Abstract
The extensive membrane network of the endoplasmic reticulum (ER) is physically juxtaposed to and functionally entwined with essentially all other cellular compartments. Therefore, the ER must sense diverse and constantly changing physiological inputs so it can adjust its numerous functions to maintain cellular homeostasis. A growing body of new work suggests that the unfolded protein response (UPR), traditionally charged with signaling protein misfolding stress from the ER, has been co-opted for the maintenance of basal cellular homeostasis. Thus, the UPR can be activated, and its output modulated, by signals far outside the realm of protein misfolding. These findings are revealing that the UPR causally contributes to disease not just by its role in protein folding but also through its broad influence on cellular physiology.
Physical and functional interconnectivity of the ER
The ER is a central coordinator of diverse cellular processes. Most notably, the ER acts as gatekeeper to the secretory pathway by folding, modifying, and exporting nascent secretory and transmembrane proteins, together encompassing ∼30% of the cellular proteome, en route to their final destinations within the endomembrane system. The ER also stores calcium for localized release by second messenger cascades. Lipogenic reactions (including those involved with synthesis of triacylglycerols, sterols, ceramides, and most cellular phospholipids) occur on the cytosolic face of the ER membrane, as does fatty acid desaturation. Enzymes involved in gluconeogenesis and specialized pathways of fatty acid oxidation are also housed at the ER. The ER forms the nuclear envelope and can contribute to biogenesis of peroxisomes, lipid droplets, and autophagic membranes. The ER makes close contacts with every other membranous structure in the cell, and these contacts likely facilitate the bidirectional transfer of lipids, calcium, and other molecules. This physical and functional interconnectivity imparts upon the ER a central role in the maintenance of numerous aspects of cellular and organismal homeostasis. Thus, disruption of ER function broadly impacts cellular function, and disruptions in other cellular processes typically redound to the ER.
Because the ER has to effectively carry out its many functions even as external conditions act upon it, it must communicate bidirectionally with other cellular signaling cascades. The organelle must be able to sense and integrate information about the nature, intensity, and duration of perturbations to it and to adjust cellular pathways accordingly to maintain homeostasis. The ER response to nutrient flux necessarily differs from its response to infection by a pathogen that usurps the secretory pathway or from its response to a differentiation stimulus that necessitates ER expansion. It is now becoming clear that the unfolded protein response (UPR), a signaling pathway from the ER to the nucleus which is best characterized as a stress response, has been drafted into the role of regulator of basal homeostasis and that the UPR provides a signaling framework into which other cellular pathways are intimately integrated. In this review, we discuss recent findings that shed light on how different physiological stimuli can activate the UPR, how intersection between the UPR and other signaling cascades allows its output to be modulated, and how the UPR might contribute to diseases beyond those that can be readily rationalized as protein folding disorders. Thus, we strive to highlight the underappreciated but emerging role of the UPR in buffering normal fluctuations in cellular state to facilitate maintenance of cellular homeostasis.
Evolutionary expansion of ER signal transduction
The observation that toxin-mediated impairment of ER protein folding leads to transcriptional induction of ER chaperones spurred the search for signaling mechanisms from the ER to the nucleus (Kozutsumi et al., 1988; Dorner et al., 1990). This stress paradigm proved to be a powerful guide in uncovering the ER signal transduction pathways that collectively became known as the UPR (for review see Ron and Walter, 2007). In yeast, the UPR is defined by a single linear pathway initiated by the ER-resident transmembrane kinase Ire1p and the downstream transcription factor Hac1p (Sidrauski et al., 1998). Transcriptional profiling has delineated the hundreds of Hac1p targets (i.e., the output of the yeast UPR; Travers et al., 2000), whereas comprehensive whole-genome analysis has identified genes that, when disrupted, activate the UPR (i.e., pathways that generate inputs for the UPR; Jonikas et al., 2009). Not unexpectedly, the inputs and outputs partially overlap and, to a first approximation, can be largely explained by a simple model in which the inputs impinge upon ER protein folding, whereas the output represents a transcriptional program designed to augment ER functionality and restore the organelle to its prestress state. In this context, the UPR acts as a closed signaling loop, with discrete and self-limited start and endpoints.
This core UPR has been markedly expanded during vertebrate evolution in three qualitatively different ways (Fig. 1). First, multiple ER-resident stress sensors in addition to IRE1 initiate complementary signaling pathways, all of which can regulate transcription. Second, UPR output is regulated by multiple means, both transcriptional and nontranscriptional. Third, many components of these UPR signaling cascades are themselves subject to regulation in trans by other cellular pathways. These evolutionary increases in complexity have greatly widened the scope of UPR signal transduction in terms of both the broader range of physiological inputs that influence UPR activity and the more nuanced outputs that are customized to cellular need. The major consequence of vastly expanded UPR signaling is the regulatory influence of cellular pathways that function well outside the realm of ER protein processing. Thus, it is increasingly clear that what began as an organelle-specific stress response has expanded into a complex signaling network that plays a central homeostatic role in normal vertebrate physiology. To provide an understanding of these newly emerging complexities of UPR regulation, we begin with an overview of the basic vertebrate UPR signaling pathways.
Figure 1.
Evolutionary expansion of the UPR allows for regulatory modulation and variable inputs and outputs. The evolutionarily conserved IRE1 arm of the UPR (green) has been expanded in metazoans by the parallel PERK and ATF6 pathways (blue). Each of these stress transducers is sensitive to protein misfolding stress in the ER, and they collectively contribute to gene regulation and UPR output. Each transducer can also interact with additional factors (orange) shared with signaling pathways responsive to other stimuli. These accessory interactions can thereby modulate the canonical stress response output contingent on cellular conditions. Physiological stimuli can further shape UPR outputs by differentially acting on the UPR at both proximal and distal steps. Two illustrative examples are depicted using red and black dashed lines. In the first example, TLR engagement activates all three UPR transducers, while simultaneously suppressing ATF4 production (Woo et al., 2009). TLR-stimulated UPR-independent pathways operate in parallel, such that TLR stimulation influences and is influenced by UPR activation to determine the final output. The second example illustrates plasma cell differentiation, where a differentiation stimulus initiates a differentiation program that includes selective activation of the ATF6-α and IRE1-α pathways of the UPR. The UPR pathways increase ER protein processing capacity, thereby facilitating the high level of antibody production and secretion that accompanies differentiation (Gass et al., 2008; Ma et al., 2010). The selectivity of UPR activation in this case precludes PERK-mediated responses, which might act at cross-purposes to differentiation.
The vertebrate UPR
The vertebrate UPR uses three ubiquitous branches defined by the ER-resident transmembrane proteins IRE1-α, PERK, and ATF6-α. Although the PERK and ATF6-α pathways also exist in simpler metazoans such as flies and worms, they appear to carry out more specialized functions than their vertebrate counterparts. During acute ER perturbation, these pathways rapidly reduce protein import into the ER by several distinct mechanisms. The best described of these is a robust and nonspecific inhibition of most mRNA translation as a consequence of eIF2-α phosphorylation by PERK (Harding et al., 2000). ER load is also attenuated by IRE1-α–mediated degradation of select ER-associated mRNAs, a process named regulated IRE1-dependent decay (RIDD; Hollien and Weissman, 2006). Finally, preemptive quality control selectively inhibits the translocation of certain proteins into the ER depending on properties of their ER targeting signal peptides (Kang et al., 2006). The possibility that these mechanisms might act in a more substrate-specific way during physiological UPR activation is conceptually appealing but has not been explored in depth.
Each UPR pathway also culminates in transcriptional regulation via unique mechanisms (for review see Ron and Walter, 2007): activated IRE1-α facilitates removal of a short intron from Xbp1 mRNA, allowing translation of full-length XBP1; activated ATF6-α is processed by regulated intramembrane proteolysis to liberate its N-terminal transcriptional activator domain; and activated PERK phosphorylates the translation initiation factor eIF2-α, which stimulates translation of ATF4. Together (and with considerable overlap among them), XBP1, ATF6-α, and ATF4 regulate genes encoding ER chaperones, ER-associated degradation factors, amino acid transport and metabolism proteins, phospholipid biosynthesis enzymes, and numerous others, including many that have no obvious direct relationship to secretory pathway function. The vertebrate IRE1-α and ATF6-α paralogues (IRE1-β and ATF6-β) add further complexity (Bertolotti et al., 2001; Thuerauf et al., 2007), as do several tissue-specific ATF6-like ER-resident transcription factors (such as CREB-H, OASIS, Luman, and others) that can be activated by regulated intramembrane proteolysis (Bailey and O’Hare, 2007). Although these paralogues and orthologues remain relatively poorly studied to date, it is clear that they can be responsive to changes in ER physiology and are increasingly considered under the general UPR umbrella (e.g., Zhang et al., 2006). Thus, even the aforementioned oversimplified description of the vertebrate UPR highlights the considerable complexity of signal transduction emanating from the ER.
