Summary
Reasons for performing study
A reverse genetics rescue system for equine influenza virus and the construction of three NS1 mutant viruses encoding carboxy-terminally truncated NS1 proteins of 73, 99 or 126 amino acids, have been previously described (Quinlivan et al., J. Virology 79, 8431–8439, 2005). These viruses are impaired in their ability to inhibit type I IFN production in vitro and are replication attenuated, thus are candidates for use as a modified live influenza virus vaccine in the horse.
Hypothesis
One or more of these mutant viruses are safe when administered to horses, and recipient horses when challenged with wild-type influenza have reduced physiological and virological correlates of disease.
Methods
Vaccination and challenge studies were done in horses, with measurement of pyrexia, clinical signs, virus shedding, and systemic pro-inflammatory cytokines.
Results
Aerosol or intranasal inoculation of horses with the viruses produced no adverse effects. Seronegative horses inoculated with the NS1-73 and NS1-126 viruses, but not the NS1-99 virus, shed detectable virus and generated significant levels of antibodies. Following challenge with wild-type influenza, horses vaccinated with NS1-126 virus did not develop fever (>38.5°C), had significantly fewer clinical signs of illness, and significantly reduced quantities of virus excreted for a shorter duration post-challenge compared to unvaccinated controls. Expression of pro-inflammatory cytokines IL-1β, IL-6, IFNγ, and TNFα was examined by quantitative RT-PCR of mRNA. Mean IL-1β and IL-6 levels were significantly higher in control animals, and were positively correlated with peak viral shedding and pyrexia on Day +2 post-challenge.
Conclusion
These data suggest the recombinant NS1 viruses are safe and effective as modified live virus vaccines against equine influenza.
Relevance
This type of reverse genetics-based vaccine can be easily updated by exchanging viral surface antigens to combat the problem of antigenic drift in influenza viruses.
Keywords: equine influenza, intranasal, NS-1, cytokine, reverse genetics, vaccine
Introduction
Influenza A virus of the H3N8 subtype circulates widely and causes outbreaks of disease in equine populations. Vaccination is the most effective method of prophylaxis against equine influenza and its complications (Paillot et al. 2006a; Park et al. 2003). Conventional equine influenza vaccines are inactivated whole virus or sub-unit preparations. However, in horses, immunity generated by natural infection is markedly different to that generated by vaccination with inactivated virus. The poor durability of the protective antibody response to these vaccines has been documented (Newton et al. 2000). Mucosal IgA is produced following natural infection but not conventional vaccination; whereas for IgG(T) (analog of mouse IgG1) the reverse is seen (Wilson et al. 2001). While there is induction of interferon-γ (IFNγ) by ISCOM vaccines (Paillot et al. 2006b) and canarypox-vectored vaccines increase the IFNγ response to challenge (Paillot et al. 2008), the antigen-specific cytotoxic T-lymphocyte (CTL) response generated after natural infection is unseen in horses vaccinated with conventional inactivated virus (Hannant and Mumford 1989).
Modified live virus (MLV) vaccines, administered intranasally, may mimic the process of natural infection better than inactivated vaccines and provide superior protection against illness. MLV vaccines are thought to induce improved cross-reactive CTL as well as humoral antibody responses (e.g. Gorse et al. 1995; Renegar and Small 1991; Tamura et al. 1990). Equine influenza virus replicates in the upper respiratory tract, thus an intranasally administered vaccine may be preferable to elicit the protective mucosal IgA response (Soboll et al. 2003b). A cold-adapted equine influenza MLV vaccine (FluAverttm IN; Heska Corp.) is safe and provides significant clinical protection at 6 months after single-dose vaccination of influenza-naïve horses (Townsend et al. 2001).
