Abstract
In this study, fluorescent metal nanoshells were synthesized as molecular imaging agent to detect single MicroRNA (miRNA) molecules in the cells positive to lung cancer. These metal nanoshells were composed of the silica spheres with the encapsulated Ru(bpy)32+ complexes as cores and the thin silver layers as shells. Compared with the silica spheres in the absence of metal, the metal nanoshells displayed the enhanced emission intensity, shortened lifetime, and extended photostability. The single-stranded probe oligonucleotides were covalently bound on the metal nanoshells to hybridize with the target miRNA-486 molecules in the cells. In was shown that with stronger emission intensity and longer lifetime, the conjugated metal nanoshells were isolated distinctly from the cellular autofluorescence on the cell images. These emission spots on the cell images were counted accurately and analyzed with a pool of cell representing the miRNA-486 expression levels in the cells. The results may reflect a genomic signal change and provide a reference to the lung cancer early diagnosis as well as other diseases.
Keywords: lung cancer, genomic aberration, MicroRNA-486 (miRNA-486), expression level, fluorescent metal nanoshell, Ru(bpy)32+ complex, fluorescence cell image, single molecule count, emission intensity, lifetime
Introduction
Lung cancer is a leading cause of cancer death through the world [1]. Traditional strategies of chest X-ray and sputum cytology have been demonstrated inefficiently to the lung tumor in early stage before spreading and becoming incurable [2]. So it is of importance to develop new diagnostic methods of early diagnosis. It is known that most lung cancers are originated from a field cancerization and characterized by molecular genetic abnormalities [3]. Fluorescence in-situ hybridization (FISH) assay may offer a potential in the early diagnosis to the lung cancer because of its visualization to the genomic aberration [4]. Generally, the DNA fragments that are derived from target genes in the cells are used as the probes.
MicroRNAs (miRNAs) are a class of small non-protein-coding RNAs [5]. They are known to control a wide range of biological functions by the expression regulation of hundreds of target genes including the cancer development and progression [6,7]. MiRNAs are recently emerged as the tissue-specific biomarkers to identify and define the cancers, including diagnose the lung cancer at early stage [8]. Thus, the development of reliable FISH techniques for the cancer-related miRNA detection in the tumor cells may provide an applicable approach in the early diagnosis of lung cancer, and it is also clinically important [9]. In this paper, miRNA-486, one of well known biomarkers to lung cancer [10], was used as the target to study in serial lung cancer positive cell lines by the FISH assay method.
In the current FISH assays, the probe molecules are generally labeled with the conventional organic fluorophores [11]. However, these organic fluorophore are known to have clear drawbacks, e.g. weak emission signal, rapid photobleaching, and strong photoblinking [12]. Most importantly, the lifetimes of organic fluorophores that take important roles in the cell imaging are ranged in 2 – 10 ns close to that of cellular autofluorescence of cell line. These features may lead to the emission signals from the probes cannot be isolated from the cellule background on the cell images, and furthermore result in a dramatic variation in intra- and inter-observer agreement. To circumvent these issues, typically, the probe molecules have to be conjugated by many organic fluorophores. However, such a treatment may reduce the hybridization capability of probe molecule in in-situ hybridization with the target and increase the possibility of mismatching in the cell. Consequently, the reduced detection accuracy may hamper the application of promising diagnostic technique in the clinical setting. Therefore, it is imperative to develop novel strategies that can be used to fluorescently label the probe molecules.
The metal nanoparticle fluorophores become attractive in the past decade [13–15]. These metal nanoparticles are developed to have sub-wavelength sizes and display typical metal plasmon resonances in visible region [16]. A near field interaction is observed to occur when an organic fluorophore is localized near the metal nanoparticle that may result in an obvious change of optical property of fluorophore including enhanced intensity, shortened lifetime, extended photostability, and reduced photoblinking [17]. In this research, we are particularly interested in the specially designed fluorescent metal nanoparticle with a core/shell structure [18]. Because the interior electric field induced by an incident light is calculated to be uniform in the core area [19–22], the fluorophores in the core can be coupled equally and efficiently so the fluorescence can be enhanced more efficiently. In addition, the metal shell may also prevent entrance of oxygen and other species that probably react with the fluorophores and thus protect the fluorophores from the possible photochemical or other reactions. The versatile surface chemistry on the metal surface can promise the binding of probe moieties on the metal nanoshells.
In this research, the silver nanoshells were synthesized with the silica cores and silver shells. For the fluorescence purpose, the Ru(bpy)32+ complexes were encapsulated into the silica cores. Even though the lifetime of the encapsulated Ru(bpy)32+ complexes was significantly shortened by the fluorophore-metal interaction, the measured lifetime was still longer than the lifetime of cellular autofluorescence. The stronger emission signal and longer lifetime may promise the identification of the metal nanoshell from the cellule background on the cell image. Using as the molecule imaging agent, the fluorescent metal nanoshells were covalently bound by multiple probe oligonucleotides on the surfaces to hybridize with the target miRNA molecules in the cell. The cell images were recorded on the confocal microscopy in the emission intensity and lifetime [23]. The expression levels of miRNA-486 in the cell lines were evaluated in term of the count numbers of emission signals of metal nanoshells on the cell images. The distribution patterns were also analyzed at the single cell level.
Materials and Methods
All reagents and spectroscopic grade solvents were used as received from Sigma-Aldrich. Nanopure water (>18.0 MΩ.cm) purified using Millipore Milli-Q gradient system was used in all experiments. A probe oligonucleotide of 5’-amino hexayl-ctcggggcagctcagtacagga-3’ was synthesized by Biopolymer Laboratory in University of Maryland at Baltimore.
Preparation of single-stranded oligonucleotide bound metal nanoshell
The silica spheres with 50 nm diameters were prepared in a modified Stöber strategy [18a]. To encapsulate the fluorophores into the silica spheres, 5 × 10−6 M Ru(bpy)32+ complex was dissolved with 1.4 × 10−2 M tetraethyl orthosilicate in the reaction solution. The silver thin nanoshells were deposited on the silica spheres layer-by-layer [24]. The metal nanoshell thickness was observed to be 10 nm. The metal nanoshells were dispersed in ethanol solution containing 10 mM hexa(ethyleneglycol)mono-11-(acetylthio)undecyl ether. The solution was stirred for 2 h at room temperature to assemble the organic monolayers on the metal shell surfaces. The metal nanoshells were recovered by centrifugation, and the solid residue was rinsed with ethanol. These organic monolayer-coated silver nanoshells were partially substituted by 4-aminothiophenol ligands via ligand exchanges [25]. Typically, the silver nanoshells (1 × 10−8 M) and 4-aminothiophenol (1 × 10−6 M) were co-dissolved in a mixing solvent of ethanol and water (v/v = 1/1), and the solution was stirred for 24 h at room temperature. The suspension was removed by centrifugation and the residue was rinsed with ethanol. The amino-ligand substituted metal nanoshells (1 × 10−8 M) and poly(ethylene glycol)bis(carboxymethyl)ether (Mn = 250, 1 × 10−5 M) were co-dissolved in water, and then an excess amount of 1-ethyl-3-(3-dimethylaminopropyl)-carbodimide hydrochloride (EDC, 2 × 10−5 M) was added in solution. The solution was stirred for 2 h at room temperature. The suspension was removed by centrifugation, and residue was rinsed with water. The residue of metal nanoshell (1 × 10−8 M) was redispersed in water containing single-stranded probe oligonucleotide (1 × 10−6 M)) and EDC (2 × 10−6 M). The solution was stirred for 5 h at room temperature, and the suspension was removed by centrifugation. The residue was rinsed with water and dispersed in 10 mM phosphate buffered saline (PBS) buffer solution with 0.05 % Tween 20 at pH 7.4.