Vertebrate diversity necessitates an expanded UPR
One reason for contrasting the yeast and vertebrate UPR pathways, besides providing essential background, is to consider why the latter has evolved such elaborate complexity when a single pathway analogous to the yeast UPR should be sufficient to up-regulate ER chaperones when protein folding is disrupted. The most plausible explanation is that an expanded UPR provides greater flexibility: a wider range of inputs can be accommodated by the multiple overlapping but distinct pathways, and the outputs of multiple, branched, and intersecting signaling pathways can be finely tuned to cellular need (Fig. 1).
This greater level of regulation in vertebrates is necessitated by the diversity of differentiated cell types, the individual requirements of these cell types for proper function, the considerably different environments in which these cells normally operate, and the different ranges in environmental parameters that these cells can expect to encounter. For example, some cell types (e.g., differentiated neurons) enjoy remarkably constant nutrient supplies, steady demands on the secretory pathway, a lack of cell division, and a minimum of morphological changes that require modification of the membrane production program. In contrast, other cells experience widely ranging flux through the secretory pathway (e.g., endocrine cells), substantial variations in the types of nutrients that are metabolized (e.g., hepatocytes), regular exposure to various toxic substances (e.g., macrophages), or widely varying oxygen tensions (e.g., alveolar cells). Thus, the normal range of physiological and environmental parameters faced by one cell type may be vastly different than that encountered by another cell type in the same organism.
This diversity among cell types means that some cells regularly adjust the functionality and capacity of their ER, whereas others maintain a comparatively constant steady-state (not unlike a typical tissue-culture cell). Consequently, UPR signaling is almost certainly used even during the course of normal organismal physiology to tune ER functionality in response to changing demands. Indeed, it is increasingly evident that in vertebrates, UPR signaling can be initiated without necessarily exceeding the protein-processing capacity of the ER, the traditional definition of ER stress. Thus, the stress paradigm, which we define for the purposes of this review as the experience of ER conditions that lie outside the physiological range of environments normally faced by that cell, probably does not accurately capture the major physiological roles of the vertebrate UPR. Although the UPR is certainly a necessary element of the protective response against pathological conditions (for review see Otsu and Sitia, 2007), here we focus on how normal physiological signaling intersects with the UPR to regulate its activation and its output, toward the goal of maintaining normal cellular function. In this view, the input is defined by the physiological stimuli that activate part or all of the UPR, whereas the output constitutes the adjustments to cellular function, mediated by both transcriptional and nontranscriptional mechanisms.
Physiological engagement of UPR pathways
To be used for physiological regulation and to have sufficient flexibility given the multitude of ER functions, the UPR must be capable of discriminating among a broad variety of activation stimuli (inputs) and initiating appropriate responses. Although positive and negative feedback loops within the UPR can intrinsically produce distinct outputs in proportion to the strength of input (Rutkowski et al., 2006), the ways in which the stress sensors are activated by physiological stimuli must necessarily be more nuanced than can be accounted for by postulating a single uniform mechanism of activation. Although client load in excess of the folding capacity of ER chaperones (in particular BiP) is generally thought of as the primary inducer of the UPR, this explanation seems limited for at least two reasons. First, misfolded proteins as the sole input would make tuning the response in all but the most limited manner quite difficult, and it would not provide for discrimination among qualitatively distinct inputs. Consistent with this idea, pharmacological perturbants that broadly impair ER protein folding activate either all three UPR pathways together or none at all if the dose is insufficient (Rutkowski et al., 2006). Second, according to a simple misfolded protein accumulation mechanism, the UPR would necessarily be reactive to altered conditions and could never be proactively induced. Although a strictly reactive system is appropriate when the need for a response is rare, unpredictable, and intermittent, it seems less desirable when requisite changes to ER functionality can be fully anticipated as part of normal physiology. Indeed, requiring ER impairment in order for the UPR to be activated in response to physiological stimuli such as differentiation cues or in preparation for cell division is an unsatisfying proposition. Therefore, it is reasonable to posit that there are many distinct physiological UPR activation states and that these represent the first step in generating diverse outputs. The following observations support this view and provide a range of interesting examples of physiological UPR activation.
Although yeast appear to use the UPR almost exclusively as a stress response, nascent physiological uses are apparent even in this context. Specifically, low-level cell cycle–dependent UPR activation under normal growth conditions suggests a stress-independent housekeeping role for the UPR, the absence of which leads to defective cytokinesis (Bicknell et al., 2007). Indeed, doubling of the ER (and indeed, of all the membranous content of the cell) is necessary in all eukaryotes, and so it is logical that this functionality of the UPR would be found even in yeast. Of course, in vertebrates, there are far more circumstances in which it would be advantageous for the UPR to anticipate impending demand on the ER and/or secretory pathway and to proactively adjust cellular physiology.
The most dramatic example of a proactive UPR is during development, when many cell types must drastically expand their ER upon differentiation. For example, the differentiation of B cells into antibody-producing plasma cells requires the UPR (Iwakoshi et al., 2003), presumably for expansion of the ER to handle the markedly increased secretory pathway load generated by up-regulated Ig synthesis. Although it seems logical that Ig induction would cause ER stress, activate the UPR, and initiate a responsive up-regulation of ER, this does not appear to be the case. Instead, differentiation-inducing stimuli lead to substantial ER expansion via activation of the IRE1-α and ATF6-α pathways before significant Ig synthesis occurs (van Anken et al., 2003). More dramatically, B cells engineered to lack Ig production nevertheless activate Xbp1 and differentiate normally (Hu et al., 2009). These findings call into question the causal role of ER stress in inducing the UPR in this context. It seems, rather, that Xbp1 (and perhaps other UPR pathways) are activated in preparation for rather than in response to secretory pathway load; in other words, a cell need not wait for dysfunction in the ER to alter its capacity. Although the mechanisms underlying UPR activation in plasma cell differentiation remain to be clarified, these observations emphasize that, depending on cellular context, UPR pathway components are capable of being activated in a selective and stress-independent fashion to regulate ER structure and function.
Proactive UPR activation may be used in several other contexts besides development. For example, diurnal induction of the UPR (and changes in its responsiveness) has been observed and is dependent on the circadian clock (Cretenet et al., 2010). Because nutritional flux and metabolism, which involve numerous facets of ER biology (see UPR pathway interactions in disease), are both predictable and typically diurnal, a mechanism to prepare the ER seems entirely appropriate. The UPR can also be triggered via signaling from the extracellular environment. For example, stimulation of thyrocytes with thyroid-stimulating hormone leads to up-regulation of ER chaperones, apparently in preparation for the increased load demanded by the need to make thyroglobulin (a large, slowly maturing secretory protein; Christis et al., 2010). Similarly, Toll-like receptors (TLRs), which signal the existence of pathogens, can modulate UPR signaling (Woo et al., 2009). This too makes logical sense because of the foreseeable increase in demands on the ER in the context of the stress of combating infection, presenting foreign antigens, and so forth. Thus, preparative UPR activation may well be protective in each of these circumstances by adjusting cellular physiology to a state more able to deal with impending environmental or physiological changes. Although this prepared state likely features a functionally and/or physically expanded ER, the ability of the UPR to regulate diverse cellular processes suggests that other functions of the UPR besides ER expansion might be as important or even more so during proactive UPR signaling.
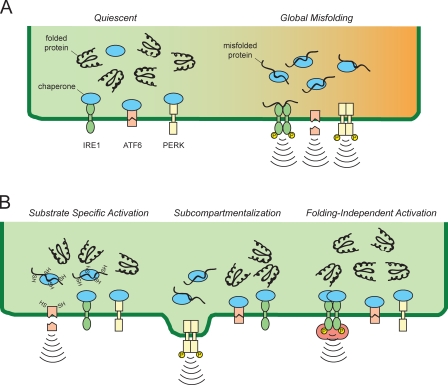
Appealing as a proactive engagement process is, there is still no mechanistic understanding of how it might work, although recent studies on activation mechanisms of the UPR stress sensors suggest some possibilities (Fig. 2). One of these is that selective physiological activation of the UPR could be achieved by a more nuanced aspect of the chaperone/client protein balance. In this view, accumulation of specific ER client proteins might trigger UPR activation even as global folding remains unperturbed. For example, B lymphocytes up-regulate numerous cell surface receptors during differentiation (Hardy and Hayakawa, 2001), and it is conceivable that one or more of these might stimulate UPR signaling in a selective manner. Although all three stress sensors are activated by global ER dysfunction, differences in their activation mechanisms could impart substrate specificity in some cases. In support of this idea, ATF6-α activation is influenced by its oxidation state, glycosylation state, and proteasome-dependent turnover (Hong et al., 2004a,b; Nadanaka et al., 2006; Yoshida et al., 2009; Fonseca et al., 2010). Thus, stimuli that preferentially alter these aspects of ER protein processing (for example, up-regulation of protein that is particularly demanding of the ER oxidation and isomerization activity) could favor ATF6-α activation more than other pathways. Pathway-selective stimulation could be further facilitated if protein folding and UPR transduction are subcompartmentalized within the ER, as has been recently suggested (Kondratyev et al., 2007).