Today, influenza MLV vaccines may be created by introducing specific mutations to viral genes resulting in attenuation while maintaining immunogenicity (Palese and Garcia-Sastre 2002). The influenza viral NS1 gene is a candidate for attenuating mutations. The influenza NS1 protein has several regulatory functions during virus infection, including antagonism of the host IFNα/β antiviral response (Donelan et al. 2003; Kochs et al. 2007). An influenza A virus lacking the NS1 gene could only efficiently replicate in IFN-incompetent systems such as STAT1−/− mice or Vero cells (Garcia-Sastre et al. 1998). Also, human influenza viruses with truncated NS1 proteins are attenuated in mice (Egorov et al. 1998) and provide protection against wild-type challenge (Talon et al. 2000).
We previously described the establishment of a reverse genetics rescue system for equine influenza virus and the construction of three recombinant equine influenza viruses with truncations in their NS1 genes (Quinlivan et al. 2005b). These viruses were impaired in their ability to inhibit IFN production in vitro and also impaired in their replication efficiency in vitro or in vivo in a murine model. Here, the potential of these NS1 mutant viruses as candidates for a live equine influenza virus vaccine was assessed in the equine model.
Materials and methods
Vaccine viruses
Three recombinant equine influenza viruses (subtype H3N8) expressing carboxy-terminally truncated NS1 proteins were tested: NS1-73, NS1-99, and NS1-126, which express respectively the first 73, 99, and 126 of the 219 amino acids of the NS1 protein (Quinlivan et al. 2005b). The vaccine viruses were propagated in 7-day-old embryonated hen eggs which have an underdeveloped IFN system. Vaccine viruses were administered to horses either by inhalation of aerosolized virus (5×106 PFU) through a mask (Trial I) or by intranasal inoculation via a canula inserted 15 cm into the nasal meatus (Trial II).
Experimental challenges
The challenge virus was the homologous wild-type equine influenza virus strain, influenza A/equine/Kentucky/5/2002 (H3N8). Challenge virus (6×107 EID50 units) was aerosolized using a DeVillbis Ultra-Neb 99 nebulizer, and pumped into a tented stall (21.5 m3) where it was inhaled by the horses for 45 minutes (Mumford et al. 1990; Townsend et al. 2001).
Other methods
Methods for clinical monitoring, serological analysis, virus shedding with quantitation and confirmation of viral genotype, inflammatory cytokine responses post challenge, and statistical analyses may be found in the Supplementary Material, at the website www.evj.co.uk/suppinfo.
Results
Trial I: Safety of NS1 mutant viruses in horses
The three prototype vaccine viruses were administered to two horses each, including both influenza-seropositive and naïve seronegative animals. The NS1-73 and NS1-126 viruses were administered to two animals each (one naïve yearling pony and one seropositive 2-year-old). The NS1-99 virus was administered to two seronegative weanling (8-months-old) ponies. None of the vaccinated animals developed fever, cough or other clinical signs in association with the vaccination, indicating that the mutant viruses were safe for administration to horses.
The seronegative horses vaccinated with NS1-73 and NS1-126 shed virus for multiple days as detected by RT-PCR and egg isolation. Gel electrophoresis of RT-PCR products of the isolated viruses confirmed the shed virus possessed the NS1-73 and NS1-126 genotype of the vaccine virus (data not shown). Neither the NS1-99 animals nor the seropositive animals vaccinated with NS1-126 and NS1-73 shed virus detectable by either method.
The two animals vaccinated with NS1-99 also failed to generate a detectable SRH antibody response post-vaccination. The naïve animals vaccinated with NS1-126 and NS1-73 were seronegative on the day of vaccination and developed an antibody response (SRH > 100mm2) by two weeks post-vaccination with a slightly stronger response seen from the NS1-126 vaccine (data not shown).