Culturing cell and in-vivo hybridization of fluorescent metal nanoshell with miRNA-486 in cell
The cell lines of human lung cancer H460, H1944, and A549 were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Of the three cell lines, A549 was known to be negative, whereas H460 and H1944 are positive to the miRNA-486 expression, according to our previous study [26]. The cells were cultured at 37°C in 5% CO2 and water-saturated atmosphere in RPMI1640 (Gibco) and F-12K (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals). The MDA-MB-231 breast cancer cells that displays positive miRNA-486 expression was purchased from ATCC. The cells medium is DMEM (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals).
The cell slides were fixed in 4% paraformaldehyde in PBS buffer for 30 min at 4°C and washed twice with 10 mM PBS buffer solution. The cell slides were treated in 10 mM PBS at 50°C for 15 min and then incubated in 10 mM PBS buffer solution with 0.05 % Tween 20 containing 1 × 10−10 M oligonucleotide-bound metal nanoshells for 24 h, and then rinsed thoroughly with 10 mM PBS solution and ethanol, respectively. Dried in air, the cell coverslips were stored at 4°C for the fluorescent imaging measurements.
Spectral, imaging, and TEM measurements
Absorption spectra were collected on a Hewlett Packard 8453 spectrophotometer. Ensemble fluorescence spectra were performed on a Cary Eclipse Fluorescence Spectrophotometer. In the preparation of Transmission electron micrograph (TEM) sample, the silica sphere or metal nanoshell sample was dispersed in ethanol at nano-molar scale. The solution was cast onto copper grids (200 mesh) with standard carbon-coated Formvar films (200–300 Å) and dried in air. The TEM images were taken with a side-entry Philips electron microscope at 120 keV. The size distributions were analyzed with Scion Image Beta Release 2 on the base on at least 100 images.
The fluorescence image measurements were performed on a time-resolved scanning confocal microscopy (MicroTime 200, PicoQuant), which consists of an inverted confocal microscope coupled to a high-sensitivity detection setup. A single mode pulsed laser diode (470 nm, 100 ps, 40 MHz) was used as the excitation source. An oil immersion objective (Olympus, 100×, 1.3NA) was used for focusing the laser light onto the sample and collecting the emission signal. The emission passed a dichroic mirror, focused onto a 75 µm pinhole for a spatial filtering to reject out-of-focus signal, and finally recorded on a single photon avalanche diode (SPAD) (SPCM-AQR-14, Perkin Elmer Inc). Bandpass filters were used to eliminate the excitation residual. The data collected with a TimeHarp 200 board were stored in the Time-Tagged Time-Resolved Mode (TTTR) that allows every detected photon with its individual timing and detection channel information.
Results and Discussion
In the current method, the silica spheres were synthesized as mono-dispersion on the size [18a] that was confirmed by TEM images. The average diameter of them was 50 nm (TEM image in Figure 1). According to the different concentrations of Ru(bpy)32+ complex in solution before and after the silica sphere formation, the concentration of Ru(bpy)32+ complex in the silica sphere could be estimated to be ca. 4 × 10−4 M. With the size of silica sphere, this value could be converted to about 120 Ru(bpy)32+ complexes in one silica sphere. The metal nanoshells were deposited on the silica spheres layer-by-layer up to 10 nm thick (TEM image in Figure 1). During the metal shell deposition on the silica spheres, no significant amount of Ru(bpy)32+ complex was tested to leak from the dispersed silica spheres into water.
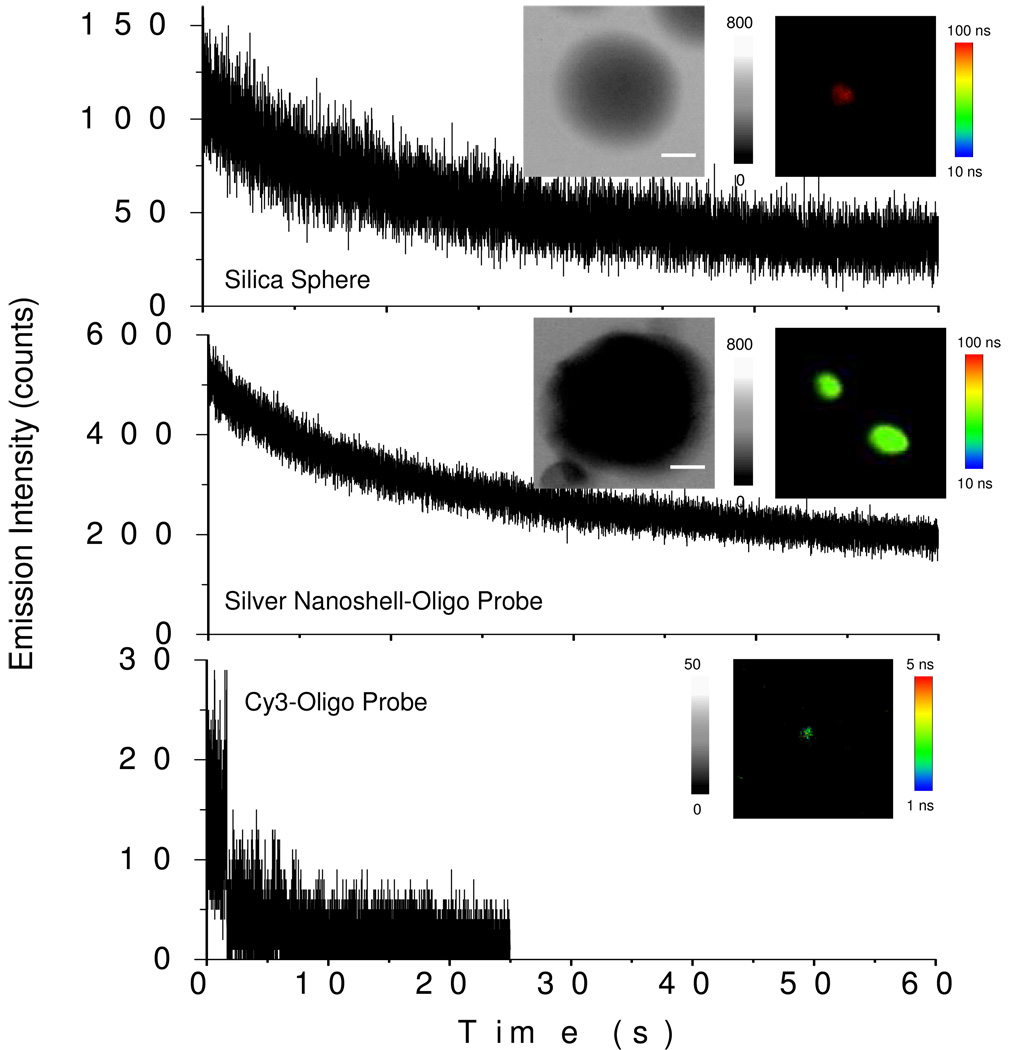
Figure 1.
