The Ste20 kinase targets the decapping enzyme Dcp2 during stress responses, allowing accumulation of Dcp2 in P-bodies and the formation of stress granules.
Abstract
Translation and messenger RNA (mRNA) degradation are important sites of gene regulation, particularly during stress where translation and mRNA degradation are reprogrammed to stabilize bulk mRNAs and to preferentially translate mRNAs required for the stress response. During stress, untranslating mRNAs accumulate both in processing bodies (P-bodies), which contain some translation repressors and the mRNA degradation machinery, and in stress granules, which contain mRNAs stalled in translation initiation. How signal transduction pathways impinge on proteins modulating P-body and stress granule formation and function is unknown. We show that during stress in Saccharomyces cerevisiae, Dcp2 is phosphorylated on serine 137 by the Ste20 kinase. Phosphorylation of Dcp2 affects the decay of some mRNAs and is required for Dcp2 accumulation in P-bodies and specific protein interactions of Dcp2 and for efficient formation of stress granules. These results demonstrate that Ste20 has an unexpected role in the modulation of mRNA decay and translation and that phosphorylation of Dcp2 is an important control point for mRNA decapping.
Introduction
The proper control of translation, mRNA degradation, and the subcellular localization of mRNAs are important aspects of the regulation of gene expression in eukaryotic cells. Eukaryotic mRNAs are typically degraded in a process initiated by deadenylation, which can lead to 3′ to 5′ degradation but often allows mRNA decapping and 5′ to 3′ degradation (Parker and Song, 2004; Garneau et al., 2007). Decapping is in competition with translation initiation (Coller and Parker, 2004) and occurs by a process involving a set of decapping activator and enhancer proteins that can inhibit translation and/or assemble a translationally repressed mRNP, which contains the decapping enzyme and is capable of decapping and 5′ to 3′ degradation (for review see Parker and Sheth, 2007).
mRNAs that are not engaged in translation can accumulate in a variety of RNA–protein granules in the cytosol. For example, untranslating mRNAs complexed with the decapping machinery accumulate in foci referred to as processing bodies (P-bodies; for reviews see Anderson and Kedersha, 2006; Eulalio et al., 2007; Parker and Sheth, 2007). P-bodies, and/or the mRNPs within them, have been implicated in mRNA decapping, nonsense-mediated decay, mRNA storage, general translation repression, µRNA-mediated repression, and viral life cycles (for reviews see Anderson and Kedersha, 2006; Eulalio et al., 2007; Parker and Sheth, 2007). Transcripts within P-bodies are in dynamic exchange with the translating pool of mRNAs and can either be degraded or can return to translation (Brengues et al., 2005; Bhattacharyya et al., 2006).
P-bodies can partially overlap in yeast (Brengues and Parker, 2007; Hoyle et al., 2007; Buchan et al., 2008) or dock in mammalian cells (Kedersha et al., 2005; Wilczynska et al., 2005), with a second cytoplasmic RNA–protein structure referred to as a stress granule. Stress granules are dynamic aggregates of untranslating mRNAs in conjunction with some translation initiation factors (e.g., eIF4E and eIF4G) and several RNA-binding proteins with the precise composition of stress granules dependent on the stress or organism examined (for reviews see Anderson and Kedersha, 2006; Buchan and Parker, 2009). Stress granules are generally not present in normal cells and form in response to defects in translation initiation, including decreased function of eIF2 or eIF4A (Kedersha et al., 2002; Dang et al., 2006; Mazroui et al., 2006), or heat shock or glucose deprivation in yeast (Brengues and Parker, 2007; Hoyle et al., 2007; Buchan et al., 2008; Grousl et al., 2009). P-bodies also increase during stress in both yeast and mammals presumably because of increases in the pool of nontranslating mRNPs (Kedersha et al., 2005; Teixeira et al., 2005; Wilczynska et al., 2005). At a minimum, the presence of P-bodies and stress granules serves as microscopic markers for pools of biochemically distinct mRNPs, although the additional properties of the larger aggregates remain to be identified.
The interaction of P-bodies and stress granules suggests that mRNA might be exchanged between these two compartments in a process involving remodeling of the composite mRNPs. Consistent with that possibility, stress granules in yeast during glucose deprivation are dependent on P-bodies for their formation and commonly form in association with preexisting P-bodies (Buchan et al., 2008). These observations led to the suggestion that, at least during glucose deprivation in yeast, mRNAs might primarily move from P-bodies to stress granules in a process that would require exchange of P-body components on the mRNA for the proteins seen associated with the mRNA in stress granules such as translation initiation factors (Buchan et al., 2008). Such a remodeling of the mRNP would be expected to impact on the fate of the transcript because transitioning from a P-body mRNP to a stress granule state would reduce the possibility of mRNA degradation and promote reentry into translation. Thus, important and unresolved issues are how individual mRNPs within P-bodies and stress granules are remodeled and how external stimuli activate signaling pathways to alter these interactions, thereby modulating translation and mRNA degradation.
Signaling pathways that activate different MAPKs control many cellular responses to external cues. In Saccharomyces cerevisae, three of the MAPK cascades are activated by the Ste20 protein kinase, which serves as a MAPKKKK. We now show that during stress, Ste20 also directly phosphorylates the decapping enzyme on Ser137. Phosphorylation of Dcp2 affects the decay of some mRNAs, is required for Dcp2 accumulation in P-bodies and specific protein interactions of Dcp2, and for efficient formation of stress granules. These results demonstrate that Ste20 has an unexpected role in the modulation of mRNA decay and translation and that phosphorylation of Dcp2 is an important control point for mRNA decapping.
Results
Dcp2 is phosphorylated during stress
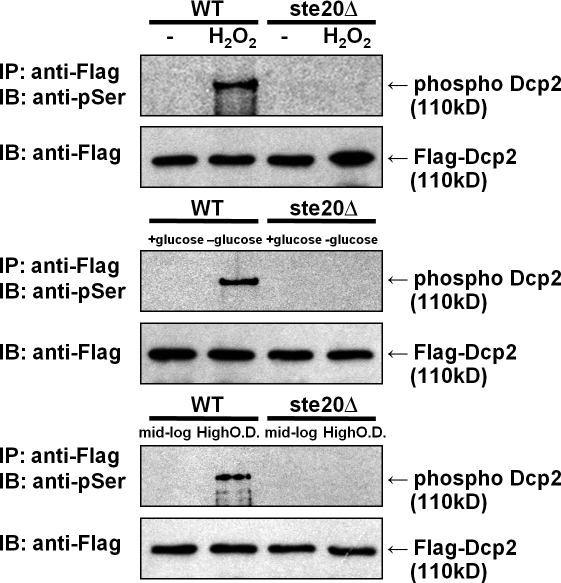
Several stresses, including glucose deprivation and growth to high cell density, lead to enhanced P-body formation and/or inhibition of mRNA degradation, suggesting that some components of the mRNA degradation machinery might be modified under these conditions (Jona et al., 2000; Benard, 2004; Teixeira et al., 2005; Greatrix and van Vuuren, 2006; Hilgers et al., 2006). Because Dcp2 is the critical catalytic component of the decapping enzyme, we examined whether Dcp2 was phosphorylated during glucose deprivation (10 min), oxidative stress induced by hydrogen peroxide (1 mM for 30 min), or at high cell density. We examined Dcp2 phosphorylation by immunopurifying Flag-tagged Dcp2 from cells with or without stress conditions and then performing a Western blot on the immunopurified material with an anti–phospho-Ser–specific antibody.
We observed that hydrogen peroxide exposure, glucose deprivation, or growth to stationary phase all led to the appearance of phosphorylated Dcp2 (Fig. 1). In contrast, in mid-log cultures, phosphorylated Dcp2 was not detected, although Dcp2 immunopurified to similar levels as during stress conditions, as judged by a Western blot (Fig. 1). These results demonstrate that Dcp2 is phosphorylated in response to hydrogen peroxide treatment, glucose deprivation, or growth to high cell density.
Figure 1.
Dcp2 is phosphorylated in response to stress in vivo. Wild-type (WT) and ste20Δ strains in the BY4741 background were transformed with Flag-Dcp2 expression plasmid (pRP983) and grown to reach OD 600 of ∼0.50–0.6. Cells were treated with 1 mM hydrogen peroxide for 30 min or deprived of glucose for 10 min. For examination at high cell density, cells were grown in synthetic media for 48 h. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody–conjugated agarose and probed with anti—phospho-Ser antibody (pSer; see Materials and methods). To detect Flag-Dcp2, the membrane was reprobed with anti-Flag antibody. IB, immunoblot.