Trial II: Vaccination and challenge
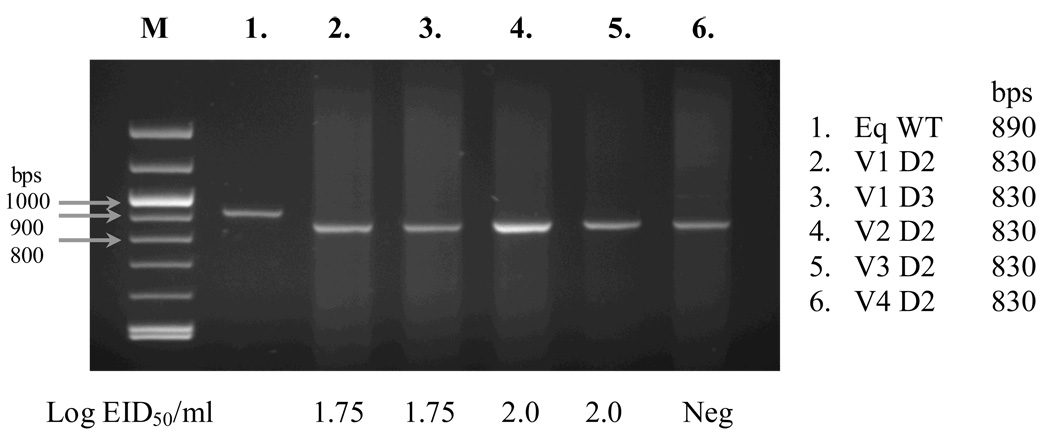
The NS1-126 virus was selected for a vaccination/challenge trial, as it was the most attenuated of the 3 recombinant viruses (Quinlivan et al. 2005b) and Trial I observations showed it to be promising in terms of immunogenicity. Six influenza-naïve yearling horses were given two doses of NS1-126 vaccine 4 weeks apart by using an intranasal canula. No clinical signs of infection were seen post-inoculation. Nasal swabs were taken after both vaccinations (V1 and V2) and NS1-126 virus was isolated from three of the vaccinated horses after V1. A fourth vaccinate was RT-PCR positive but no infectious virus was isolated. None excreted detectable virus after V2. The genotype of excreted virus was confirmed by RT-PCR (Figure 1).
Fig. 1. Trial II viral shedding post-vaccination.
RT-PCR of NS segments of (1) wild-type (WT) equine (Eq) influenza virus and virus shed from vaccinate animals (2–6, animals V1–V4) on days 2 and 3 (D2/3) post-vaccination. Sizes of the DNA markers (M) are indicated at the left, and lengths of the RT-PCR products are shown at the right. The WT equine influenza NS segment is 890bps long whereas the vaccine virus NS1-126 is 830bps long. The log10 EID50/ml values for the corresponding nasal secretions are indicated below the figure. Neg = negative.
Four weeks after V2, 5 of these vaccinated horses (the 6th, chosen at random, was omitted for lack of space) and 3 influenza-naïve yearling horses as unvaccinated controls, were challenged with wild type virus. Horses were exposed to the aerosolized challenge virus in two groups of 4, each group including both vaccinates and controls.
SRH antibody responses
All animals were seronegative on the day of V1, and controls remained seronegative throughout the vaccination period (Supplementary Material, Table 3). Four weeks later on the day of V2, all vaccinates had seroconverted (Table 3). On the day of challenge (four weeks after V2) all vaccinates were seropositive but there were no further increases in response to the second vaccination. At one week post-challenge one vaccinate exhibited an anamnestic response. At two weeks post-challenge the three control horses and 4/5 of vaccinates had seroconverted to equine influenza.
Febrile responses post-challenge
Mean rectal temperatures were calculated for the 5 NS1-126 vaccinated animals and the 3 unvaccinated control horses for Day −1 to Day +8 post-challenge (Fig. 2). There was a significant effect of day post-challenge (p=0.03) and vaccination status p<0.0001) on rectal temperatures for 8 days post-challenge with a significant interaction between day and status (p=0.049). None of the vaccinated animals developed fever >38.5°C after challenge with wild type equine influenza virus. In contrast, all 3 of the control animals had fever, which was a statistically significant difference between the two groups (p=0.009).
Fig. 2. Trial II febrile response post-challenge.
Mean rectal temperatures ± SD were taken for the day prior to (−1), the day of (0) and for 8 days post-challenge. From Day 6 onwards, mean rectal temperatures of controls, but not vaccinates, are presumed to be artificially reduced by instigation of antibiotic/anti-inflammatory therapy.
