Abstract
Background
Many risk factors have been associated with cancer, such as age, family history, race, smoking, high-fat diet, and poor nutrition. It is important to reveal the molecular changes related to risk factors that could facilitate early detection, prevention, and overall control of cancer.
Methods
We selected six cancer-specific methylated genes that have previously been reported in primary tumors and have also been detected in different bodily fluids of cancer patients. Here, we used quantitative fluorogenic real-time methylation-specific PCR in plasma DNA samples for the detection of methylation changes from an asymptomatic population who do not have any known cancer.
Results
The promoter methylation frequencies of the studied genes were as follows: APC (7%), CCND2 (22%), GSTP1 (2%), MGMT (9%), RARβ2 (29%), and P16 (3%). Promoter methylation of at least one of the genes analyzed was observed in ~46% (72 of 157) of the samples by binary dichotomization. Promoter hypermethylation of at least two genes was detected in 17% (26 of 157) of the samples. RARβ2 methylation was observed in 45% of subjects who had a high-fat diet in contrast with those who had a low-fat diet (23%; P = 0.007).
Discussion
Our findings may help to elucidate early methylation changes that may lead to cancer development. These methylation changes could be due to exposure to risk factors and may be useful for cancer prevention measures such as changes in lifestyle. Longitudinal follow-up of a high-risk population is needed to understand the association of methylation of candidate genes in cancer development.
Introduction
Cancer incidence and mortality rates vary worldwide, including by race, gender, and age (1). In the United States, in 2008, 1,437,180 individuals, or 3,937 persons per day, were expected to be diagnosed with cancer; and about 565,650 persons, or more than 1,500 per day, were expected to die from cancer (2). African Americans have higher cancer incidence and mortality rates than any other racial and ethnic group in the United States. The disparities in cancer mortality are much greater than those in incidence, in part, because African Americans have the shortest survival rate due to later stage at diagnosis and less access to appropriate and timely treatment (3). The causes of this disparity are complex and interrelated but likely arise from socioeconomic disparities in work, wealth, income, education, housing, overall standard of living, prevention, early detection and treatment services, and the effect of racial discrimination on all these factors (4). Over the past 3 decades, research efforts have come to the conclusion that most cancers are multifactorial (i.e., they are probably related to the interactions of multiple genetic, epigenetic, and environmental risk factors). Risk factors that have been associated with cancer include race (3), smoking status (5), age (6), family cancer history (7), poverty (8), alcohol consumption (9), and high-fat diet (10).
Epigenetic changes are defined as all meiotically and mitotically heritable changes in gene expression that are not coded in the DNA sequence itself and deregulate the mechanisms, such as transcriptional control, which lead to the inappropriate silencing or activation of cancer-associated genes (11). It is known that certain environmental factors induce epigenetic changes. These changes will establish a gene expression pattern by altering the epigenetic state of the genome (12, 13). Both global hypomethylation and gene-specific promoter hypermethylation are associated with cancer (14, 15). Several studies have shown that these epigenetic changes are an early event in carcinogenesis and are present in the precursor lesions of a variety of cancers, including lung (16), thyroid (17), and colon (18).
Cancer development occurs through the accumulation of genetic and epigenetic changes that allow cells to behave independently from the tight network of controls that regulate the homeostatic balance between cell proliferation and cell death (19). The interaction of external factors with one or more internal factors drives the genetic and epigenetic changes, increasing the probability of developing cancer (20). Recent studies have provided convincing evidence that methylation in the promoter region is frequently associated with “gene silencing”; that is, the gene is not expressed in the presence of methylation but is expressed in its absence (21). Most importantly, promoter methylation of these genes can be detected in bodily fluids with high sensitivity and specificity (22–25).
Before Watson and Crick described the helical structure of the DNA, Mandel and Metais in 1947 (26) discovered DNA in plasma and serum by using a simple perchloric acid precipitation method. Previous reports paved a way to find circulating DNA in higher concentrations among patients with cancer compared with cancer-free patients (27, 28). Stroun et al. (29) showed that circulating free DNA was derived from tumor cells and certain characteristics of tumor DNA were present in circulating DNA. Other studies reported the presence of methylated DNA in the bodily fluids of patients with various types of malignancies and the absence of it in normal subjects (22–25).
The goal of this study is to determine the level of promoter hypermethylation in the plasma DNA of established cancer-related genes (APC, CCND2, GSTP1, MGMT, RARβ2, and P16) and to test their association with several potential risk factors in cancer-free individuals. We anticipate that in the future this information can be used to stratify high-risk groups by a molecular approach and thereby contribute to cancer prevention and control. To our knowledge, this is the first study to evaluate the association between lifestyle factors and CpG island methylation of genes in cancer-free subjects.