Ste20 is required for Dcp2 phosphorylation
To identify protein kinases that potentially phosphorylate Dcp2, we focused on protein kinases activated during stress. Previous work has shown that oxidative stress activates the yeast MAPK pathway, including Ste20, Ste11, Ste7, Fus3, and Kss1. (Staleva et al., 2004). Moreover, a genetic screen identified Ste20 as being important in stress granule formation in yeast (unpublished data). Thus, we hypothesized that Ste20 might be responsible for the phosphorylation of Dcp2 during stress, which we tested by examining whether Dcp2 was phosphorylated during stress in a ste20Δ strain.
We observed that after hydrogen peroxide treatment, glucose deprivation, or growth to high cell density, the ste20Δ strain showed reduced phosphorylation of Dcp2 as compared with a wild-type strain (Fig. 1). Thus, Ste20 either directly phosphorylates Dcp2 or is required for stress-induced phosphorylation of Dcp2 in Saccharomyces cerevisiae by activating a downstream kinase. However, strains lacking the Ste11 and Ste7 proteins, which are downstream of Ste20 in its canonical MAPK pathway, still show phosphorylation of Dcp2 during glucose deprivation (Fig. S1), arguing that Ste20 might directly phosphorylate Dcp2.
Ste20 can directly phosphorylate Dcp2
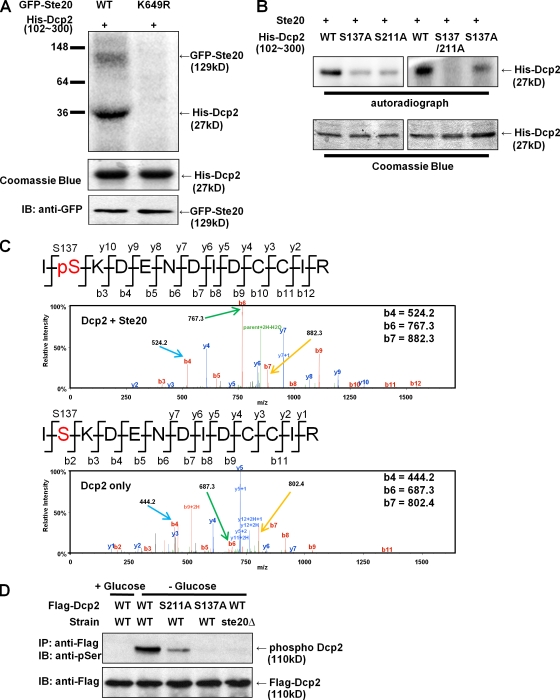
To determine whether Ste20 could directly phosphorylate Dcp2, we immunopurified Ste20 from yeast and determined whether it could phosphorylate recombinant Dcp2 in vitro. In these experiments, a wild-type or kinase-dead mutant allele of Ste20 tagged with GFP at its genomic locus (Ahn et al., 2005) was immunoprecipitated with an anti-GFP antibody. The resulting immunopellet was mixed with the Dcp2 catalytic domain (amino acids 102–300) purified from Escherichia coli in the presence of radioactive ATP. If Ste20 is capable of directly phosphorylating Dcp2, Dcp2 phosphorylation should be observed and should be dependent on the kinase activity of Ste20.
Incubation of Dcp2 102–300 with the wild-type Ste20 immunopellet, but not Ste20 kinase-dead allele (K649R), led to the labeling of an ∼130-kD band, which is likely to be autophosphorylation of Ste20-GFP, and a band running at ∼35 kD (Fig. 2 A). Although this band is larger than the expected size of Dcp2 102–300 (27 kD with tags included), this band comigrates with the major Coomassie-stained band in Dcp2 102–300 preparations, which we have verified by excision of the band followed by mass spectroscopy to be Dcp2 102–300. Additional data that the phosphorylated ∼35-kD migrating band is Dcp2 102–300 is the detection of phosphopeptides from Dcp2 after kinasing with Ste20 (see below). These results argue that Ste20 can directly phosphorylate Dcp2, which is confirmed by demonstrating that Ste20 purified from E. coli was also able to phosphorylate Dcp2 102–300 (Fig. S1 B).
Figure 2.
Ste20 phosphorylates Dcp2 in vitro and in vivo. (A) GFP-tagged wild-type (WT) or kinase-dead Ste20 (K649R) integrated at the genomic locus was immunoprecipitated with anti-GFP antibody. The resulting immunopellets were incubated with Dcp2 catalytic domain (residues 102–300; pRP1211) purified from E. coli in the presence of radioactive ATP for 30 min at 30°C. The reaction mixture was subjected to SDS-PAGE and autoradiograph. (B) Dcp2 S137 and/or S211 residue was mutated to an alanine residue with site-directed mutagenesis (pRP1678, pRP1684, or pRP1685). The recombinant protein was purified from E. coli and mixed with Ste20 purified from yeast for in vitro phosphorylation analysis. (C) His-tagged Dcp2 (102–300) was incubated with (top) or without (bottom) GST-Ste20 purified from E. coli (pRP2135) in the presence of nonradioactive ATP. The resulting reaction mixtures were digested with trypsin for MS analysis. Product b and y ions were indicated in the peptide sequences and mass spectrum, respectively. Key m/z values were indicated to compare mass shift after phosphorylation. The b4, b5, and b6 peptides, which show a shift corresponding to phosphorylation, are marked with colored arrows. (D) Plasmids expressing Dcp2 wild type, S137A, or S211A (pRP983, pRP1676, or pRP1682, respectively) were transformed into wild-type or ste20Δ strains. Cells were grown in mid-log phase and exposed to glucose deprivation stress. Cell lysates were examined to detect phosphorylated Dcp2 by SDS-PAGE and immunoblot (IB) assay.
Ser137 is a target for Dcp2 phosphorylation in yeast
To determine the significance of Dcp2 phosphorylation, we desired to identify the Ser and/or threonine residues where yeast Dcp2 was phosphorylated and then use genetic approaches to address the function of phosphorylation. Because the phosphorylation sites in vitro were localized in the catalytic domain of yeast Dcp2 (residues 102–300), we examined this portion of Dcp2 for possible Ste20 phosphorylation sites based on comparison with the sites mapped in histone H2B and Ste11 (Wu et al., 1995; Ahn et al., 2005). This analysis identified Ser137 (S137) and 211 (S211) as possible Ste20 phosphorylation sites. We tested this possibility by mutating these sites to alanine either individually or in combination and examining their phosphorylation in vitro with immunopurified Ste20 from yeast or affinity-purified Ste20 from E. coli. We observed that the extent of Dcp2 phosphorylation by Ste20 in vitro was reduced after either the S137A or S211A mutations (Fig. 2 B and Fig. S1 B). Moreover, the double-mutant S137A, S211A, showed almost a complete loss to phosphorylation in vitro (Fig. 2 B and Fig. S1 B). We interpret these results to indicate that both S137 and S211 can serve as sites of Ste20 phosphorylation in vitro.
To verify that Dcp2 was phosphorylated by Ste20 on Ser137, we phosphorylated Dcp2 in vitro with recombinant Ste20 purified from E. coli and analyzed the products by mass spectrometry (MS; see Materials and methods). We observed phosphorylated peptide fragments corresponding to phosphorylation on Ser137 when Dcp2 was incubated with Ste20 and ATP (Fig. 2 C). This provides direct evidence that Ste20 phosphorylates Dcp2 on Ser137.
In vivo, the specificity of phosphorylation might be influenced by additional factors. Thus, we examined how the S137A and S211A mutations affected the phosphorylation of Dcp2 in yeast during glucose deprivation. We observed that the S137A mutant was no longer phosphorylated, whereas the S211A mutant showed reduced phosphorylation (Fig. 2 D). All proteins were equivalently immunopurified based on Western analysis for the Flag epitope fused to Dcp2. We also observed that the S137A mutant showed reduced phosphorylation after hydrogen peroxide treatment, whereas the S211A mutant was phosphorylated to levels similar to wild-type Dcp2 (unpublished data). These observations argue that S137 is required for phosphorylation in vivo by Ste20 and is likely to be the major site of phosphoryl group addition by Ste20 in cells. However, there may be additional sites, including S211A, that are phosphorylated in some stresses. Additional evidence that S137 is the key site for phosphorylation during stress is the phenotypes of a charge-mimetic allele at this position (see below).