Clinical scores post-challenge
Clinical scores were assessed as described in the Supplementary Materials (Table 1). There were statistically significant differences between NS1-126 vaccinated animals and controls in clinical scores for coughing (p=0.017), nasal discharge (p=0.02) and abnormal lung sounds (p=0.022) (Fig. 3). All three control horses exhibited coughing, nasal discharge, and abnormal lung sounds, while one animal also had elevated respiration rate on Day +6. Two controls developed signs severe enough to trigger instigation of antibiotic/anti-inflammatory therapy on Day +5 and Day +7, respectively, in accordance with the veterinary care protocol approved by the University of Kentucky IACUC . In contrast, one vaccinate had a cough and abnormal lung sounds on Day +3 only, two vaccinates had slight nasal discharge for one day only, and another had mild abnormal lung sounds on Days +1 and +6.
Fig. 3. Trial II clinical scores post-challenge.
The clinical signs scoring index is found in Supplementary Materials online (Table 1). (a) Mean ± SD clinical signs recorded on the day prior to (−1), the day of (0) and for 8 days post-challenge. (b) Mean ± SD clinical scores for each sign recorded from days 1 to 8 post-challenge. Nasal Dx, nasal discharge. Resp, respiration.
Viral shedding post-challenge
Viral nucleic acid and infectious virus in nasal secretions were quantified by qRT-PCR (Fig. 4a) and EID50 analysis (Fig. 4b) respectively for eight days post-challenge. Viral RNA was detected in all 3 control horses (100%) from Days +2 to +8, compared to 3/5 of vaccinated animals (60%). The maximum amount in any swab from a control horse was 109 copies/ml RNA, compared to the maximum in any vaccinate of 6×107 copies/ml.
Fig. 4. Trial II viral shedding post-challenge.
Quantification of viral RNA (a) and infectious virus (b) shed in nasal secretions for eight days post-challenge as determined by qRT-PCR and EID50 analysis respectively. Error bars show SD.
All three control horses shed detectable infectious virus for 5 consecutive days from Day +2 to +6 post-challenge, with a maximum of 5.6×105 EID50/ml shed by a control horse on Day +4 (Fig. 4b). In contrast, 3/5 of vaccinated animals shed detectable infectious virus, with duration of shedding reduced to 1–3 days. The greatest amount shed by a vaccinated animal was 1.8×103 EID50/ml on Day +4 post-challenge. PCR detected viral RNA in samples where infectious virus was not detected (Fig.4), which we attribute to its much greater sensitivity.
Vaccination with NS1-126 mutant virus significantly reduced the total amount of viral RNA (p=0.017) and infectious virus (p<0.0001) detected in swabs post-challenge. Vaccination also significantly reduced the duration of viral shedding post-challenge; mean of 7 ± 0.0 versus 3.2 ± 1.4 days (p=0.049) by qRT-PCR, or 5.3 ± 0.6 versus 1.2 ± 1.3 days (p=0.002) by EID50. On the day of peak virus shedding (Day +3), vaccination resulted in mean reductions of 106 copies/ml RNA or 103 EID50/ml of infectious virus.
Inflammatory cytokine mRNA production
RT-PCR analysis of pro-inflammatory cytokine expression following challenge (Fig. 5) revealed that mean IL-1β and IL-6 mRNA levels were significantly heightened in controls compared to vaccinates on Day +2 (both p<0.001), correlating with observed rectal temperatures. TNFα mRNA levels in controls peaked on Day +3, however there were no significant differences between controls and vaccinates. While there was a significant difference in IFNγ production over time (P=0.048), there were no significant differences between vaccinates and controls.
Fig. 5. Pro-inflammatory cytokine mRNA induction post-challenge.
Cytokine mRNA expression in peripheral blood cells was determined by quantitative RT-PCR, using primers/probes as described in Supplementary Materials online(Table 2). Y-axes are relative quantity (RQ) with the average of the Day -1 samples for each cytokine being used as the calibrator. Days post-challenge are shown at bottom. Panels (clockwise from upper left) show mean RQ +/− SEM for IL-1β, IL-6, IFNγ, and TNFα, respectively.