The time trace profiles silica sphere, silver nanoshell, and Cy3-labeled probe oligonucleotide. The insets include the respective TEM images with the scale bars of 20 nm and typical single nanoparticle/molecule fluorescence image recorded in the emission intensity and lifetime. The scales of diagrams were 5 × 5 µm and the resolutions were 100 x 100 pixel with an integration of 0.6 ms/pixel.
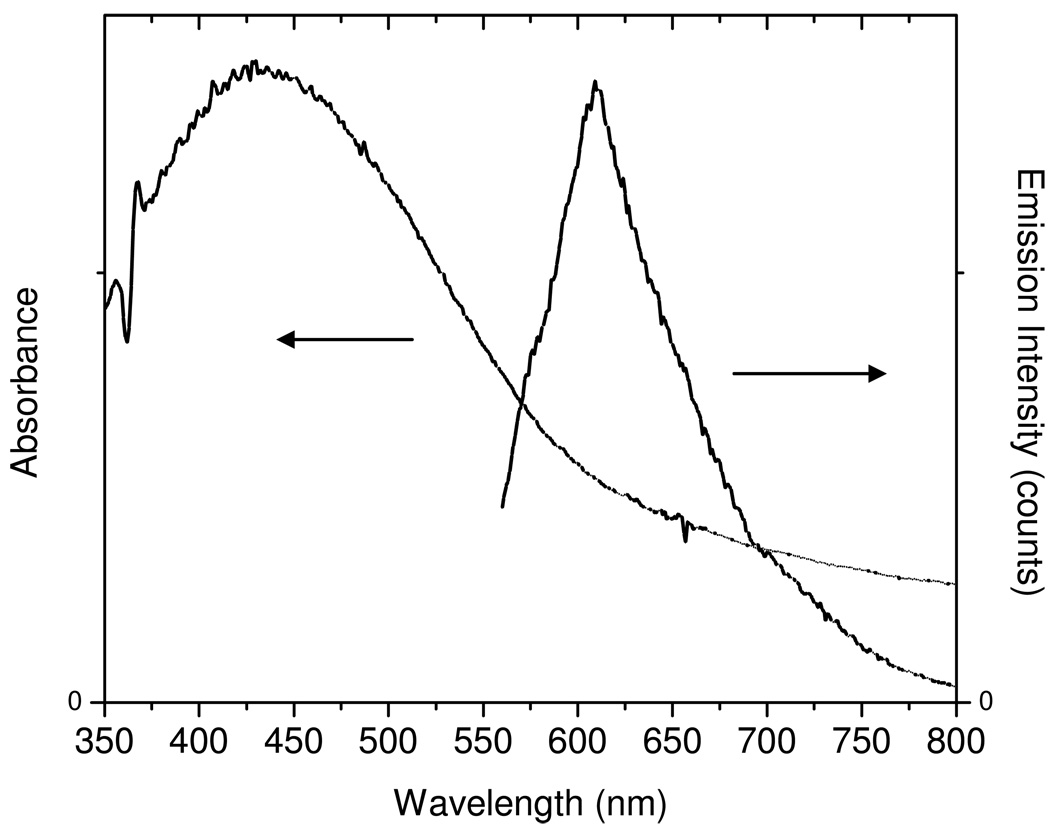
The silver nanoshells displayed an absorbance of plasmon resonance at 400 nm (Figure 2), consistent with our previous observation [18]. Upon excitation at 470 nm, the encapsulated Ru(bpy)32+ complexes in the metal nanoshells displayed an emission maximum at 605 nm (Figure 2), close to that of silica spheres in the absence of metal. The optical properties of single metal nanoshells were monitored on the scanning confocal microscopy. The time trace profiles of both the silica sphere and metal nanoshell were compared in Figure 1 expressing the progressive decays of emission intensity due to the presences of multiple fluorophores in the single nanoparticle. Even though the amount of fluorophore in one nanoparticle was not significantly altered, the metal nanoshells were detected to display the enhanced emission intensity up to 6-fold and the shortened lifetime from approx. 600 ns (silica sphere) to 45 ns (silver nanoshell). The enhanced emission intensity is due to the near-field interaction of fluorophore with metal [17]. This interaction can increase the intrinsic decay rate of fluorophore near the metal surface leading to the shortened lifetime. We also noticed that the shortened lifetime of metal nanoshells was still much longer than that of cellular autofluorescence of cell line (2 – 10 ns). With the enhanced emission intensity and longer lifetime, we thus believe that the emission signals from the metal nanoshells could be isolated distinctly from the cellule autofluorescence on the cell image.
Figure 2.
Absorbance and ensemble fluorescence spectra of metal nanoshell with the Ru(bpy)32+ complex encapsulation in 10 mM PBS solution. The emission spectrum was determined upon the excitation at 450 nm.
To compare with the nanoparticle fluorophores, the single Cy3-labeled oligonucleotide with the same sequence of probe oligonucleotide was also tested, displaying a typical single-step photobleaching of single molecule fluorophore and much weaker emission intensity under the same irradiation conditions.