Consequences of Dcp2 phosphorylation
The aforementioned results argued that Dcp2 is phosphorylated on S137A during stress by Ste20. To determine the role of Dcp2 phosphorylation, we examined the consequences of mutations that either prevent phosphorylation (S137A) or are charge mimetic (S137E) on mRNA decapping, the formation of P-bodies, and stress granules. In addition, because Ste20 affects Dcp2 phosphorylation, we also examined the effects of a ste20Δ in these assays. Strikingly, we observed that dcp2Δ strains expressing the dcp2-S137A allele grew more slowly than strains expressing the wild-type or S137E allele (see Discussion). This indicates that phosphorylation of Dcp2 is required for optimal growth rate. More detailed experiments to understand the role of Dcp2 phosphorylation are described in Materials and methods.
Dcp2 phosphorylation does not generally affect decapping
Because Dcp2 is the decapping enzyme, we first asked whether alteration of the phosphorylation site affected its catalytic activity. S137 is located adjacent to, but does not overlap, the active site of Dcp2 (She et al., 2006, 2008; Deshmukh et al., 2008). Given this, we purified the catalytic domain of Dcp2 from E. coli (residues 102–300) either as wild-type or with the S137A or S137E mutations and assayed the catalytic ability of this protein in vitro with a cap-labeled substrate based on the MFA2 mRNA. We observed that the S137A or S137E mutation did not substantially alter the decapping activity of Dcp2 in vitro (Fig. S2). Based on this, we suggest that phosphorylation does not directly inhibit or stimulate Dcp2 enzymatic activity.
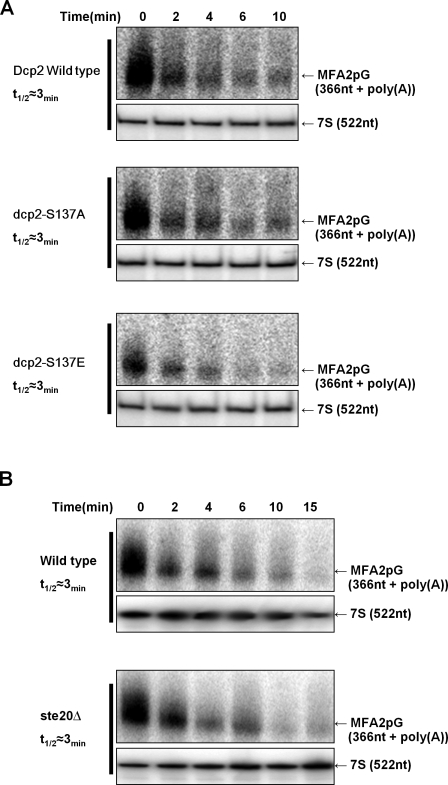
To examine the effects of these lesions on decapping in vivo, we examined the decay of the MFA2pG reporter mRNA, which is under control of the GAL promoter, in dcp2Δ strains transformed with plasmids expressing Dcp2 wild type, dcp2-S137A, or dcp2-S137E. We examined mRNA decay during both mid-log growth, in which Dcp2 is generally not phosphorylated and mRNA decay is normal, and during glucose deprivation, in which Dcp2 is phosphorylated and mRNA decay is inhibited, primarily by a block to deadenylation (Hilgers et al., 2006).
We observed that the decay rate of the MFA2pG mRNA was largely unaffected by the S137A or S137E alleles of Dcp2 in both mid-log cultures and during glucose deprivation (Fig. 3 A and Fig. S3). However, we did observe that the total levels of mRNA were consistently reduced in the S137E strain for unknown reasons (≤40% mRNA compared with wild type). Nevertheless, the main implication is that Dcp2 phosphorylation on S137 does not globally alter mRNA decay in vivo. Similarly, we observed that the decay of the MFA2pG reporter was the same in ste20Δ and wild-type strains both in mid-log and stress conditions (Fig. 3 B and Fig. S3). We interpret these results to indicate that phosphorylation of Dcp2 does not globally alter mRNA decay, although it remains possible that Dcp2 phosphorylation affects the decapping of a subset of mRNAs (see below).
Figure 3.
Dcp2 phosphorylation is not required for mRNA decay. (A) dcp2Δ strain (yRP1358) expressing Gal-MFA2pG mRNA was transformed with centromere plasmids expressing either wild-type, S137A, or S137E Dcp2-GFP (pRP1275, pRP1677, or pRP1680, respectively; Coller and Parker, 2005; this study). Cells were grown in synthetic media containing galactose, and transcription was repressed by changing to glucose media. At each time point, cells were harvested, and total RNA was analyzed by Northern blotting. (B) BY4741 wild-type or ste20Δ was transformed with GAL-MFA2pG plasmid and grown in minimal media with galactose. MFA2pG transcription was blocked by glucose addition, and MFA2pG mRNA levels were examined over time by Northern blotting.
Dcp2 phosphorylation affects its localization in P-bodies
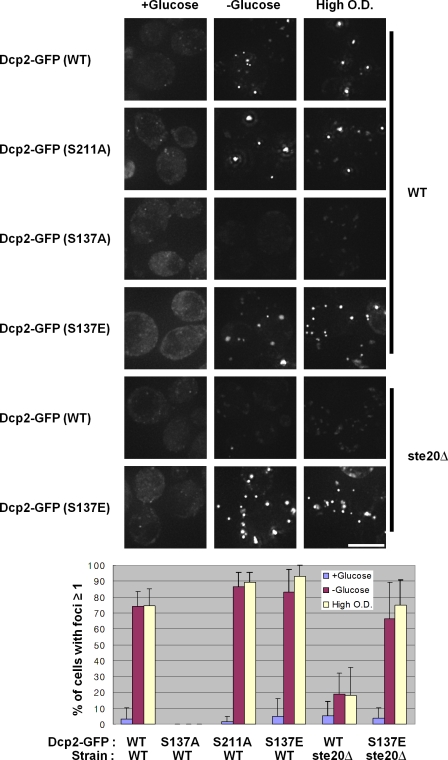
We also investigated the effect of Dcp2 phosphorylation on the subcellular location of Dcp2. During stresses such as glucose deprivation and high cell density, Dcp2 accumulates in P-bodies (Teixeira et al., 2005). We transformed Dcp2-GFP expression plasmids either with or without the S137A, S137E, or S211A mutations in wild-type strains and examined the subcellular location of Dcp2 in mid-log cultures as well as those exposed to glucose deprivation and grown to stationary phase.
We observed that the Dcp2-S137A-GFP failed to accumulate in P-bodies during glucose deprivation and growth to high cell density (Fig. 4). In contrast, the Dcp2 wild-type, Dcp2-S211A, and Dcp2-S137E proteins all accumulated in P-bodies. The failure of Dcp2-S137A proteins to accumulate in P-bodies is not because of changes in its expression levels (Fig. S4). These observations argue that phosphorylation of Dcp2 is required for its efficient accumulation in P-bodies.
Figure 4.
Dcp2 phosphorylation is necessary for its accumulation in P-bodies. A wild-type (WT) strain (BY4741) was transformed with wild-type, S137A, S137E, or S211A Dcp2-GFP (pRP1683) plasmids. Cells were grown in synthetic media to mid-log and deprived of glucose for 10 min or grown to high cell density before microscopic examination on a microscope. A ste20Δ strain in the BY4741 background was transformed with wild-type or S137E Dcp2-GFP plasmid and exposed to glucose deprivation or high OD stress. Cells having at least one Dcp2 foci were counted by ImageJ for quantification (see Materials and methods). Error bars indicate SD. Bar, 5 µm.
If Dcp2 phosphorylation is required for its accumulation in P-bodies, strains lacking Ste20, which affects Dcp2 phosphorylation, should also show a defect in the accumulation of Dcp2 in P-bodies. Moreover, if this defect is largely caused by the loss of Dcp2 phosphorylation, then a charge-mimetic allele of Dcp2 would be predicted to restore Dcp2 accumulation in P-bodies in a ste20Δ strain. To test these predictions, we transformed a ste20Δ strain with GFP-tagged versions of either wild-type Dcp2 or the different mutant alleles and examined their location during mid-log growth and after glucose deprivation.
This experiment revealed the following important observations. First, we observed that the accumulation of the wild-type Dcp2-GFP protein in P-bodies was reduced in ste20Δ strains (Fig. 4). The accumulation of Dcp2-GFP in P-bodies in the ste20Δ was not reduced as much as the accumulation of the Dcp2-S137A protein in a wild-type strain. This suggests that additional kinases might be able to phosphorylate Dcp2 at low levels. Consistent with this possibility, we observed that after long exposures, low levels of phosphorylated Dcp2 could be detected in the ste20Δ strain during glucose deprivation (unpublished data). Nevertheless, the reduction in Dcp2-GFP accumulation in P-bodies in the ste20Δ strain provides additional evidence that phosphorylation of Dcp2 promotes its accumulation in P-bodies during stress.