Discussion
Influenza viruses undergo antigenic drift whereby variant strains arise from an accumulation of point mutations in the viral surface glycoproteins haemagglutinin (HA) and neuraminidase (NA) (Palese and Shaw 2007). Therefore vaccines need to be regularly updated with relevant strains (Daly et al. 2004). Generation of vaccine viruses using reverse genetics holds a unique advantage in that the virus is created entirely from cloned plasmid DNA (Fodor et al. 1999; Neumann et al. 1999). This allows for rapid creation of updated vaccine strains as only the relevant surface antigens (HA and NA) would need to be replaced in a master strain.
A previous study of live equine influenza viruses with carboxy-terminally truncated NS1 proteins showed that these viruses were attenuated in vitro and in vivo in a mouse model (Quinlivan et al. 2005b). In either seropositive or seronegative horses (Trial I) these viruses did not produce any signs of influenza illness or adverse side effects. Antibody responses and patterns of viral shedding post-vaccination in NS1-126 and NS1-73 vaccinated animals were indicative of viral replication in the host.
Trial II showed that vaccination using the NS1-126 mutant virus resulted in clinical protection from challenge with wild type equine influenza virus, with significantly reduced clinical signs and significant reductions in quantity and duration of viral shedding, compared to control animals. Comparable results have been reported using NS1-truncated swine influenza viruses inoculated into pigs (Richt et al. 2006; Solorzano et al. 2005), and NS1-truncated human influenza virus in macaques (Baskin et al. 2007).
The protection afforded by traditional inactivated vaccines correlates with levels of circulating anti-HA antibody (Morley et al. 1995; Newton et al. 2000). However, natural infection induces relatively longer-term immunity which is not entirely dependent on anti-HA antibodies (Hannant et al. 1988). Also, protection status with live influenza vaccines probably involves other mediators of the immune system such as CTL and local mucosal immunity. Vaccination with MLV and DNA based equine influenza vaccines provided clinical protection despite low antibody responses to vaccination (Chambers et al. 2001; Lunn et al. 1999; Soboll et al. 2003a; Townsend et al. 2001). Minimal antibody production has also been observed with human cold-adapted vaccine virus (Edwards et al. 1994).
There is limited information on cytokine expression in influenza-infected horses (Quinlivan et al. 2007). In the present study, there were significant differences between NS1 vaccinated and control animals in expression of pro-inflammatory cytokines post-challenge. Mean IL-1β and IL-6 levels were higher in controls, peaking on Day +2 post-challenge, which correlated with peak pyrexia and peak viral shedding. These findings are similar to previous findings for influenza in horses (Wattrang et al. 2003) and also humans (Hayden et al. 1998) and pigs (Van Reeth et al. 1998). IL-6 peaks were also seen in the NS1-126 vaccinated horses; however, these did not correlate with pyrexia and their trigger is unclear. Our study did not reveal clear trends of TNFα or IFNγ expression associated with vaccination status.
The present data indicate the safety and effectiveness of recombinant NS1 viruses for vaccination against equine influenza. This approach may have practical benefits. The reverse genetics based vaccine holds unique advantages in that surface antigens can be updated relatively easily to combat antigenic drift, the attenuation strategy produces genetically stable vaccine viruses, and there is also potential for delivery of immunostimulating genes (e.g., cytokines) within the vaccine virus (Ferko et al. 2006). Future studies should focus on duration of protection, protection against challenge with heterologous influenza viruses, and elucidation of optimal schedules and dosages to elicit maximal protection.
