A newly discovered interaction between LL5s, laminins, and integrins reveals how the extracellular matrix directs microtubule polarity in epithelial tissues.
Abstract
LL5β has been identified as a microtubule-anchoring factor that attaches EB1/CLIP-associating protein (CLASP)–bound microtubule plus ends to the cell cortex. In this study, we show that LL5β and its homologue LL5α (LL5s) colocalize with autocrine laminin-5 and its receptors, integrins α3β1 and α6β4, at the basal side of fully polarized epithelial sheets. Depletion of both laminin receptor integrins abolishes the cortical localization of LL5s, whereas LL5 depletion reduces the amount of integrin α3 at the basal cell cortex. Activation of integrin α3 is sufficient to initiate LL5 accumulation at the cell cortex. LL5s form a complex with the cytoplasmic tails of these integrins, but their interaction might be indirect. Analysis of the three-dimensional distribution of microtubule growth by visualizing EB1-GFP in epithelial sheets in combination with RNA interference reveals that LL5s are required to maintain the density of growing microtubules selectively at the basal cortex. These findings reveal that signaling from laminin–integrin associations attaches microtubule plus ends to the epithelial basal cell cortex.
Introduction
Appropriate organization of microtubule networks is critical for maintaining cell structure and polarity (Goode et al., 2000; Rodriguez et al., 2003). Because microtubules are dynamic polymers that exhibit repeated stochastic growth and shortening, local regulation of their dynamic properties and anchoring of their ends to specific sites within cells are central to the generation of polarized microtubule arrays (for reviews see Kirschner and Mitchison, 1986; Desai and Mitchison, 1997).
To date, it has become clear that an intriguing class of microtubule-associated factors, plus end–tracking proteins (+TIPs; Schuyler and Pellman, 2001), which specifically accumulate at microtubule plus ends and are typified by EB1 family proteins, play important roles in setting up the well-organized microtubule network by linking microtubule ends to various cellular structures (Mimori-Kiyosue and Tsukita, 2003; Lansbergen and Akhmanova, 2006). Some of the +TIPs such as CLIP-associating proteins (CLASPs), actin cross-linking family 7 (ACF7; also known as MACF1), and adenomatous polyposis coli (APC) tumor suppressor protein localize to the cell cortex near migrating cell edges and attach EB1-positive microtubule plus ends to these structures (Kodama et al., 2003; Etienne-Manneville et al., 2005; Mimori-Kiyosue et al., 2005, 2007). Efforts to study the molecular mechanisms of association of CLASPs with the cell cortex identified LL5β as their direct binding partner participating in the cortical microtubule attachment (Lansbergen et al., 2006).
LL5β, also known as pleckstrin homology (PH)–like domain, family B, member 2 (PHLDB2), is a member of the LL5 protein family, which consists of LL5α, -β, and -γ (or PHLDB1, -2, and -3). LL5γ is a short protein carrying only a PH domain; it has no similarities with LL5α and -β in other regions, including the CLASP-binding region. In contrast, LL5α and -β show considerable similarity in the CLASP-binding region, PH domain, and the part adjacent to the PH domain (Fig. 1 A). LL5α was first identified in a rat pituitary cDNA library and named after the clone number (Levi et al., 1993), whereas LL5β was identified in database searches for proteins possessing a PH domain that contained a putative phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3)–binding motif (Dowler et al., 2000). LL5β does indeed bind to PtdIns(3,4,5)P3, and also to the cytoskeletal adaptor γ-filamin (Dowler et al., 2000; Paranavitane et al., 2003). Previously, LL5β has been detected at skeletal neuromuscular junctions, where it was shown to play a role in the clustering of acetylcholine receptors (Kishi et al., 2005).
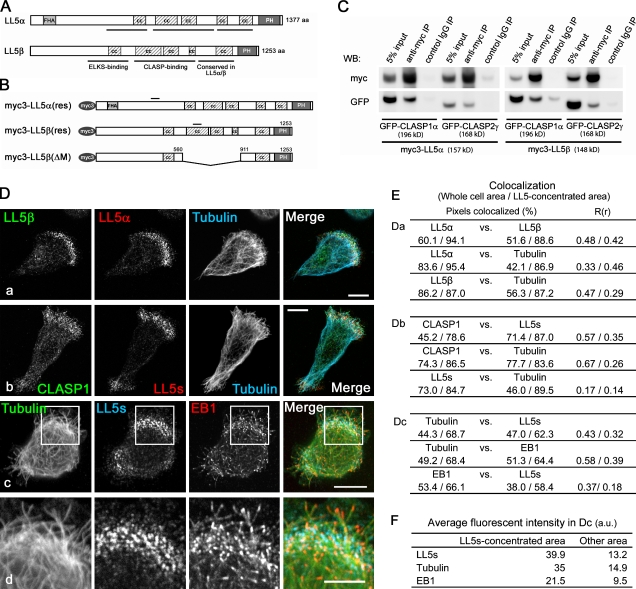
Figure 1.
Comparison of LL5α and -β functions in MCF-10A cells. (A) Domain compositions of LL5α and -β. FHA, forkhead-associated domain; cc, coiled-coil domain; PH, PH domain. (B) Myc3-tagged constructs used for rescue experiments. Bars indicate silent mutations conferring siRNA resistance. For live cell imaging, fluorescent protein-tagged versions were used. (C) Extracts from HEK293T cells ectopically expressing myc3-tagged LL5s and GFP-tagged CLASPs were subjected to immunoprecipitation (IP) with an anti-myc antibody and analyzed by Western blotting (WB). (D) At 3 h after seeding on coverslips, MCF-10Aeco cells were fixed and stained with the indicated antibodies. LL5s exhibit similar distributions at the migrating edges of cells and colocalize with CLASPs (a and b). Microtubule plus ends visualized with EB1 (c) are concentrated in the region with CLASP/LL5 accumulation. The boxed areas in c are magnified in d. (E) The colocalized fluorophores in D (a–c) were quantified. The percentages of colocalized pixels and the Pearson’s correlation coefficients in colocalized volume (R(r)) between the two color channels inside the whole cell area and the LL5-concentrated area are presented. (F) The mean fluorescence intensities of tubulin, LL5s, and EB1 staining in D (c) were compared between the LL5-concentrated area and other area. Bars: (D, a–c) 10 µm; (D, d) 5 µm.
For a long time, it has been known that the microtubules in epithelial cells are aligned along the apicobasal axis with their plus ends facing toward the basal side (Bacallao et al., 1989). However, the molecular mechanisms underlying the polarization and organization of these microtubule networks remain to be clarified, and it is therefore interesting to examine the involvement of +TIPs in this process. In vivo, the basal surfaces of epithelial cells adhere to the underlying basement membrane (BM) comprised of type IV collagens, proteoglycans, and laminins, and cell adhesion to the ECM is one of the primary extrinsic cues for generating apicobasal polarity (for reviews see Rodriguez-Boulan and Nelson, 1989; Schoenenberger and Matlin, 1991; Yeaman et al., 1999).
Laminin-5 (also known as laminin-332, kalinin, nicelin, epiligrin, and radsin), which contains α3, β3, and γ2 chains, is one of the BM components secreted by epithelial cells themselves, whereas integrins play central roles in the adhesion of cells to the BM and subsequent signal transduction across the plasma membrane (Hintermann and Quaranta, 2004; Katayama and Sekiguchi, 2004). Integrins are heterodimeric transmembrane proteins consisting of α and β subunits, with the α subunits contributing to their ligand-binding specificities (Hemler, 1999; for reviews see van der Flier and Sonnenberg, 2001; Hynes, 2002). Epithelia are known to express the laminin receptor integrins α3β1 and α6β4 (Katayama and Sekiguchi, 2004). Integrin α3β1 is involved in epithelial cell migration, whereas integrin α6β4 stabilizes epithelial cell–BM attachment (Nguyen et al., 2000b).
In the present study, we used the noncancerous human mammary epithelial cell line MCF-10A to investigate the mechanisms that determine the cortical distribution of LL5β and its homologue LL5α (LL5s). We show that LL5s are colocalized with laminin-5 deposition and laminin receptor integrins in cultured cells as well as in epithelial tissues. We show that the localization of LL5s relies on laminin receptor integrins and that LL5s stabilize the localization of integrin α3 at the basal cortex. We also demonstrate that activation of integrin α3 is sufficient to initiate LL5 accumulation and that LL5s associate with the cytoplasmic tails of these laminin receptor integrins. Finally, we describe the LL5-dependent three-dimensional distribution of microtubules in fully polarized epithelial sheets.
Results
CLASP-binding ability of LL5s and their subcellular distribution
MCF-10A cells, which comprise a nontransformed human mammary epithelial cell line derived by spontaneous immortalization of human breast epithelial cells from a patient with fibrocystic disease (Soule et al., 1990), express LL5β as well as LL5α, a close homologue of LL5β (Fig. 1 A and Fig. S1 A). LL5α and -β have similar domain compositions, including several coiled-coil regions and a PH domain. In addition, LL5α has an N-terminal forkhead-associated domain. LL5α was similar to LL5β in its CLASP-binding ability (Fig. 1 C).
During active cell spreading shortly after cell attachment to glass, LL5α showed a similar distribution pattern to that of LL5β and colocalized with CLASP1 at sites where microtubules were significantly concentrated (Fig. 1, D–F; and Video 1), which is consistent with previous observations (Lansbergen et al., 2006). Previously, using total internal reflection fluorescence (TIRF) microscopy, we showed that in HeLa cells, CLASP1 is located in the close vicinity of glass substrates (Mimori-Kiyosue et al., 2005). Growing microtubule ends visualized with EB1 (Mimori-Kiyosue et al., 2000) were also concentrated at sites exhibiting LL5 accumulation (Fig. 1, D [c and d], E, and F), which is in agreement with our previous observations that demonstrated that EB1-positive growing microtubule ends are frequently trapped at the CLASP-accumulating cortical regions (Mimori-Kiyosue et al., 2005).