A second important observation was that the charge-mimetic dcp2-S137E allele rescued the defect in Dcp2 accumulation seen in the ste20Δ strain (Fig. 4). This observation strongly argues that the defect in Dcp2 accumulation in P-bodies in the ste20Δ strain is caused by the failure of Dcp2 to get phosphorylated.
A third observation was that the dcp2-S137E mutant did not accumulate above normal levels in P-bodies during mid-log growth (Fig. 4). This argues that phosphorylation of Dcp2 is not sufficient by itself to induce large P-bodies, although Dcp2 phosphorylation is necessary during stress responses for the accumulation of Dcp2 in P-bodies.
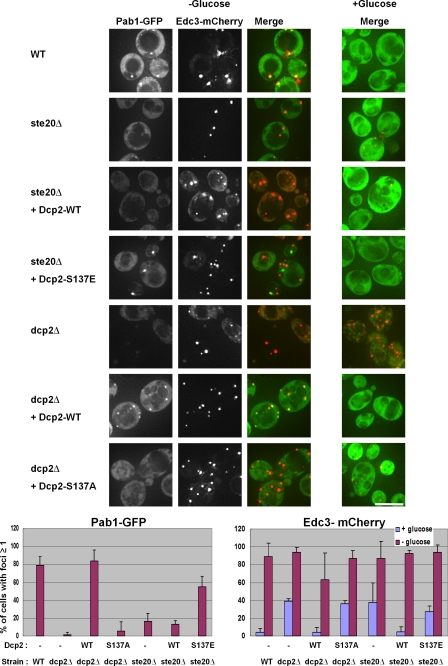
Dcp2 phosphorylation affects stress granule formation but not P-body formation
The aforementioned observations indicated that phosphorylation of Dcp2 was required for its accumulation in P-bodies during stress. This could be because phosphorylation of Dcp2 is required for P-bodies to form or because Dcp2 phosphorylation specifically affects Dcp2 accumulation in P-bodies. Given this, we examined how P-bodies formed in the various Dcp2 alleles as well as in ste20Δ strains using Edc3-mCherry as a marker of P-bodies. Moreover, because recent results suggest that P-bodies promote the formation of stress granules in yeast (Buchan et al., 2008), we also examined how the Dcp2-S137A and -S137E alleles and the ste20Δ affected stress granule formation in the same experiments using a Pab1-GFP fusion protein as a marker of yeast stress granules (Buchan et al., 2008). Thus, either wild-type, ste20Δ, or various dcp2 mutant strains were transformed with a centromere plasmid expressing Edc3-mCherry (a P-body marker) and Pab1-GFP (a stress granule marker) protein fusions and their subcellular location examined with and without glucose deprivation. These experiments revealed the following points.
First, we observed that dcp2Δ strains expressing the dcp2-S137A allele still produced P-bodies as judged by the accumulation of Edc3-mCherry (Fig. 5). Moreover, as seen previously, dcp2Δ strains also formed robust P-bodies during glucose deprivation and formed enhanced P-bodies during mid-log growth, presumably caused by a defect in mRNA decapping (Sheth and Parker, 2003; Teixeira and Parker, 2007). These results demonstrate that neither Dcp2 phosphorylation nor Dcp2 itself is required for P-body formation per se and, therefore, demonstrates that phosphorylation of Dcp2 is required for Dcp2 accumulation in P-bodies.
Figure 5.
Dcp2 phosphorylation affects stress granule but not P-body formation. Wild-type (WT; BY4742), ste20Δ (yRP2547), or dcp2Δ (yRP1358) strains were transformed with Pab1-GFP/Edc3-mCherry plasmid (pRP1658; Buchan et al., 2008), grown to OD 600 of ∼0.5–0.6 in synthetic media, and deprived of glucose for 10 min before the localization of Pab1p-GFP and Edc3p-mCherry were examined with a microscope. In addition, the dcp2Δ strain was transformed with wild-type or S137A Flag-Dcp2 plasmid (pRP983 or pRP1676) before glucose deprivation. The ste20Δ strain containing Pab1-GFP/Edc3-mCherry plasmids was transformed with wild-type or S137E Flag-Dcp2 (pRP983 or pRP1679). Images were quantified as in Fig. 4 and in Materials and methods. Error bars indicate SD. Bar, 5 µm.
Second, we observed that Dcp2 wild-type strains efficiently formed stress granules, whereas the dcp2Δ and dcp2-S137A strains showed reduced accumulation of stress granules (Fig. 5, Pab1-GFP). These results suggest that Dcp2 and its phosphorylation are required for optimal stress granule formation. We also observed that ste20Δ strains showed reduced stress granule formation during glucose deprivation as compared with wild-type cells, although P-bodies formed normally in the ste20Δ strain (Fig. 5). This observation argues that Ste20 enhances stress granule formation either through phosphorylation of Dcp2 and/or by phosphorylation of additional proteins. The observed changes in Pab1 accumulation were not caused by reduction of protein expression (Fig. S4).
The requirement for Dcp2 phosphorylation for stress granule formation suggests that the requirement for Ste20 in stress granule formation is at least in part caused by phosphorylation of Dcp2. This predicts that the dcp2-S137E allele would restore stress granule formation in the ste20Δ strain. To test this possibility, we expressed the wild-type or dcp2-S137E alleles in a ste20Δ strain and examined P-body and stress granule formation in response to glucose deprivation. We observed that the ste20Δ strain expressing the Dcp2-S137E protein showed partially restored stress granule formation as compared with the wild-type strain, whereas ste20Δ strains expressing exogenous wild-type Dcp2 still showed a defect in stress granule formation (Fig. 5). The ability of the charge-mimetic form of Dcp2 to restore stress granule formation in the ste20Δ strain provides additional evidence that phosphorylated Dcp2 promotes stress granule formation and also provides strong evidence that at least part of the role of Ste20 in stress granule formation is to phosphorylate Dcp2.
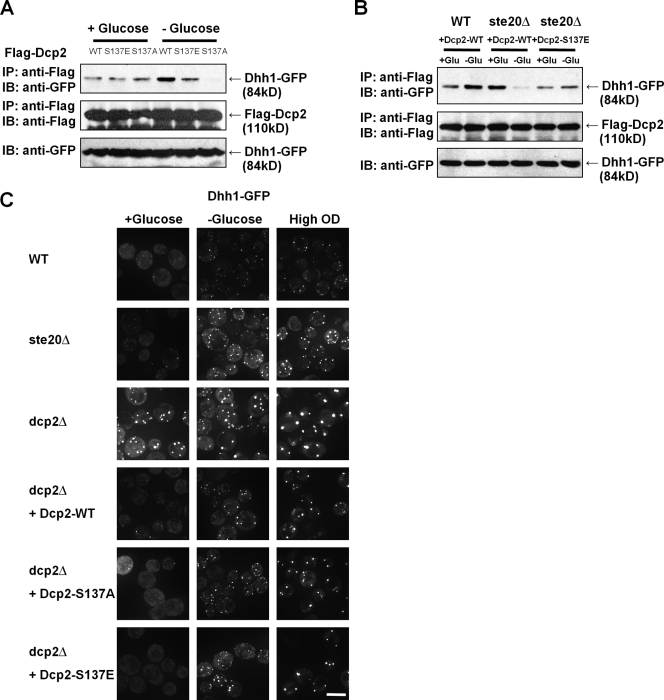
Dcp2 phosphorylation is required to maintain Dhh1–Dcp2 interactions during glucose deprivation
One possible mechanism by which Dcp2 phosphorylation promoted stress granule formation is that Dcp2 phosphorylation alters protein–protein interactions within mRNPs accumulating in P-bodies, and thereby leads to mRNP transitions that transform an mRNA into a stress granule mRNP. This possibility is also raised by the observation that stress granule formation in S. cerevisiae is promoted by preexisting P-bodies (Buchan et al., 2008). Interestingly, the Dhh1 protein, which interacts with Dcp2 (Decker et al., 2007) and localizes to P-bodies, is required for optimal stress granule formation (Buchan et al., 2008). This suggested a possible mechanism whereby Dcp2 phosphorylation might alter the Dhh1–Dcp2 interaction and thereby affect the ability of Dhh1 to promote stress granule formation. Given this, we examined the coimmunoprecipitation of wild-type, S137A, and S137E Dcp2 variants with Dhh1 in wild-type or ste20Δ strains.