Materials and Methods
Study Population
We conducted a cross-sectional study on the association between selected methylation markers and specific risk factors for cancer in a community-based sample of mostly African American men. We recruited a convenience sample of men through a variety of community-based venues, including a local market, churches, and health fairs, and a variety of other activities. Eligible participants were age 40 y and above, without a known history of a symptomatic disease, including cancer. In our cross-sectional study, 534 cancer-free subjects were recruited. Among the 534 individuals, 262 are male and were selected for this study. However, 105 were excluded from the analysis for the following reasons: 81 did not consent to a blood draw, 14 did not have amplifiable DNA extracted from their plasma samples, and 10 did not have enough plasma available. The remaining sample for this analysis included 149 African Americans, 2 Asians, 1 Hispanic/Latino, 4 Caucasians, and 1 “other.” We obtained plasma samples from the participants and tested for a panel of markers that have been previously shown to have high methylation frequencies in prostate cancer. The participants gave written informed consent and responded to a standardized questionnaire administered by a trained interviewer during a face-to-face interview. The content of the interview includes questions on the following areas: socioeconomic factors (e.g., income, education, and parental education); access to care (medical insurance, economic barriers, transportation); cultural beliefs (mistrust, religiosity); psychosocial factors (depression, self-esteem, social support); cancer screening practices; and related behavioral risk and protective factors, including smoking, alcohol consumption, and dietary habits. Further demographic characteristics are detailed in Table 1.
Table 1.
Demographic characteristics of the studied population
| Characteristic | No. subjects (%) |
|---|---|
| Age | |
| 40–49 | 99 (63) |
| 50–90 | 58 (37) |
| Smoking | |
| None | 36 (23) |
| Former and current | 121 (77) |
| Income | |
| $0–49,999 | 131 (83) |
| $50,000+ | 14 (9) |
| Unknown | 12 (8) |
| Family cancer history | |
| None in family | 86 (55) |
| Anyone in family | 71 (45) |
| Diet and nutrition | |
| Low fat | 108 (69) |
| High fat | 47 (30) |
| Unknown | 2 (1) |
| CO | |
| <6 ppm | 60 (38) |
| >6 ppm | 81 (52) |
| Unknown | 16 (10) |
| Environmental tobacco smoke | |
| None | 8 (5) |
| Child | 22 (14) |
| Adult | 23 (15) |
| Child + adult | 95 (60) |
| Unknown | 9 (6) |
| None and adult only | 31 (21) |
| Child and child + adult | 117 (79) |
| Alcohol consumption | |
| None and former | 131 (83) |
| Current | 26 (17) |
| Diabetes | |
| No diabetes | 149 (95) |
| Diabetes | 8 (5) |
Carbon Monoxide Measurement
A single-breath carbon monoxide (CO) level measurement was used to immediately assess and confirm smoking status and severity of the smoking. Following tobacco smoke inhalation, CO displaces oxygen in the erythrocyte to form carboxyhemoglobin, a biological indicator of the amount of CO in the body. In this form, CO has a half-life (t1/2) of about 6 to 16 h and may remain in the blood for up to 24 h (30). However, measuring carboxyhemoglobin levels is not always possible due to the invasive nature of collecting blood samples. Studies have reported that the level of CO in end expiratory breath shows a close relationship to blood carboxyhemoglobin level (31). Hence, measuring breath CO levels can provide an immediate, noninvasive, and effective way of confirming a subject’s smoking status. Reported breath CO level in nonsmokers ranges from 0 to 6 parts per million (ppm), whereas CO level of smokers will be >6 ppm.
The portable Breath CO-Carbon Monoxide Monitor (Vitalographs, Inc.) was used to measure both alveolar and environmental CO levels. The environmental (interview site/background) CO level was measured before measuring volunteer’s lung CO level. After recording the highest CO level obtained for the interview site, a sampling-T and new disposable mouthpiece were mounted on the monitor. When the display showed 000 +2, the volunteer was asked to take a deep breath, hold it as long as he can, and then to breathe out slowly and gradually through the mouthpiece over a period of 20 s. The highest level displayed on the specimen collection form was recorded. The volunteer was asked to provide three expiratory breaths, and the average of the three measures was taken. Pearson correlation coefficients among each of the measures show acceptable variability.
Blood Sample Collection
After obtaining the informed consent, a 20 mL blood sample was collected into three tubes: 3 mL into a Vacutainer tube (BD Diagnostics) containing K3 EDTA 7.5% solution, 7 mL into a Vacutainer tube without anticoagulant, and 10 mL into a Vacutainer tube containing K3 EDTA 7.5% solution. The blood samples were transported to the laboratory within 2 h of being drawn. The Vacutainer containing 3 mL of blood was used for DNA isolation from lymphocytes. The Vacutainer tube containing 7 mL was used to obtain serum. The plasma was obtained by centrifuging 10 mL of blood at 800 × g for 10 min at 4°C. An equal volume of plasma (~1 mL) was aliquoted into three 1.8 mL vials (VWR International) and stored at −80°C until used. For this study, 1 mL of plasma was used for DNA extraction.
DNA Extraction
DNA from plasma was extracted as previously described (24). Briefly, DNA was obtained from 1 mL of plasma by digestion with 50 μg/mL proteinase K (Boehringer Mannheim) in the presence of 1% SDS at 48°C for 2 d followed by phenol/chloroform extraction and ethanol precipitation and finally dissolved in 20 μL of LoTE (2.5 mmol/L EDTA and 10 mmol/L Tris-HCl) and stored at −20°C until used.
Sodium Bisulfite Conversion of DNA Extracted from Plasma
The EpiTect Bisulfite kit (Qiagen, Inc.), was used to convert unmethylated cytosines to uracil in DNA extracted from plasma according to the manufacturer’s instructions. Converted DNA was stored at −80°C until used.