Figure 2.
Potential activation mechanisms for the UPR during physiological stimulation. (A) Global protein misfolding stress in the ER results in activation of all three UPR sensors. ER chaperones (blue) can maintain UPR signal transducers in an inactive state during quiescent conditions. Unfolded proteins can activate the stress transducers by indirect chaperone titration or direct transducer binding. The global balance between chaperone reserve and ER folding clients might represent the stimulus for physiological activation of the UPR, with parallel signaling pathways modifying the activity and/or abundance of phosphatases, proteases, nucleases, etc. that determine net UPR output. However, it is likely that at least some physiological stimuli are capable of activating UPR pathways selectively by several hypothetical mechanisms. (B) First, a UPR sensor could be preferentially sensitive to the folding status of certain types of substrates or specific environmental conditions (e.g., redox status; left). Second, spatial restriction of UPR transducers and/or individual substrates might lead to localized preferential activation (middle). And third, stimulated dimerization of IRE1-α or PERK in trans (or enforced ectopic transit of ATF6-α to the Golgi, where it is activated) could activate limbs of the UPR independently of the ER folding status. These or other hypothetical mechanisms might be used during different physiological stimuli, and they need not be mutually exclusive.
Another possible mechanism for proactive UPR induction, not mutually exclusive with the first, is that UPR activation can occur independently of protein folding. As proof of principle, artificial dimerization of PERK and IRE1-α was shown to allow their stress-independent activation (Papa et al., 2003; Lu et al., 2004). Thus, one might imagine physiological stimuli that achieve the same outcome, for instance, the existence of endogenous factors or even small molecule ligands that can scaffold sensor self-association. This idea is supported by the observation that the flavonol quercetin can stimulate dimerization and activation of yeast Ire1p (Wiseman et al., 2010). It is important to emphasize that both of these possibilities are purely hypothetical at this point. The mechanisms of sensor activation, even during classical ER stress, remain unresolved, and mechanisms both dependent and independent of direct binding to misfolded proteins by the proximal stress sensors have been proposed (Credle et al., 2005; Zhou et al., 2006; Oikawa et al., 2009).
In addition to proactive UPR engagement preceding dramatic cell-shaping exogenous stimuli such as differentiation cues, the UPR is also likely to be activated to maintain cellular function in the face of normal fluctuations of a cell’s physiological state. Such a basal function is evident from the observation that UPR activation is apparent in some cells and tissues of normal mice under normal conditions. For example, a GFP reporter for Xbp1 activation was observed in situ in both pancreas and muscle (Iwawaki et al., 2004). Similar reporters applied to several other cell types (mammalian B and T cells [Brunsing et al., 2008], dendritic cells [Hayashi et al., 2007], and placenta [Iwawaki et al., 2009] among others) provide further support for Xbp1 splicing under various stress-free physiological settings. Although less extensively characterized, activated forms of other UPR pathways components, including ATF4, ATF6, and PERK, have been detected in various tissues of unchallenged mice (Harding et al., 2000; Yang et al., 2004; Lee et al., 2008; Wang et al., 2009), despite the fact that these forms are not usually detected in cultured cells that have not been subjected to acute stress. Thus, it is likely that these transcription factors are contributing to the basal regulation of gene expression in vivo, although their precise roles are currently poorly defined.
Physiological roles for the UPR have been challenging to study because of early lethality in animals with constitutive deletions in UPR components (IRE1-α, XBP1, and PERK). However, conditional gene deletion studies are overcoming this problem and providing insights into the situations in which UPR signaling is important during normal physiology. Mice in which Xbp1 was ablated in the adult liver show no evidence for ER stress or a general impairment of the secretory pathway but show a marked reduction in serum cholesterol, triglyceride, and free fatty acid (Lee et al., 2008). The expression of a subset of lipogenic genes was reduced in these animals, suggesting that XBP1 contributes to lipid homeostasis in the liver through the regulation of these genes. The gut-specific IRE1-α paralogue IRE1-β also regulates lipid metabolism, as the RIDD activity of IRE1-β regulates chylomicron production in the intestine (Iqbal et al., 2008). Similarly, eIF2-α maintains homeostasis in pancreatic β cells; inducible ablation of eIF2-α phosphorylation in the adult pancreas leads to rapid and severe β-cell dysfunction and diabetes, accompanied by unregulated protein translation in these cells (Back et al., 2009). Finally, acutely knocking down hepatic ATF6-α expression using an adenoviral vector results in elevated expression of gluconeogenic genes and hyperglycemia, effects which have been attributed to competition between ATF6-α and the metabolic transcriptional regulator CREB for interaction with the coregulator CRTC2 (Wang et al., 2009). Collectively, these examples provide prima facie evidence that all three pathways of the UPR are active to one extent or another in at least certain tissues under normal conditions. Thus, the UPR, in addition to being crucial to sensing and responding to presumably rare and sporadic protein misfolding stress, is physiologically engaged by many other cellular pathways (developmental, cell surface signaling, circadian, and others). These results suggest that the UPR, or at least elements of it, are frequently or perhaps even continually activated in some cell types as a means of fine-tuning cellular conditions in real time.
UPR functionality is context dependent
Because physiological uses of the UPR occur in rather diverse cellular contexts, its output is likely to be tailored to the stimulus. This raises several key questions. How broad is the range of UPR outputs that can be achieved by the cell? Is this range simply quantitative (i.e., a strong vs. weak response), or is UPR output substantively and qualitatively diverse? And how would context-dependent diversity in outputs be generated?
The fact that signal transduction pathways can generate different outputs depending on cellular context is well established in many systems. An obvious example would be MAPK signaling, whose effects on downstream gene expression depend greatly on the stimulus that induces it, the complement of modifier proteins expressed in a given cell type, and the activity of parallel signaling pathways (Shaul and Seger, 2007). An important requirement in such systems is that key components of a signaling cascade are sensitive to trans-acting modifiers that can influence their abundance or activity. Because such modifiers may vary depending on conditions or cell type, responsiveness of the signaling pathway would be variable. Thus, the physiological, environmental, genetic, developmental, and historical state of the cell would determine the context within which a signaling pathway acts, which in turn would determine the output generated in response to that signal. The following examples illustrate how these principles are beginning to emerge in the field of UPR regulation.
The ability of cells to produce alternate outputs during UPR activation is most clearly illustrated by the observation that excessive ER stress (that which a cell cannot successfully ameliorate via UPR activation) leads to programmed cell death, an outcome which is obviously incompatible with most instances of physiological UPR activation. A physiological UPR must rely on mechanisms whereby adaptive UPR signaling can be maintained (potentially even perpetually) while apoptotic signaling is suppressed. One UPR-mediated event of particular importance is up-regulation of the transcription factor CHOP by ATF4 (Zinszner et al., 1998). Activation of the UPR by mild stress promotes long-term up-regulation of ER chaperones, which are quite stable at the protein and mRNA levels. This presumably helps to improve ER folding and blunt the UPR, whereas rapid degradation of proapoptotic components such as CHOP helps the cell escape death. However, when ER stress is severe, this mitigating action of the UPR is insufficient (Rutkowski et al., 2006). Conversely, it has been suggested that cells must attenuate protective features of the UPR, most notably IRE1-α activation, to execute apoptosis when stress is severe (Lin et al., 2007). These alternate fates are an intrinsic consequence of the way that the UPR is structured, allowing cells to naturally discriminate strong from mild stresses. However, the importance of proapoptotic UPR signaling components such as CHOP in potentiating cell death suggests that they might also be regulated in trans at other points along the signaling axes that promotes their production.
One possible means of suppressing CHOP in trans, for example, would be to maintain PERK in a quiescent state during UPR activation. Such selective inactivation would sacrifice the benefits that PERK activation provides to the cell, including ATF4-mediated up-regulation of amino acid biosynthetic pathways and redox defenses. Nonetheless, this appears to be exactly the mechanism used during plasma cell differentiation, during which CHOP is not robustly up-regulated (Gass et al., 2002), a finding which is subsequently attributed to the quiescence of PERK (Gass et al., 2008; Ma et al., 2010). In that context, ATF6-α and IRE1-α must be activated to up-regulate ER chaperones and expand the ER, yet PERK activation is undesirable. This is in part because of PERK’s effects on CHOP and also because PERK activation would lead to translational repression, which would subvert the increased Ig synthesis needed as the cells differentiate.