In wild-type strains, we observed that during mid-log, Dhh1 coimmunoprecipitated with Dcp2, and this coimmunoprecipitation was unaffected by the S137E or S137A alleles. This indicates that Dcp2 can interact with Dhh1 independent of the phosphorylation status of Dcp2 during mid-log growth. In contrast, during glucose deprivation, we observed that the S137A allele showed reduced ability to coimmunoprecipitate Dhh1, although it was expressed at normal levels (Fig. 6 A). Similarly, in ste20Δ strains, Dhh1 and Dcp2 coimmunoprecipitated during mid-log cultures but showed reduced interaction during glucose deprivation (Fig. 6 B). Strikingly, the S137E allele of Dcp2 could restore the interaction with Dhh1 during glucose deprivation in the ste20Δ strain. These observations argue that the interaction between Dhh1 and Dcp2 is altered during stress either directly or indirectly and that phosphorylation of Dcp2 is required to maintain the interaction of Dhh1 and Dcp2 under stress conditions.
Figure 6.
Dcp2 phosphorylation is required for its interaction with Dhh1p during glucose deprivation. (A) Wild-type (WT; BY4742) strains were transformed with expression plasmid of Dhh1-GFP (pRP 1151; Coller et al., 2001) controlled by its own promoter and centromeric replication origin. Again, these strains were cotransformed with Flag-tagged wild-type (pRP983), S137A (pRP1676), or S137E (pRP1679) Dcp2 plasmids. Cells were grown in synthetic media to reach OD 600 of ∼0.5–0.6 with or without glucose deprivation for 10 min. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody–conjugated beads, and copurified Dhh1p-GFP was detected with anti-GFP antibody after SDS-PAGE. (B) Wild-type or ste20Δ (yRP2547) strains was cotransformed with plasmids expressing Dhh1-GFP and plasmids expressing Flag-tagged wild-type or S137E Dcp2 as indicated. (C) Wild-type, ste20Δ, or dcp2Δ (yRP1358) strains were transformed with a plasmid expressing Dhh1-GFP, or in some cases, the dcp2Δ strain was cotransformed with plasmids expressing Dcp2-WT, S137A, or S137E as indicated. Dhh1 localization was examined with a microscope with or without stress. IB, immunoblot. Bar, 5 µm.
The requirement for Dcp2 phosphorylation to maintain interactions with Dhh1 and to assemble into P-bodies suggested two possible models by which these events could be occurring, which can be distinguished by the subcellular location of Dhh1 during stress. In one model, stress induces translation repression, forming an initial P-body containing Edc3 and Dhh1 (as well as other proteins), then phosphorylated Dcp2 would be recruited to this complex, which predicts that Dhh1 accumulation in P-bodies would be independent of Dcp2 phosphorylation. In an alternative model, the order of assembly would be formation of a P-body containing Edc3 (as well as other proteins), which then recruits phosphorylated Dcp2, leading to the recruitment of Dhh1, which predicts that Dhh1 accumulation in P-bodies would be dependent of Dcp2 phosphorylation. Thus, we examined the subcellular location of Dhh1-GFP in various mutants affecting Dcp2 phosphorylation with or without stress.
Consistent with earlier results (Teixeira and Parker, 2007), we observed that Dhh1 accumulated in P-bodies during mid-log in a dcp2Δ strain. More importantly, we observed that Dhh1-GFP was also present in P-bodies in dcp2-S137A and ste20Δ strains (Fig. 6 C). This indicates that Dhh1 recruitment into P-bodies is independent of Dcp2 or its phosphorylation status and is therefore upstream of Dcp2 recruitment into P-bodies.
Dcp2 phosphorylation affects the expression and decay of certain mRNAs
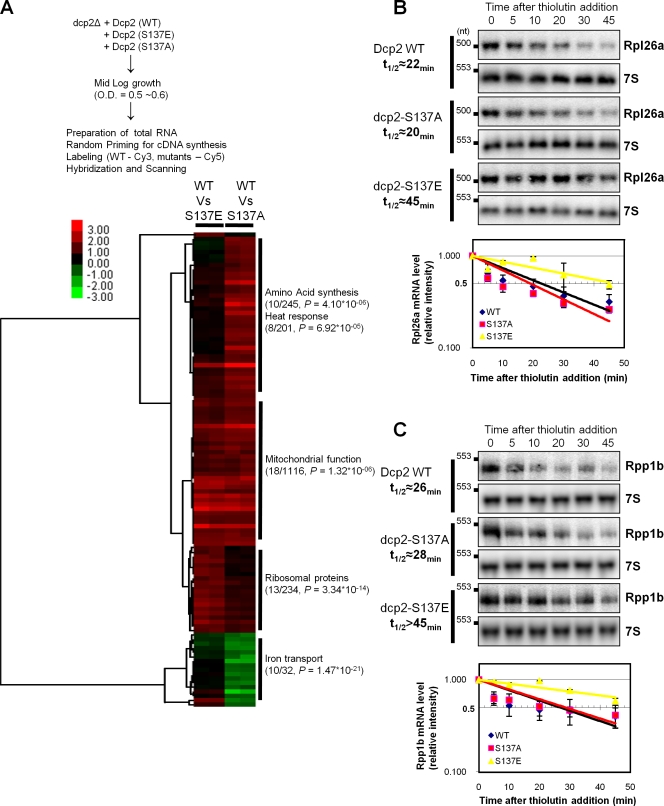
The aforementioned results raised the possibility that phosphorylation of Dcp2 might affect the decay of some, but not all, transcripts. To identify mRNAs whose degradation might be affected by Dcp2 phosphorylation, we performed microarray analysis comparing the dcp2-S137E and dcp2-S137A alleles with wild-type Dcp2, which led to several important observations.
Most importantly, in dcp2-S137E cells, no mRNAs were down-regulated more than twofold, and 40 mRNAs out of ∼6,200 were up-regulated more than twofold, which suggests that Dcp2 phosphorylation stabilizes a subset of mRNAs. Strikingly, the mRNAs up-regulated in the dcp2-S137E stain were overrepresented in ribosomal protein mRNAs (Fig. 7 A), suggesting that Dcp2 phosphorylation led to preferential stabilization of this class of mRNAs. Moreover, by direct measurement of mRNA decay rates, we validated that two ribosomal protein mRNAs (Rpl26a and Rpp1b) showed slower rates of mRNA degradation in the dcp2-S137E strain as compared with either wild-type or dcp2-S137A strains (Fig. 7, B and C). Collectively, these results argue that phosphorylation of Dcp2 at S137 stabilizes a subclass of mRNAs enriched in ribosomal mRNAs.
Figure 7.
Dcp2 phosphorylation is required for the expression and degradation of certain mRNAs. (A) The schematic of expression microarray analysis and gene clustering of 109 genes, which are up-regulated or down-regulated more than twofold in dcp2-S137E or 137A allele. Number of genes found in the microarray, total genes involved in specific gene ontology, and p-values were described. (B and C) Dcp2-WT, dcp2-S137A, or dcp2-S137E strains (yRP2680, yRP2681, or yRP2682, respectively) were grown in synthetic media containing to reach mid-log. And the cells were incubated with 5 µg/ml thiolutin for the indicated times, and total RNA was prepared at each time point for Northern blot analysis of Rpl26a or Rpp1b mRNA. The relative intensity of each band was quantified for a half-life measurement with two independent experiments.
Our microarray results also revealed other alterations in mRNA levels in response to alterations at S137. We observed that there was a class of mRNAs, preferentially enriched in mitochondrial function (Fig. 7 A), that were increased in both the dcp2-S137E and dcp2-S137A strains as compared with wild-type and, therefore, may be mRNAs whose degradation is normally enhanced by S137. We also observed a class of mRNAs, preferentially enriched in mRNAs involved in amino acid synthesis or heat response, which were unregulated in the dcp2-S137A strain and unaffected in the dcp2-S137E strain. Finally, we observed a class of mRNAs enriched in iron transporters, which were unaffected in the dcp2-S137E strain but down-regulated in the dcp2-S137A strain. Collectively, these results indicate that modification of Dcp2 on S137 impacts the levels of several mRNAs by both direct and indirect mechanisms.