Methylation Analysis
Bisulfite-modified DNA was used as a template for fluorescence-based real-time PCR, as previously described (22). Amplification reactions were carried out in triplicate in a final volume of 20 μL containing 1 μL of bisulfite-modified DNA; 600 nmol/L concentrations of forward and reverse primers; 200 nmol/L probe; 0.6 units of platinum Taq polymerase (Invitrogen); 200 μmol/L concentrations each of dATP, dCTP, dGTP, and dTTP; and 6.7 mmol/L MgCl2. Primers and probes were designed to specifically amplify the promoters of the six genes of interest and the promoter of a reference gene, β-actin; primer and probe sequences and annealing temperatures are provided in Supplementary Table S1. Amplifications were carried out using the following profile: 95°C for 3 min followed by 50 cycles at 95°C for 15 s and 60°C for 1 min. Amplification reactions were carried out in 384-well plates in a 7900 sequence detector (Perkin-Elmer Applied Biosystems) and analyzed by a sequence detector system (SDS 2.3; Applied Biosystems). Each plate included subjects’ DNA samples, positive (in vitro methylated leukocyte DNA) and negative (normal leukocyte DNA or DNA from a known unmethylated cell line) controls, and multiple water blanks. Leukocyte DNA from a healthy individual was methylated in vitro with excess SssI methyltransferase (New England Biolabs, Inc.) to generate completely methylated DNA, and serial dilutions (90–0.009 ng) of this DNA were used to construct a calibration curve for each plate. All samples were within the range of sensitivity and reproducibility of the assay based on the amplification of the internal reference standard (threshold cycle value for β-actin of 40). The relative level of methylated DNA for each gene in each sample was determined as a ratio of methylation-specific PCR–amplified gene to β-actin (reference gene) and then multiplied by 1,000 for easier tabulation (average value of triplicates of the gene of interest divided by the average value of triplicates of β-actin × 1,000). The samples were categorized as unmethylated or methylated based on the sensitivity of the assay. We considered methylated and unmethylated by dichotomization. Any normalized methylation values above the cut point value (here 0) were considered as methylated.
Gene Selection
A total of 6 genes (GSTP1, APC, CCND2, MGMT, P16, and RARβ2) were selected for promoter methylation status analysis. The panel included genes reported as targets for epigenetic silencing in human cancer.
Risk Factors
We examined the association between several potential risk factors and promoter methylation at each of the six genes. Age was analyzed both as a continuous variable, ranging from 40 to 86, and as a dichotomous variable, ranging from 40 to 49 and 50 and above. Income was analyzed as a dichotomous variable under $50,000 per year (per household) and ≥$50,000. The cutoff for age and income was determined empirically. A smoker was defined as an individual who reported having smoked ≥100 cigarettes in his lifetime. Family cancer history was dichotomized to anyone with “brothers, sisters, parents, children, or other close family members” having/had cancer and those who have not. Diet and nutrition were characterized as a dichotomous variable based on consumption of red meat, processed meat, whole-milk products, and fried foods. Individuals who ate two or more of these food groups on at least a daily basis were considered high-fat consumers and individuals who ate less were considered low-fat consumers. The CO level found in the breath was considered a measure of current smoke exposure through either personal smoking or environmental exposure. Levels ranging from 0 to 6 ppm were considered non-exposed and levels of ≥6 ppm were considered as exposed. Childhood environmental tobacco smoke exposure was dichotomized based on the response to the question whether the individual was exposed or not in the home where they grew up. Alcohol use was dichotomized with never users and former users as one group and current users as the other group. A physician diagnosis of diabetes was also a binary variable. We decided for the empirical categorization based on our findings.
Statistical Analysis
We examined the relationship between promoter methylation frequencies of each individual gene and total numbers of methylated genes with the hypothesized risk factors, first by χ2. Because of the skewed distribution of methylation levels, we also tested with nonparametric rank sum tests. The categorization of methylation status into binary variables (1 = methylated and 0 = no methylation) was also used to test the relationship using χ2 and logistic regression. An association was considered statistically significant with P value of <0.05. All statistical tests were two-sided. Data analyses were done with Stata version 9.1.
Results
The characteristics of the study participants, including age, income, race, smoking status, family cancer history, diabetes, diet and nutrition, CO level, environmental tobacco smoke, and alcohol consumption, are summarized in Table 1.
Frequency of Methylation in Plasma Samples by Quantitative Methylation-Specific PCR
We tested the promoter methylation status of six genes that have been previously associated with cancer (GSTP1, APC, CCND2, MGMT, P16, and RARβ2).
The frequencies observed were 3 of 157 (2%) for GSTP1, 11 of 157 (7%) for APC, 34 of 157 (22%) for CCND2, 14 of 157 (9%) for MGMT, 4 of 157 (3%) for P16, and 46 of 157 (29%) for RARβ2. Approximately 46% of the subjects showed promoter methylation of at least one gene, 16.5% for two or more genes, 7% for three or more genes, and 2% for four or more genes. The promoter methylation frequencies of these six individual genes are listed in Table 2.
Table 2.