CHOP expression could also be regulated downstream of PERK activation, preserving the benefits of the latter. Such pathway suppression was recently described during stimulation of cells by TLRs (Woo et al., 2009). These receptors, which signal the presence of pathogens, activate the UPR, perhaps in preparation for pathogen-mediated stress or as a consequence of increased ER load caused by cytokine induction. Remarkably, while triggering the UPR, TLR signaling simultaneously suppresses CHOP production at a step after eIF2-α phosphorylation. This selective suppression permits cells to survive the stress of responding to pathogens by taking advantage of some, but not all, facets of the UPR. Indeed, because PERK suppression occurs downstream of eIF2-α, the adaptive translational suppression can take place without the apoptotic consequences. Thus, the aforementioned examples make it clear that a benefit of the vertebrate UPR, with its multiple pathways and multiple steps in each pathway, is that the response’s output can be finely modulated based on the points at which it is regulated by intrinsic and extrinsic factors.
In addition to the dichotomy between adaptation and apoptosis, other more nuanced outputs of the UPR can also be regulated. The basic principle of this regulation typically is based on the sharing of components between the UPR and other signaling pathways. This kind of cross talk can bidirectionally impact the output of both signaling pathways, allowing each to be modified by the other. Different mechanisms of pathway intersection have been described for each of the three primary limbs of the UPR. For example, the activity of ATF6-α is influenced by its interaction with the transcriptional coregulator CRTC2 (Wang et al., 2009). This interaction links UPR activity to the status of cellular metabolic pathways because CRTC2 also serves as a coactivator for CREB, a transcription factor which (among many things) is responsive to glucagon signaling. Therefore, UPR activation influences the metabolic state by sequestering CRTC2 through ATF6-α, thereby altering CREB activity. Conversely, concurrent glucagon stimulation would tend to titrate CRTC2 away from ATF6-α, potentially regulating UPR output. As another example of regulation of ATF6-α transcriptional output in trans, the ATF6-α influence on BiP expression is counteracted by HDAC1 (histone deacetylase 1)-mediated repression (Baumeister et al., 2009). Thus, the balance between UPR activation and external regulation of the histone machinery in a physiologically activated UPR is expected to control the ultimate output.
The PERK and IRE1 pathways also intersect with non-UPR signaling cascades. For PERK, a key point of intersection is eIF2-α phosphorylation, which can be regulated by multiple kinases and phosphatases (Wek et al., 2006). Several physiological inputs, including nutritional status, hypoxia, hormone stimulation, infection, and others, influence eIF2-α phosphorylation both directly through other eIF2-α kinases and indirectly. Notable among these kinases is PKR because it can be activated by signals that overlap with PERK activation such as viral infection and nutrient sensing (Baltzis et al., 2004; Nakamura et al., 2010). PERK also has been shown to phosphorylate, and thus activate, the NRF2 transcription factor, linking PERK activation to a pathway more classically considered important in redox sensing (Cullinan et al., 2003; Cullinan and Diehl, 2004). Thus, the effect of PERK during UPR activation is integrated with multiple other cellular conditions to shape the downstream output.
For IRE1, activity may be controlled in part by interactions with BCL-2 family members (Hetz et al., 2006; Lisbona et al., 2009) or PI3 kinase (Park et al., 2010; Winnay et al., 2010). IRE1 also interacts with TRAF2 to influence signaling by its substrates, including JNK, ASK1, and perhaps nuclear factor κB (NF-κB; Urano et al., 2000; Nishitoh et al., 2002; Kaneko et al., 2003), suggesting that IRE1 might serve as a surprisingly broad signal transduction platform (Hetz and Glimcher, 2009). As with ATF6-α, XBP1 activity can be regulated directly at the level of its interaction with target genes. Myogenic differentiation induces Xbp1 splicing, and the subset of DNA sequences to which XBP1 can bind in that context overlaps only partially with those genes regulated by XBP1 during classical ER stress (Acosta-Alvear et al., 2007). This specificity in output likely reflects the influence of other transcriptional regulators specific to the differentiation process that are capable of modulating XBP1 activity. Similarly, TLR stimulation can lead to IRE1-α/XBP1 activation, giving rise to a pattern of gene expression quite distinct from the stress paradigm (Martinon et al., 2010). Again, these interacting partners for eIF2-α, IRE1, XBP1, etc. are not themselves static, and their abundance and activity can affect the output of UPR activation. Thus, all three UPR pathways are subject to modulation in trans by a variety of mechanisms involving rather diverse signaling pathways.
Finally, UPR output might also be regulated via the abundance or activity of key UPR signaling molecules. Whether or how this abundance varies among cell types or historical events (e.g., whether the UPR was recently activated) is not well studied. However, modulating the expression of each of the key UPR molecules by small degrees (say, 10 or 20%) could have a significant combined effect on the sensitivity and output of the response. Notably, ATF6-α, IRE1-α, PERK, and ATF4 are all themselves transcriptionally regulated by UPR activation, and the relative amounts of the key UPR components could tune the response. For example, XBP1 protein translated from unspliced Xbp1 mRNA has been shown to antagonize transcriptional activation by the protein translated from the spliced mRNA (Tirosh et al., 2006; Yoshida et al., 2006). Thus, the steady-state ratio between these two forms of the XBP1 protein, controlled by the sum of regulation of Xbp1 mRNA expression and splicing, controls transcriptional output.
These and yet other mechanisms for modulating UPR activity at multiple points along each UPR branch illustrate the remarkably rich regulatory potential of the vertebrate UPR, which allows the response to achieve fine context specificity in ways that are only beginning to be uncovered. Thus, the UPR is capable of eliciting a rather wide range of qualitatively and quantitatively distinct outputs that are molded by both the nature of the activating inputs and the context within which the UPR pathways operate. Understanding more precisely the scope of UPR functional diversity, as well as the detailed mechanisms by which that diversity is generated, represents a major challenge for the future.
Causal roles for the UPR in disease
Given the centrality of the ER to many cellular and organismal functions, it is quite likely to be associated with a variety of diseases. Indeed, certain markers of UPR activation have been observed in many disease states, which is not surprising because the signaling pathways of the UPR appear to respond to many stimuli and because the outputs of UPR signaling are so diverse. These diseases include cancer, diabetes, neurodegeneration, infectious disease, autoimmunity, and many others (for review see Zhao and Ackerman, 2006). However, these associations are almost impossible to interpret from the standpoint of causality. In fact, given the interconnectedness of many cellular pathways and the ways cells interact within a complex organ system or organism, UPR activation in a given disease could well be a secondary or even tertiary event that is only distantly related to the underlying cause of disease. In this sense, UPR activity is simply a marker, and not necessarily a proximal one, of generally altered cellular function. A more relevant question is to ask under what circumstances ER stress and the UPR more directly contribute to disease.
The UPR would most obviously contribute to diseases caused by direct impairment of protein folding, of which there are three general classes. In the first class, a partial or complete defect in a UPR signaling component is anticipated to cause disease because the cells would be far less able to adjust some facet of their ER functionality to changing cellular conditions. This impairment would constrain the scope of physiological conditions to which the UPR could then effectively respond, and organs and cell types that place the greatest burden on that UPR component would be the first to evince pathology. According to this model, the organs most often affected would include the pancreas and liver, which are subject to constitutively heavy traffic through the secretory pathway and whose activities must be modulated constantly to meet fluctuating nutritional and metabolic needs. Indeed, PERK loss of function in humans causes Wolcott-Rallison syndrome, a disease characterized by defects in many highly secretory cell types, including pancreatic β cells. Mice with analogous deficiencies in either PERK or eIF2-α phosphorylation develop pancreatic β-cell failure because they cannot tolerate the burden of regulated insulin production and secretion (Harding et al., 2001; Scheuner et al., 2001; Zhang et al., 2002). Even more subtle deficiencies such as Xbp1 heterozygosity or a single nonphosphorylatable eIF2-α allele lead to phenotypes when the animals are challenged with a high-fat diet that imposes greater physiological demands on the liver and pancreas (among other tissues; Özcan et al., 2004; Scheuner et al., 2005).
The converse of diseases arising from impaired UPR signaling is cancer, where UPR hyperactivation in tumor cells can exacerbate disease by protecting them from cell death. Alterations in UPR function are not typically found in screens for driver mutations producing oncogenic transformation (e.g., Jonkers and Berns, 1996; Collier et al., 2005), but altered UPR activity nonetheless appears to be important for progression and prognosis of at least some cancers (Carrasco et al., 2007; Lee, 2007; Schewe and Aguirre-Ghiso, 2008). In such cases, UPR involvement is not caused by genetic lesion of a UPR component but by positive feedback between the transformation process and UPR activation. It is thought that altered conditions of the transformed cell, including rapid growth, high metabolic demand, and hypoxia, activate the UPR. This may favor adaptation, protection from cell death, and continued growth, thereby furthering transformation. The apparent ability of proteasome inhibitors to tip the balance of UPR activation toward apoptosis in multiple myeloma (Lee et al., 2003) hints at the importance of a properly regulated UPR, including its death-promoting activities, in cancer prevention.