Comparison of the microtubule-anchoring activities of LL5s
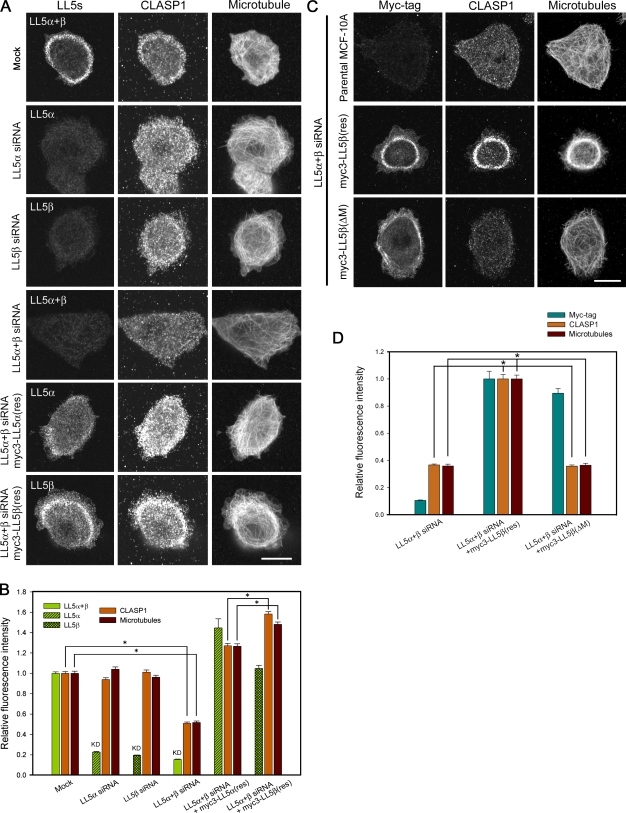
The microtubule-anchoring activities of LL5s were analyzed by RNAi and rescue experiments in the MCF-10Aeco cell line, a subclone of MCF-10A which is susceptible to retrovirus-mediated gene transfer (Fig. S1 D and Fig. S2, A, B, and E). RNAi-mediated depletion of LL5s separately or together revealed that the attachment of CLASP-bound microtubules to the basal cell cortex was affected only when both LL5s were depleted simultaneously, whereas individual LL5α or -β knockdown had no effect (Fig. 2, A and B). Rescue experiments showed that the expression of myc3-LL5β(res) increased the densities of CLASP1 and microtubules more efficiently than myc3-LL5α(res), although the expression level of myc3-LL5α(res) was higher than that of myc3-LL5β(res) (Fig. 2 B). Importantly, myc3-LL5β(ΔM) lacking the CLASP-binding region failed to recruit CLASP and microtubules to the cell cortex, although this mutant was still able to associate with the plasma membrane (Fig. 2, C and D). The expression levels of CLASPs were not affected by LL5 depletion (unpublished data). All of these observations indicate that LL5s have redundant roles in microtubule regulation, although LL5β is more potent than LL5α.
Figure 2.
Microtubule-anchoring activity of LL5s. (A and B) Effects of LL5 depletion on the distributions of CLASP1 and microtubules at the basal cortex. Parental MCF-10Aeco cells or cells expressing myc3-LL5α(res) or myc3-LL5β(res) were transfected with the indicated siRNAs, seeded on coverslips, fixed, and stained for LL5s (LL5α alone, LL5β alone, or LL5α+β), CLASP1, and microtubules. (A) Images of the basal cortex were collected using a confocal microscope. (B) The fluorescence intensities of LL5s, CLASP1, and microtubules at the lamellipodia were analyzed and plotted. The data were transformed into a relative fluorescence index. The fluorescence intensities of the mock control were set as 1. The results are presented as means ± SEM (*, P < 0.01 vs. the mock control). (C and D) LL5β recruits microtubules to the basal cortex through its middle region. (C) Under LL5-depleted conditions, parental MCF-10Aeco cells or cells expressing myc3-LL5β(res) or myc3-LL5β(ΔM) were fixed and stained for myc tag, CLASP1, and microtubules. (D) The fluorescence intensities of myc tag, CLASP1, and microtubules at the lamellipodia were analyzed and plotted. The data were transformed into a relative fluorescence index. The fluorescence intensities of the myc3-LL5β–expressing cells were set as 1. Myc3-LL5β(ΔM) is unable to recruit CLASP1/microtubules. The results are presented as means ± SEM (*, P < 0.01 vs. the LL5α+β siRNA + myc3-LL5β(res) sample). Bars, 10 µm.
LL5s are localized at cell adhesion sites containing laminin-5 deposition
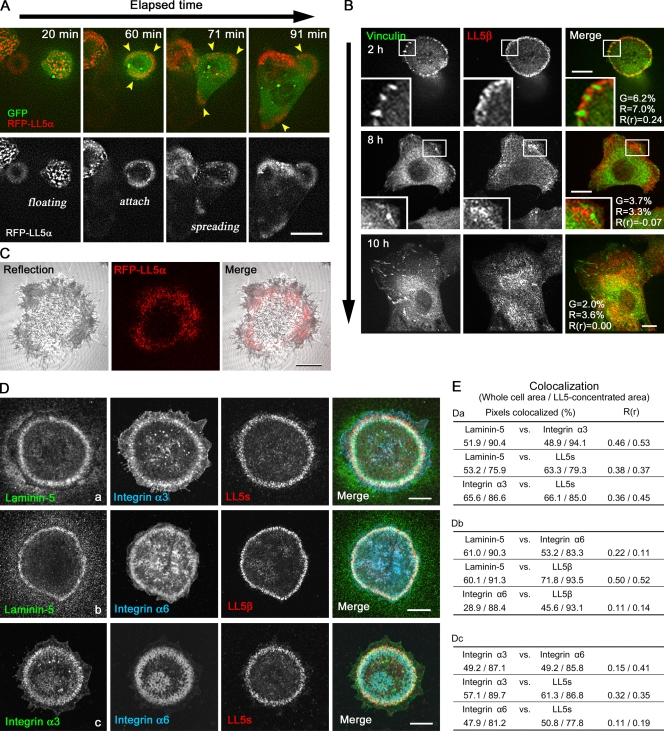
To explore the mechanisms of LL5 recruitment to the cell cortex, we carefully analyzed their dynamic behavior and distribution in comparison to other subcellular structures. The distribution of LL5s was strictly restricted to the substratum-attached basal cortex (Video 2). Live cell imaging revealed that accumulation of LL5s was initiated upon cell–substratum attachment (Fig. 3 A and Video 3). Cortical patches of LL5s appeared soon after cell attachment to glass, and these structures were dynamically relocated upon outgrowth of lamellipodia. After the cells became fully extended, LL5s were no longer concentrated in patches at the cell periphery but were instead randomly distributed throughout the basal plasma membrane (Fig. 3 B and Video 4). In line with previous observations (Lansbergen et al., 2006), LL5s never colocalized with focal adhesions (FAs), which are the major cell adhesion structures (Fig. 3 B). However, interference reflection microscopy revealed that RFP-LL5α was distributed at cell–substratum attachment sites visualized as gray zones (Fig. 3 C), which have previously been termed “close contacts” (Carter et al., 1990). From these observations, we hypothesized that cell–substratum attachment may trigger the accumulation of LL5s at the basal cell cortex.
Figure 3.
Subcellular localization of LL5s in MCF-10A cells. (A) Selected images from a time-lapse video of MCF-10Aeco cells expressing RFP-LL5α and GFP (Video 1). GFP allows visualization of the cell shape. The images were collected soon after cell seeding. Accumulation of RFP-LL5α (arrowheads) is initiated upon cell attachment to the substratum. (B) Comparison of the distributions of LL5β and FAs visualized with antivinculin antibodies. Representative images of cells stained for LL5β and vinculin at 2 (top), 8 (middle), and 10 (bottom) h after seeding are shown. Insets are magnified images of the boxed areas. The percentage of colocalized pixels and the Pearson’s correlation coefficients (R(r)) between vinculin (G) and LL5β (R) channels are presented in the figures. (C) Comparison of the distributions of RFP-LL5α and cell–substratum attachment sites visualized by interference reflection microscopy (grayscale). (D and E) Colocalization of LL5s with laminin-5 deposition and laminin receptor integrins. (D) Autocrine laminin-5 is secreted and deposited in a flower pattern on the glass surface by the cells. (E) The colocalization of fluorophores in D (a–c) was quantified inside the whole cell area and the LL5-concentrated area. Colocalization analyses are similar to Fig. 1 E. Bars: (A and B) 10 µm; (C and D) 5 µm.
To identify the factors that initiate the accumulation of LL5s, we screened for ECM proteins with a similar distribution pattern to LL5s. Among the ECM proteins examined, including fibronectin and collagen I and IV, we found that laminin-5 colocalized with LL5 patches in MCF-10Aeco cells (Fig. 3, D and E; and not depicted for ECMs other than laminin-5 because they exhibit no colocalization with LL5s). Mass spectrometry analysis revealed that laminin-5 was the dominant ECM protein secreted and deposited on the substrate by MCF-10Aeco cells (Table I). Migrating MCF-10A cells have previously been shown to secrete laminin-5 (Stahl et al., 1997; Goldfinger et al., 1999). Indeed, live imaging of GFP-fused laminin-γ2 revealed that it was deposited on the glass surface during cell migration (Fig. S1 E and Video 5). The scattering of LL5 patches along the basal cortex (Fig. 3 B) coincided with the broadening of laminin-5 deposition on the entire glass surface (not depicted).
Table I.
Mass spectrometry analysis of ECM proteins deposited by MCF-10A cells
| Gene symbol | Protein name | Amino acid residues | Mascot scorea | Number of total peptides detected |
| LAMA3 | Laminin subunit α3 chain (laminin-5 α3 subunit) | 1713 | 715 | 113 |
| LAMB3 | Laminin subunit β3 chain (laminin-5 β3 subunit) | 1170 | 441 | 69 |
| LAMC2 | Laminin subunit γ2 chain (laminin-5 γ2 subunit) | 1193 | 388 | 106 |
| LAMB1 | Laminin subunit β1 chain | 1786 | 79 | 6 |
| LAMC1 | Laminin subunit γ1 chain | 1609 | 96 | 3 |
| AGRN | Agrin | 2026 | 67 | 3 |
| CYR61 | CYR61 (cysteine-rich angiogenic inducer 61) | 381 | 64 | 4 |
| THBS1 | Thrombospondin 1 (THBS1) | 1170 | 135 | 6 |
| VCAN | Versican, proteoglycan PG-M (V3) | 655 | 47 | 2 |
| FGB | Fibrinogen, β chain | 483 | 49 | 1 |
Factors indicated by boldface are laminin-5 components. MCF-10Aeco cells grown in serum-free MEGM were detached from culture plates with PBS containing 10 mM EDTA. The proteins remaining on the plates were lysed in SDS sample buffer, separated by SDS-PAGE, and analyzed by mass spectrometry. Only ECM proteins with clear mass spectra with scores of P < 0.05 are listed. Laminin-5 subunits are detected as the major ECM components of MCF-10Aeco cells.