Promoter methylation frequency for the six genes analyzed in the studied population
| Gene | Methylation positive % (no. methylation positive/no. total cases) |
|---|---|
| APC | 7 (11/157) |
| CCND2 | 22 (34/157) |
| GSTP1 | 2 (3/157) |
| MGMT | 9 (14/157) |
| RARβ2 | 29 (46/157) |
| P16 | 3 (4/157) |
Association of Plasma DNA Hypermethylation Profiles with Cancer Risk Factors
Several demographic parameters considered to be risk factors (age, smoking status, family cancer history, diabetes, diet and nutrition, CO level, environmental tobacco smoke, and alcohol) were compared with the methylation frequency. CCND2 and RARβ2 have higher methylation frequencies compared with other genes (Table 2).
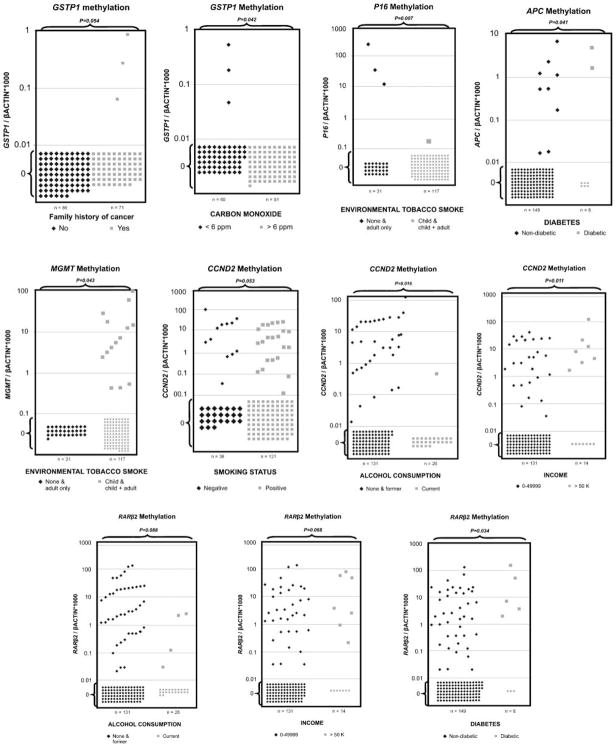
Associations between variables and gene promoter methylation were observed for every gene with at least one of the analyzed parameters. The bivariate analysis data are shown in Table 3, with χ2 analysis. Briefly, RARβ2 promoter methylation was positively associated with high-fat consumers and diabetes (P = 0.007 and P = 0.034, χ2 test, respectively). Promoter methylation levels and frequencies of RARβ2 in plasma DNA of high-and low-fat consumers are shown in Fig. 1. Representative scatter plots showing the relationship of promoter methylation values of tested genes with different cancer risk factors are shown in Fig. 2. GSTP1 promoter methylation frequency was significantly correlated with family history of cancer and CO levels <6 ppm (P = 0.054 and P = 0.042, χ2 test, respectively; Fig. 2; Table 3). APC promoter methylation frequency was significantly correlated with diabetes (P = 0.041, χ2 test; Fig. 2; Table 3). MGMT methylation frequency was significantly correlated with childhood environmental tobacco smoke whereas P16 was correlated with adult (or no) exposition (P = 0.043 and P = 0.007, χ2 test, respectively; Fig. 2; Table 3). CCND2 promoter methylation was significantly higher in the never smokers, nonalcoholic, and higher income group (P = 0.053, P = 0.016, and P = 0.011, χ2 test, respectively; Fig. 2; Table 3). We also did rank sum test to emphasize the strength and veracity of the findings and the data are shown in Supplementary Table S2.
Table 3.
Correlation of variables by exposure type associated with gene-specific promoter methylation
| Variables | APC | CCND2 | GSTP1 | MGMT | RARβ2 | P16 |
|---|---|---|---|---|---|---|
| P (χ2)* | P (χ2) | P (χ2) | P (χ2) | P (χ2) | P (χ2) | |
| Age | 0.544 | 0.822 | 0.181 | 0.921 | 0.715 | 0.616 |
| Nonsmoking | 0.722 | 0.053 | 0.665 | 0.889 | 0.306 | 0.921 |
| Income (>$50,000) | 0.187 | 0.011 | 0.567 | 0.06 | 0.068 | 0.507 |
| Family cancer history | 0.203 | 0.884 | 0.054 | 0.454 | 0.673 | 0.846 |
| Diet and nutrition (high fat) | 0.819 | 0.998 | 0.249 | 0.645 | 0.007 | 0.814 |
| CO (<6 ppm) | 0.621 | 0.542 | 0.042 | 0.553 | 0.529 | 0.132 |
| Environmental tobacco smoke | 0.315 | 0.353 | 0.368 | 0.043 | 0.25 | 0.007 |
| Nonalcohol | 0.125 | 0.016 | 0.43 | 0.321 | 0.088 | 0.367 |
| Diabetes | 0.041 | 0.814 | 0.685 | 0.715 | 0.034 | 0.639 |
NOTE: Referent group for each variable: age 1 (40–49) and 2 (50 and 90); current smokers; $0 to $49,999; no cancer in family; low fat; ≥6 ppm; no environmental tobacco smoke exposure; current alcohol; no diabetes.
P ≤ 0.05 was considered statistically significant.