The third class of diseases likely to be causally linked to the UPR and ER stress encompasses those directly involving globally compromised ER function, including protein misfolding, maintenance of calcium homeostasis, redox balance, etc. (for review see Zhao and Ackerman, 2006). Here, the inciting event would be a chronic additional load on the ER, resulting in activation of the UPR to remediate the stress by increasing the functional capacity of the ER. At some point, the capacity to respond would be overwhelmed, resulting eventually in cell death. That such disease states are caused mainly by UPR-mediated cell death is supported by the fact that in some cases, CHOP ablation at least partially rescues the phenotype (Silva et al., 2005; Pennuto et al., 2008; Song et al., 2008; Namba et al., 2009; Thorp et al., 2009). Diseases in this class could be caused by mutations in secretory pathway client proteins or in the ER protein folding machinery.
A noteworthy observation from patients with such diseases (and analogous mouse models) is that even an inciting event that occurs universally (or widely) often leads to considerable cell type–specific pathology (e.g., Zhao et al., 2005). This underscores the notion that the cellular context in which a perturbation occurs shapes the capacity to adapt to or absorb that perturbation. Similarly, because the capacity of the ER to carry out its various functions, and that of the UPR to respond, can change during development and aging (Naidoo, 2009), disease phenotypes are often temporally regulated and age dependent. Neurodegenerative disorders are classical examples of age-related diseases in which the UPR has been implicated (Paschen and Mengesdorf, 2005). Although no definitive causal relationship has been established between Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, and other such disorders and the UPR, it is tempting to speculate that neurons, which enjoy privilege from most environmental fluctuations that challenge other cell types, might be especially sensitive to progressive perturbations such as proteotoxicity.
UPR pathway interactions in disease
Beyond the predictable disease-causing effects of deficiencies in either UPR components or protein folding, how else might pathological states be caused by the UPR? Is it possible that UPR activation in response to a given stimulus can be beneficial under one set of conditions but detrimental under another depending on the context in which activation occurs? Pathogenesis by such a mechanism would not involve a specific lesion or deficiency but rather a more nuanced system-wide change in how a complex set of homeostatic pathways interact and are organized. To illustrate this idea, we consider the role of UPR signaling in liver metabolism and insulin action, which has been a topic of several recent studies.
Nutrient availability fluctuates constantly in response to feeding and fasting cycles. Among the many consequences of this flux is the secretion of insulin by pancreatic β cells, an event which has many downstream consequences in insulin target tissues. At least two of these consequences are capable of influencing the ER (Fig. 3). First, insulin signaling up-regulates the central nutrient-sensing mammalian target of rapamycin (mTOR) pathway (Hietakangas and Cohen, 2009). Activation of mTOR favors anabolism, including increased protein synthesis. Second, insulin signaling stimulates both lipogenesis and cholesterologenesis by activating SREBP-1c and SREBP2 in the liver to facilitate the production of very low-density lipoprotein (VLDL) in the ER (Laplante and Sabatini, 2009). Thus, both mTOR activity and VLDL biogenesis would be expected to increase load on the ER, which necessitates activation of the UPR to increase ER protein processing capacity (Özcan et al., 2004, 2008; Su et al., 2009). Consistent with this idea, the UPR is activated in the liver during and after eating, and this activation is ameliorated by the inhibition of mTOR (Oyadomari et al., 2008; Pfaffenbach et al., 2010). Although the stimulus of feeding can clearly be sensed by the cell, it can hardly be thought of as a stress in a conventional sense because simply eating a meal is unlikely to grossly impair ER function.
Figure 3.
UPR-mediated feedback inhibition of insulin signaling. Nutritional intake upon feeding stimulates insulin release, which initiates at least two signaling pathways in the liver (green) that impact the ER in distinct ways. First, enhanced protein synthesis caused by mTOR activation increases general substrate load in the ER. Second, activation of SREBP pathways stimulate production of lipoproteins such as VLDL, whose components are assembled in the ER. These (and possibly other) effects activate the UPR, which can inhibit insulin action by multiple feedback mechanisms (orange). Although this feedback inhibition likely serves a beneficial role during the postprandial state, it could become deleterious during chronic overnutrition. IR, insulin receptor; IRS, IR substrate.
UPR signaling in the context of insulin action encompasses several pathways of negative feedback that, although perhaps beneficial or largely irrelevant for the short-term nutrient fluctuations that occur during feeding and fasting cycles, may become detrimental in the context of chronic overnutrition. Among these pathways, activation of JNK by IRE1-α (and possibly PKR; Nakamura et al., 2010) is particularly interesting because JNK phosphorylates the insulin receptor substrate (IRS-1), which then inhibits signaling through the insulin receptor (Tanti and Jager, 2009). Insulin signaling is further attenuated by UPR-mediated autophagic destruction of the insulin receptor (Zhou et al., 2009). JNK can also activate proinflammatory cytokines, as can NF-κB, another transcription factor which can also be activated by the UPR (Zhang and Kaufman, 2008). UPR activation can also suppress VLDL secretion through the production of CHOP, which, in addition to promoting cell death during severe ER stress, can antagonize the regulation of metabolic gene expression (Rutkowski et al., 2008). In the context of normal feeding and fasting cycles, these signaling pathways likely contribute to normal feedback inhibition of insulin action at multiple points along the cascade (Fig. 3). However, when nutritional excess and relatively constant insulin signaling is the norm, they may contribute to a chronically altered state. In this context, these normally beneficial UPR activities could lead to insulin resistance (as a consequence of persistent IRS-1 phosphorylation and insulin receptor degradation), elevated cytokine levels (via both JNK and NF-κB), fatty liver (by inhibition of VLDL secretion), and other UPR-mediated effects on metabolism (Hotamisligil, 2010). Some of these effects (e.g., elevated cytokine levels) could lead to further UPR activation, setting up a positive feedback loop that sustains unalleviated UPR activation, inflammation, and chronic insulin resistance.
Indeed, this aforementioned sequence of events centered on UPR signaling is emerging as a key pathogenic mechanism in models of type 2 diabetes. Although many aspects of this model remain to be rigorously established, the model is consistent with several studies in which the UPR or ER functionality is experimentally manipulated. For example, JNK deficiency leads to protection from diet-induced insulin resistance (Hirosumi et al., 2002), whereas constitutive mTOR activation causes marked insulin resistance (Shah et al., 2004). With respect to the ER, mice heterozygous for Xbp1 are unable to reestablish ER homeostasis as efficiently, and they develop increased insulin resistance more readily than wild-type animals in response to overnutrition, concomitant with increased JNK activation (Özcan et al., 2004). Conversely, chemical chaperones or BiP overexpression improve insulin sensitivity, perhaps caused by dampened signaling via UPR pathways that initiate the aforementioned positive feedback loops (Özcan et al., 2006; Kammoun et al., 2009).
Together, the aforementioned studies illustrate that the integration of UPR pathways with metabolic pathways, at the levels of both signaling and at the level of transcriptional control, makes UPR outputs highly dependent on the cellular context. This is because each metabolic state (e.g., fed, fasted, high fat, low fat, etc.) will, in addition to regulating the UPR, also modulate independent signaling cascades with which the UPR can then interact. Thus, the connections between the UPR and metabolism can be understood as autoregulatory mechanisms that are normally beneficial and work to mutually promote ER homeostasis and a suitable cellular energy balance. These connections between pathways then become deleterious only in special contexts, such as chronic overnutrition, that are unlikely to have been commonly encountered during the evolution of UPR pathways. Thus, unameliorated ER stress (i.e., misfolded protein accumulation beyond ER processing capacity) may not feature prominently in such diseases; instead, the pathological events are likely caused by physiological UPR activation in combination with altered environmental context.
In a broader sense, many disease states in which UPR activation is implicated may not necessarily reflect misfolded protein stress, given that this is not the sole means of UPR activation. UPR-associated disease states may instead reflect the appropriate engagement of UPR pathways by physiological inputs, but within a cellular or environmental context where the pathway outputs are not adaptive to the organism, thus contributing to the pathology. Therefore, understanding the nuances of the interactions of UPR pathways with the cellular context will continue to be crucial in dissecting the pathogenesis of complex, chronic, multifactorial diseases such as metabolic syndrome, inflammation, neurodegeneration, cancer, and others.
Future aims and challenges
Although the UPR was originally identified as a response to acute ER perturbation and clearly serves that function in physiologically meaningful ways, it is now evident that UPR signaling pathways are enmeshed with cellular physiology in more complex and subtle ways. We have highlighted several observations illustrating that inputs into the UPR are considerably more diverse and nuanced than solely protein misfolding stress and that UPR outputs are highly complex, quite malleable, and strongly influenced by cellular context. The considerable flexibility in the way these pathways are engaged allows them to be deployed under a rather wide range of physiological conditions that intersect in one way or another with ER homeostasis. Conversely, the same complex set of UPR pathway interactions, while providing considerable regulation and flexibility, also carries greater opportunity for misappropriation in a variety of pathological contexts. We are only just glimpsing the mechanisms that allow a core set of UPR pathways to be differentially responsive in a context-dependent manner to regulate diverse outputs.