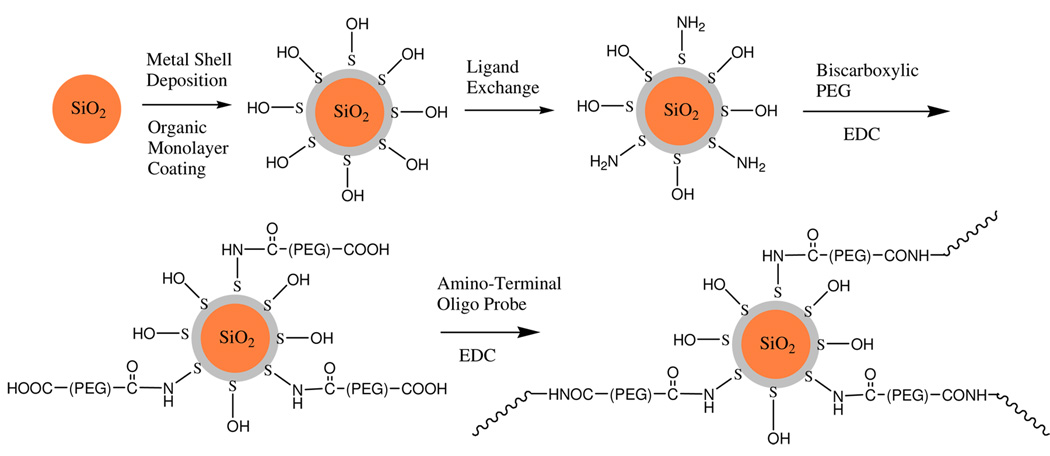
To improve the chemical stability, the surfaces of metal nanoshells were coated by the organic monolayers via sulfur-metal interactions. The optical properties of metal nanoshells were influenced insignificantly by the surface-coated organic monolayers. In addition, we also expected that the poly(ethylene glycol)-like ligands on the metal nanoshells could reduce the resistances of them in the penetration through the cell membranes and increase the hybridization with the miRNA targets in the cells. To furthermore reduce the influence to the hybridization from the bulky nanoparticles, a flexible poly(ethylene glycol) linker of ca. 10 nm long was specially designed to insert between the probe oligonucleotide and metal surface that was accomplished via three-step surface reactions. Firstly, the metal nanoshells were partially substituted by the amino-terminal ligands via ligand exchange [25], and then covalently bound by poly(ethylene glycol)bis(carboxymethyl)ether via condensation [26]. Finally, the single-stranded probe oligonucleotides were covalently bound on the metal nanoshells by an additional condensation. We could not provide direct evidence to demonstrate the occurrences of these surface reactions. Alternatively, in the same strategy, Cy5.5-labeled oligonucleotides that have the same sequences and 5’-amino-terminations were bound on the metal nanoshells without the encapsulated Ru(bpy)32+ complexes in the cores. Upon the excitation at 680 nm, the metal nanoshells that were treated with the Cy5.5-labeled oligonucleotides displayed a fluorescence maximum at 705 nm. This fact indirectly demonstrated the occurrence of surface reactions. Using the Cy5.5-oligonucleotide metal nanoshell complexes, we estimated the loading number of oligonucleotide on the metal nanoshell. Typically, the metal nanoshell was dispersed in 10 mM PBS buffer solution. The concentration of metal nanoshell could be estimated from the absorbance of plasmon resonance [17]. Subsequently, several drops of 0.1 M NaCN aqueous solution were added. With a progressive disappearing of plasmon resonance in solution, the metal nanoshells were regarded to remove by the dissolution with NaCN [14]. As a result, the Cy5.5-labeled oligonucleotides were released from the metal nanoshells into solution. The concentration of Cy5.5-labeled oligonucleotides in solution was determined by the emission intensity. By a molar ratio of Cy5.5-labeled oligonucleotide over metal nanoshell, the loading number of Cy5.5-labeled oligonucleotide on the metal nanoshell was estimated to be 13. Since the probe oligonucleotides were bound on the silver nanoshells in the same strategy, we feel that the loading number of probe oligonucleotide on the silver nanoshell should remain to be 13.
This loading number is an important parameter because it can influence the hybridization properties of metal nanoshell with the target miRNA in the cell. The metal nanoshell with a high loading number may lead to a strong hybridization capability but also increase the opportunity of mismatching in the hybridization, and consequently the accuracy of detection will be reduced. In addition, since the oligonucleotide has the negative charges, the metal nanoshell with a high loading number may result in surplus charges on the surface that resist the penetration through the cell membrane and decrease the mobility in the cell. To reduce the influence from the conjugated oligonucleotides to the metal nanoshell, we selectively loaded very few probe oligonucleotides of 13 on one metal nanoshell in this research. The optimization to the metal nanoshells regarding with the loading number and its influence to the hybridization capability and mismatching is being carried out in our laboratory.
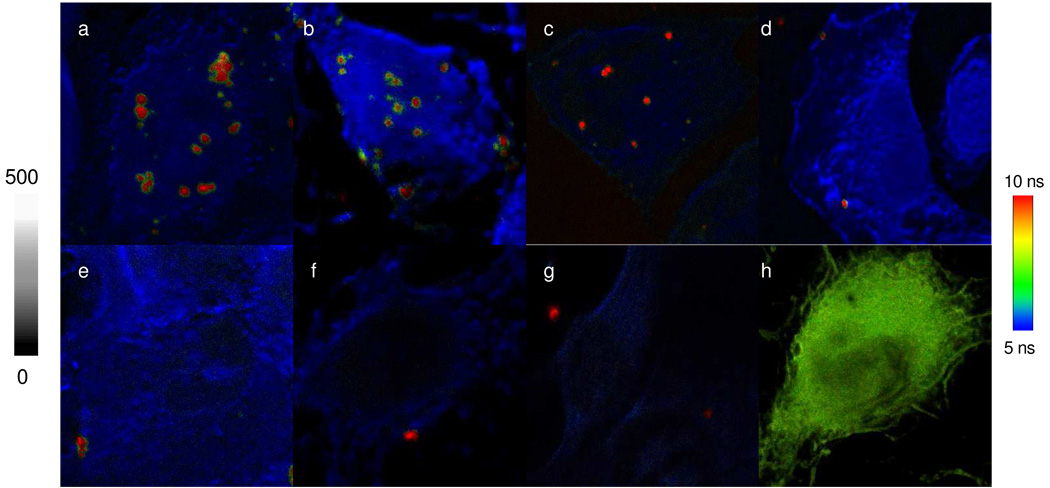
The cell lines were incubated with the metal nanoshell probes. Because the metal nanoshells were significantly larger than the typical oligonucleotide probes and the hybridizations of them with the target miRNAs were expected to meet stronger steric hindrances in the cells, the incubations were extended up to 24 h. Three positive to lung cancer cell lines (H460, H1944, and MDA-MB-231) and one negative (A549) were used in the experiments. After incubated, the cell lines were washed thoroughly with the buffer solution and ethanol to remove the metal nanoshells other than the hybridizations with the target miRNAs in the cells. The fluorescent cell images were recorded on the confocal microscopy in the emission intensity and lifetime. Representative cell images were presented in Figure 3. Because of stronger intensity and longer lifetime, the emission signals from the metal nanoshells could be isolated distinctly from the cellular autofluorescence on the collected cell images and the emission spots could be counted accurately at the single miRNA level. We also noticed that there was no significant emission signal from the metal nanoshell out of the cell image, reflecting that the observed metal nanoshells were mostly conjugated on the cells. To confirm that they were hybridized indeed with the miRNAs in the cells, in the same strategy, the cell lines were also incubated with the fluorescent metal nanoshells without the conjugated probe oligonucleotide as negative controls. The typical cell images collected from H460, H1944, and MDA-MB-231 were also given in Figure 3, showing significantly reduced emission signals in comparison with the corresponding cell images.
Figure 3.
Representative fluorescence images from the single cells that were incubated by the metal nanoshell with the single-stranded probe oligonucleotide conjugation (up panel) (a) H460, (b) H1944, (c) MDA-MB-231, and (d) A549 or incubated by the metal nanoshell without the probe oligonucleotide conjugation (bottom panel): (e) H460, (f) H1944, and (g) MDA-MB-231. Images (a) to (g) have the emission intensity bar from 0 to 500 and the lifetime bar from 5 to 10 ns. Image (h) represents the cell image of H1944 hybridized with Cy3-labeled oligonucleotide probe. Image (h) has the emission intensity bar from 0 to 200 and the lifetime bar from 1 to 4 ns. The scales of (a) and (e) diagrams are 20 × 20 µm and others are 50 × 50 µm. The resolutions of diagrams are an integration of 0.6 ms/pixel.
