1989. Yeast nuclear envelope proteins cross react with an antibody against mammalian pore complex proteins. J. Cell Biol. 108:2059–2067 10.1083/jcb.108.6.2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A., Desai A., Oegema K. 2007. A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 178:43–56 10.1083/jcb.200701139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailer S.M., Siniossoglou S., Podtelejnikov A., Hellwig A., Mann M., Hurt E. 1998. Nup116p and nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor gle2p. EMBO J. 17:1107–1119 10.1093/emboj/17.4.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N., Snay-Hodge C., Pasteau F., Dagher S., Cole C.N., Doye V. 1998. Functional characterization of a Nup159p-containing nuclear pore subcomplex. Mol. Biol. Cell. 9:3475–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsel A., Viklund H., Hennerdal A., Elofsson A. 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37:W465–468 10.1093/nar/gkp363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S.G., Partridge J.R., Whittle J.R., Schwartz T.U. 2009. The nuclear pore complex has entered the atomic age. Structure. 17:1156–1168 10.1016/j.str.2009.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial H.J., Rout M.P., Giddings T.H., Winey M. 1998. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 143:1789–1800 10.1083/jcb.143.7.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw J.M., Krutchinsky A.N., Zhang W., Chait B.T., Matunis M.J. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158:915–927 10.1083/jcb.200206106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo M.A., Hetzer M.W. 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18:456–466 10.1016/j.tcb.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo M.A., Anderson D.J., Richard E., Hetzer M.W. 2006. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 312:440–443 10.1126/science.1124196 [DOI] [PubMed] [Google Scholar]
- Dawson T.R., Lazarus M.D., Hetzer M.W., Wente S.R. 2009. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J. Cell Biol. 184:659–675 10.1083/jcb.200806174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyn Kops A., Guthrie C. 2001. An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport. EMBO J. 20:4183–4193 10.1093/emboj/20.15.4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHoratius C., Silver P.A. 1996. Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol. Biol. Cell. 7:1835–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vit M.J., Waddle J.A., Johnston M. 1997. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell. 8:1603–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Williams R., Alber F., Eswar N., Chait B.T., Rout M.P., Sali A. 2006. Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 103:2172–2177 10.1073/pnas.0506345103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V., Hurt E. 1997. From nucleoporins to nuclear pore complexes. Curr. Opin. Cell Biol. 9:401–411 10.1016/S0955-0674(97)80014-2 [DOI] [PubMed] [Google Scholar]
- Doye V., Wepf R., Hurt E.C. 1994. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 13:6062–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G., Casella J.F., Gautier R., Boehmer T., Schwartz T.U., Antonny B. 2007. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 14:138–146 10.1038/nsmb1194 [DOI] [PubMed] [Google Scholar]
- Fernandez-Martinez J., Rout M.P. 2009. Nuclear pore complex biogenesis. Curr. Opin. Cell Biol. 21:603–612 10.1016/j.ceb.2009.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fevrier B., Vilette D., Archer F., Loew D., Faigle W., Vidal M., Laude H., Raposo G. 2004. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. USA. 101:9683–9688 10.1073/pnas.0308413101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming D., Sarges P., Stelter P., Hellwig A., Böttcher B., Hurt E. 2009. Two structurally distinct domains of the nucleoporin Nup170 cooperate to tether a subset of nucleoporins to nuclear pores. J. Cell Biol. 185:387–395 10.1083/jcb.200810016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsch L.C., Dockendorff T.C., Cole C.N. 1995. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 129:939–955 10.1083/jcb.129.4.939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J., Mari M., De Mazière A., Reggiori F. 2008. A cryosectioning procedure for the ultrastructural analysis and the immunogold labelling of yeast Saccharomyces cerevisiae. Traffic. 9:1060–1072 10.1111/j.1600-0854.2008.00753.x [DOI] [PubMed] [Google Scholar]