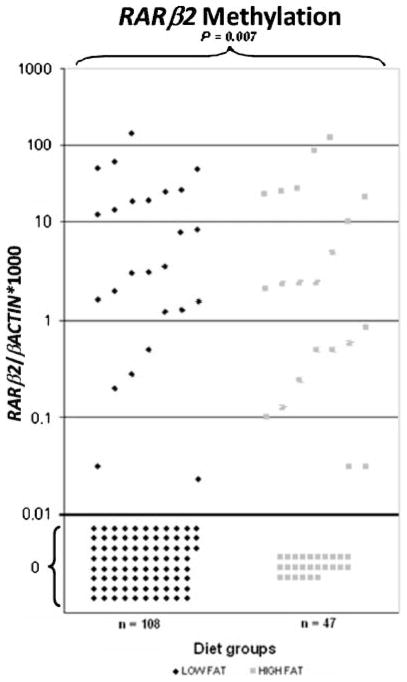
Figure 1.
Methylation levels and frequencies of RARβ2 in plasma DNA of high- and low-fat consumers. Calculation of the RARβ2 gene to β-actin ratios was based on the fluorescence emission intensity values for both the genes obtained by quantitative methylation-specific real-time PCR analysis. The obtained ratios were multiplied by 1,000 for easier tabulation. Values designated as 0.01 are zero values, which cannot be plotted correctly on a log scale. RARβ2 methylation was found in plasma DNA of 25 of 108 (23%) low-fat consumers and in plasma DNA of 21 of 47 (45%) high-fat consumers (P = 0.007). The number of subjects is represented by dots. We can observe 25 dots above 0.1 (methylated) versus 83 below 0.01 (unmethylated) for the low-fat consumers, whereas 21 dots show methylated (above 0.01) and 26 dots show unmethylated values (below 0.01) for the high-fat consumer group.
Figure 2.
Representative scatter plots of methylation values of tested genes with different high-risk variables. Methylation levels of GSTP1, APC, CCND2, MGMT, P16, and RARβ2 in plasma DNA of different groups. Calculation of the gene of interest: ratios were based on the fluorescence emission intensity values for both the gene of interest and β-actin obtained by quantitative real-time PCR analysis. The obtained ratios were multiplied by 1,000 for easier tabulation. Values designated as 0.01 are zero values, which cannot be plotted correctly on a log scale.
All of the variables of interest (age, income, race, smoking status, family cancer history, diabetes, diet and nutrition, CO level, environmental tobacco smoke, and alcohol consumption) were then entered in a logistic regression model. In this analysis, CpG methylation of genes (yes/no status) APC, GSTP1, MGMT, and P16 was not associated with any risk factors (data not shown). However, the association between CCND2 methylation and the absence of alcohol consumption was statistically significant (P = 0.05; odds ratio, 0.12; 95% confidence interval, 0.015–1.003) and a positive association between RARβ2 methylation and smokers approached significance (P = 0.066; odds ratio, 0.33; 95% confidence interval, 0.101–1.076). The multivariate analysis was done, but the results are not shown because none achieved statistical significance (data not shown).
Discussion
Epigenetic changes provide a potential explanation for how environmental factors modify the risk for common diseases among individuals (13). It is established that inactivation by promoter methylation of certain genes is one of the hallmarks of developing cancer. Methylation of cancer-related genes has been found in some cancer-free individuals, raising several questions, such as “What is the prevalence of these methylated genes in cancer-free individuals?,” “Could the presence of methylation signal of any of the cancer-related gene heighten the risk of developing cancer in the future and thus be used to identify people who should be monitored more closely?,” and “What are the individual, environmental, and behavioral factors that lead to methylation of these genes?”
In a cross-sectional study, we selected a cancer-free population with known demographic, lifestyle, and other associated factors that are risk factors for developing cancer, such as diet, family history of cancer, and environmental exposure. We hypothesized that this population would have a high prevalence of promoter methylation in certain genes that have been linked to cancer (GSTP1, APC, CCND2, MGMT, P16, and RARβ2; refs. 17, 22–25, 32). It was possible to detect promoter methylation of these genes in plasma DNA samples, and indeed, 46% of the participants were found to have methylation of at least one of six genes we tested. To determine whether methylation in plasma DNA of this cancer-free population is due to exposure to specific risk factors, rather than just a normal occurrence, future research needs to compare the methylation patterns among individuals at varying levels of risk for developing cancer. If methylation patterns vary by risk level, it may mean that detection of promoter methylation of these genes in plasma specimens could provide a novel, minimally invasive approach to screen high-risk populations long before any symptoms of cancer become established. Longitudinal follow-up of a cancer-free cohort with methylation and without methylation needs to be done to evaluate the usefulness of DNA methylation in plasma as a screening tool or as an aid in assessing the need for preventive measures.
Previous studies (18, 33) have reported the detection of genetic and epigenetic alterations in matched samples from tumor tissue and plasma in patients with lung, head and neck, kidney, colon, and pancreatic cancers. The circulating plasma DNA is presumably shed from the original primary tumor or preneoplastic lesions. The promoter methylation of the studied genes has also been detected in bodily fluids of cancer patients, whereas no or less frequent methylation was detected in age-matched controls (22–25). However, thus far, little is known about the relevance of the detection of promoter methylation of certain genes in plasma of cancer-free subjects and how these epigenetic alterations are related to exposures of risk factors that are related to cancer. Recently, in a nested case-control study of an extremely high-risk cohort of developing lung cancer, Belinsky et al. (16) reported that simultaneous methylation of three or more of the six genes they tested was associated with a 6.5-fold increased risk of developing cancer. Both the sensitivity and the specificity were 64% of the later study. They prospectively examined a large panel of genes for their ability to predict lung cancer and showed the promise of gene promoter hypermethylation in sputum as a molecular marker for identifying people at high risk for cancer development.