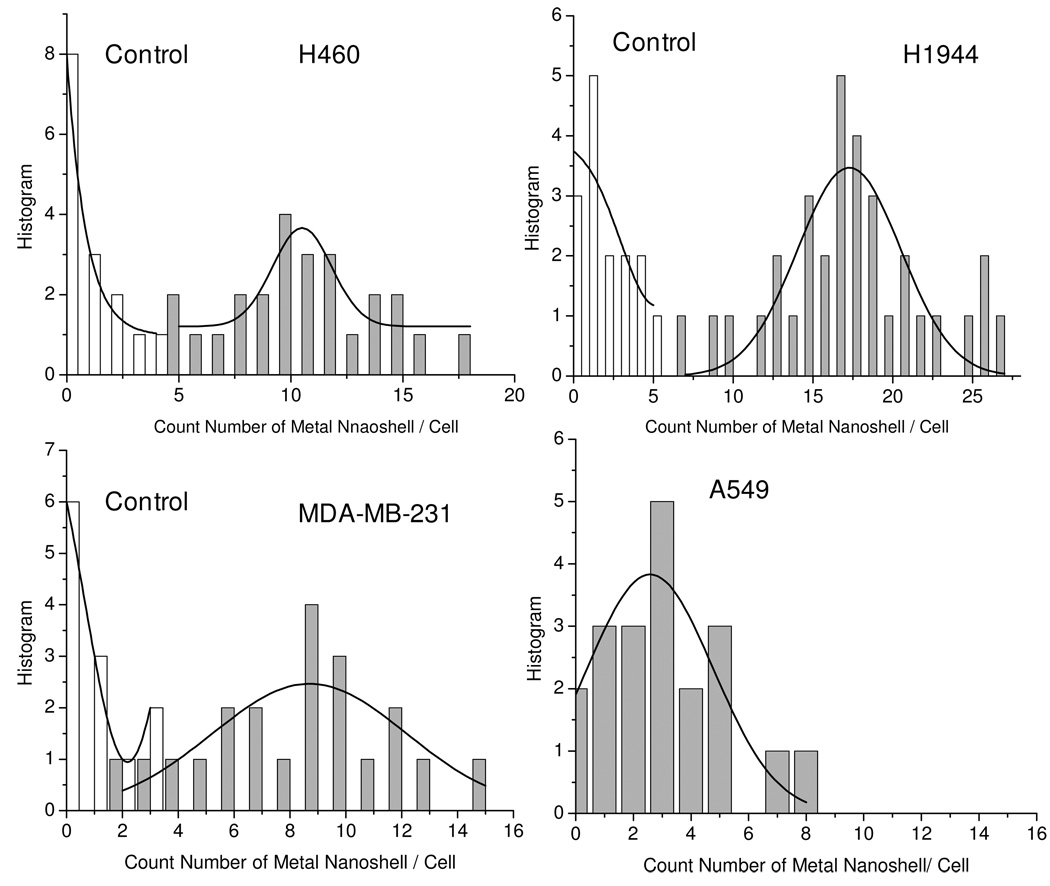
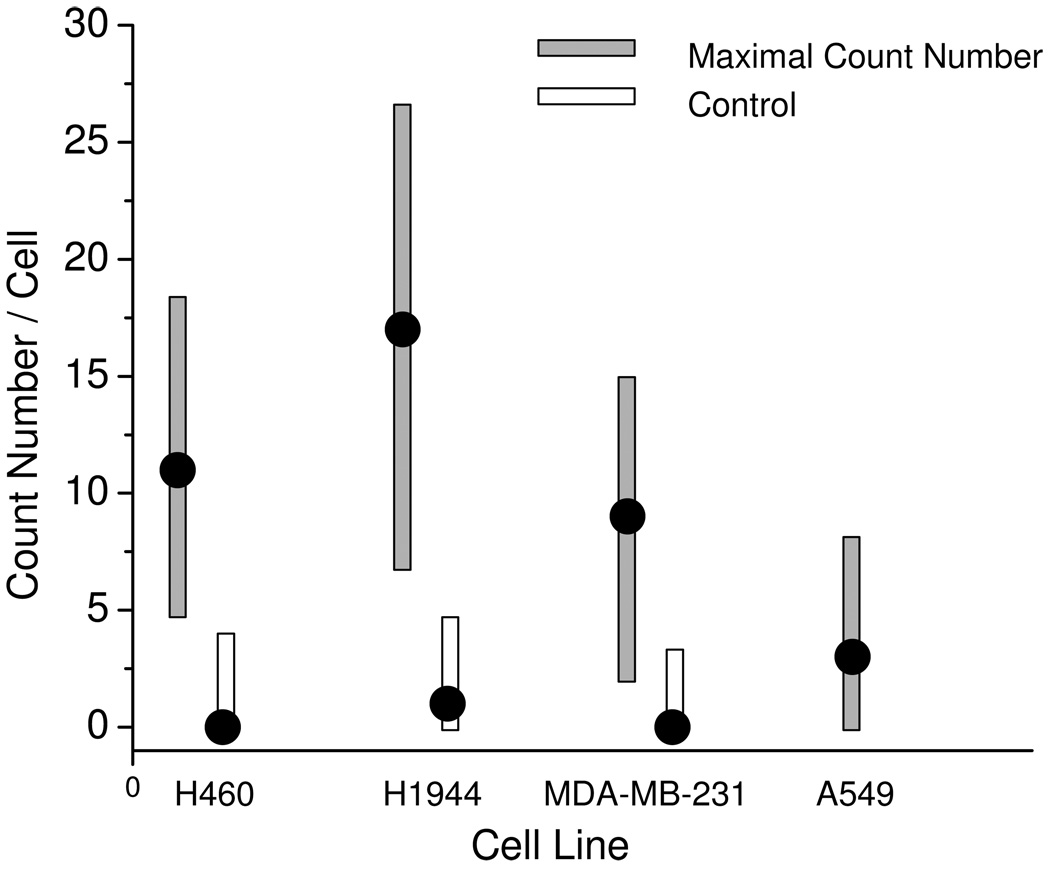
To isolate the emission signals more clearly and count the numbers more accurately, the collected cell images were subsequently treated with Picoquant Analytical software and OriginPro7.0 software. Firstly, the cell image was transferred using Picoquant Analytical software. The emissions with the lifetimes shorter than 10 ns were removed using OriginPro7.0 software and then the residual data were recovered to the cell images. Consequently, only the emissions with the lifetime longer than 10 ns were clearly distinct on the image to count the number of the metal nanoshell. For each cell sample, a population of more than twenty cell pools was preceded and the number distributions were presented in Figure 4. It was shown that to the cancer positive H460, H1944, and MDA-MB-231 cells, the maximal numbers were ranged in 9 – 17 (Figure 5); whereas the number of cancer negative cell A549 was only 3 reflecting the negative cell has a low miRNA-486 expression. We also notice that in three types of cancer positive cells, H1944 expresses the highest miRNA-486 level. The cell images collected from the controlled samples were also analyzed showing the maximal numbers less than 5 (Figure 4), significantly lower than those achieved on the corresponding cells. All maximal values from the cell samples and controls in Figure 4 were presented in Figure 5 with the error bars. Although the value from MDA-MB-231 displays a slight overlapping with that from the control, the differences are distinct turning out that cell and control results can be separated clearly when using the fluorescent metal nanoshell as the molecule imaging agent to detect the single molecule miRNA in the cell.