Significance threshold of proteins was set at P < 0.05.
LL5s are colocalized with the laminin receptor integrins α3β1 and α6β4
During initial spreading, the cells formed a flower pattern of laminin-5 by remodeling previously deposited molecules, and this process has been reported to be dependent on the laminin receptor integrin α3 (Carter et al., 1991; Baker et al., 1996; deHart et al., 2003). Integrin α3β1 was previously shown to be localized at close contacts (Carter et al., 1990). Epithelia express the laminin receptor integrins α3β1 and α6β4, the ligand specificities of which rely on their α subunits (for reviews see van der Flier and Sonnenberg, 2001; Hynes, 2002). Therefore, we investigated the distribution of laminin-5 deposition and integrins α3 and α6 in comparison to LL5s. Both integrins α3 and α6 exist as two variants, a and b, which have different cytoplasmic domains. In MCF-10A cells, both integrins α3 and α6 were mainly represented by the variant a, together with a small amount of variant b of integrin α6 (Fig. S1 C).
As expected, LL5s frequently colocalized with these integrins in the laminin-5 flower patterns (Fig. 3, D and E). Colocalization analysis revealed that LL5 distribution displayed a stronger overlap with integrin α3 than with integrin α6. Note that LL5s were significantly concentrated on the laminin-5 deposition, whereas integrins were detectable diffusely also at the other plasma membrane sites (Fig. 3, D and E), suggesting that LL5s are colocalized with activated pools of integrins associated with laminin-5.
These patchy structures slightly resembled the hemidesmosome-like stable anchoring contacts formed by integrin α6β4 in vitro (Carter et al., 1991). However, LL5s did not significantly colocalize with either keratin filaments or plectin, which are the intrinsic components of hemidesmosomes (unpublished data). We conclude that LL5s localize to migratory laminin–integrin adherent complexes that are different from stable anchoring contacts.
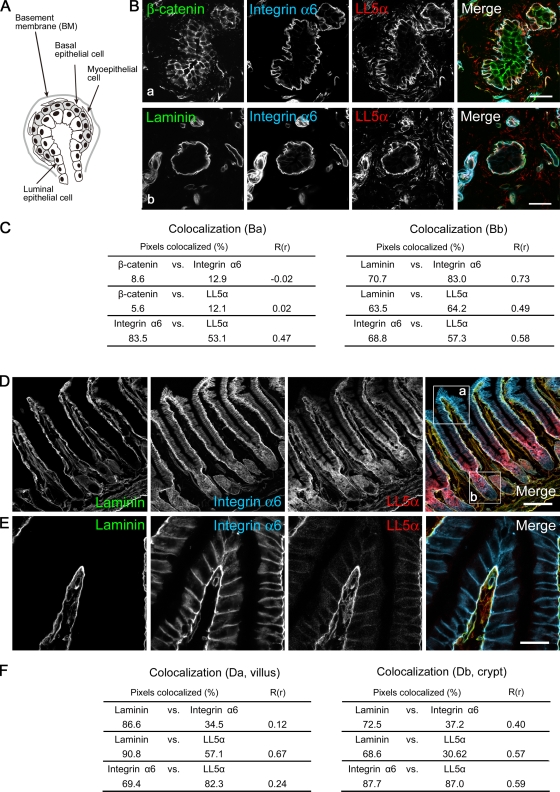
To examine the physiological relationship between LL5s and laminin, we investigated their tissue distributions. Laminin-5 was previously detected in the BMs underlying various epithelial tissues and glands such as the skin, lung, small intestine, stomach, and kidney (Mizushima et al., 1998). Consistent with these data, LL5α colocalized with laminin as well as integrin α6 at the basal cell membranes in the mammary gland (Fig. 4, A–C). In the small intestine, which represents a typical columnar epithelial tissue, LL5α also accumulated at the basal surface (Fig. 4, D–F). LL5β showed distribution patterns that were similar to LL5α (unpublished data).
Figure 4.
Localization of LL5α in mouse tissues. (A) Schematic diagram of a lobule from a mammary gland based on an example from a previous paper (Debnath et al., 2003). The mammary epithelium possesses a polarized architecture surrounding a hollow lumen, which is surrounded by an inner layer of luminal epithelial cells and an outer layer of myoepithelial and basal epithelial cells. (B) Immunofluorescence staining of mammary tissues with the indicated antibodies. LL5α is localized to the basal cortex of the outer epithelium attached to laminin-containing BMs (b) but not to the β-catenin–positive lateral membranes (a). (C) Colocalization analysis similar to Fig. 1 E. Entire square areas, including the gland structure, were analyzed. (D and E) Immunofluorescence staining of the small intestine. LL5α is localized to the basal cell cortex attached to the laminin BMs. The tip region of a villus is magnified in E. (F) Colocalization analysis similar to Fig. 1 E. Boxed areas a (villus) and b (crypt) in D were analyzed. LL5β showed a similar distribution pattern to LL5α both in mammary glands and intestines (not depicted). Bars: (B) 20 µm; (D) 100 µm; (E) 50 µm.
LL5 localization depends on laminin receptor integrins
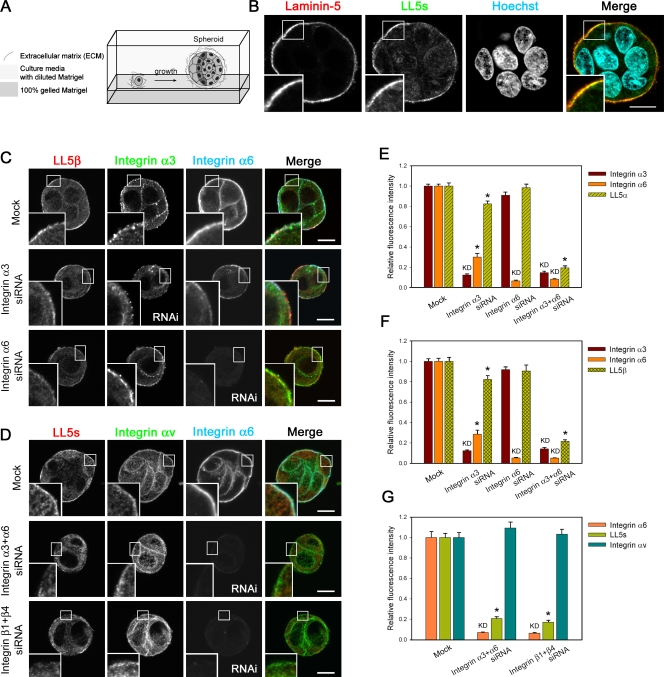
The aforementioned observations raised the possibility that laminin receptor integrins may be involved in LL5 localization not only in monolayer cell cultures but also in three-dimensional epithelial tissues. To examine this possibility, we used an RNAi strategy and a spheroid culture system in Matrigel (Fig. 5 A). We chose this method because inhibition of the laminin-5–integrin pathway in MCF-10A cells in conventional monolayer cultures induces detachment of the cells from the substratum, thereby prohibiting cytological analyses. In addition, we used fluorescence confocal imaging of spheroid cross sections to allow precise measurements of protein accumulation along the basal cell cortex.
Figure 5.
Distribution of LL5s in MCF-10A spheroids and its dependency on laminin receptor integrins. (A) Schematic diagram of the overlay method for three-dimensional culture of MCF-10A cells in Matrigel (Debnath et al., 2003). MCF-10Aeco cells seeded onto a solid bed as a single-cell suspension proliferate and start to form clusters (spheroids) in three-dimensional culture. (B) Representative confocal microscopic images of MCF-10Aeco spheroids. Spheroids at day 3 were fixed and immunostained with antibodies against LL5s and laminin-5. Nuclei were stained with Hoechst 33342. The outer surface of the spheroids covered with laminin-5–positive BM is the basal side of the cells. (C and D) Confocal images of MCF-10Aeco spheroids treated with the indicated siRNAs and stained for LL5s and integrins. Integrin αv is known to localize to FAs. Note that knockdown of integrin β1 + β4 is estimated by the integrin α6 signals, owing to the limited availability of primary antibodies from different host species for simultaneous staining. The reduction of integrin α6 after integrin β1 + β4 knockdown was confirmed in independent experiments (Fig. S4). In integrin α3 knockdown cells, integrin α6β4 is also partially reduced for unknown reasons, but LL5s are still retained at the basal cortex. (B–D) Insets are magnified images of the boxed areas. (E–G) Quantitative analyses of the fluorescent signals at the basal cortex in the spheroids. For each condition, >35 cells were analyzed. The data were transformed into a relative fluorescence index, and the values of the mock control were set as 1. LL5 localization is inhibited when integrins α3 + α6 or β1 + β4 are knocked down. Note that integrin αv is unaffected by knockdown of integrins α3 + α6 or β1 + β4, whereas the signals for LL5s are abolished. The results are presented as means ± SEM (*, P < 0.01 vs. the mock control). KD, protein that was knocked down. Bars, 10 µm.
In MCF-10A spheroids grown in Matrigel, LL5s, laminin-5, and laminin receptor integrins were all distributed at the basal side of the cells (Fig. 5 B), similar to their distributions in tissues. When integrin α3 or α6 was separately knocked down, we observed only an ∼20% decrease in LL5 accumulation at the basal cell cortex (Fig. 5, C, E, and F; and Fig. S2 C). Furthermore, knockdown of either integrin β1 or β4, which form heterodimers with integrin α3 or α6, respectively, had only subtle effects on the LL5 distribution (Fig. S2 D and Fig. S3). However, when integrins α3 and α6 were simultaneously depleted, LL5s were dramatically reduced at the basal cortex, whereas integrin αv, an FA component, was unaffected (Fig. 5, D–G). To confirm the specificity of this RNAi effect, we performed simultaneous knockdown of integrins β1 and β4. Because the activity and stability of integrin α and β subunits are mutually dependent, knockdown of either subunit resulted in a reduction of its partner subunit at the basal cortex (Fig. S4). In cells depleted of integrins β1 and β4, the cortical distribution of integrins α3 and α6 as well as LL5s was abolished (Fig. 5, D and G). These findings indicate that the localization of LL5s largely relies on integrins α3β1 and α6β4. In addition, we observed that ACF7 and APC, other microtubule-stabilizing proteins, remained at the basal cortex after knockdown of integrin α3β1 or α6β4 (unpublished data), suggesting the presence of several independent pathways that anchor microtubules to the epithelial basal cortex.