Supplementary Material
Acknowledgements
Partial support of this work (A.G-S and P.P.) was provided by NIH grants UO1AI070469, HHSN2662000700010C, U54 AI057158-04, and 1 UC19 AI062623-023, by NIH training grant AI007647 (D.Z.) and by USDA/CSREES Project No. KY014028, a project of the Kentucky Agricultural Experiment Station (publication no. 08-014-020). Work done at the Irish Equine Centre was funded by the Department of Agriculture and Food under the National Development Plan. Michelle Quinlivan was funded by the Irish Research Council for Science, Engineering and Technology. The authors wish to thank Lynn Tudor, Lynn Ennis and the UK Veterinary Science farm crew, and Rachel Kenna of the Virology Unit, Irish Equine Centre.
References
- Baskin CR, Bielefeldt-Ohmann H, Garcia-Sastre A, Tumpey TM, Van Hoeven N, Carter VS, Thomas MJ, Proll S, Solorzano A, Billharz R, Fornek JL, Thomas S, Chen CH, Clark EA, Murali-Krishna K, Katze MG. Functional genomic and serological analysis of the protective immune response resulting from vaccination of macaques with an NS1-truncated influenza virus. J Virol. 2007;81:11817–11827. doi: 10.1128/JVI.00590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TM, Holland RE, Tudor LR, Townsend HG, Cook A, Bogdan J, Lunn DP, Hussey S, Whitaker-Dowling P, Youngner JS, Sebring RW, Penner SJ, Stiegler GL. A new modified live equine influenza virus vaccine: phenotypic stability, restricted spread and efficacy against heterologous virus challenge. Equine Vet J. 2001;33:630–636. doi: 10.2746/042516401776249291. [DOI] [PubMed] [Google Scholar]
- Daly JM, Yates PJ, Newton JR, Park A, Henley W, Wood JL, Davis-Poynter N, Mumford JA. Evidence supporting the inclusion of strains from each of the two co-circulating lineages of H3N8 equine influenza virus in vaccines. Vaccine. 2004;22:4101–4109. doi: 10.1016/j.vaccine.2004.02.048. [DOI] [PubMed] [Google Scholar]
- Donelan NR, Basler CF, Garcia-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KM, Dupont WD, Westrich MK, Plummer WD, Jr, Palmer PS, Wright PF. A randomized controlled trial of cold-adapted and inactivated vaccines for the prevention of influenza A disease. J Infect Dis. 1994;169:68–76. doi: 10.1093/infdis/169.1.68. [DOI] [PubMed] [Google Scholar]
- Egorov A, Brandt S, Sereinig S, Romanova J, Ferko B, Katinger D, Grassauer G, Alexandrova G, Katinger H, Muster T. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J Virol. 1998;72:6437–6441. doi: 10.1128/jvi.72.8.6437-6441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferko B, Kittel C, Romanova J, Sereinig S, Katinger H, Egorov A. Live attenuated influenza virus expressing human interleukin-2 reveals increased immunogenic potential in young and aged hosts. J Virol. 2006;80:11621–11627. doi: 10.1128/JVI.01645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Gorse GJ, Campbell MJ, Otto EE, Powers DC, Chambers GW, Newman FK. Increased anti-influenza A virus cytotoxic T cell activity following vaccination of the chronically ill elderly with live attenuated or inactivated influenza virus vaccine. J Infect Dis. 1995;172:1–10. doi: 10.1093/infdis/172.1.1. [DOI] [PubMed] [Google Scholar]
- Hannant D, Mumford JA. Cell mediated immune responses in ponies following infection with equine influenza virus (H3N8): the influence of induction culture conditions on the properties of cytotoxic effector cells. Vet Immunol Immunopathol. 1989;21:327–337. doi: 10.1016/0165-2427(89)90040-8. [DOI] [PubMed] [Google Scholar]
- Hannant D, Mumford JA, Jessett DM. Duration of circulating antibody and immunity following infection with equine influenza virus. Vet Rec. 1988;122:125–128. doi: 10.1136/vr.122.6.125. [DOI] [PubMed] [Google Scholar]