Aberrant methylation of CpG islands in the promoter regions has gained importance in the initiation of cancer and can be used as biomarkers for early detection of cancer (22, 24, 34). Promoter methylation of several genes has been reported previously in nonmalignant tissues, sputum, and serum DNA of smokers (35). Detection of CpG island promoter hypermethylation in plasma DNA could be a marker of disease (an early neoplastic effect), exposure (a biological effect of any risk factor), or both. The presence and extent of CpG island promoter hypermethylation in a cancer-free subject may reflect chronic exposure to known or still unidentified carcinogenic risk factors. Longitudinal studies need to assess the risk of developing precursor lesions associated with the presence of methylation of specific genes, and the assessment of additional genes may help to further elucidate this relationship, perhaps identifying high-risk individuals who could benefit from more intensive standard evaluation that could eventually detect cancer earlier.
One of the most interesting statistically significant associations observed in this study was between the RARβ2 promoter methylation and high fatty food intake (defined by consumption of red meat, processed meat, whole-milk products, or fried foods more than twice a day, every day). This gene encodes retinoic acid receptor β, a member of the thyroid-steroid hormone receptor superfamily of nuclear transcriptional regulators. It binds retinoic acid, the biologically active form of vitamin A, which mediates cellular signaling in embryonic morphogenesis, cell growth, and differentiation. This gene is inactivated by promoter methylation in many types of cancer (17, 24, 36). Because high intake of fat has been related to a large number of cancers (37–39), and RARβ2 methylation silencing is a common feature of malignancy, one can speculate that this association is related to preneoplastic changes. Environmental and dietary factors in animals and humans inevitably affect epigenetic patterns, although a clear-cut causal relationship has yet to be established. The major obstacle in establishing such relationship is the fact that environmental and dietary factors induce molecular changes that are most likely subtle and cumulative, resulting into a quantitative manifestation over a long period of time (20).
Cyclin D2 is one of the D-type cyclins that function as rate-limiting controllers of G1 to S phase during the cell cycle. Inactivation of CCND2 associated with aberrant promoter hypermethylation was reported in different cancer types (32, 40). In this study, we found a high frequency of methylation of CCND2 (22%) in our cancer-free population. Interestingly CCND2 methylation frequency is significantly higher in never smokers compared with smokers. Although tobacco smoking is known to cause lung and other cancers, these diseases are still reported in those who have never smoked and are considered as different diseases in never smokers (41), suggesting that there may be different pathways that need to be altered in smokers and nonsmokers. For each situation, CCND2 methylation in never smokers may indicate the subjects who are high risk for developing nonsmoking-related cancer, such as prostate cancer, and may also indicate that this alteration belongs to a nonsmoking pathway of carcinogenesis. There are contradictory data for MGMT methylation in smokers (42) and never smokers (43). Our findings show a positive correlation between MGMT methylation and environmental tobacco smoke. These results are consistent with Liu et al.’s (42) study, which showed that tumors from male smokers have more MGMT promoter methylation compared with tumors from male nonsmokers (50% versus 10%; P < 0.05). In our data, P16 methylation frequency was significantly correlated with adult environmental tobacco smoke. Recent studies showed that aberrant P16 promoter methylation is an early and critical event in the non–small cell lung carcinoma development (44), and it has been observed in serum and sputum of chronic smokers without clinical disease (45). P16 inactivation by promoter hypermethylation may be a useful predictive marker of lung cancer development in high-risk subjects such as heavy smokers.
We observed a significant association between GSTP1 methylation and family cancer history. Many studies have shown the association of presence of methylation and family history of colon cancer (46), supporting the concept of a shared etiology, such as genetic predisposition (47). The biological relevance of these epigenetic alterations associated with other parameters, such as diabetes, is not yet clear. Further studies in cancer patients who present this condition may elucidate the relevance of these alterations and their correlation with carcinogenesis.
There are several limitations in our study. (a) Our population is consisted of only 157 individuals. A larger sample would give us more power to detect relationships between promoter methylation and relevant risk factors. (b) The study population was recruited through convenience sampling. Although this limits our findings, the method of participant selection would have no effect on the association between specific risk factors and promoter methylation. (c) Our study is restricted to males. (d) This is a cross-sectional study. Although we can report associations between methylation and risk factors, we cannot infer causal relationships, as we do not know the temporal relationship between the exposures of interest and promoter methylation. This temporal relationship will need to be examined in a longitudinal study with a larger cohort. (e) Our diet measurement is based on participant’s self-report that may not be accurate. Animal studies with accurate diet consumption report may help to elucidate the role of fatty food on RARβ2 methylation; however, animal studies may not accurately reflect results in humans.