LL5s affect the localization of integrin α3
Next, we examined the effect of LL5 depletion on the integrin distribution. LL5 knockdown reduced the accumulation of integrin α3 at the basal cell cortex (Fig. 6, A and B). Although depletion of LL5α or -β alone reduced integrin α3 by up to ∼40% (Fig. 6, C and D), simultaneous knockdown of both proteins had a greater effect (Fig. 6 B). Consistent results were obtained with two different integrin α3 antibodies directed against different regions of the protein (a pAb against the cytoplasmic region and an mAb [P1B5] against the extracellular domain), indicating that the reduction of the integrin α3 signal after LL5 depletion was unlikely to be caused by epitope masking by conformational changes or binding to other molecules (Fig. 6, C and D). Expression of an LL5β rescue construct alone restored the localization of integrin α3 (Fig. 6 B). Although these results suggested that LL5α and -β have redundant roles in integrin α3 localization, the finding that the amount of LL5α was increased to 180% in LL5β knockdown cells (Fig. S2 A) implied that LL5α is less potent than LL5β in regulating integrin α3. Regarding integrin α6, we did not observe any LL5 knockdown–specific reduction of its cortical accumulation (unpublished data). These observations suggest that the basal localizations of LL5s and integrin α3 are interdependent.
Figure 6.
LL5s regulate α3 integrin localization in MCF-10A spheroids. (A and B) LL5s stimulate recruitment of integrin α3 to the basal cell cortex. (A) Confocal images of MCF-10Aeco spheroids treated with the indicated siRNAs and stained for LL5s, myc tag, and integrin α3 (pAb) are shown. Insets are magnified images of the boxed areas. (B) The fluorescent signals at the basal cortex were analyzed and plotted. For each condition, >137 cells were analyzed. The fluorescence intensity of the mock control was set as 1. (C and D) Knockdown of either LL5α or -β reduces the localization of integrin α3 at the basal cortex in MCF-10Aeco spheroids. For each condition, >60 cells were analyzed. To detect integrin α3, two different antibodies recognizing different regions of the integrin (a pAb against the cytoplasmic region and an mAb [P1B5] against the extracellular domain) were used in consideration of possible epitope masking by conformational changes or binding to other molecules. The changes in the integrin α3 signals are the same for the pAb and mAb. (B–D) The results are presented as means ± SEM (*, P < 0.01 vs. the mock control). KD, protein that was knocked down. Bars, 10 µm.
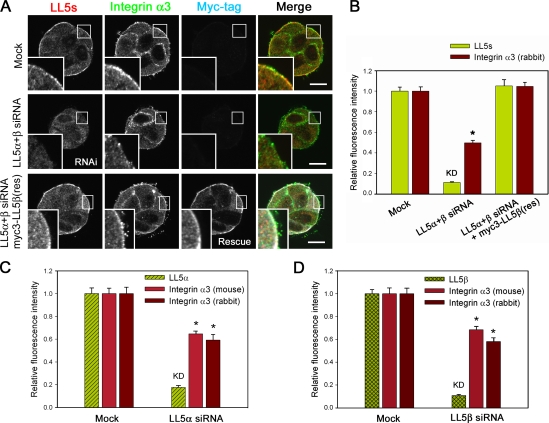
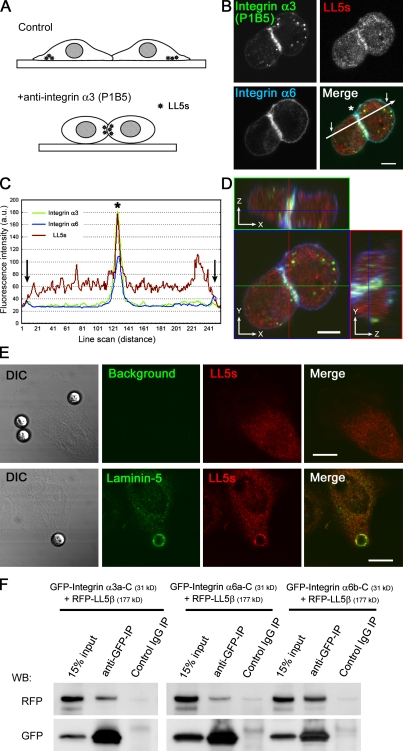
Interactions between integrins and LL5s
The aforementioned results suggested that signaling dependent on laminin–integrin coupling is required for LL5 localization. This idea was further explored using an integrin α3–activating antibody (clone P1B5), which binds to the extracellular domain of integrin α3 and prevents its binding to laminin-5 but still activates the integrin. Previously, this antibody was reported to induce artificial integrin-dependent cell aggregation (Fig. 7 A; Symington et al., 1993). In the presence of this antibody, LL5s were ectopically accumulated at the artificially induced cell–cell attachment sites together with integrins α3 and α6 (Fig. 7, B–D). Moreover, attachment of laminin-5–coated microbeads to the cell surface induced the accumulation of LL5s around the beads (Fig. 7 E). These observations demonstrate that integrin activation mediated by laminin-5 is sufficient for recruitment of LL5s to the cell cortex.
Figure 7.
Integrin α3 activation initiates LL5 accumulation and LL5–integrin α3 association. (A–D) An activating antibody against integrin α3 (clone P1B5) induces cell aggregation (A; Symington et al., 1993) and LL5 accumulation at the cell cortex. (B) Suspended cells (1 × 106) were preincubated with 5 µg of anti–integrin α3 antibody P1B5 in 1 ml MEGM, cultured on coverslips for 3 h, fixed, and stained with the indicated antibodies. (C) The relative fluorescence intensities along the white line in B were plotted. The arrows and asterisk indicate the plasma membranes and cell attachment sites, respectively. (D) Z-stack images of cells were collected with an LSM510 confocal microscope. All the fluorescent signals show significant accumulation at the artificial cell attachment sites. (E) Laminin-5–conjugated beads induce the accumulation of LL5s. Cells were stained with the indicated antibodies. DIC, differential interference contrast. (F) Extracts from HEK293T cells ectopically expressing RFP-LL5β and GFP-tagged cytoplasmic tails of integrins α3 (variant a) and α6 (variant a and b) were subjected to immunoprecipitation (IP) with an anti-GFP antibody and analyzed by Western blotting (WB). Bars: (B and D) 5 µm; (E) 10 µm.
Next, we investigated whether LL5s physically associate with integrins. To detect LL5–integrin interactions, we performed pull-down assays using RFP-LL5β and the GFP-fused cytoplasmic tails of integrins α3 (variant a) and α6 (variant a and b), which are expressed in MCF-10A cells (Fig. S1 C), in HEK293T cells. As shown in Fig. 7 F, both the integrin α3 and α6 fragments were precipitated with RFP-LL5β, indicating that they can indeed form a complex. Subsequently, we examined their direct interaction by yeast two-hybrid analysis and in vitro GST pull-down assays using four LL5β fragments covering the entire length of LL5β and the cytoplasmic tails of integrins α3, α6, αv, and β1a or GFP-fused LL5s purified from HEK293 cells and the same integrin fragments fused to GST, respectively. We observed no positive results for any of the combinations tested (unpublished data). All of these results indicate that LL5s can form complexes with laminin receptor integrins through their cytoplasmic tails, although their interactions may be indirect.
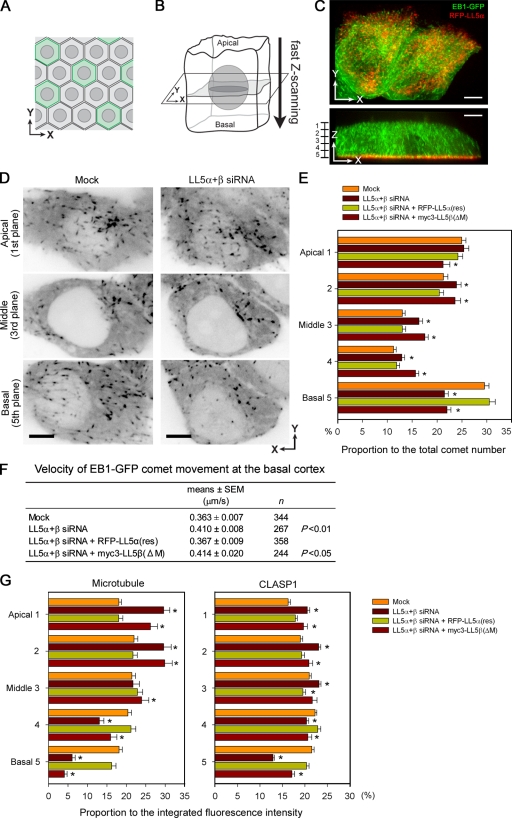
Microtubule-anchoring activity of LL5s in polarized epithelial sheets
Our present findings suggest that LL5s may affect the unique orientation of microtubules in polarized epithelia. Therefore, we examined whether the aforementioned laminin–integrin-dependent localization of LL5s is involved in the regulation of microtubule plus end dynamics at the epithelial basal cortex. To investigate this aspect, we set up a system to analyze the microtubule dynamics three-dimensionally in polarized epithelial cells using EB1-GFP, a marker for growing microtubule ends (Fig. 1 D, c and d), because direct visualization of individual microtubule filaments in three-dimensional space is technically difficult. MCF-10Aeco cells expressing EB1-GFP together with RFP-LL5α were co-cultured with nonfluorescent parental cells to highlight individual cells within confluent monolayers on noncoated glass-bottomed dishes (Fig. 8 A), and images were acquired by fast scanning along the z axis (Fig. 8 B and Video 6). A representative three-dimensional reconstituted image of a cell and two-dimensional images in different planes are shown in Fig. 8 (C and D) and Video 7.
Figure 8.