Discussion
Ste20 modulates mRNA decay by phosphorylation of Dcp2
Our observations indicate that Dcp2 is phosphorylated by Ste20 during certain stresses. Specifically, during glucose deprivation, hydrogen peroxide exposure or growth to high cell density, immunopurified Dcp2 reacts with antibody against phospho-Ser in a manner dependent on Ste20 (Fig. 1). Moreover, immunopurified or recombinant Ste20 can phosphorylate the catalytic region of Dcp2 in vitro on Ser137 as confirmed by MS analysis (Fig. 2). Phosphorylation of Dcp2 by Ste20 is primarily on Ser137 because mutation of Ser137 to alanine reduces phosphorylation of Dcp2 by Ste20 in vitro and in vivo (Fig. 2 B, Fig. S1 B, and Fig. S2 D). In addition, substitution of Ser137 with a charge-mimetic allele, S137E, suppresses some of the defects seen in a ste20Δ strain (Figs. 4 and 5). These results reveal an unexpected function of Ste20, modifying the decapping enzyme, and demonstrate that activation of Ste20 leads to a bifurcated response, whereby phosphorylation of a downstream MAPK cascade leads to transcriptional regulation (Herskowitz, 1995), and Ste20 directly impinges on molecules involved in posttranscriptional control, thereby affecting the rates of translation and/or decay of certain mRNAs.
An unresolved issue is the cellular compartment wherein Ste20 phosphorylates Dcp2. Ste20 shuttles between the nucleus and cytoplasm, and during oxidative stress accumulates in the nucleus (Ahn et al., 2005). Because Dcp2 also shuttles between the nucleus and cytosol (Grousl et al., 2009), it could be that Ste20 phosphorylates Dcp2 in the nucleus. However, because some Ste20 remains in the cytosol during oxidative stress (Ahn et al., 2005) and we have not observed accumulation of Ste20 in the nucleus during glucose deprivation or growth to high cell concentrations (unpublished data), it could be that Dcp2 is phosphorylated by cytoplasmic Ste20.
Dcp2 phosphorylation affects its accumulation in P-bodies
Several observations indicate that phosphorylation of Dcp2 by Ste20 affects its accumulation in P-bodies and its protein–protein interactions. First, the S137A allele of Dcp2 fails to accumulate in P-bodies during stress, although P-bodies form as judged by Edc3p accumulation (Figs. 4 and 5). Second, ste20Δ strains still form P-bodies during glucose deprivation, as judged by accumulation of Edc3, but fail to recruit Dcp2 to these complexes (Fig. 5). Third, the ability of Dcp2 to accumulate in P-bodies in the ste20Δ strain can be restored by the charge-mimetic S137E allele of Dcp2 (Fig. 4). Fourth, the dcp2-S137A allele in wild-type cells and the wild-type Dcp2 in ste20Δ strains show reduced coimmunoprecipitation with Dhh1 during glucose deprivation (Fig. 6). Moreover, the reduced interaction of Dcp2 and Dhh1 in the ste20Δ strain can be restored by the S137E allele (Fig. 6). Thus, phosphorylation of Dcp2 increases its coimmunoprecipitation with Dhh1 at least during glucose deprivation. These results indicate that the phosphorylation of Dcp2 is likely to change some aspect of its protein–protein interactions, thereby affecting the assembly of an mRNP containing the decapping enzyme.
One surprising observation was that the Dhh1–Dcp2 interaction is normal during mid-log growth when Dcp2 is not phosphorylated, yet requires Dcp2 to be phosphorylated during glucose deprivation to maintain the interaction. This could be because phosphorylation of Dcp2 is required to counterbalance additional modifications to Dhh1 or Dcp2 during stress that directly reduces their interaction. Alternatively, during stress, Dcp2 might interact with other factors that compete for Dhh1 binding to Dcp2 and that phosphorylation decreases the binding to allow Dhh1 to bind more tightly.
Dcp2 phosphorylation promotes stress granule formation
Several observations indicate that phosphorylation of Dcp2 by Ste20 affects the formation of stress granules. First, strains lacking Dcp2 or Ste20 show reduced numbers and fainter stress granules during glucose deprivation (Fig. 5). This is not a result of defects in decapping per se because dcp1Δ strains, which are as defective as dcp2Δ strains in decapping, show increased numbers of stress granules during glucose deprivation (Buchan et al., 2008). Second, strains with the dcp2-S137A allele show a defect in forming stress granules (Fig. 5). Third, the defect in stress granule formation in a ste20Δ strain can be partially suppressed by expression of the Dcp2-S137E protein. This latter observation indicates that at least part of the defect in stress granule formation of the ste20Δ strain is the result of a failure to phosphorylate Dcp2. These observations demonstrate that Dcp2 is not just a decapping enzyme but plays a second role in stress granule formation in a manner dependent on its phosphorylation. This also provides additional evidence that stress granule formation during glucose deprivation in yeast is dependent on P-bodies (Buchan et al., 2008).
Our results suggest that Ste20 might also impact stress granule formation through additional targets. This possibility is based on the observations that although the ste20Δ strain shows dramatically decreased stress granule formation during glucose deprivation, a dcp2Δ strain only has partially reduced stress granules. Moreover, the dcp2-S137E allele only partially rescued the ste20Δ defect in stress granule formation. Another possible target for Ste20 affecting stress granule formation is eIF-4E, which was identified as a substrate for Ste20 in a genome-wide screen (Ptacek et al., 2005).
An unresolved issue is how phosphorylated Dcp2 promotes stress granule formation. One possibility is that when mRNAs are exiting translation, they can either assemble an mRNP capable of accumulation into stress granules or an mRNP capable of assembling into P-bodies, and that phosphorylated Dcp2 limits the accumulation of the P-body type of mRNP, thereby promoting stress granule formation. However, three observations make this possibility unlikely. First, recent results indicate that P-bodies promote the assembly of stress granules in yeast and are not in competition with stress granule formation (Buchan et al., 2008). Second, we observe that dcp2Δ strains are also defective in stress granule formation (Fig. 5), which is inconsistent with Dcp2 functioning in competition with stress granule formation. Third, we observe that dcp2-S137A and dcp2Δ strains not only show reduced stress granules, they show increased size and intensity of P-bodies in mid-log as judged by Edc3. Given this, the simplest interpretation is that phosphorylation of Dcp2 increases the formation of stress granules by affecting remodeling of mRNPs within P-bodies and thereby promoting the transition of some mRNPs from P-bodies into stress granules, perhaps by affecting Dhh1 function, which is required for optimal stress granule assembly (Buchan et al., 2008). This model provides a possible explanation for why dcp2-S137A strains grow poorly (unpublished data). In the absence of Dcp2 phosphorylation, some mRNAs will be unable to exit P-bodies and reenter translation, which is consistent with the small P-body formation in S137A strains even during mid-log growth (Fig. 5).
Dcp2 phosphorylation stabilizes some mRNAs encoding ribosomal proteins
Several observations argue that phosphorylation of Dcp2 modulates the levels and/or decay of some, but not all, yeast mRNAs. Most importantly, microarray analysis of the dcp2-S137E allele revealed an increase in a set of mRNAs enriched in ribosomal proteins (Fig. 7 A). Moreover, direct measurement of the mRNA decay rates of some of these mRNAs revealed that they were more stable in the dcp2-S137E strain as compared with either wild-type or dcp2-S137A strains (Fig. 7, B and C). However, this effect is limited to a subset of mRNA conditions because the levels of many mRNAs were not changed in the different Dcp2 alleles, and phosphorylation of Dcp2 did not affect the decay of the MFA2pG reporter mRNA (Fig. 3). To date, we have only been able to examine the effects of Dcp2 phosphorylation during mid-log growth because during the stresses we have used to date, mRNA deadenylation is also inhibited, and bulk mRNA is stabilized (Hilgers et al., 2006), thus obscuring our ability to measure the effect of Dcp2 phosphorylation on decapping of the total population of mRNAs. However, it is likely that the role of Dcp2 phosphorylation under stress would be to stabilize the subpopulation of mRNAs that are already deadenylated. Because we only analyzed mRNAs that changed greater than twofold in our microarray experiments, it is possible that additional mRNAs are affected by Dcp2 phosphorylation to a smaller degree. Thus, phosphorylation of Dcp2 stabilizes a subset of mRNAs, particularly those involved in ribosome biogenesis. Because the mRNAs involved in ribosome biogenesis represent a large fraction of the total mRNA in the cell, this suggests that during stress, Dcp2 phosphorylation contributes to the formation of an mRNP that can selectively promote the remodeling of this class of mRNPs from a P-body state to a stress granule complex. Such a remodeling would be expected to stabilize the mRNA by reducing its interactions with the decapping complex and possibly also promote its subsequent reentry into translation. Thus, posttranslational modification of the decapping enzyme by Ste20 plays an important role in modulating the fate of cytoplasmic mRNAs.