- Hayden FG, Fritz RS, Lobo MC, Alvord WG, Strober W, Straus E. Local and systemic cytokine responses during experimental human influenza A virus infection. J Clin Invest. 1998;101:643–649. doi: 10.1172/JCI1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochs G, Garcia-Sastre A, Martinez-Sobrido L. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol. 2007;81:7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn DP, Soboll G, Schram BR, Quass J, McGregor MW, Drape RJ, Macklin MD, McCabe DE, Swain WF, Olsen CW. Antibody responses to DNA vaccination of horses using the influenza virus hemagglutinin gene. Vaccine. 1999;17:2245–2258. doi: 10.1016/s0264-410x(98)00496-4. [DOI] [PubMed] [Google Scholar]
- Morley PS, Hanson LK, Bogdan JR, Townsend HG, Appleton JA, Haines DM. The relationship between single radial hemolysis, hemagglutination inhibition, and virus neutralization assays used to detect antibodies specific for equine influenza viruses. Vet Microbiol. 1995;45:81–92. doi: 10.1016/0378-1135(94)00105-6. [DOI] [PubMed] [Google Scholar]
- Mumford EL, Traub-Dargatz JL, Salman MD, Collins JK, Getzy DM, Carman J. Monitoring and detection of acute viral respiratory tract disease in horses. J Am Vet Med Assoc. 1998;213:385–390. [PubMed] [Google Scholar]
- Mumford JA, Hannant D, Jessett DM. Experimental infection of ponies with equine influenza (H3N8) viruses by intranasal inoculation or exposure to aerosols. Equine Vet J. 1990;22:93–98. doi: 10.1111/j.2042-3306.1990.tb04217.x. [DOI] [PubMed] [Google Scholar]
- Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton JR, Townsend HG, Wood JL, Sinclair R, Hannant D, Mumford JA. Immunity to equine influenza: relationship of vaccine-induced antibody in young Thoroughbred racehorses to protection against field infection with influenza A/equine-2 viruses (H3N8) Equine Vet J. 2000;32:65–74. doi: 10.2746/042516400777612116. [DOI] [PubMed] [Google Scholar]
- Paillot R, Grimmett H, Elton D, Daly JM. Protection, systemic IFNgamma, and antibody responses induced by an ISCOM-based vaccine against a recent equine influenza virus in its natural host. Vet Res. 2008;39:21. doi: 10.1051/vetres:2007062. [DOI] [PubMed] [Google Scholar]
- Paillot R, Hannant D, Kydd JH, Daly JM. Vaccination against equine influenza: Quid novi? Vaccine. 2006a doi: 10.1016/j.vaccine.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Paillot R, Kydd JH, Sindle T, Hannant D, Edlund Toulemonde C, Audonnet JC, Minke JM, Daly JM. Antibody and IFN-gamma responses induced by a recombinant canarypox vaccine and challenge infection with equine influenza virus. Vet Immunol Immunopathol. 2006b doi: 10.1016/j.vetimm.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Palese P, Garcia-Sastre A. Influenza vaccines: present and future. J Clin Invest. 2002;110:9–13. doi: 10.1172/JCI15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P, Shaw ML. Orthomyxoviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields' Virology. 5th edn. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007. pp. 1647–1690. [Google Scholar]
- Park AW, Wood JL, Newton JR, Daly J, Mumford JA, Grenfell BT. Optimising vaccination strategies in equine influenza. Vaccine. 2003;21:2862–2870. doi: 10.1016/s0264-410x(03)00156-7. [DOI] [PubMed] [Google Scholar]
- Quinlivan M, Nelly M, Prendergast M, Breathnach C, Horohov D, Arkins S, Chiang YW, Chu HJ, Ng T, Cullinane A. Pro-inflammatory and antiviral cytokine expression in vaccinated and unvaccinated horses exposed to equine influenza virus. Vaccine. 2007;25:7056–7064. doi: 10.1016/j.vaccine.2007.07.059. [DOI] [PubMed] [Google Scholar]