Microtubule organization in polarized monolayers of MCF-10Aeco cells. (A and B) Schematic diagram of live cell imaging in a polarized epithelial monolayer. The cells were maintained on uncoated inert glass substrates. (A) Fluorescent cells are surrounded by nonfluorescent cells. (B) Three-dimensional images of whole cells were collected by fast z scanning along the apicobasal axis using a Revolution XD system. (C) Three-dimensional distribution of EB1-GFP comets in polarized monolayers. See also Videos 6 and 7. (D) Representative images of EB1-GFP at the apical, middle, and basal cell planes in control and LL5 knockdown cells. The contrast of the images is inverted. (E) Analysis of the EB1-GFP comet distribution along the apicobasal axis under the indicated conditions. The z sections were divided into five equal parts as shown in the left of C, and the number of EB1-GFP comets in each part was counted. For each condition, >53 cells were analyzed, whereas n = 30 for LL5β(ΔM). In LL5-depleted cells, basal EB1-GFP comets are specifically reduced. (F) Velocity of EB1-GFP comet movement at the basal cortex. The conditions showing statistically significant results with P < 0.01 or P < 0.05 versus the mock control are indicated in the table. (G) The three-dimensional distributions of the fluorescence intensities of microtubules (left) and CLASP1 (right) in polarized epithelial monolayers were analyzed using conventional confocal microscopy. Five equally spaced confocal sections of 1-µm thickness were collected from the apical to basal sides of polarized MCF-10Aeco cells under the indicated conditions. For quantitative analysis of the fluorescent signals in each confocal section, ROIs were selected by avoiding centrosomes and Golgi regions, where microtubules are concentrated independent of LL5s. For microtubule and CLASP1 staining, n > 43 and n > 32, respectively. (E and G) The results are presented as means ± SEM (*, P < 0.01 vs. the mock control). Bars, 5 µm.
In polarized MCF-10Aeco cells, analysis of the three-dimensional distribution of EB1-GFP revealed that microtubule growth frequently occurred near the apical and basal cell surfaces (Fig. 8, D and E; and Videos 7, 8, and 9). After LL5s were knocked down, the number of growing microtubules along the basal cortex was selectively reduced, whereas it was increased in other cell regions (Fig. 8, D and E). The effects of LL5 depletion were rescued by full-length LL5α but not by LL5β(ΔM) lacking the CLASP-binding region (Fig. 8 E). We used LL5α for the rescue experiment because the ability of exogenous LL5β to increase microtubule density was too strong, as shown in Fig. 2, and affected the overall distribution of the microtubules, thus making interpretation of the data complicated.
Next, we measured the velocity of EB1-GFP comet movement along the basal cortex (Fig. 8 F and Video 10). After LL5 depletion, the velocity of EB1-GFP movement, or the microtubule growth speed, was mildly but significantly increased, indicating that the remaining microtubules became more dynamic in this cell area, which is similar to what we demonstrated in our previous analyses in HeLa cells (Mimori-Kiyosue et al., 2005).
Furthermore, we confirmed the distributions of microtubules and CLASP1 in immunostained MCF-10Aeco cells using conventional confocal microscopy. As shown in Fig. 8 G, the intensities of both microtubule and CLASP1 staining were reduced at the basal cortex and increased in the upper part of the cells. All of these observations indicated that LL5s were needed to maintain a dense array of microtubules targeting the basal side of epithelial cells.
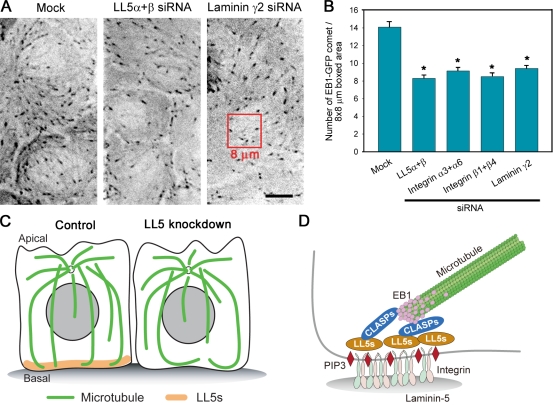
Finally, we confirmed the involvement of the laminin–integrin pathway in LL5-mediated microtubule regulation by depleting laminin-5 or its receptor integrins in EB1-GFP–expressing MCF-10Aeco cells cultured on collagen-coated glass substrates, representing conditions in which cell attachment was maintained in the absence of these molecules. Without laminin or integrins, the number of growing microtubules visualized with EB1-GFP comets along the basal cortex was reduced, which is very similar to the effect observed after LL5 depletion (Fig. 9, A and B), supporting the importance of these molecules in LL5 localization and microtubule organization (Fig. 9, C and D).
Figure 9.
Laminin–integrin signaling activates the LL5/CLASP/EB1–microtubule anchoring pathway. (A and B) Analysis of EB1-GFP comet numbers at the basal cell cortex in cells treated with the indicated siRNAs. The cells were maintained on collagen IV–coated glass substrates to retain siRNA-treated cells. (A) Representative images are shown. (B) The number of EB1-GFP comets in the 8 × 8–µm boxed area was counted. For each condition, n > 31. The results are presented as means ± SEM (*, P < 0.01 vs. the mock control). (C) Microtubule organization in MCF-10A cells estimated from the observations obtained in this study. LL5s attach microtubule ends to the basal cortex and suppress their dynamicity. Without LL5 activities, the microtubule-stabilizing activity at the basal cortex is reduced, and once microtubules reach the basal surface, they remain dynamic. (D) Schematic diagram of the molecular link between laminin-5 and microtubule plus ends. Bar, 5 µm.
Discussion
Visualization of microtubule plus ends in three dimensions
In this study, we analyzed the three-dimensional distribution of microtubule plus ends in polarized columnar epithelial cells using EB1-GFP and high-speed confocal microscopy. Previously, we have analyzed microtubule dynamics in two dimensions by visualizing both microtubules and EB1-GFP in strongly flattened HeLa cells and demonstrated that the parameters of EB1-GFP motility are good indicators of the overall microtubule dynamics (Mimori-Kiyosue et al., 2005; Lansbergen et al., 2006), which is in agreement with many other studies (for review see Morrison, 2007). In this study, we expanded EB1-GFP imaging into three-dimensional epithelial cells, in which individual microtubule filaments were hard to distinguish especially along the apicobasal axis, and showed that EB1-GFP is useful to detect the trends of microtubule organization also in this system. Thus, imaging of microtubule plus end markers emerges as a very valuable approach to examine microtubule organization in thick specimens, including living animals (Norden et al., 2009).
Laminin–integrin-dependent localization of LL5s
Using the aforementioned technique in combination with conventional confocal microscopy and three-dimensional cell culture, we explored the mechanisms that determine the localization of LL5s, microtubule-anchoring factors, in MCF-10A human mammary epithelial cells and found that their association with the basal cell cortex was dependent on laminin-5–mediated non-FA cell–substrate attachments. Furthermore, we found that accumulation of LL5s at the basal cortex was initiated by activation of integrin α3β1 in an interdependent manner and that their localization was maintained by both integrins α3β1 and α6β4. We also found that LL5s form complexes with these integrins, although the interactions might be indirect. Because LL5β is known to associate with γ-filamin, an integrin-binding protein (Paranavitane et al., 2003), we tested the possibility of the involvement of γ-filamin and another integrin adaptor, talin, in the LL5 localization but found no indications for their direct contribution (unpublished data). Our findings suggest that there could be unknown factors acting together with the cytoplasmic tails of integrins to determine the distribution of LL5s at the basal cell cortex. The precise molecular mechanisms mediating the basal localization of LL5s should be further clarified in future studies.
Basal localization of LL5s via coincidence sensing
The PH domain of LL5s has been known to bind to PtdIns(3,4,5)P3 (Dowler et al., 2000; Paranavitane et al., 2003). Because it was previously shown that cell adhesion to laminin generates higher levels of activity of phosphoinositide 3-kinase and its downstream effector Akt than adhesion to collagen or fibronectin (Nguyen et al., 2000a; Gu et al., 2002), signaling via laminin–integrin coupling could be an ideal cue to initiate the formation of microtubule attachment sites via LL5s. In a previous study, the LL5β PH domain tagged with GFP was localized to the plasma membrane but was not restricted to the basal cortex and instead was abundantly present in entire areas of the plasma membrane (Lansbergen et al., 2006), indicating a requirement for another localization determination factor. In the present study, we found that the basal accumulation of LL5s was dependent on laminin receptor integrins. It is likely that LL5s localize selectively at the basal cortex by acting as detectors of coincidence localization signals, similar to other PH domain–containing proteins (for review see Carlton and Cullen, 2005), downstream of both phosphoinositide signaling and integrin signaling.
Interactions between microtubules and cell–substratum adhesions
The association of microtubules with cellular adhesions was first described in migrating fish fibroblasts, in which microtubule ends targeted to FAs were visualized by vinculin staining using live cell fluorescence microscopy (Kaverina et al., 1998). It has also been reported that fibronectin-mediated stimulation of FAK induces localized stabilization of microtubules by Rho signaling in migrating mouse fibroblasts (Palazzo et al., 2004). TIRF imaging revealed that microtubule ends come into close proximity (<100 nm) of the plasma membrane at FAs (Krylyshkina et al., 2003). Following this observation, we found that the microtubule plus ends in HeLa cells are anchored in the close vicinity of the plasma membrane through complexes with EB1/CLASP/LL5β using TIRF microscopy, although LL5β was not colocalized with FAs (Mimori-Kiyosue et al., 2005; Lansbergen et al., 2006). In the present study, we have revealed that LL5s are accumulated at non-FA cell–substratum adhesion sites composed of deposited laminin-5 and laminin receptor integrins. Taking these findings into consideration, it is likely that microtubules can be targeted to different types of cell–substratum adhesion sites through distinct pathways. However, because microtubule targeting to FAs has been shown to regulate FA turnover and thereby facilitate cell migration in concert with the endocytic process (Small and Kaverina, 2003; Ezratty et al., 2005; Matsui et al., 2008), it is possible that microtubules anchored by LL5s could affect laminin-mediated cell adhesion and migration in epithelia.
LL5s attach microtubules to the basal surface of polarized epithelial sheets
In this study, using a highly sensitive fast-scanning fluorescence microscope system and EB1-GFP imaging/analysis, we collected images of the entire cell volumes inside the epithelial sheet with sufficient quality to count the number of EB1-GFP comets and analyzed their three-dimensional distribution. Using this technique in combination with conventional confocal microscopy, we demonstrated that in fully polarized epithelial monolayers, LL5s are localized at the basal surfaces and act as microtubule-anchoring factors by recruiting CLASP/EB1-positive microtubule plus ends to these structures.