Materials and methods
Yeast strains, growth conditions, plasmids, and oligonucleotides
The list of strains, plasmids, and oligonucleotides used in this work are shown in Tables S1, S2, and S3, respectively. Strains were grown at 30°C in a shaking water incubator in yeast/extract/peptone medium or synthetic medium supplemented with the appropriate amino acid drop out solutions and 2% glucose. All site-directed mutagenesis was performed using standard protocols, and resulting plasmids were verified by sequencing (Wang and Malcolm, 1999).
Microscopic analysis
Cells were grown to OD 600 of ∼0.5–0.6 in synthetic media with 2% glucose. For glucose deprivation, cells were centrifuged and quickly washed with synthetic medium lacking glucose. Pellets were resuspended in synthetic medium lacking glucose and incubated for 10 min in a shaking water bath at 30°C. Images were collected by a deconvolution microscope (Deltavision RT; Applied Precision) with a UPlan S Apo 100× 1.4 NA objective (Olympus). 512 × 512–pixel files were acquired with a camera (CoolSNAP HQ; Photometrics) by 1 × 1 binning. Z-series images were compiled with maximum intensity projections using ImageJ (National Institutes of Health). Each image was blindly randomized and quantified as described previously (Buchan et al., 2008). The counting and measurement of foci size were performed using ImageJ with smoothing, thresholding, and analyze particle functions. Quantitation datasets represent the analysis of at least two independent experiments with at least 50 cells. The details for quantitation are shown in Table S4.
Western blot analysis
Western blots were performed following standard protocols. The following antibodies and their sources were used: anti–phospho-Ser antibody (BD), anti-Flag antibody (Sigma-Aldrich), anti-GFP antibody (Covance), or anti-actin antibody (Abcam). Goat anti–mouse HRP (Thermo Fisher Scientific) was used as a secondary antibody.
mRNA decay assay
For in vivo mRNA decay assay, cells were grown to reach OD 600 of ∼0.5–0.6 in synthetic medium supplemented with either 2% glucose or 2% galactose at 30°C. Transcription was blocked by transferring cells to 4% glucose media (when grown in galactose) by adding tetracycline (when using a tet-off reporter) or by adding thiolutin (when probing Rpl26a or Rpp1b; Sigma-Aldrich). Total RNA was extracted and probed with oRP140 (Muhlrad et al., 1994; Caponigro and Parker, 1995), 100 (Caponigro et al., 1993), 1479, or 1480. The resulting images were acquired with a phosphoimager (Molecular Dynamics) and quantified using 7S RNA as a loading control (Caponigro et al., 1993). Images are representative of two independent experiments.
Phosphorylation assays
To analyze phosphorylation of Dcp2 in vitro, the catalytic core of Dcp2 (amino acids 102–300) and Ste20 were purified from E. coli as previously described (Ahn et al., 2005; She et al., 2006). Ste20 was purified from yeast using either Ste20-TAP or Ste20-GFP fusion proteins (Ahn et al., 2005). The TAP purification method was used as described previously (Rigaut et al., 1999). To immunopurify the GFP-tagged protein, cell lysates were incubated with anti-GFP antibody (AnaSpec) for 2 h, and IgG-conjugated Sepharose bead (GE Healthcare) was used to pull down antibody (Ryoo et al., 2004). The resulting immunopellet was washed three times with lysis buffer and mixed with purified recombinant proteins and radioactive ATP in the kinase reaction buffer as described previously (Ryoo et. al., 2004).
To analyze phosphorylation in vivo, cells were grown in synthetic media to mid-log (or stationary phases) and harvested with or without described stresses. Cells were lysed, and Flag-Dcp2 (Dunckley and Parker, 1999) was purified as previously described (Beckham et al., 2007). Immunopellets were run on SDS-PAGE and subjected to Western analysis with antibody against phospho-Ser. Images are representative of three independent experiments
Microarray analysis
Expression microarray analysis was performed as described previously (Capaldi et al., 2008). dcp2Δ strain was transformed with expression plasmid of Dcp2-GFP wild-type, S137A, or S137E mutant. Cells were grown in synthetic complete media to reach OD 600 of ∼0.5–0.6 and frozen in dry ice. Total RNA was purified by phenol/chloroform and chloroform extraction. 40 µg total RNA was converted into cDNA by SuperScript III (Invitrogen) with 40 µg random primer (N9) and amino allyl UTP (Sigma-Aldrich) and purified with the use of gel extraction kit (QIAGEN). 5 µg purified DNA was labeled with NHS ester Cy3 or Cy5 (GE Healthcare) by incubating in sodium bicarbonate for 5 h, and the free dye was removed by gel extraction kit. 500 ng Cy3- or Cy5-labeled cDNA was hybridized to a microarray with 6,200 60-base probes (G4140A arrays; Agilent Technologies) in hybridization buffer (Agilent Technologies) for 16 h at 65°C in rotation chamber. The arrays were washed and scanned with the use of a scanner (4000B; Axon). The resulting images were analyzed with GenePix, and the all of the files were uploaded to Stanford Microarray Database. The further clustering work was performed with Cluster 3.0 (Eisen Laboratory) and Java Tree view. Two independent microarray analyses were performed (Table S5).
Liquid chromatography tandem MS
Mapping of phosphorylation on Dcp2 in vitro was performed as follows. First, His-Dcp2 (102–300) was incubated with or without GST-Ste20 purified from E. coli for 2 h at 37°C. The reaction mixtures were digested in 100 mM ammonium bicarbonate by trypsin (10 µg/ml) at 37°C overnight as previously described (Flannery et al., 1989). Liquid chromatography MS/MS analyses of trypsin-digested proteins were performed using a linear quadrupole-ion trap mass spectrometer (LTQ; Thermo Fisher Scientific) equipped with an HPLC system (Paradigm MS4; Michrom), an autosampler (AS3000; SpectraSystems), and a nanoelectrospray source as described previously (Andon et al., 2002; Lantz et al., 2007). Tandem MS spectra of peptides were analyzed with a program that allows the correlation of experimental tandem MS data with theoretical spectra generated from known protein sequences (TurboSEQUEST version 3.1; Thermo Fisher Scientific; Eng et al., 1994). The peak list (DTA files) for the search was generated by Bioworks (version 3.1; Thermo Fisher Scientific). Parent peptide mass error tolerance, fragment ion mass tolerance, and criteria used for preliminary positive peptide identification are the same as previously described (Cooper et al., 2003; Qian et al., 2005). All matched peptides were confirmed by visual examination of the spectra. All spectra were searched against an S. cerevisiae database (downloaded October 3, 2009; National Center for Biotechnology Information) and the primary sequence of DCP2. At the time of the search, the S. cerevisiae protein database from contained 64,422 entries. The results were also validated using XTandem, another search engine (Craig and Beavis, 2004), and with Scaffold, a program that relies on various search engine results (i.e., Sequest, XTandem, and MASCOT) and that uses Bayesian statistics to reliably identify more spectra (Keller et al., 2002; Nesvizhskii et al., 2003).
Online supplemental material
Fig. S1 shows Dcp2 phosphorylation during glucose deprivation in wild type or various deletion strains of MAPK pathway in yeast and in vitro phosphorylation assay results with Ste20 purified from E. coli. Fig. S2 shows in vitro decapping activity of wild-type or charge-mimetic Dcp2. Fig. S3 shows the decay rate of MFA2pG during glucose deprivation in wild type, Dcp2 mutant alleles, or the Ste20 deletion strain. Fig. S4 shows expression level of Dcp2-GFP or Pab1-GFP in various alleles with Western blot analysis. Tables S1–S5 detail properties of yeast strains, plasmids, and oligonucleotides as well as quantitation of microscopic results and raw data of microarray analysis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912019/DC1.
Acknowledgments
We thank C. David Allis for yeast strains and expression plasmid, J. Ross Buchan and Carolyn Decker for critical review of the manuscript, Anne Webb for assistance with blind scoring of microscopic images, Carl Boswell for assistance with microscopy, Andrew Capaldi for the support of microarray analysis, George Tsaprailis for MS analysis, and the Parker laboratory for support and discussions. Mass spectrometric data were acquired by the Arizona Proteomics Consortium supported by NIEHS (grant ES06694 to the SWEHSC), National Institutes of Health/NCI (grant CA023074 to the AZCC), and the BIO5 Institute of the University of Arizona. This work was supported by the Howard Hughes Medical Institute and the National Institutes of Health (grant R37GM45443).