Figure 4.
The distributions of count numbers from the hybridized metal nanoshells in H460, (b) H1944, (c) MDA-MB-231, and (d) A549 cells as well as their corresponding controls. The count numbers were achieved from the analysis to a population of more than twenty cell pools.
Figure 5.
Comparisons of maximal count numbers from the conjugated metal nanoshells on H460, H1944, MDA-MB-231, and A549 as well as their corresponding controls. The error bars were included.
We have considered using the metal nanoshell with the scrambled oligonucleotide loading as the control in the experiment. However, the metal nanoshells without the oligonucleotide conjugation were finally selected because the scrambled oligonucleotides on the metal nanoshell were regarded to be able to increase the mismatching opportunity in the hybridizations with the DNAs or RNAs in the cells. We also intended to compare the single organic fluorophore with the metal nanoshell on the cell imaging. H1944 was incubated with Cy3-labeled oligonucleotide probe and the cell images were collected in the same strategy (Figure 3h). The result reveals that the emission signals from the single molecule Cy3 fluorophores cannot be isolated from the cellular autofluorescence on the collected cell image because of their weak emission intensity and close lifetime to the cellular autofluorescence.
It is interesting to notice that during the cell imaging, even as the same type of cell, the collected images are observed with somehow differences on the cell structures, which are probably due to the different formation conditions of the tested cells on the coverslip. The metal nanoshells do not always display the same lifetimes on the cells images that can be attributed to the cellular autofluorescence. It is known that the cellular autofluorescence has the lifetime below 10 ns, shorter than that of metal nanoshell. The apparent lifetimes of metal nanoshells on the cell images are collected in the combination of the cellular autofluorescence as shorter-lifetime component and the metal nanoshell as longer-lifetime component. When the emission from the metal nanoshell is not altered significantly, a strong autofluorescence may increase the shorter-lifetime component leading to a shortened apparent lifetime of the observed metal nanoshell whereas a weaker autofluorescence may reduce the shorter-lifetime component leading to a lengthened apparent lifetime. It is observed that the autofluorescence of cell is not always homogeneous on the single cell image and also changes from cell to cell individuals, so the emission lifetimes from the conjugated metal nanoshells may fluctuate to some extent on the collected cell image.
We also concerted the detection accuracy that is probably influenced by two possible factors probably in this research. First is the steric hindrance from the bulky metal nanoshell. In fact, it is always a challenge using the nanoparticles as probes in the actual hybridization with the targets in the cell in the competition with the free single-stranded oligonucleotide [28]. However, as described above, the presence of multiple oligonucleotides on the metal nanoshell as well as the long and flexible linkers between the probe oligonucleotides and metal surfaces can compensate to some extent the loss of hybridization capability. On the other hand, trailed by the fluorescent metal nanoshell, the probe oligonucleotides are not required to label with multiple organic fluorophores, so the probability of mismatching can be reduced. The second factor is from the resolution of confocal microscopy. A regular confocal microscopy has a resolution of 200 µm on the x–y dimension larger than the size of metal nanoshell. As a result, the emission signals from two or more metal nanoshells with the close localizations cannot be recorded accurately.
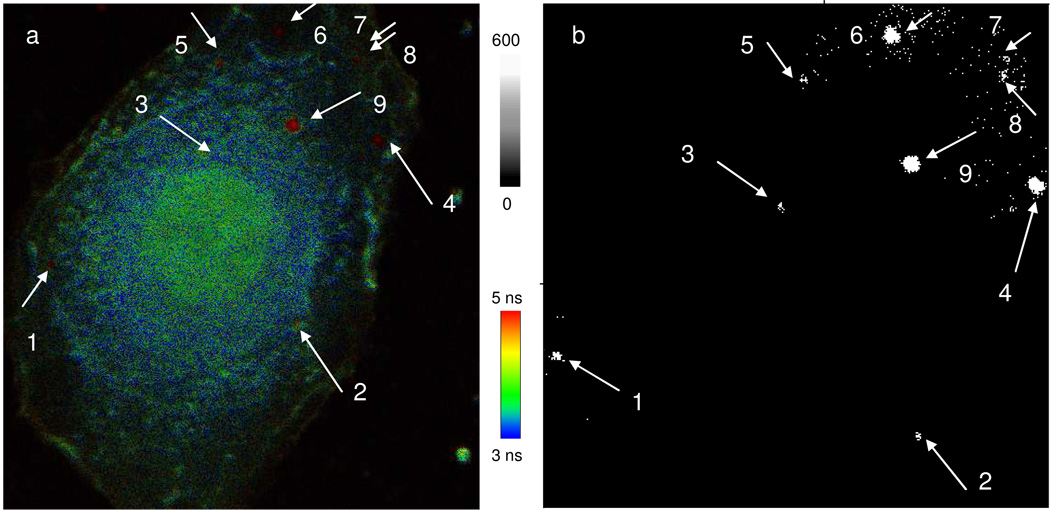
Besides the expression level, we were interested in the localizations of target miRNA-486 molecules in the cells. H1944 was incubated with the metal nanoshell probe, and then fluorescently labeled with Texas Red on the plasma membrane and with YOYO-1 on the nucleus. In this research, we pursued the locations of miRNAs in the cell, and the cell imaging was carried out by one-step scanning with the lifetime factor. Texas Red can display the lifetime of 4.5 ns and YOYO-1 displays the lifetime of 3.6 ns, so the distinct membrane (surrounding yellow) and nucleus (center blue and green) were shown on a representative image (Figure 6a). Although the emission from stained Texas Red and YOYO-1 became much stronger than the cellular autofluorescence of cell, the emission signals from the metal nanoshells could be approximately separated on the cell image when they were localized out of nucleus because of their longer lifetimes. The cell images were treated in the same strategy of emission spot counting, in which the emission signals with the lifetime shorter than 10 ns were removed and the residues with the lifetime longer than 10 ns were uniformly set to be one by OriginPro7.0 software. The uniformed data were converted into the image (Figure 6b), showing the clearly isolated emission signals of the silver nanoshells that represented the locations of the metal nanoshells in the cell. Because of strong fluorescence from YOYO-1, the emission from the metal nanoshells near the nucleus became invisible. Nerveless, the observation may provide direct evidence for the miRNA expression out of nucleus area. This result in reasonable because according to reports, some mature miRNA-486 molecules can be indeed expressed in the ribosome or/and cytoplasm [29,30]. This observation is also important to understand the function of miRNA in the occurrence of lung cancer and offer a potential to innovate the therapy in the treatments to the patients.