- Quinlivan M, Zamarin D, Garcia-Sastre A, Cullinane A, Chambers T, Palese P. Attenuation of equine influenza viruses through truncations of the NS1 protein. J Virol. 2005b;79:8431–8439. doi: 10.1128/JVI.79.13.8431-8439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renegar KB, Small PA., Jr Passive transfer of local immunity to influenza virus infection by IgA antibody. J Immunol. 1991;146:1972–1978. [PubMed] [Google Scholar]
- Richt JA, Lekcharoensuk P, Lager KM, Vincent AL, Loiacono CM, Janke BH, Wu WH, Yoon KJ, Webby RJ, Solorzano A, Garcia-Sastre A. Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J Virol. 2006;80:11009–11018. doi: 10.1128/JVI.00787-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboll G, Horohov DW, Aldridge BM, Olsen CW, McGregor MW, Drape RJ, Macklin MD, Swain WF, Lunn DP. Regional antibody and cellular immune responses to equine influenza virus infection, and particle mediated DNA vaccination. Vet Immunol Immunopathol. 2003a;94:47–62. doi: 10.1016/s0165-2427(03)00060-6. [DOI] [PubMed] [Google Scholar]
- Soboll G, Nelson KM, Leuthner ES, Clark RJ, Drape R, Macklin MD, Swain WF, Olsen CW, Lunn DP. Mucosal co-administration of cholera toxin and influenza virus hemagglutinin-DNA in ponies generates a local IgA response. Vaccine. 2003b;21:3081–3092. doi: 10.1016/s0264-410x(03)00161-0. [DOI] [PubMed] [Google Scholar]
- Solorzano A, Webby RJ, Lager KM, Janke BH, Garcia-Sastre A, Richt JA. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J Virol. 2005;79:7535–7543. doi: 10.1128/JVI.79.12.7535-7543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon J, Salvatore M, O Neill RE, Nakaya Y, Zheng H, Muster T, García-Sastre A, Palese P. Influenza A and B viruses expressing altered NS1 proteins: A vaccine approach. Proc Natl Acad Sci U S A. 2000;97:4309–4314. doi: 10.1073/pnas.070525997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Funato H, Hirabayashi Y, Kikuta K, Suzuki Y, Nagamine T, Aizawa C, Nakagawa M, Kurata T. Functional role of respiratory tract haemagglutinin-specific IgA antibodies in protection against influenza. Vaccine. 1990;8:479–485. doi: 10.1016/0264-410x(90)90250-p. [DOI] [PubMed] [Google Scholar]
- Toulemonde EC, Daly J, Sindle T, Guigal PM, Audonnet JC, Minke JM. Efficacy of a recombinant equine influenza vaccine against challenge with an American lineage H3N8 influenza virus responsible for the 2003 outbreak in the United Kingdom. Vet Rec. 2005;156:367–371. doi: 10.1136/vr.156.12.367. [DOI] [PubMed] [Google Scholar]
- Townsend HG, Penner SJ, Watts TC, Cook A, Bogdan J, Haines DM, Griffin S, Chambers T, Holland RE, Whitaker-Dowling P, Youngner JS, Sebring RW. Efficacy of a cold-adapted, intranasal, equine influenza vaccine: challenge trials. Equine Vet J. 2001;33:637–643. doi: 10.2746/042516401776249354. [DOI] [PubMed] [Google Scholar]
- Van Reeth K, Nauwynck H, Pensaert M. Bronchoalveolar interferon-alpha, tumor necrosis factor-alpha, interleukin-1, and inflammation during acute influenza in pigs: a possible model for humans? J Infect Dis. 1998;177:1076–1079. doi: 10.1086/517398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattrang E, Jessett DM, Yates P, Fuxler L, Hannant D. Experimental infection of ponies with equine influenza A2 (H3N8) virus strains of different pathogenicity elicits varying interferon and interleukin-6 responses. Viral Immunol. 2003;16:57–67. doi: 10.1089/088282403763635456. [DOI] [PubMed] [Google Scholar]
- Wilson WD, Mihalyi JE, Hussey S, Lunn DP. Passive transfer of maternal immunoglobulin isotype antibodies against tetanus and influenza and their effect on the response of foals to vaccination. Equine Vet J. 2001;33:644–650. doi: 10.2746/042516401776249435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.