However, in the absence of the LL5s, many microtubules still remained normally oriented toward the basal cortex after LL5 knockdown, as shown in Fig. 8. Some of them grow toward the basal side stochastically without being attached firmly to the cortex and exhibit random growth/shortening dynamics. But because we also observed that ACF7 and APC, other microtubule-stabilizing proteins, remained at the basal cortex after knockdown of integrin α3β1 or α6β4 (unpublished data), this suggested the presence of other microtubule-anchoring mechanisms at the epithelial basal cortex. Previously, the involvement of APC in the nucleation and organization of acentrosomal microtubules at the epithelial basal cortex has been described (Reilein et al., 2005). The redundancy and/or interrelationship between multiple microtubule-anchoring mechanisms should be further clarified in the future. However, from the viewpoint of the overall organization of epithelial microtubules, the fact that the populations of microtubules and EB1-GFP comets were increased in the upper parts of the cells after LL5 depletion suggests that the majority of the microtubule-nucleating factors, which frequently reside around the centrosomes and Golgi structures, are maintained at the apical side. This indicates the possibility that the concentration of microtubule-nucleating factors at the apical side occurs first and independently of the LL5-mediated regulation of basal microtubules.
Physiological meaning of basal microtubule anchoring
What is the physiological meaning of microtubule attachment to the basal cortex? Apicobasal microtubule alignment could be beneficial for the directional transport of specific cell components by plus end–directed motors. However, to date, no specific cargoes transported exclusively toward the basal cell cortex have been reported, although apical and basolateral targeting is well documented (Rodriguez-Boulan et al., 2005). In this regard, it is noteworthy that during avian development, laminin BM integrity is controlled by the stability of the basal microtubules (Nakaya et al., 2008). Consequently, it would be interesting if LL5s were involved in the trafficking of BM components. However, we observed that knockdown of LL5s did not inhibit laminin-5 deposition, at least under our experimental conditions. This finding is consistent with a previous report that integrins α3β1 and α1β4 are not essential for the deposition of laminin-5 into the BM, as evaluated using integrin-null animal models (DiPersio et al., 2000). However, further analyses of the microtubule attachment mechanisms and their importance at the epithelial basal cortex in more physiological environments will provide important insights into the biological significance of microtubule–ECM communication during epithelial morphogenesis as well as development.
Materials and methods
Cells and gene transfer
Human kidney epithelial cell line HEK293T cells were maintained in DME (Invitrogen) supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C under a 5% CO2 atmosphere. Plasmid transfections were performed using the Effectene transfection reagent (QIAGEN) according to the manufacturer’s instructions. Nontumorigenic human mammary epithelial MCF-10A cells (American Type Culture Collection) were grown in mammary epithelial cell growth medium (MEGM; Sanko-Junyaku Co.) supplemented with 100 ng/ml cholera toxin, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C under a 5% CO2 atmosphere. Retrovirus-mediated gene transfer was performed as previously described (Akagi et al., 2003). Parental MCF-10A cells were first infected with an amphotropic retroviral vector expressing a murine ecotropic retrovirus receptor. This procedure rendered MCF-10A cells susceptible to subsequent infection with ecotropic viral vectors (MCF-10Aeco cell line). For ecotropic retrovirus-mediated gene transfer, cDNAs were inserted into pCX4 series retroviral vectors carrying different drug-selection markers. The infected cells were selected in media containing appropriate drugs.
Three-dimensional culture of MCF-10A cells
Three-dimensional cultures were performed as described previously (Debnath et al., 2003). In brief, for spheroid formation, siRNA-treated cells (1 × 104 cells) in MEGM containing 2% Matrigel were seeded as single-cell suspensions into 4-well chamber slides (Nunc) coated with growth factor–reduced Matrigel (BD) at 24 h after transfection. The cells were cultured in the Matrigel for 2 d. For branching morphogenesis assays (Stahl et al., 1997), cells (2 × 105 or 4 × 105) in the medium containing 2% Matrigel were seeded as single-cell suspensions into 4-well chamber slides coated with growth factor–reduced Matrigel.
Expression constructs
GFP- or RFP-fused LL5β expression constructs were described previously (Lansbergen et al., 2006). GFP-fused LL5α was generated by inserting an LL5α cDNA into the pEGFP-C vector. To express C-terminal fragments of integrin α3 and α6 in cells, PCR-amplified fragments were generated using human integrin α3 variant a cDNA (OriGene) and MCF-10A first strand cDNA as templates. siRNA-resistant mutated LL5α and -β constructs (rescue; represented as LL5α(res) and LL5β(res), respectively) were PCR engineered by introducing eight silent substitutions into the target site of LL5α siRNA (resulting in the sequence 5′-TGGTGGCCATCAGCCTGAGTGAATA-3′) and seven silent substitutions into the target site of LL5β siRNA (resulting in the sequence 5′-GGAGAAGGAAATCCTAGACCACTTA-3′) and subcloned into pCX4bsr and pCX4hyg, respectively. An LL5β mutant lacking the CLASP-binding region (aa 561–910; LL5β(ΔM)) was generated by PCR engineering. To add N-terminal tags, a sequence encoding EGFP (Takara Bio Inc.), TagRFP (Evrogen), mKO (monomeric Kusabira orange; MBL), or three repeats of myc tag (myc3) was inserted at the 5′ ends of the LL5 cDNAs. GFP–α-tubulin (Takara Bio Inc.) and EB1-GFP (Mimori-Kiyosue et al., 2000) were subcloned into pCX4puro. A laminin-γ2 cDNA (Thermo Fisher Scientific) was subcloned into pCX4puro, and a C-terminal GFP tag was added.
Transfection of siRNAs
Synthetic stealth siRNAs (Invitrogen) were transfected using the HiPerFect transfection reagent (QIAGEN) according to the manufacturer’s instructions. siRNAs were directed against the following target sites: LL5α, 5′-TGGTGGCCATCAGCCTGAGTGAATA-3′; LL5β, 5′-GGAGAAGGAGATTTTGGATCATCTA-3′; integrin α3, 5′-ATAAACCAAAGGCAGAGCCACTGGG-3′; integrin α6, 5′-ATACATTGTCGTCTCCACATCCCTC-3′; integrin β1, 5′-TACACTTACAGACACCACACTCGCA-3′; integrin β4, 5′-TGAACATCTCGTCTGTGCAGTAGGC-3′; and laminin-γ2, 5′-TGTCCAGCTTGCTATAATCAAGTGA-3′. The siRNAs were transfected at a concentration of 100 nM. The knockdown efficiency was analyzed by Western blotting (Fig. S2). To retain laminin-γ2– or integrin-depleted cells on the culture plates, the siRNA-treated cells were seeded onto collagen-coated plates (Iwaki).
Antibodies
To generate mAbs against LL5α and -β, fragments of mouse LL5α and -β corresponding to aa 703–951 (FLJ00141) and aa 563–916, respectively, were fused to a GST tag using the pGEX-5X-2 vector, expressed in Escherichia coli, purified with GST-affinity resin (GE Healthcare), and used to immunize Armenian hamsters. Hybridomas were produced by standard methods (BMR Division of Sunplanet Co. Ltd.), and their products were assayed by ELISA, Western blotting, and immunofluorescence staining using GFP-fused human LL5α or -β as controls to select LL5α– and LL5β-specific clones (clones #223 and #843, respectively). The characterization of the antibodies is shown in Fig. S1 (A and B). Rabbit anti-CLASP1 pAbs (Akhmanova et al., 2001), rabbit anti-LL5β pAb (Lansbergen et al., 2006), and mouse anti-LL5β mAb (provided by J. Sanes, Harvard University, Cambridge, MA; Kishi et al., 2005) were described previously. The following primary antibodies were purchased from the indicated sources: mouse anti–integrin α3 mAb (clone P1B5; Millipore), mouse anti–integrin α3 mAb (clone ASC-1; Millipore), rabbit anti–integrin α3 pAb (Millipore), rat anti–integrin α6 mAb (clone GoH3; Abcam), mouse anti–integrin β1 mAb (clone P5D2; Millipore), mouse anti–integrin β1 mAb (BD), mouse anti–integrin β4 mAb (clone M126; Abcam), mouse anti–integrin αv mAb (clone P2W7; Santa Cruz Biotechnology, Inc.), rabbit anti–laminin-5 (Abcam), mouse anti-epiligrin (clone P3E4; Millipore), mouse anti-epiligrin (clone P3H9-2; Millipore), mouse anti–laminin-5 (γ2 subunit) mAb (clone D4B5; Millipore), rabbit antilaminin pAb (Sigma-Aldrich), mouse antivinculin mAb (Sigma-Aldrich), rabbit anti–β-catenin pAb (Sigma-Aldrich), mouse anti-EB1 mAb (BD), anti-MACF1 (ACF7) mAb (Abnova), mouse anti-p150glued mAb (BD), mouse anti–α-tubulin mAb (DM1A; Sigma-Aldrich), FITC-conjugated anti–α-tubulin mAb (DM1A; Sigma-Aldrich), rat anti–α-tubulin mAb (YL1/2; Abcam), rabbit anti-GFP pAb (Millipore), mouse anti-GFP mAb (Millipore), rabbit anti-DsRed antibody (denoted as an anti-RFP antibody in the text; Takara Bio Inc.), mouse anti-myc mAb (clone 4A6; Millipore), and HRP-conjugated goat anti-myc pAb (Abcam). As secondary antibodies, Cy2-, FITC-, rhodamine red–X-, and Cy5-conjugated anti–mouse, anti–rabbit, anti–rat, or anti–Armenian hamster antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc.
Immunofluorescence staining
For conventional two-dimensional cultures, the cells were seeded on uncoated coverslips as described previously (Mimori-Kiyosue et al., 2005). Fixed and immunostained samples were mounted in ProLong Antifade reagent (Invitrogen) or PermaFluor (Beckman Coulter). Cells cultured in Matrigel were fixed with a 1:1 mixture of methanol/acetone at −20°C for 10 min. After washing in PBS, the cells were permeabilized with 0.5% Triton X-100 in PBS, blocked with immunofluorescence buffer (3% BSA, 0.05% Tween 20, 0.04% sodium azide, and PBS), and sequentially incubated with primary and secondary antibodies. The stained cells were mounted with ProLong Gold Antifade reagent (Invitrogen). Tissue samples from C57BL6/J strain mice (8 wk of age) were prepared by perfusion fixation and processed as described previously (Nakatani et al., 2004).