It is clear that if we are to fully appreciate the range of UPR regulation in physiology and pathophysiology, we will need systems and methods that differ qualitatively from those used to elucidate the core stress-initiated pathways. First, it will be necessary to develop model systems for experimentally inducing and studying physiological activation of the UPR. Because the inputs that initiate the UPR are diverse, it is likely that different specific experimental systems will be needed to understand the range of mechanisms by which each UPR component can be selectively activated or selectively suppressed. Second, we will need tools for manipulating (turning on, turning off, and tuning) individual UPR pathways with higher spatial and temporal resolution. An attractive system in this respect is the chemical genetic tools that have been developed that allow exogenously expressed PERK and IRE1-α to be specifically activated in the absence of stress (Papa et al., 2003; Lu et al., 2004). In the case of IRE1-α, such activation allows for dissociation of its Xbp1 splicing and RIDD-stimulating activities (Han et al., 2009). Such chemical genetic strategies, in combination with tissue-specific transgene expression (or, more optimally, knockin expression), would allow highly precise experimental manipulation of UPR pathways to test their roles in complex physiology. This will be the next stage of refinement beyond conditional knockouts, the current state of the art, needed to probe causal relationships between the UPR and specific physiological and pathological states.
Paralleling the implementation of better methods for inducing UPR pathways will be the development of in vivo biosensors for monitoring both UPR activation and the various UPR-regulated functions, including protein folding, maintenance of calcium homeostasis, and lipid synthesis. The development of a transgenic mouse expressing an IRE1-responsive fluorescent indicator is an important first step; biosensors that sensitively report on other UPR pathways at points both proximal and distal to UPR activation will also be necessary. Likewise, probes for assessing ER calcium handling and, more recently, the ER folding environment (Merksamer et al., 2008) have been developed for use in cultured cells and simple organisms; these must now be adapted for use in vivo in vertebrates. Biosensors of this sort will be necessary in part because, in most tissues, UPR pathways will likely be activated to a very modest extent, such that detection will require amplification. Once such methods are developed, it will be possible to use them to follow the ebb and flow of UPR pathway activation in vivo in response to stimuli such as feeding and fasting cycles.
Finally, the interconnection between the UPR and other cellular signaling pathways will be best understood when studied as an integrated whole. Fortunately, the methods available for quantitative and global analysis of cellular responses to alterations in homeostasis are expanding exponentially. The ability to analyze whole “omes” (transcriptome, proteome, metabolome, and epigenome) in combination with sophisticated computational tools for integrating the vast amounts of data generated by these analyses will allow us to visualize the UPR less as series of linear pathways with discrete endpoints and more as a contextually dependent contributor to a complex and dynamic signaling network that maintains cellular homeostasis. Indeed, such tools are already being applied to simple organisms (Jonikas et al., 2009), and it is only a matter of time before they can be applied to the vertebrate UPR as well.
Acknowledgments
We thank C. Blaumueller for comments on the manuscript.
Work in the laboratory of D.T. Rutkowski is supported by the National Institutes of Health (grant DK084058) and the Carver Trust Medical Research Initiative. Work in the laboratory of R.S. Hegde is supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development at the National Institutes of Health.
Footnotes
Abbreviations used in this paper:
- mTOR
- mammalian target of rapamycin
- NF-κB
- nuclear factor κB
- RIDD
- regulated IRE1-dependent decay
- TLR
- Toll-like receptor
- UPR
- unfolded protein response
- VLDL
- very low-density lipoprotein
References
- Acosta-Alvear D., Zhou Y., Blais A., Tsikitis M., Lents N.H., Arias C., Lennon C.J., Kluger Y., Dynlacht B.D. 2007. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell. 27:53–66 10.1016/j.molcel.2007.06.011 [DOI] [PubMed] [Google Scholar]
- Back S.H., Scheuner D., Han J., Song B., Ribick M., Wang J., Gildersleeve R.D., Pennathur S., Kaufman R.J. 2009. Translation attenuation through eIF2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metab. 10:13–26 10.1016/j.cmet.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D., O’Hare P. 2007. Transmembrane bZIP transcription factors in ER stress signaling and the unfolded protein response. Antioxid. Redox Signal. 9:2305–2321 10.1089/ars.2007.1796 [DOI] [PubMed] [Google Scholar]
- Baltzis D., Qu L.K., Papadopoulou S., Blais J.D., Bell J.C., Sonenberg N., Koromilas A.E. 2004. Resistance to vesicular stomatitis virus infection requires a functional cross talk between the eukaryotic translation initiation factor 2alpha kinases PERK and PKR. J. Virol. 78:12747–12761 10.1128/JVI.78.23.12747-12761.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister P., Dong D., Fu Y., Lee A.S. 2009. Transcriptional induction of GRP78/BiP by histone deacetylase inhibitors and resistance to histone deacetylase inhibitor-induced apoptosis. Mol. Cancer Ther. 10.1158/1535-7163.MCT-08-1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A., Wang X., Novoa I., Jungreis R., Schlessinger K., Cho J.H., West A.B., Ron D. 2001. Increased sensitivity to dextran sodium sulfate colitis in IRE1beta-deficient mice. J. Clin. Invest. 107:585–593 10.1172/JCI11476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell A.A., Babour A., Federovitch C.M., Niwa M. 2007. A novel role in cytokinesis reveals a housekeeping function for the unfolded protein response. J. Cell Biol. 177:1017–1027 10.1083/jcb.200702101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunsing R., Omori S.A., Weber F., Bicknell A., Friend L., Rickert R., Niwa M. 2008. B- and T-cell development both involve activity of the unfolded protein response pathway. J. Biol. Chem. 283:17954–17961 10.1074/jbc.M801395200 [DOI] [PubMed] [Google Scholar]
- Carrasco D.R., Sukhdeo K., Protopopova M., Sinha R., Enos M., Carrasco D.E., Zheng M., Mani M., Henderson J., Pinkus G.S., et al. 2007. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 11:349–360 10.1016/j.ccr.2007.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christis C., Fullaondo A., Schildknegt D., Mkrtchian S., Heck A.J., Braakman I. 2010. Regulated increase in folding capacity prevents unfolded protein stress in the ER. J. Cell Sci. 123:787–794 10.1242/jcs.041111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier L.S., Carlson C.M., Ravimohan S., Dupuy A.J., Largaespada D.A. 2005. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 436:272–276 10.1038/nature03681 [DOI] [PubMed] [Google Scholar]
- Credle J.J., Finer-Moore J.S., Papa F.R., Stroud R.M., Walter P. 2005. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 102:18773–18784 10.1073/pnas.0509487102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretenet G., Le Clech M., Gachon F. 2010. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 11:47–57 10.1016/j.cmet.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Cullinan S.B., Diehl J.A. 2004. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J. Biol. Chem. 279:20108–20117 10.1074/jbc.M314219200 [DOI] [PubMed] [Google Scholar]
- Cullinan S.B., Zhang D., Hannink M., Arvisais E., Kaufman R.J., Diehl J.A. 2003. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 23:7198–7209 10.1128/MCB.23.20.7198-7209.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner A.J., Wasley L.C., Raney P., Haugejorden S., Green M., Kaufman R.J. 1990. The stress response in Chinese hamster ovary cells. Regulation of ERp72 and protein disulfide isomerase expression and secretion. J. Biol. Chem. 265:22029–22034 [PubMed] [Google Scholar]
- Fonseca S.G., Ishigaki S., Oslowski C.M., Lu S., Lipson K.L., Ghosh R., Hayashi E., Ishihara H., Oka Y., Permutt M.A., Urano F. 2010. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J. Clin. Invest. 120:744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass J.N., Gifford N.M., Brewer J.W. 2002. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 277:49047–49054 10.1074/jbc.M205011200 [DOI] [PubMed] [Google Scholar]
- Gass J.N., Jiang H.Y., Wek R.C., Brewer J.W. 2008. The unfolded protein response of B-lymphocytes: PERK-independent development of antibody-secreting cells. Mol. Immunol. 45:1035–1043 10.1016/j.molimm.2007.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Lerner A.G., Vande Walle L., Upton J.P., Xu W., Hagen A., Backes B.J., Oakes S.A., Papa F.R. 2009. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 138:562–575 10.1016/j.cell.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Bertolotti A., Zeng H., Ron D. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 5:897–904 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zeng H., Zhang Y., Jungries R., Chung P., Plesken H., Sabatini D.D., Ron D. 2001. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol. Cell. 7:1153–1163 10.1016/S1097-2765(01)00264-7 [DOI] [PubMed] [Google Scholar]