- Hodge C.A., Choudhary V., Wolyniak M.J., Scarcelli J.J., Schneiter R., Col C.N. 2010. Integral membrane proteins Brr6 and Apq12 link assembly of the nuclear pore complex to lipid homeostasis in the endoplasmic reticulum. J. Cell Sci. 123:141–151 10.1242/jcs.055046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., Kozlov M.M., Rapoport T.A., Prinz W.A. 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 319:1247–1250 10.1126/science.1153634 [DOI] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Kampmann M., Blobel G. 2009. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat. Struct. Mol. Biol. 16:782–788 10.1038/nsmb.1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsani K.R., Karess R.E., Dostatni N., Doye V. 2008. In vivo dynamics of Drosophila nuclear envelope components. Mol. Biol. Cell. 19:3652–3666 10.1091/mbc.E07-11-1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Melén K., Osterberg M., von Heijne G. 2006. A global topology map of the Saccharomyces cerevisiae membrane proteome. Proc. Natl. Acad. Sci. USA. 103:11142–11147 10.1073/pnas.0604075103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E., Morozova K.N., Voeltz G.K., Allen T.D., Goldberg M.W. 2007. Reticulon 4a/NogoA locates to regions of high membrane curvature and may have a role in nuclear envelope growth. J. Struct. Biol. 160:224–235 10.1016/j.jsb.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Hetzer M.W. 2008. Reorganization of the nuclear envelope during open mitosis. Curr. Opin. Cell Biol. 20:669–677 10.1016/j.ceb.2008.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C.K., Giddings T.H., Jr, Winey M. 2004. A novel allele of Saccharomyces cerevisiae NDC1 reveals a potential role for the spindle pole body component Ndc1p in nuclear pore assembly. Eukaryot. Cell. 3:447–458 10.1128/EC.3.2.447-458.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C.K., Delmar V.A., Forbes D.J. 2006. Topology of yeast Ndc1p: predictions for the human NDC1/NET3 homologue. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 288:681–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A., Felberbaum R., Hochstrasser M. 2007. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell Biol. 178:813–827 10.1083/jcb.200702154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeillet S., Palancade B., Cartron M., Thierry A., Richard G.-F., Dujon B., Doye V., Nicolas A. 2005. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair (Amst.) 4:459–468 10.1016/j.dnarep.2004.11.010 [DOI] [PubMed] [Google Scholar]
- Loïodice I., Alves A., Rabut G., Van Overbeek M., Ellenberg J., Sibarita J.B., Doye V. 2004. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell. 15:3333–3344 10.1091/mbc.E03-12-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid A.S., Mancuso J., Cande W.Z., Weis K. 2006. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J. Cell Biol. 173:361–371 10.1083/jcb.200506199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makio T., Stanton L.H., Lin C.C., Goldfarb D.S., Weis K., Wozniak R.W. 2009. The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J. Cell Biol. 185:459–473 10.1083/jcb.200810029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J., Güttinger S., Hawryluk-Gara L.A., Panté N., Mall M., Galy V., Haselmann U., Mühlhäusser P., Wozniak R.W., Mattaj I.W., et al. 2006. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell. 22:93–103 10.1016/j.molcel.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Miao M., Ryan K.J., Wente S.R. 2006. The integral membrane protein Pom34p functionally links nucleoporin subcomplexes. Genetics. 172:1441–1457 10.1534/genetics.105.052068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R., Watkins J.L., Wente S.R. 1996. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol. Biol. Cell. 7:1921–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E., Stanton L.H., Madrid A.S., Kieselbach T., Weis K. 2009. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol. 185:475–491 10.1083/jcb.200810030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P., Albright C., Robbins P.W. 1988. Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J. Biol. Chem. 263:17499–17507 [PubMed] [Google Scholar]
- Palancade B., Zuccolo M., Loeillet S., Nicolas A., Doye V. 2005. Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol. Biol. Cell. 16:5258–5268 10.1091/mbc.E05-06-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B., Liu X., Garcia-Rubio M., Aguilera A., Zhao X., Doye V. 2007. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell. 18:2912–2923 10.1091/mbc.E07-02-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poullet P., Carpentier S., Barillot E. 2007. myProMS, a web server for management and validation of mass spectrometry-based proteomic data. Proteomics. 7:2553–2556 10.1002/pmic.200600784 [DOI] [PubMed] [Google Scholar]