Figure 6.
A: representative fluorescence cell images of H1944. The cell lines were first incubated with the metal nanoshell probes to hybridize with the target miRNA 486 molecules in the cells. The cell lines were furthermore stained with 10 nM 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine triethyammonium salt and 10 nM YOYO-1 iodide for 10 min. In such a treatment, the cell membranes were fluorescently labeled with Texas Red and the cell nucleuses were fluorescently labeled with YOYO-1. The scale of diagram is 50 × 50 µm. The resolution is an integration of 0.6 ms/pixel. B: the collected emission data with the lifetime shorter than 10 ns were removed. The residual data with the lifetime longer than 10 ns were uniformly set to be one and converted into the image. The arrows and numbers on the images represent the locations of metal nanoshells in the cell.
The novel nanoparticle fluorophores were developed to detect the single miRNA molecule in the cells positive to the lung cancer in this report. Thus, it is important to compare this method with other traditional methods. In fact, numerous techniques can be used to determine the miRNA expression including northern blot and RNA-primed microarray [31–33]. However, these techniques require the lysis of a population of cells and thus do not allow cell-to-cell variations in miRNA expression to be elucidated. FISH assay can be also practically used to test the targets in the cell [34]. However, it is known that a typical miRNA is 19–25 nucleotides in length, shorter than most FISH probe, so only less number of fluorophore is conjugated on the probe. Consequently, the emission intensity is lower and the detection sensitivity is lower. Actually, only few reports appear on the detections to the high expression miRNA using FISH assay. MiRNA-486, probe in this research, is known to have a low expression in the cells. To our knowledge, there is not a conventional method to test miRNA 486 in the cells. One of our authors in this paper has tried to determine the expression of miRNA 486 in the cells by RT-qPCR method [35]. The samples were collected from the sputum of lung cancer patients. The total RNA isolated from one sputum sample was diluted in diethylpyrocarbonate water to serial concentrations. The result revealed to be capable of detecting as low as 10 copies of the target genes. Lower number could not be detected accurately. In addition, the RT-qPCR method could not provide direct observation to the locations of miRNAs in the cells. In this report, using the fluorescent metal nanoshells as probes, we have achieved the emission signals from the hybridized metal nanoshells in the cells less than 10. The locations of them were also observed. Although the achieved results are needed to be furthermore evaluated, we feel that the metal nanoshell-based FISH assay can provide a powerful tool to detect the low expression molecules in the cells and map their spatial localizations at the tissue and cellular level, or even to the subcellular level.
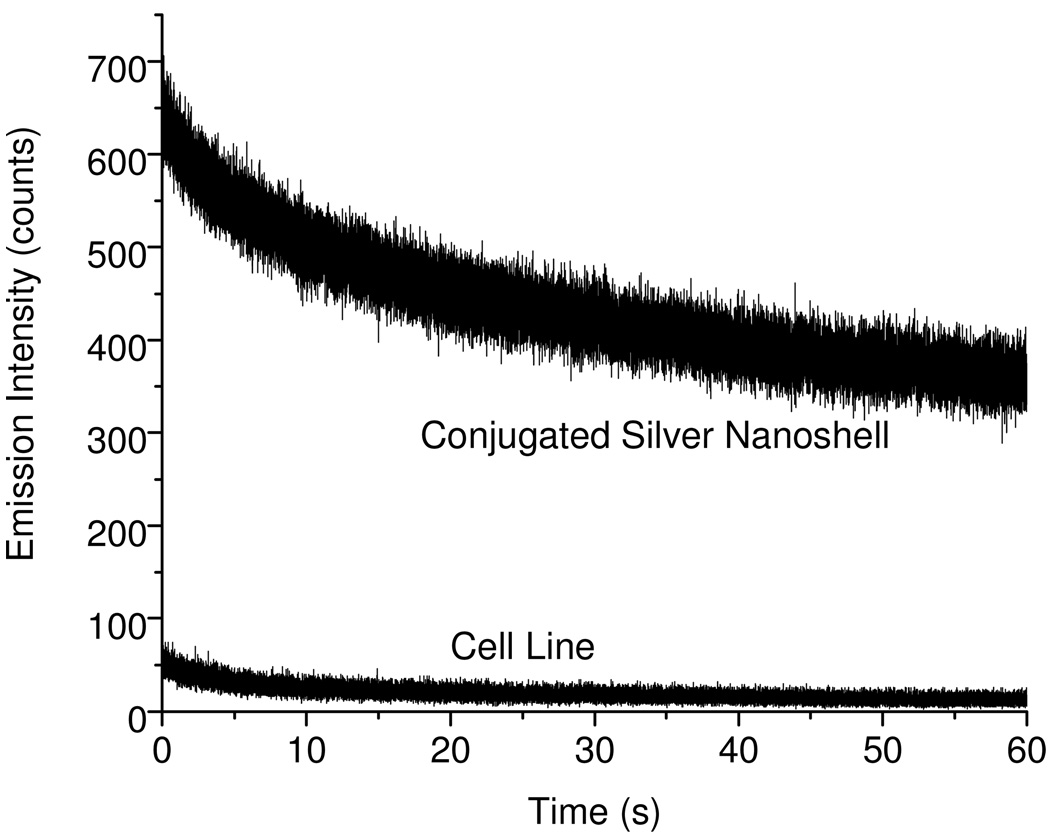
The photostability of fluorophore may be improved by the near-field interaction between the fluorophore and metal nanostructure [17]. It was noticed on the time trace profiles collected from the emission signals of silica sphere and silver nanoshell (Figure 1) that within 60 seconds the emission intensity from the silica sphere decays to ¼ whereas that from the silver nanoshell decays to ½ only under the same irradiation conditions, indicating that the metal nanoshell may increase the photostability of encapsulated Ru(bpy)32+ complexes approx. 2-fold. We also evaluated the long lifetime emission signal from the metal nanoshell by comparing with the short time cellular autofluorescence (Figure 7), showing that the emission intensity from the metal nanoshell decayed to only 1/2 within 60 s whereas the cellular autofluorescence eliminated completely within 20 s. This result indicates the metal nanoshells may remain luminescent even though the cellular autofluorescence is completely eliminated by the laser irradiation.
Figure 7.
Emission decays from the conjugated metal nanoshell and typical cellular autofluorescence on the cell image under the same irradiation conditions.
Nerveless, the metal nanoshell can be used as molecular imaging agent to hybridize with the target miRNAs in the cells for the cell imaging, and the collected expression levels of miRNA-486 in the cells may distinct the lung cancer cell line that is important to evaluate the occurrence of lung cancer and diagnose in earl stage to the patients.
Scheme 1.
Preparation of metal shell/silica core structure with the Ru(bpy)32+ complex encapsulation and surface reaction to bind with the single-stranded oligonucleotide on the metal nanoshell.