- Hardy R.R., Hayakawa K. 2001. B cell development pathways. Annu. Rev. Immunol. 19:595–621 10.1146/annurev.immunol.19.1.595 [DOI] [PubMed] [Google Scholar]
- Hayashi A., Kasahara T., Iwamoto K., Ishiwata M., Kametani M., Kakiuchi C., Furuichi T., Kato T. 2007. The role of brain-derived neurotrophic factor (BDNF)-induced XBP1 splicing during brain development. J. Biol. Chem. 282:34525–34534 10.1074/jbc.M704300200 [DOI] [PubMed] [Google Scholar]
- Hetz C., Glimcher L.H. 2009. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol. Cell. 35:551–561 10.1016/j.molcel.2009.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C., Bernasconi P., Fisher J., Lee A.H., Bassik M.C., Antonsson B., Brandt G.S., Iwakoshi N.N., Schinzel A., Glimcher L.H., Korsmeyer S.J. 2006. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 312:572–576 10.1126/science.1123480 [DOI] [PubMed] [Google Scholar]
- Hietakangas V., Cohen S.M. 2009. Regulation of tissue growth through nutrient sensing. Annu. Rev. Genet. 43:389–410 10.1146/annurev-genet-102108-134815 [DOI] [PubMed] [Google Scholar]
- Hirosumi J., Tuncman G., Chang L., Görgün C.Z., Uysal K.T., Maeda K., Karin M., Hotamisligil G.S. 2002. A central role for JNK in obesity and insulin resistance. Nature. 420:333–336 10.1038/nature01137 [DOI] [PubMed] [Google Scholar]
- Hollien J., Weissman J.S. 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 313:104–107 10.1126/science.1129631 [DOI] [PubMed] [Google Scholar]
- Hong M., Li M., Mao C., Lee A.S. 2004a. Endoplasmic reticulum stress triggers an acute proteasome-dependent degradation of ATF6. J. Cell. Biochem. 92:723–732 10.1002/jcb.20118 [DOI] [PubMed] [Google Scholar]
- Hong M., Luo S., Baumeister P., Huang J.M., Gogia R.K., Li M., Lee A.S. 2004b. Underglycosylation of ATF6 as a novel sensing mechanism for activation of the unfolded protein response. J. Biol. Chem. 279:11354–11363 10.1074/jbc.M309804200 [DOI] [PubMed] [Google Scholar]
- Hotamisligil G.S. 2010. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 140:900–917 10.1016/j.cell.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.C., Dougan S.K., McGehee A.M., Love J.C., Ploegh H.L. 2009. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 28:1624–1636 10.1038/emboj.2009.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J., Dai K., Seimon T., Jungreis R., Oyadomari M., Kuriakose G., Ron D., Tabas I., Hussain M.M. 2008. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 7:445–455 10.1016/j.cmet.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakoshi N.N., Lee A.H., Vallabhajosyula P., Otipoby K.L., Rajewsky K., Glimcher L.H. 2003. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4:321–329 10.1038/ni907 [DOI] [PubMed] [Google Scholar]
- Iwawaki T., Akai R., Kohno K., Miura M. 2004. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat. Med. 10:98–102 10.1038/nm970 [DOI] [PubMed] [Google Scholar]
- Iwawaki T., Akai R., Yamanaka S., Kohno K. 2009. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc. Natl. Acad. Sci. USA. 106:16657–16662 10.1073/pnas.0903775106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas M.C., Collins S.R., Denic V., Oh E., Quan E.M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J.S., Schuldiner M. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 323:1693–1697 10.1126/science.1167983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J., Berns A. 1996. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim. Biophys. Acta. 1287:29–57 [DOI] [PubMed] [Google Scholar]
- Kammoun H.L., Chabanon H., Hainault I., Luquet S., Magnan C., Koike T., Ferré P., Foufelle F. 2009. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J. Clin. Invest. 119:1201–1215 10.1172/JCI37007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M., Niinuma Y., Nomura Y. 2003. Activation signal of nuclear factor-kappa B in response to endoplasmic reticulum stress is transduced via IRE1 and tumor necrosis factor receptor-associated factor 2. Biol. Pharm. Bull. 26:931–935 10.1248/bpb.26.931 [DOI] [PubMed] [Google Scholar]
- Kang S.W., Rane N.S., Kim S.J., Garrison J.L., Taunton J., Hegde R.S. 2006. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 127:999–1013 10.1016/j.cell.2006.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratyev M., Avezov E., Shenkman M., Groisman B., Lederkremer G.Z. 2007. PERK-dependent compartmentalization of ERAD and unfolded protein response machineries during ER stress. Exp. Cell Res. 313:3395–3407 10.1016/j.yexcr.2007.07.006 [DOI] [PubMed] [Google Scholar]
- Kozutsumi Y., Segal M., Normington K., Gething M.J., Sambrook J. 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 332:462–464 10.1038/332462a0 [DOI] [PubMed] [Google Scholar]
- Laplante M., Sabatini D.M. 2009. An emerging role of mTOR in lipid biosynthesis. Curr. Biol. 19:R1046–R1052 10.1016/j.cub.2009.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Iwakoshi N.N., Anderson K.C., Glimcher L.H. 2003. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. USA. 100:9946–9951 10.1073/pnas.1334037100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Scapa E.F., Cohen D.E., Glimcher L.H. 2008. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 320:1492–1496 10.1126/science.1158042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.S. 2007. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 67:3496–3499 10.1158/0008-5472.CAN-07-0325 [DOI] [PubMed] [Google Scholar]
- Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B., Shokat K.M., Lavail M.M., Walter P. 2007. IRE1 signaling affects cell fate during the unfolded protein response. Science. 318:944–949 10.1126/science.1146361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisbona F., Rojas-Rivera D., Thielen P., Zamorano S., Todd D., Martinon F., Glavic A., Kress C., Lin J.H., Walter P., et al. 2009. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol. Cell. 33:679–691 10.1016/j.molcel.2009.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P.D., Jousse C., Marciniak S.J., Zhang Y., Novoa I., Scheuner D., Kaufman R.J., Ron D., Harding H.P. 2004. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 23:169–179 10.1038/sj.emboj.7600030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Shimizu Y., Mann M.J., Jin Y., Hendershot L.M. 2010. Plasma cell differentiation initiates a limited ER stress response by specifically suppressing the PERK-dependent branch of the unfolded protein response. Cell Stress Chaperones. 15:281–293 10.1007/s12192-009-0142-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F., Chen X., Lee A.H., Glimcher L.H. 2010. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat. Immunol. 11:411–418 10.1038/ni.1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merksamer P.I., Trusina A., Papa F.R. 2008. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell. 135:933–947 10.1016/j.cell.2008.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadanaka S., Yoshida H., Mori K. 2006. Reduction of disulfide bridges in the lumenal domain of ATF6 in response to glucose starvation. Cell Struct. Funct. 31:127–134 10.1247/csf.06024 [DOI] [PubMed] [Google Scholar]
- Naidoo N. 2009. The endoplasmic reticulum stress response and aging. Rev. Neurosci. 20:23–37 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Furuhashi M., Li P., Cao H., Tuncman G., Sonenberg N., Gorgun C.Z., Hotamisligil G.S. 2010. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 140:338–348 10.1016/j.cell.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T., Tanaka K., Ito Y., Ishihara T., Hoshino T., Gotoh T., Endo M., Sato K., Mizushima T. 2009. Positive role of CCAAT/enhancer-binding protein homologous protein, a transcription factor involved in the endoplasmic reticulum stress response in the development of colitis. Am. J. Pathol. 174:1786–1798 10.2353/ajpath.2009.080864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitoh H., Matsuzawa A., Tobiume K., Saegusa K., Takeda K., Inoue K., Hori S., Kakizuka A., Ichijo H. 2002. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 16:1345–1355 10.1101/gad.992302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D., Kimata Y., Kohno K., Iwawaki T. 2009. Activation of mammalian IRE1alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp. Cell Res. 315:2496–2504 10.1016/j.yexcr.2009.06.009 [DOI] [PubMed] [Google Scholar]
- Otsu M., Sitia R. 2007. Diseases originating from altered protein quality control in the endoplasmic reticulum. Curr. Med. Chem. 14:1639–1652 10.2174/092986707780830952 [DOI] [PubMed] [Google Scholar]
- Oyadomari S., Harding H.P., Zhang Y., Oyadomari M., Ron D. 2008. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 7:520–532 10.1016/j.cmet.2008.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan U., Cao Q., Yilmaz E., Lee A.H., Iwakoshi N.N., Ozdelen E., Tuncman G., Görgün C., Glimcher L.H., Hotamisligil G.S. 2004. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 306:457–461 10.1126/science.1103160 [DOI] [PubMed] [Google Scholar]