- Rabut G., Lénárt P., Ellenberg J. 2004. Dynamics of nuclear pore complex organization through the cell cycle. Curr. Opin. Cell Biol. 16:314–321 10.1016/j.ceb.2004.04.001 [DOI] [PubMed] [Google Scholar]
- Rexach M. 2009. Piecing together nuclear pore complex assembly during interphase. J. Cell Biol. 185:377–379 10.1083/jcb.200904022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., Chait B.T. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651 10.1083/jcb.148.4.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.J., McCaffery J.M., Wente S.R. 2003. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 160:1041–1053 10.1083/jcb.200209116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.J., Zhou Y., Wente S.R. 2007. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol. Biol. Cell. 18:886–898 10.1091/mbc.E06-06-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli J.J., Hodge C.A., Cole C.N. 2007. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J. Cell Biol. 178:799–812 10.1083/jcb.200702120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R., Hitomi M., Ivessa A.S., Fasch E.-V., Kohlwein S.D., Tartakoff A.M. 1996. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol. Cell. Biol. 16:7161–7172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcheprova Z., Baldi S., Frei S.B., Gonnet G., Barral Y. 2008. A mechanism for asymmetric segregation of age during yeast budding. Nature. 454:728–734 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Voss C., Rist J.M., Hu J., Rapoport T.A., Prinz W.A., Voeltz G.K. 2008. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283:18892–18904 10.1074/jbc.M800986200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N., Roberts P., Gu Z., Spitz L., Tabb M.M., Nomura M., Goldfarb D.S. 1996. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 135:329–339 10.1083/jcb.135.2.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., Hurt E. 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17:6449–6464 10.1093/emboj/17.22.6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot J.W., Geuze H.J. 2007. Cryosectioning and immunolabeling. Nat. Protoc. 2:2480–2491 10.1038/nprot.2007.365 [DOI] [PubMed] [Google Scholar]
- Stade K., Ford C.S., Guthrie C., Weis K. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 90:1041–1050 10.1016/S0092-8674(00)80370-0 [DOI] [PubMed] [Google Scholar]
- Stavru F., Hülsmann B.B., Spang A., Hartmann E., Cordes V.C., Görlich D. 2006. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 173:509–519 10.1083/jcb.200601001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel G., Rout M.P. 1995. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. J. Cell Biol. 131:19–31 10.1083/jcb.131.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus L.C., Dawson T.R., Rexer D.J., Ryan K.J., Wente S.R. 2010. Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization. Mol. Biol. Cell. 21:1072–1087 10.1091/mbc.E09-07-0615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Carmo-Fonseca M., Hurt E.C. 1991. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 10:573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E.J., Wente S.R. 2006. Dynamic nuclear pore complexes: life on the edge. Cell. 125:1041–1053 10.1016/j.cell.2006.05.027 [DOI] [PubMed] [Google Scholar]
- Vasu S., Shah S., Orjalo A., Park M., Fischer W.H., Forbes D.J. 2001. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155:339–354 10.1083/jcb.200108007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., Rapoport T.A. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Vollmar F., Hacker C., Zahedi R.P., Sickmann A., Ewald A., Scheer U., Dabauvalle M.C. 2009. Assembly of nuclear pore complexes mediated by major vault protein. J. Cell Sci. 122:780–786 10.1242/jcs.039529 [DOI] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. 2009. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 25:1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S.R., Blobel G. 1993. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol. 123:275–284 10.1083/jcb.123.2.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.S., Strittmatter S.M. 2007. The reticulons: a family of proteins with diverse functions. Genome Biol. 8:234 10.1186/gb-2007-8-12-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel U., Doye V., Tekotte H., Wepf R., Grandi P., Hurt E.C. 1996. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J. Cell Biol. 133:1141–1152 10.1083/jcb.133.6.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]