In summary, this cross-sectional study suggests that it is possible to detect cancer-related gene hypermethylation in plasma of a cancer-free population, and it may be related to the exposure to certain risk factors or may represent a preneoplastic alteration. A large longitudinal study of a cancer-free population needs to be conducted to elucidate the effect of exposure to specific risk factors on the methylation of these genes and ultimately establish the use of methylation markers as a valid method for early cancer detection. In such a study, individuals with methylated genes could be tracked to see whether they develop cancer at a higher rate than those without methylation of these genes. Those subjects who had no methylated genes at baseline could be tracked to see if they developed them over time and which factors and behaviors play a role in causing the methylation. Because methylation of certain genes occurs in the very early stages of tumorigenesis, detection of methylation in plasma specimens could provide a low invasive approach to screen high-risk population long before any symptoms of any cancer are established.
Supplementary Material
Acknowledgments
Grant support: Flight Attendant Medical Research Institute Young Clinical Scientist Award, International Association for the Study of Lung and Career development award from Specialized Program of Research Excellence in Cervical Cancer grant P50 CA098252 (M.O. Hoque); Flight Attendant Medical Research Institute Young Clinical Scientist Award (S. Begum); Cigarette Restitution Fund of Maryland; and National Cancer Institute grant U54 CA091409. This abstract was awarded the AACR-MEG (Molecular Epidemiology Group) Scholar-in-Training Award at the 100th American Association for Cancer Research Annual Meeting (2009).
Footnotes
Note: Supplementary data for this article are available at Cancer Epidemiology, Biomarkers & Prevention Online (http://cebp.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
M.O. Hoque is a paid consultant to Oncomethylome Sciences, SA.
References
- 1.Ward E, Jemal A, Cokkinides V, et al. Cancer disparities by race/ethnicity and socioeconomic status. CA Cancer J Clin. 2004;54:78–93. doi: 10.3322/canjclin.54.2.78. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Shavers VL, Brown ML. Racial and ethnic disparities in the receipt of cancer treatment. J Natl Cancer Inst. 2002;94:334–57. doi: 10.1093/jnci/94.5.334. [DOI] [PubMed] [Google Scholar]
- 4.Ghafoor A, Jemal A, Cokkinides V, et al. Cancer statistics for African Americans. CA Cancer J Clin. 2002;52:326–41. doi: 10.3322/canjclin.52.6.326. [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer GP, Denissenko MF, Olivier M, et al. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–51. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 6.Issa JP. CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol. 2000;249:101–18. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- 7.Johns LE, Houlston RS. A systematic review and meta-analysis of familial colorectal cancer risk. Am J Gastroenterol. 2001;96:2992–3003. doi: 10.1111/j.1572-0241.2001.04677.x. [DOI] [PubMed] [Google Scholar]
- 8.Ward-Smith P. The effects of poverty on urologic health. Urol Nurs. 2007;27:445–6. [PubMed] [Google Scholar]
- 9.Hashibe M, Brennan P, Benhamou S, et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99:777–89. doi: 10.1093/jnci/djk179. [DOI] [PubMed] [Google Scholar]
- 10.Popkin BM. Understanding global nutrition dynamics as a step towards controlling cancer incidence. Nat Rev Cancer. 2007;7:61–7. doi: 10.1038/nrc2029. [DOI] [PubMed] [Google Scholar]
- 11.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 12.Feil R. Environmental and nutritional effects on the epigenetic regulation of genes. Mutat Res. 2006;600:46–57. doi: 10.1016/j.mrfmmm.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 13.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:S245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J Nutr. 2002;132:2424S–29S. doi: 10.1093/jn/132.8.2424S. [DOI] [PubMed] [Google Scholar]
- 15.Momparler RL. Cancer epigenetics. Oncogene. 2003;22:6479–83. doi: 10.1038/sj.onc.1206774. [DOI] [PubMed] [Google Scholar]
- 16.Belinsky SA, Liechty KC, Gentry FD, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–44. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 17.Hoque MO, Rosenbaum E, Westra WH, et al. Quantitative assessment of promoter methylation profiles in thyroid neoplasms. J Clin Endocrinol Metab. 2005;90:4011–8. doi: 10.1210/jc.2005-0313. [DOI] [PubMed] [Google Scholar]
- 18.Esteller M, Sparks A, Toyota M, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–71. [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 20.Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22:91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- 21.Hoque MO, Kim MS, Ostrow KL, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res. 2008;68:2661–70. doi: 10.1158/0008-5472.CAN-07-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoque MO, Begum S, Topaloglu O, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst. 2006;98:996–1004. doi: 10.1093/jnci/djj265. [DOI] [PubMed] [Google Scholar]
- 23.Hoque MO, Begum S, Topaloglu O, et al. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 2004;64:5511–7. doi: 10.1158/0008-5472.CAN-04-0799. [DOI] [PubMed] [Google Scholar]