- Özcan U., Yilmaz E., Özcan L., Furuhashi M., Vaillancourt E., Smith R.O., Görgün C.Z., Hotamisligil G.S. 2006. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 313:1137–1140 10.1126/science.1128294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan U., Özcan L., Yilmaz E., Düvel K., Sahin M., Manning B.D., Hotamisligil G.S. 2008. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol. Cell. 29:541–551 10.1016/j.molcel.2007.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa F.R., Zhang C., Shokat K., Walter P. 2003. Bypassing a kinase activity with an ATP-competitive drug. Science. 302:1533–1537 10.1126/science.1090031 [DOI] [PubMed] [Google Scholar]
- Park S.W., Zhou Y., Lee J., Lu A., Sun C., Chung J., Ueki K., Ozcan U. 2010. The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat. Med. 16:429–437 10.1038/nm.2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen W., Mengesdorf T. 2005. Endoplasmic reticulum stress response and neurodegeneration. Cell Calcium. 38:409–415 10.1016/j.ceca.2005.06.019 [DOI] [PubMed] [Google Scholar]
- Pennuto M., Tinelli E., Malaguti M., Del Carro U., D’Antonio M., Ron D., Quattrini A., Feltri M.L., Wrabetz L. 2008. Ablation of the UPR-mediator CHOP restores motor function and reduces demyelination in Charcot-Marie-Tooth 1B mice. Neuron. 57:393–405 10.1016/j.neuron.2007.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffenbach K.T., Nivala A.M., Reese L., Ellis F., Wang D., Wei Y., Pagliassotti M.J. 2010. Rapamycin inhibits postprandial-mediated X-box-binding protein-1 splicing in rat liver. J. Nutr. 140:879–884 10.3945/jn.109.119883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8:519–529 10.1038/nrm2199 [DOI] [PubMed] [Google Scholar]
- Rutkowski D.T., Arnold S.M., Miller C.N., Wu J., Li J., Gunnison K.M., Mori K., Sadighi Akha A.A., Raden D., Kaufman R.J. 2006. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 4:e374 10.1371/journal.pbio.0040374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D.T., Wu J., Back S.H., Callaghan M.U., Ferris S.P., Iqbal J., Clark R., Miao H., Hassler J.R., Fornek J., et al. 2008. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev. Cell. 15:829–840 10.1016/j.devcel.2008.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R.J. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 7:1165–1176 10.1016/S1097-2765(01)00265-9 [DOI] [PubMed] [Google Scholar]
- Scheuner D., Vander Mierde D., Song B., Flamez D., Creemers J.W.M., Tsukamoto K., Ribick M., Schuit F.C., Kaufman R.J. 2005. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat. Med. 11:757–764 10.1038/nm1259 [DOI] [PubMed] [Google Scholar]
- Schewe D.M., Aguirre-Ghiso J.A. 2008. ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proc. Natl. Acad. Sci. USA. 105:10519–10524 10.1073/pnas.0800939105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah O.J., Wang Z., Hunter T. 2004. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 14:1650–1656 10.1016/j.cub.2004.08.026 [DOI] [PubMed] [Google Scholar]
- Shaul Y.D., Seger R. 2007. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim. Biophys. Acta. 1773:1213–1226 10.1016/j.bbamcr.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Sidrauski C., Chapman R., Walter P. 1998. The unfolded protein response: an intracellular signalling pathway with many surprising features. Trends Cell Biol. 8:245–249 10.1016/S0962-8924(98)01267-7 [DOI] [PubMed] [Google Scholar]
- Silva R.M., Ries V., Oo T.F., Yarygina O., Jackson-Lewis V., Ryu E.J., Lu P.D., Marciniak S.J., Ron D., Przedborski S., et al. 2005. CHOP/GADD153 is a mediator of apoptotic death in substantia nigra dopamine neurons in an in vivo neurotoxin model of parkinsonism. J. Neurochem. 95:974–986 10.1111/j.1471-4159.2005.03428.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Scheuner D., Ron D., Pennathur S., Kaufman R.J. 2008. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Invest. 118:3378–3389 10.1172/JCI34587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q., Tsai J., Xu E., Qiu W., Bereczki E., Santha M., Adeli K. 2009. Apolipoprotein B100 acts as a molecular link between lipid-induced endoplasmic reticulum stress and hepatic insulin resistance. Hepatology. 50:77–84 10.1002/hep.22960 [DOI] [PubMed] [Google Scholar]
- Tanti J.F., Jager J. 2009. Cellular mechanisms of insulin resistance: role of stress-regulated serine kinases and insulin receptor substrates (IRS) serine phosphorylation. Curr. Opin. Pharmacol. 9:753–762 10.1016/j.coph.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Thorp E., Li G., Seimon T.A., Kuriakose G., Ron D., Tabas I. 2009. Reduced apoptosis and plaque necrosis in advanced atherosclerotic lesions of Apoe-/- and Ldlr-/- mice lacking CHOP. Cell Metab. 9:474–481 10.1016/j.cmet.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuerauf D.J., Marcinko M., Belmont P.J., Glembotski C.C. 2007. Effects of the isoform-specific characteristics of ATF6 alpha and ATF6 beta on endoplasmic reticulum stress response gene expression and cell viability. J. Biol. Chem. 282:22865–22878 10.1074/jbc.M701213200 [DOI] [PubMed] [Google Scholar]
- Tirosh B., Iwakoshi N.N., Glimcher L.H., Ploegh H.L. 2006. Rapid turnover of unspliced Xbp-1 as a factor that modulates the unfolded protein response. J. Biol. Chem. 281:5852–5860 10.1074/jbc.M509061200 [DOI] [PubMed] [Google Scholar]
- Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., Walter P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 101:249–258 10.1016/S0092-8674(00)80835-1 [DOI] [PubMed] [Google Scholar]
- Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H.P., Ron D. 2000. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 287:664–666 10.1126/science.287.5453.664 [DOI] [PubMed] [Google Scholar]
- van Anken E., Romijn E.P., Maggioni C., Mezghrani A., Sitia R., Braakman I., Heck A.J. 2003. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 18:243–253 10.1016/S1074-7613(03)00024-4 [DOI] [PubMed] [Google Scholar]
- Wang Y., Vera L., Fischer W.H., Montminy M. 2009. The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature. 460:534–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R.C., Jiang H.Y., Anthony T.G. 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34:7–11 10.1042/BST0340007 [DOI] [PubMed] [Google Scholar]
- Winnay J.N., Boucher J., Mori M.A., Ueki K., Kahn C.R. 2010. A regulatory subunit of phosphoinositide 3-kinase increases the nuclear accumulation of X-box-binding protein-1 to modulate the unfolded protein response. Nat. Med. 16:438–445 10.1038/nm.2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman R.L., Zhang Y., Lee K.P., Harding H.P., Haynes C.M., Price J., Sicheri F., Ron D. 2010. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol. Cell. 38:291–304 10.1016/j.molcel.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C.W., Cui D., Arellano J., Dorweiler B., Harding H., Fitzgerald K.A., Ron D., Tabas I. 2009. Adaptive suppression of the ATF4-CHOP branch of the unfolded protein response by toll-like receptor signalling. Nat. Cell Biol. 11:1473–1480 10.1038/ncb1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H.C., Schinke T., Li L., Brancorsini S., Sassone-Corsi P., Townes T.M., et al. 2004. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 117:387–398 10.1016/S0092-8674(04)00344-7 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Oku M., Suzuki M., Mori K. 2006. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 172:565–575 10.1083/jcb.200508145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Uemura A., Mori K. 2009. pXBP1(U), a negative regulator of the unfolded protein response activator pXBP1(S), targets ATF6 but not ATF4 in proteasome-mediated degradation. Cell Struct. Funct. 34:1–10 10.1247/csf.06028 [DOI] [PubMed] [Google Scholar]
- Zhang K., Kaufman R.J. 2008. From endoplasmic-reticulum stress to the inflammatory response. Nature. 454:455–462 10.1038/nature07203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D.T., Back S.H., Kaufman R.J. 2006. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 124:587–599 10.1016/j.cell.2005.11.040 [DOI] [PubMed] [Google Scholar]
- Zhang P., McGrath B., Li S., Frank A., Zambito F., Reinert J., Gannon M., Ma K., McNaughton K., Cavener D.R. 2002. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell. Biol. 22:3864–3874 10.1128/MCB.22.11.3864-3874.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Ackerman S.L. 2006. Endoplasmic reticulum stress in health and disease. Curr. Opin. Cell Biol. 18:444–452 10.1016/j.ceb.2006.06.005 [DOI] [PubMed] [Google Scholar]
- Zhao L., Longo-Guess C., Harris B.S., Lee J.W., Ackerman S.L. 2005. Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat. Genet. 37:974–979 10.1038/ng1620 [DOI] [PubMed] [Google Scholar]
- Zhou J., Liu C.Y., Back S.H., Clark R.L., Peisach D., Xu Z., Kaufman R.J. 2006. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA. 103:14343–14348 10.1073/pnas.0606480103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Zhang J., Fang Q., Liu M., Liu X., Jia W., Dong L.Q., Liu F. 2009. Autophagy-mediated insulin receptor down-regulation contributes to endoplasmic reticulum stress-induced insulin resistance. Mol. Pharmacol. 76:596–603 10.1124/mol.109.057067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H., Stevens J.L., Ron D. 1998. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12:982–995 10.1101/gad.12.7.982 [DOI] [PMC free article] [PubMed] [Google Scholar]