Fluorescence microscopy and image analysis
Confocal imaging was performed using a confocal laser-scanning microscope system (Fluoview FV1000; Olympus) driven by FV10-ASW software (Olympus) equipped with an inverted microscope (IX81; Olympus) and a Plan-Apochromat 100× NA 1.40 oil immersion or 10× NA 0.40 objective (Olympus) or a confocal laser-scanning microscope (LSM510; Carl Zeiss, Inc.) driven by LSM510 software (version 2.3; Carl Zeiss, Inc.) equipped with an inverted microscope (Axiovert 100M; Carl Zeiss, Inc.) and a Plan-Apochromat 63× NA 1.40 or 100× NA 1.40 oil immersion objective (Carl Zeiss, Inc.). Reflection interference microscopy was performed using the LSM510 system. Live cell imaging was performed using a microscope system (DeltaVision Core; Applied Precision) driven by SoftWoRx software (Applied Precision) equipped with an IX70 microscope with a Plan-Apochromat 100× NA 1.40 oil immersion objective (Olympus), a cooled charge-coupled device camera (CoolSNAP HQ2; Photometrics) and a CO2 incubator (Tokai Hit Co., Ltd.) for live cell culture, or a Revolution XD system driven by IQ software (Andor Technology) equipped with an IX81 inverted microscope with a UPlanSApo 100× NA 1.40 oil immersion objective (Olympus), an electron-multiplying charge-coupled device camera (iXon DU-888; Andor Technology), a confocal spinning disk (CSU22; Yokogawa), a piezo-Z stage, a laser combiner with an acousto-optic tunable filter and a CO2 incubator for live cell culture. For live cell imaging, cells were seeded on uncoated or collagen IV (BD)–coated glass-based dishes (Iwaki) in MEGM at 37°C under a 5% CO2 atmosphere. To establish polarized epithelial sheets, cells were seeded on glass-bottomed dishes (2–3 × 106 cells/35-mm diameter dish) and cultured for 24–36 h.
As for the images collected with a DeltaVision system, out of focus signals were removed using the three-dimensional deconvolution technique included in the system. Quantification and analyses of fluorescent signals were performed using ImageJ (National Institutes of Health), MetaMorph (MDS Analytical Technologies), or Imaris 6.4 (Bitplane AG) software. For quantitative analysis of the fluorescent signals at the basal cortex in confocal cross section images of spheroids, the entire length of the circumferential region of the spheroids was manually selected to obtain the mean fluorescent signal intensity, and the mean intensity of the background staining in the cytoplasm was subtracted. For quantitative analysis of the fluorescent signals in confocal images, the mean fluorescent signal intensity in an appropriate region of interest (ROI) was analyzed. Measurement of microtubule growth parameters from EB1-GFP videos was performed manually using the MetaMorph software, and the microtubule growth rates were measured by kymograph analysis (Mimori-Kiyosue et al., 2005). The statistical significance of differences was assessed using a two-sample t test. Colocalization analysis was performed using the ImarisColoc software (Bitplane AG). The ROI was selected by the threshold masking function, and the intensity values for the equivalent pixel positions inside the ROI were compared. The percentage of colocalized pixels and Pearson’s correlation coefficient in colocalized volume (R(r)) between the two color channels were obtained on a pixel by pixel basis. For tissues, entire areas of pictures or cropped regions were analyzed.
The images were processed using Photoshop software (Adobe Systems Inc.), and the adjustments of brightness, contrast, and gamma settings were applied to the whole image. The pictures of series of immunolabeling samples under different conditions were collected with same machine settings and processed equally using Photoshop software. For Fig. 9 A, an unsharp masking was used to sharpen the images. The time-lapse images were processed using MovieMaker and Media Convert software installed on an O2 computer (Silicon Graphics, Inc.) or ImageJ to generate composite videos.
Immunoprecipitation, Western blotting, and microbead assays
For immunoprecipitation, cells were lysed with radioimmunoprecipitation assay buffer (Cell Signaling Technology) containing a protease inhibitor cocktail (Sigma-Aldrich) and immunoprecipitated with protein G–agarose (Invitrogen). To prepare total cell lysates, cells were lysed in SDS sample buffer containing a protease inhibitor cocktail (Sigma-Aldrich). The samples were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and probed with appropriate antibodies. Proteins were visualized using ECL Plus Western blotting detection reagents (GE Healthcare) and a LuminoImager (LAS-3000; Fujifilm). The band densities were analyzed with the Multi Gauge software (version 3.1; Fujifilm). Laminin-5–conjugated microbeads were prepared using a PolyLink protein-coupling kit (Polyscience) with an anti–laminin-5 mAb and MCF-10A–conditioned medium. The images were processed using Photoshop software, and the adjustments of brightness, contrast, and gamma settings were applied to the whole image.
Mass spectrometry analysis
To collect ECM proteins deposited by MCF-10A cells, the cells were cultured for 3 d and dissociated from the culture dishes with 10 mM EDTA. The proteins remaining on the dishes were lysed in SDS sample buffer, separated by SDS-PAGE, and subjected to negative staining. Protein identification was performed by mass spectrometric analysis as previously described (Tabata et al., 2007). The mass spectrometric data files were searched against the National Center for Biotechnology Information nonredundant human database using Mascot 2.1 (Matrix Science).
Online supplemental material
Fig. S1 shows the characterization of human mammary epithelial cell line MCF-10A cells, subclones derived from MCF-10A cells, and antibodies against LL5s. Fig. S2 shows the characterization of siRNA tools. Fig. S3 shows the effects of integrin β1 or β4 subunit depletion on the LL5 distribution. Fig. S4 shows the mutual effects of integrin α and β subunit depletion on their localizations at the basal cortex. Video 1 shows that microtubules are attached to the basal cortex where mKO-LL5β is accumulated. Video 2 shows a three-dimensional representation of the distributions of RFP-LL5α and microtubules in a single living cell. Video 3 shows the behavior of RFP-LL5α during cell attachment and spreading on the substratum. Video 4 shows behaviors of RFP-LL5α and microtubules in living cells undergoing random migration. Videos 5 and 6 show the deposition of laminin-γ2–GFP on the glass surface. Video 7 shows z-axis scanning of EB1-GFP in a living cell within a polarized epithelial monolayer. Video 8 shows a three-dimensional representation of the distributions of RFP-LL5α and EB1-GFP in a live cell within a polarized epithelial monolayer. Video 9 shows time-lapse videos of EB1-GFP at the apical, middle, and basal planes of a living cell within a polarized epithelial monolayer. Video 10 shows time-lapse videos of EB1-GFP at the basal cortex of living cells within a polarized epithelial monolayer. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200910095/DC1.
Acknowledgments
We are grateful to Dr. M. Takeichi (RIKEN Center for Developmental Biology, Chuo-ku, Kobe, Japan) for critical reading of the manuscript and advice. We also thank Drs. K. Sekiguchi (Osaka University, Suita, Osaka, Japan), W. Ikeda (Kobe University, Chuo-ku, Kobe), and T. Otsuka (University of Yamanashi, Kofu, Yamanashi, Japan) for helpful discussions and advice, K. Kato (RIKEN Center for Developmental Biology) for editing a QuickTime video, H. Ogasawara (KAN Research Institute, Inc., Chuo-ku, Kobe, Japan) for excellent technical support, E. Fuchs (The Rockefeller University, New York, NY) for the rabbit anti-ACF7 pAb, and J. Sanes for the mouse anti-LL5β mAb. A part of image acquisition and analysis was performed at the Center for Developmental Biology Imaging Facility.
This study was supported by the Netherlands Organization for Scientific Research ZonMw-TOP and ALW-VICI grants to A. Akhmanova.
Footnotes
Abbreviations used in this paper:
- ACF7
- actin cross-linking family 7
- APC
- adenomatous polyposis coli
- BM
- basement membrane
- CLASP
- CLIP-associating protein
- FA
- focal adhesion
- MEGM
- mammary epithelial cell growth medium
- PH
- pleckstrin homology
- PtdIns(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- ROI
- region of interest
- +TIP
- plus end–tracking protein
- TIRF
- total internal reflection fluorescence
References
- Akagi T., Sasai K., Hanafusa H. 2003. Refractory nature of normal human diploid fibroblasts with respect to oncogene-mediated transformation. Proc. Natl. Acad. Sci. USA. 100:13567–13572 10.1073/pnas.1834876100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A., Hoogenraad C.C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B.M., De Zeeuw C.I., Grosveld F., Galjart N. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 104:923–935 10.1016/S0092-8674(01)00288-4 [DOI] [PubMed] [Google Scholar]
- Bacallao R., Antony C., Dotti C., Karsenti E., Stelzer E.H., Simons K. 1989. The subcellular organization of Madin-Darby canine kidney cells during the formation of a polarized epithelium. J. Cell Biol. 109:2817–2832 10.1083/jcb.109.6.2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.E., DiPasquale A.P., Stock E.L., Quaranta V., Fitchmun M., Jones J.C. 1996. Morphogenetic effects of soluble laminin-5 on cultured epithelial cells and tissue explants. Exp. Cell Res. 228:262–270 10.1006/excr.1996.0325 [DOI] [PubMed] [Google Scholar]
- Carlton J.G., Cullen P.J. 2005. Coincidence detection in phosphoinositide signaling. Trends Cell Biol. 15:540–547 10.1016/j.tcb.2005.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter W.G., Wayner E.A., Bouchard T.S., Kaur P. 1990. The role of integrins α2β1 and α3β1 in cell-cell and cell-substrate adhesion of human epidermal cells. J. Cell Biol. 110:1387–1404 10.1083/jcb.110.4.1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter W.G., Ryan M.C., Gahr P.J. 1991. Epiligrin, a new cell adhesion ligand for integrin alpha 3 beta 1 in epithelial basement membranes. Cell. 65:599–610 10.1016/0092-8674(91)90092-D [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S.K., Brugge J.S. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 30:256–268 10.1016/S1046-2023(03)00032-X [DOI] [PubMed] [Google Scholar]