- 24.Hoque MO, Feng Q, Toure P, et al. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol. 2006;24:4262–9. doi: 10.1200/JCO.2005.01.3516. [DOI] [PubMed] [Google Scholar]
- 25.Hoque MO, Topaloglu O, Begum S, et al. Quantitative methylation-specific polymerase chain reaction gene patterns in urine sediment distinguish prostate cancer patients from control subjects. J Clin Oncol. 2005;23:6569–75. doi: 10.1200/JCO.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Mandel P, Metais P. Les acides nucleiques du plasma sanguin chez l’homme. CR Acad Sci Paris. 1948;142:241–43. [PubMed] [Google Scholar]
- 27.Leon SA, Ehrlich GE, Shapiro B, Labbate VA. Free DNA in the serum of rheumatoid arthritis patients. J Rheumatol. 1977;4:139–43. [PubMed] [Google Scholar]
- 28.Hoque MO, Lee J, Begum S, et al. High-throughput molecular analysis of urine sediment for the detection of bladder cancer by high-density single-nucleotide polymorphism array. Cancer Res. 2003;63:5723–6. [PubMed] [Google Scholar]
- 29.Stroun M, Anker P, Maurice P, et al. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46:318–22. doi: 10.1159/000226740. [DOI] [PubMed] [Google Scholar]
- 30.Ahijevych KL, Tyndale RF, Dhatt RK, Weed HG, Browning KK. Factors influencing cotinine half-life during smoking abstinence in African American and Caucasian women. Nicotine Tob Res. 2002;4:423–31. doi: 10.1080/1462220021000018452. [DOI] [PubMed] [Google Scholar]
- 31.Jarvis MJ, Belcher M, Vesey C, Hutchison DC. Low cost carbon monoxide monitors in smoking assessment. Thorax. 1986;41:886–7. doi: 10.1136/thx.41.11.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brait M, Begum S, Carvalho AL, et al. Aberrant promoter methylation of multiple genes during pathogenesis of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2786–94. doi: 10.1158/1055-9965.EPI-08-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez R, Silva JM, Sanchez A, et al. Microsatellite alterations and TP53 mutations in plasma DNA of small-cell lung cancer patients: follow-up study and prognostic significance. Ann Oncol. 2000;11:1097–104. doi: 10.1023/a:1008305412635. [DOI] [PubMed] [Google Scholar]
- 34.Ibanez de Caceres I, Battagli C, Esteller M, et al. Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res. 2004;64:6476–81. doi: 10.1158/0008-5472.CAN-04-1529. [DOI] [PubMed] [Google Scholar]
- 35.Zochbauer-Muller S, Lam S, Toyooka S, et al. Aberrant methylation of multiple genes in the upper aerodigestive tract epithelium of heavy smokers. Int J Cancer. 2003;107:612–6. doi: 10.1002/ijc.11458. [DOI] [PubMed] [Google Scholar]
- 36.Jeronimo C, Henrique R, Hoque MO, et al. Quantitative RARβ2 hypermethylation: a promising prostate cancer marker. Clin Cancer Res. 2004;10:4010–4. doi: 10.1158/1078-0432.CCR-03-0643. [DOI] [PubMed] [Google Scholar]
- 37.Crowe FL, Key TJ, Appleby PN, et al. Dietary fat intake and risk of prostate cancer in the European Prospective Investigation into Cancer and Nutrition. Am J Clin Nutr. 2008;87:1405–13. doi: 10.1093/ajcn/87.5.1405. [DOI] [PubMed] [Google Scholar]
- 38.Johnson IT, Belshaw NJ. Environment, diet and CpG island methylation: epigenetic signals in gastrointestinal neoplasia. Food Chem Toxicol. 2008;46:1346–59. doi: 10.1016/j.fct.2007.09.101. [DOI] [PubMed] [Google Scholar]
- 39.Linos E, Willett WC, Cho E, Colditz G, Frazier LA. Red meat consumption during adolescence among premenopausal women and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2146–51. doi: 10.1158/1055-9965.EPI-08-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeronimo C, Henrique R, Hoque MO, et al. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res. 2004;10:8472–8. doi: 10.1158/1078-0432.CCR-04-0894. [DOI] [PubMed] [Google Scholar]
- 41.Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol. 2007;25:561–70. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Lan Q, Siegfried JM, Luketich JD, Keohavong P. Aberrant promoter methylation of p16 and MGMT genes in lung tumors from smoking and never-smoking lung cancer patients. Neoplasia. 2006;8:46–51. doi: 10.1593/neo.05586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pulling LC, Divine KK, Klinge DM, et al. Promoter hypermethylation of the O6-methylguanine-DNA methyltransferase gene: more common in lung adenocarcinomas from never-smokers than smokers and associated with tumor progression. Cancer Res. 2003;63:4842–8. [PubMed] [Google Scholar]
- 44.Topaloglu O, Hoque MO, Tokumaru Y, et al. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10:2284–8. doi: 10.1158/1078-0432.ccr-1111-3. [DOI] [PubMed] [Google Scholar]
- 45.Belinsky SA, Grimes MJ, Casas E, et al. Predicting gene promoter methylation in non-small-cell lung cancer by evaluating sputum and serum. Br J Cancer. 2007;96:1278–83. doi: 10.1038/sj.bjc.6603721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frazier ML, Xi L, Zong J, et al. Association of the CpG island methylator phenotype with family history of cancer in patients with colorectal cancer. Cancer Res. 2003;63:4805–8. [PubMed] [Google Scholar]
- 47.Chan AO, Issa JP, Morris JS, Hamilton SR, Rashid A. Concordant CpG island methylation in hyperplastic polyposis. Am J Pathol. 2002;160:529–36. doi: 10.1016/S0002-9440(10)64872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.