ACKNOWLEDGMENT
This research was supported by grants from NIH and American Cancer Society (JZ: EB009509, JR: HG-002655, HG005090, EB006521, FJ: American Cancer Society Research Scholar Grant, NCI CA-135382, CA-137742, and CA-133956).
REFERENCES
- 1.Greenlee RT, Murray T, Wingo PA. CA. Cancer. J. Clin. 2000;5:7. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- 2.Mountain C. Chest. 1997;6:1710. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 3.(a) Chatterjee SK, Zetter BR. Future Oncol. 2005;1:37. doi: 10.1517/14796694.1.1.37. [DOI] [PubMed] [Google Scholar]; (b) Colby TV, Wistuba II, Gazdar A. Adv. Anat. Pathol. 1998;5:205. doi: 10.1097/00125480-199807000-00001. [DOI] [PubMed] [Google Scholar]
- 4.(a) Thunnissen FB. J. Clin. Pathol. 2003;11:805. doi: 10.1136/jcp.56.11.805. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Varella-Garcia M, Kittelson J, Schulte AP, Vu KO, Wolf HJ, Zeng C, Hirsch FR, Byers T, Kennedy T, Miller YE, Keith RL, Franklin WA. Cancer Detect Prev. 2004;28:244. doi: 10.1016/j.cdp.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Science. 2001;294:853. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 6.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Cell. 2006;9:189. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 7.Ortholan C, Puissegur MP, Ilie M, Barbry P, Mari B, Hofman P. Curr. Med. Chem. 2009;16:1047. doi: 10.2174/092986709787581833. [DOI] [PubMed] [Google Scholar]
- 8.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Cancer Res. 2004;64:3753. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 9.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, Gemma A, Kudoh S, Croce CM, Harrisa CC. PNAS. 2009;106:12085. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Z, Zhao Y, Tian T, Liang J, Jin G, Shen H. Biomedicine Pharmacotherapy. Volume 63(Issue 5):322–322. [Google Scholar]
- 11.(a) Jiang F, Li RY, Wang HJ, Caraway NP. Neoplaisa. 2004;2:212. [Google Scholar]; (b) Li R, Wang H, Bekele BN, Katz RL, Stass SA, Jiang F. Oncogene. 2006;25:2628. doi: 10.1038/sj.onc.1209289. [DOI] [PubMed] [Google Scholar]
- 12.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd ed. New York: Springer Published; 2006. [Google Scholar]
- 13.El-Sayed MA. Acc. Chem. Res. 2004;37:326. doi: 10.1021/ar020204f. [DOI] [PubMed] [Google Scholar]
- 14.Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 15.Lal S, Clare SE, Halas NJ. Acc. Chem. Res. 2008;41:1842. doi: 10.1021/ar800150g. [DOI] [PubMed] [Google Scholar]
- 16.(a) Hayat MA, editor. Colloidal Gold: Principles, Methods, and Applications. San Diego: Academic Press; 1991. [Google Scholar]; (b) Feldheim DL, Foss CA. Metal Nanoparticles. New York: Synthesis, Characterization and Applications, Marcel Dekker, Inc.; 2002. [Google Scholar]
- 17.(a) Lakowicz JR. Anal. Biochem. 2001;298:1. doi: 10.1006/abio.2001.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lakowicz JR. Anal. Biochem. 2005;337:171. doi: 10.1016/j.ab.2004.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Zhang J, Fu Y, Lakowicz JR. J. Phys. Chem. C. 2007;111:1955. doi: 10.1021/jp063996u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang J, Gryczynski I, Gryczynski Z, Lakowicz JR. J. Phys. Chem. B. 2006;110:8986. doi: 10.1021/jp057032z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Enderlein J. Phys. Chem. Chem. Phys. 2004;4:2780. [Google Scholar]; (b) Enderlein J. Appl. Phys. Letter. 2002;80:315. [Google Scholar]
- 20.(a) Wang H, Goodrich GP, Tam F, Oubre C, Nordlander P, Halas NJ. J. Phys. Chem. B. 2005;109:11083. doi: 10.1021/jp051466c. [DOI] [PubMed] [Google Scholar]; (b) Nehl CL, Grady NK, Goodrich GP, Tam F, Halas NJ, Hafner JH. Nano Lett. 2004;4:2355. [Google Scholar]
- 21.Olson TY, Schwartzberg AM, Orme CA, Talley CE, O'Connell B, Zhang JZ. J. Phys. Chem. C. 2008;112:6319. [Google Scholar]
- 22.(a) Hubert C, Rumyantseva A, Lerondel G, Grand J, Kostcheev S, Billot L, Vial A, Bachelot R, Royer P, Chang S-h, Gray SK, Wiederrecht GP, Schatz GC. Nano Lett. 2005;5:615. doi: 10.1021/nl047956i. [DOI] [PubMed] [Google Scholar]; (b) Alvarez-Puebla RA, Ross DJ, Nazri G-A, Aroca RF. Langmuir. 2005;21:10504. doi: 10.1021/la051645l. [DOI] [PubMed] [Google Scholar]
- 23.(a) Zhang J, Fu Y, Liang D, Nowaczyk K, Zhao RY, Lakowicz JR. Nano Lett. 2008;8:1179. doi: 10.1021/nl080093z. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang J, Fu Y, Liang D, Zhao RY, Lakowicz JR. Anal. Chem. 2009;81:883. doi: 10.1021/ac801932m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Fu Y, Lakowicz JR. J. Phys. Chem. C. 2009;113:19404. doi: 10.1021/jp906742q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Templeton AC, Wuelfing WP, Murray RW. Acc. Chem. Res. 2000;33:27. doi: 10.1021/ar9602664. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Fu Y, Chowdhury M, Lakowicz JR. J. Phys. Chem. C. 2007;111:11784. doi: 10.1021/jp067887r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Y, Todd NW, Liu Z, Zhan M, Fang H, Peng H, Alattar M, Deepak J, Stass SA, Jiang F. Lung Cancer. 2009;87:1728. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R, Todd N, Stass S, Jiang F. J. Nanosci. Nanotech. 2007;10:1. doi: 10.1166/jnn.2007.920. [DOI] [PubMed] [Google Scholar]
- 29.Politz JCR, Zhang F, Pederson T. PNAS. 2006;103:18957. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.(a) O'Connell MA, Keegan LP. Nature Struc. & Mol. Biol. 2006;13:3. doi: 10.1038/nsmb0106-3. [DOI] [PubMed] [Google Scholar]; (b) Leuschner PJF, Martinez J. Methods. 2007;43:105. doi: 10.1016/j.ymeth.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Thomas JR, Hergenrother PJ. Chem. Rev. 2008;108:1172. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 32.Kaur H, Babu BR, Maiti S. Chem. Rev. 2007;107:4672. doi: 10.1021/cr050266u. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu J, Tsourkas A. Nucleic Acids Res. 2009;37:e100. doi: 10.1093/nar/gkp482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu Z, Fang H, Zhang J, Katz RL, Jiang F. International Journal of Cancer. 2010 doi: 10.1002/ijc.25289. printed online. [DOI] [PMC free article] [PubMed] [Google Scholar]