- deHart G.W., Healy K.E., Jones J.C. 2003. The role of alpha3beta1 integrin in determining the supramolecular organization of laminin-5 in the extracellular matrix of keratinocytes. Exp. Cell Res. 283:67–79 10.1016/S0014-4827(02)00028-9 [DOI] [PubMed] [Google Scholar]
- Desai A., Mitchison T.J. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83–117 10.1146/annurev.cellbio.13.1.83 [DOI] [PubMed] [Google Scholar]
- DiPersio C.M., van der Neut R., Georges-Labouesse E., Kreidberg J.A., Sonnenberg A., Hynes R.O. 2000. alpha3beta1 and alpha6beta4 integrin receptors for laminin-5 are not essential for epidermal morphogenesis and homeostasis during skin development. J. Cell Sci. 113:3051–3062 [DOI] [PubMed] [Google Scholar]
- Dowler S., Currie R.A., Campbell D.G., Deak M., Kular G., Downes C.P., Alessi D.R. 2000. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 351:19–31 10.1042/0264-6021:3510019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S., Manneville J.B., Nicholls S., Ferenczi M.A., Hall A. 2005. Cdc42 and Par6–PKCζ regulate the spatially localized association of Dlg1 and APC to control cell polarization. J. Cell Biol. 170:895–901 10.1083/jcb.200412172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty E.J., Partridge M.A., Gundersen G.G. 2005. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 7:581–590 10.1038/ncb1262 [DOI] [PubMed] [Google Scholar]
- Goldfinger L.E., Hopkinson S.B., deHart G.W., Collawn S., Couchman J.R., Jones J.C. 1999. The alpha3 laminin subunit, alpha6beta4 and alpha3beta1 integrin coordinately regulate wound healing in cultured epithelial cells and in the skin. J. Cell Sci. 112:2615–2629 [DOI] [PubMed] [Google Scholar]
- Goode B.L., Drubin D.G., Barnes G. 2000. Functional cooperation between the microtubule and actin cytoskeletons. Curr. Opin. Cell Biol. 12:63–71 10.1016/S0955-0674(99)00058-7 [DOI] [PubMed] [Google Scholar]
- Gu J., Fujibayashi A., Yamada K.M., Sekiguchi K. 2002. Laminin-10/11 and fibronectin differentially prevent apoptosis induced by serum removal via phosphatidylinositol 3-kinase/Akt- and MEK1/ERK-dependent pathways. J. Biol. Chem. 277:19922–19928 10.1074/jbc.M200383200 [DOI] [PubMed] [Google Scholar]
- Hemler M.E. 1999. Integrins. Guidebook to the Extracellular Matrix, Anchor, and Adhesion Proteins. Second edition Kreis T., Vale R., Oxford University Press, New York: 196–212 [Google Scholar]
- Hintermann E., Quaranta V. 2004. Epithelial cell motility on laminin-5: regulation by matrix assembly, proteolysis, integrins and erbB receptors. Matrix Biol. 23:75–85 10.1016/j.matbio.2004.03.001 [DOI] [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Katayama M., Sekiguchi K. 2004. Laminin-5 in epithelial tumour invasion. J. Mol. Histol. 35:277–286 10.1023/B:HIJO.0000032359.35698.fe [DOI] [PubMed] [Google Scholar]
- Kaverina I., Rottner K., Small J.V. 1998. Targeting, capture, and stabilization of microtubules at early focal adhesions. J. Cell Biol. 142:181–190 10.1083/jcb.142.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M., Mitchison T. 1986. Beyond self-assembly: from microtubules to morphogenesis. Cell. 45:329–342 10.1016/0092-8674(86)90318-1 [DOI] [PubMed] [Google Scholar]
- Kishi M., Kummer T.T., Eglen S.J., Sanes J.R. 2005. LL5β: a regulator of postsynaptic differentiation identified in a screen for synaptically enriched transcripts at the neuromuscular junction. J. Cell Biol. 169:355–366 10.1083/jcb.200411012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama A., Karakesisoglou I., Wong E., Vaezi A., Fuchs E. 2003. ACF7: an essential integrator of microtubule dynamics. Cell. 115:343–354 10.1016/S0092-8674(03)00813-4 [DOI] [PubMed] [Google Scholar]
- Krylyshkina O., Anderson K.I., Kaverina I., Upmann I., Manstein D.J., Small J.V., Toomre D.K. 2003. Nanometer targeting of microtubules to focal adhesions. J. Cell Biol. 161:853–859 10.1083/jcb.200301102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G., Akhmanova A. 2006. Microtubule plus end: a hub of cellular activities. Traffic. 7:499–507 10.1111/j.1600-0854.2006.00400.x [DOI] [PubMed] [Google Scholar]
- Lansbergen G., Grigoriev I., Mimori-Kiyosue Y., Ohtsuka T., Higa S., Kitajima I., Demmers J., Galjart N., Houtsmuller A.B., Grosveld F., Akhmanova A. 2006. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev. Cell. 11:21–32 10.1016/j.devcel.2006.05.012 [DOI] [PubMed] [Google Scholar]
- Levi L., Hanukoglu I., Raikhinstein M., Kohen F., Koch Y. 1993. Cloning of LL5, a novel protein encoding cDNA from a rat pituitary library. Biochim. Biophys. Acta. 1216:342–344 [DOI] [PubMed] [Google Scholar]
- Matsui C., Kaieda S., Ikegami T., Mimori-Kiyosue Y. 2008. Identification of a link between the SAMP repeats of adenomatous polyposis coli tumor suppressor and the Src homology 3 domain of DDEF. J. Biol. Chem. 283:33006–33020 10.1074/jbc.M800420200 [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Tsukita S. 2003. “Search-and-capture” of microtubules through plus-end-binding proteins (+TIPs). J. Biochem. 134:321–326 10.1093/jb/mvg148 [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Shiina N., Tsukita S. 2000. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10:865–868 10.1016/S0960-9822(00)00600-X [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., Akhmanova A. 2005. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168:141–153 10.1083/jcb.200405094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Matsui C., Sasaki H., Tsukita S. 2007. Adenomatous polyposis coli (APC) protein regulates epithelial cell migration and morphogenesis via PDZ domain-based interactions with plasma membranes. Genes Cells. 12:219–233 10.1111/j.1365-2443.2007.01045.x [DOI] [PubMed] [Google Scholar]
- Mizushima H., Koshikawa N., Moriyama K., Takamura H., Nagashima Y., Hirahara F., Miyazaki K. 1998. Wide distribution of laminin-5 gamma 2 chain in basement membranes of various human tissues. Horm. Res. 50:7–14 10.1159/000053118 [DOI] [PubMed] [Google Scholar]
- Morrison E.E. 2007. Action and interactions at microtubule ends. Cell. Mol. Life Sci. 64:307–317 10.1007/s00018-007-6360-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani T., Mizuhara E., Minaki Y., Sakamoto Y., Ono Y. 2004. Helt, a novel basic-helix-loop-helix transcriptional repressor expressed in the developing central nervous system. J. Biol. Chem. 279:16356–16367 10.1074/jbc.M311740200 [DOI] [PubMed] [Google Scholar]
- Nakaya Y., Sukowati E.W., Wu Y., Sheng G. 2008. RhoA and microtubule dynamics control cell-basement membrane interaction in EMT during gastrulation. Nat. Cell Biol. 10:765–775 10.1038/ncb1739 [DOI] [PubMed] [Google Scholar]
- Nguyen B.P., Gil S.G., Carter W.G. 2000a. Deposition of laminin 5 by keratinocytes regulates integrin adhesion and signaling. J. Biol. Chem. 275:31896–31907 10.1074/jbc.M006379200 [DOI] [PubMed] [Google Scholar]
- Nguyen B.P., Ryan M.C., Gil S.G., Carter W.G. 2000b. Deposition of laminin 5 in epidermal wounds regulates integrin signaling and adhesion. Curr. Opin. Cell Biol. 12:554–562 10.1016/S0955-0674(00)00131-9 [DOI] [PubMed] [Google Scholar]
- Norden C., Young S., Link B.A., Harris W.A. 2009. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 138:1195–1208 10.1016/j.cell.2009.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo A.F., Eng C.H., Schlaepfer D.D., Marcantonio E.E., Gundersen G.G. 2004. Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science. 303:836–839 10.1126/science.1091325 [DOI] [PubMed] [Google Scholar]
- Paranavitane V., Coadwell W.J., Eguinoa A., Hawkins P.T., Stephens L. 2003. LL5beta is a phosphatidylinositol (3,4,5)-trisphosphate sensor that can bind the cytoskeletal adaptor, gamma-filamin. J. Biol. Chem. 278:1328–1335 10.1074/jbc.M208352200 [DOI] [PubMed] [Google Scholar]
- Reilein A., Yamada S., Nelson W.J. 2005. Self-organization of an acentrosomal microtubule network at the basal cortex of polarized epithelial cells. J. Cell Biol. 171:845–855 10.1083/jcb.200505071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez O.C., Schaefer A.W., Mandato C.A., Forscher P., Bement W.M., Waterman-Storer C.M. 2003. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5:599–609 10.1038/ncb0703-599 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Nelson W.J. 1989. Morphogenesis of the polarized epithelial cell phenotype. Science. 245:718–725 10.1126/science.2672330 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Kreitzer G., Müsch A. 2005. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol. 6:233–247 10.1038/nrm1593 [DOI] [PubMed] [Google Scholar]
- Schoenenberger C.A., Matlin K.S. 1991. Cell polarity and epithelial oncogenesis. Trends Cell Biol. 1:87–92 10.1016/0962-8924(91)90035-8 [DOI] [PubMed] [Google Scholar]
- Schuyler S.C., Pellman D. 2001. Microtubule “plus-end-tracking proteins”: The end is just the beginning. Cell. 105:421–424 10.1016/S0092-8674(01)00364-6 [DOI] [PubMed] [Google Scholar]
- Small J.V., Kaverina I. 2003. Microtubules meet substrate adhesions to arrange cell polarity. Curr. Opin. Cell Biol. 15:40–47 10.1016/S0955-0674(02)00008-X [DOI] [PubMed] [Google Scholar]
- Soule H.D., Maloney T.M., Wolman S.R., Peterson W.D., Jr., Brenz R., McGrath C.M., Russo J., Pauley R.J., Jones R.F., Brooks S.C. 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50:6075–6086 [PubMed] [Google Scholar]
- Stahl S., Weitzman S., Jones J.C. 1997. The role of laminin-5 and its receptors in mammary epithelial cell branching morphogenesis. J. Cell Sci. 110:55–63 [DOI] [PubMed] [Google Scholar]
- Symington B.E., Takada Y., Carter W.G. 1993. Interaction of integrins alpha 3 beta 1 and alpha 2 beta 1: potential role in keratinocyte intercellular adhesion. J. Cell Biol. 120:523–535 10.1083/jcb.120.2.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T., Sato T., Kuromitsu J., Oda Y. 2007. Pseudo internal standard approach for label-free quantitative proteomics. Anal. Chem. 79:8440–8445 10.1021/ac701628m [DOI] [PubMed] [Google Scholar]
- van der Flier A., Sonnenberg A. 2001. Function and interactions of integrins. Cell Tissue Res. 305:285–298 10.1007/s004410100417 [DOI] [PubMed] [Google Scholar]
- Yeaman C., Grindstaff K.K., Nelson W.J. 1999. New perspectives on mechanisms involved in generating epithelial cell polarity. Physiol. Rev. 79:73–98 [DOI] [PubMed] [Google Scholar]