Footnotes
Abbreviations used in this paper:
- MS
- mass spectrometry
- P-body
- processing body
References
- Ahn S.H., Cheung W.L., Hsu J.Y., Diaz R.L., Smith M.M., Allis C.D. 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 120:25–36 10.1016/j.cell.2004.11.016 [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. 2006. RNA granules. J. Cell Biol. 172:803–808 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andon N.L., Hollingworth S., Koller A., Greenland A.J., Yates J.R., III, Haynes P.A. 2002. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics. 2:1156–1168 [DOI] [PubMed] [Google Scholar]
- Beckham C.J., Light H.R., Nissan T.A., Ahlquist P., Parker R., Noueiry A. 2007. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J. Virol. 81:9759–9768 10.1128/JVI.00844-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard L. 2004. Inhibition of 5′ to 3′ mRNA degradation under stress conditions in Saccharomyces cerevisiae: from GCN4 to MET16. RNA. 10:458–468 10.1261/rna.5183804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. 2006. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb. Symp. Quant. Biol. 71:513–521 10.1101/sqb.2006.71.038 [DOI] [PubMed] [Google Scholar]
- Brengues M., Parker R. 2007. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell. 18:2592–2602 10.1091/mbc.E06-12-1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 310:486–489 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 36:932–941 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., Muhlrad D., Parker R. 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183:441–455 10.1083/jcb.200807043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi A.P., Kaplan T., Liu Y., Habib N., Regev A., Friedman N., O’Shea E.K. 2008. Structure and function of a transcriptional network activated by the MAPK Hog1. Nat. Genet. 40:1300–1306 10.1038/ng.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G., Parker R. 1995. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 9:2421–2432 10.1101/gad.9.19.2421 [DOI] [PubMed] [Google Scholar]
- Caponigro G., Muhlrad D., Parker R. 1993. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 13:5141–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J., Parker R. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73:861–890 10.1146/annurev.biochem.73.011303.074032 [DOI] [PubMed] [Google Scholar]
- Coller J., Parker R. 2005. General translational repression by activators of mRNA decapping. Cell. 122:875–886 10.1016/j.cell.2005.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J.M., Tucker M., Sheth U., Valencia-Sanchez M.A., Parker R. 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 7:1717–1727 10.1017/S135583820101994X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B., Eckert D., Andon N.L., Yates J.R., III, Haynes P.A. 2003. Investigative proteomics: identification of an unknown plant virus from infected plants using mass spectrometry. J. Am. Soc. Mass Spectrom. 14:736–741 10.1016/S1044-0305(03)00125-9 [DOI] [PubMed] [Google Scholar]
- Craig R., Beavis R.C. 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 20:1466–1467 10.1093/bioinformatics/bth092 [DOI] [PubMed] [Google Scholar]
- Dang Y., Kedersha N., Low W.K., Romo D., Gorospe M., Kaufman R., Anderson P., Liu J.O. 2006. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281:32870–32878 10.1074/jbc.M606149200 [DOI] [PubMed] [Google Scholar]
- Decker C.J., Teixeira D., Parker R. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179:437–449 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh M.V., Jones B.N., Quang-Dang D.U., Flinders J., Floor S.N., Kim C., Jemielity J., Kalek M., Darzynkiewicz E., Gross J.D. 2008. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol. Cell. 29:324–336 10.1016/j.molcel.2007.11.027 [DOI] [PubMed] [Google Scholar]
- Dunckley T., Parker R. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18:5411–5422 10.1093/emboj/18.19.5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J.K., McCormack A.L., Yates J.R., III 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8:9–22 10.1038/nrm2080 [DOI] [PubMed] [Google Scholar]
- Flannery A.V., Beynon R.J., Bond J.S. 1989. Proteolysis of Proteins for Sequencing Analysis and Peptide Mapping. Proteolytic Enzymes: A Practical Approach. Beynon R.J, Bond J.S., Oxford University Press, Oxford/New York: 259 pp [Google Scholar]
- Garneau N.L., Wilusz J., Wilusz C.J. 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8:113–126 10.1038/nrm2104 [DOI] [PubMed] [Google Scholar]
- Greatrix B.W., van Vuuren H.J. 2006. Expression of the HXT13, HXT15 and HXT17 genes in Saccharomyces cerevisiae and stabilization of the HXT1 gene transcript by sugar-induced osmotic stress. Curr. Genet. 49:205–217 10.1007/s00294-005-0046-x [DOI] [PubMed] [Google Scholar]
- Grousl T., Ivanov P., Frýdlová I., Vasicová P., Janda F., Vojtová J., Malínská K., Malcová I., Nováková L., Janosková D., et al. 2009. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 122:2078–2088 10.1242/jcs.045104 [DOI] [PubMed] [Google Scholar]
- Herskowitz I. 1995. MAP kinase pathways in yeast: for mating and more. Cell. 80:187–197 10.1016/0092-8674(95)90402-6 [DOI] [PubMed] [Google Scholar]
- Hilgers V., Teixeira D., Parker R. 2006. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae.RNA. 12:1835–1845 10.1261/rna.241006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle N.P., Castelli L.M., Campbell S.G., Holmes L.E., Ashe M.P. 2007. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 179:65–74 10.1083/jcb.200707010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jona G., Choder M., Gileadi O. 2000. Glucose starvation induces a drastic reduction in the rates of both transcription and degradation of mRNA in yeast. Biochim. Biophys. Acta. 1491:37–48 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., Anderson P. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 13:195–210 10.1091/mbc.01-05-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- Lantz R.C., Lynch B.J., Boitano S., Poplin G.S., Littau S., Tsaprailis G., Burgess J.L. 2007. Pulmonary biomarkers based on alterations in protein expression after exposure to arsenic. Environ. Health Perspect. 115:586–591 10.1289/ehp.9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R., Sukarieh R., Bordeleau M.E., Kaufman R.J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 17:4212–4219 10.1091/mbc.E06-04-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Decker C.J., Parker R. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′—>3′ digestion of the transcript. Genes Dev. 8:855–866 10.1101/gad.8.7.855 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- Parker R., Sheth U. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell. 25:635–646 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Parker R., Song H. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11:121–127 10.1038/nsmb724 [DOI] [PubMed] [Google Scholar]
- Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., et al. 2005. Global analysis of protein phosphorylation in yeast. Nature. 438:679–684 10.1038/nature04187 [DOI] [PubMed] [Google Scholar]
- Qian W.J., Liu T., Monroe M.E., Strittmatter E.F., Jacobs J.M., Kangas L.J., Petritis K., Camp D.G., II, Smith R.D. 2005. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J. Proteome Res. 4:53–62 10.1021/pr0498638 [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032 10.1038/13732 [DOI] [PubMed] [Google Scholar]
- Ryoo K., Huh S.H., Lee Y.H., Yoon K.W., Cho S.G., Choi E.J. 2004. Negative regulation of MEKK1-induced signaling by glutathione S-transferase Mu. J. Biol. Chem. 279:43589–43594 10.1074/jbc.M404359200 [DOI] [PubMed] [Google Scholar]
- She M., Decker C.J., Chen N., Tumati S., Parker R., Song H. 2006. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 13:63–70 10.1038/nsmb1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M., Decker C.J., Svergun D.I., Round A., Chen N., Muhlrad D., Parker R., Song H. 2008. Structural basis of dcp2 recognition and activation by dcp1. Mol. Cell. 29:337–349 10.1016/j.molcel.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 300:805–808 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staleva L., Hall A., Orlow S.J. 2004. Oxidative stress activates FUS1 and RLM1 transcription in the yeast Saccharomyces cerevisiae in an oxidant-dependent manner. Mol. Biol. Cell. 15:5574–5582 10.1091/mbc.E04-02-0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Parker R. 2007. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 18:2274–2287 10.1091/mbc.E07-03-0199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 11:371–382 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Malcolm B.A. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. Biotechniques. 26:680–682 [DOI] [PubMed] [Google Scholar]
- Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. 2005. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 118:981–992 10.1242/jcs.01692 [DOI] [PubMed] [Google Scholar]
- Wu C., Whiteway M., Thomas D.Y., Leberer E. 1995. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 270:15984–15992 10.1074/jbc.270.27.15984 [DOI] [PubMed] [Google Scholar]