Abstract
Damage to the genetic material can affect cellular function in many ways. Therefore, maintenance of the genetic integrity is of primary importance for all cells. Upon DNA damage, cells respond immediately with proliferation arrest and repair of the lesion or apoptosis. All these consequences require recognition of the lesion and transduction of the information to effector systems. The accomplishment of DNA repair, but also of cell cycle arrest and apoptosis furthermore requires protein-protein interactions and the formation of larger protein complexes. More recent research shows that the formation of many of these aggregates depends on post-translational modifications. In this article, we have summarized the different cellular events in response to a DNA double strand break, the most severe lesion of the DNA.
Keywords: DNA double strand breaks, homologous recombination, non-homologous endjoining, cell cycle arrest, post-translational modifications.
INTRODUCTION
The integrity of cellular DNA is threatened every day by exogenous factors like UV-light, ionizing radiation or environmental and therapeutic chemicals, as well as by endogenous conditions such as DNA replication, meiotic recombination or programmed rearrangements [reviewed in: 1-4]. These DNA lesions can result in somatic mutations, cell death, genomic instability and carcinogenesis. Accordingly, maintenance of genomic integrity represents one of the most important challenges for cellular organisms. To ensure functionality despite recurring insults, cells have developed several strategies to handle DNA lesions which are in principle prohibition of further cell divisions, repair of the lesions and elimination of cells with damaged DNA.
The repair of DNA lesions is a complex process. Moreover, diverse damages of the DNA that result from exposure to various adverse agents are fixed by different repair programs. Single strand breaks and damaged nucleotides that can result from oxidation, alkylation, hydrolysis, deamination or exposure to ionizing rays are replaced through base excision repair (BER), a mechanism that uses the sister strand as a template to direct de novo synthesis of the damaged strand around the lesion [reviewed in: 5]. Photoproducts and bulky DNA adducts that result e.g. from covalent binding of chemicals to DNA bases or exposure to UV-light are fixed by the nucleotide excision repair (NER) pathway [reviewed in: 6]. The mismatch repair (MMR) pathway, on the other hand, eliminates mispaired nucleotides that can arise during DNA replication and recombination [reviewed in: 7]. Ionizing radiation and replication in the presence of DNA lesions lead to the generation of DNA double strand breaks (DSBs), which are mainly fixed by two different repair mechanisms: homologous recombination (HR) and non-homologous endjoining (NHEJ).
While relatively rare, DSBs are the most severe DNA lesion since repair is difficult und the original state is seldom fully reconstituted. Moreover, we still understand only little about the response to DSBs, although a large number of the proteins that are involved have been identified. However, it is still ill-defined how these proteins are recruited to the site of the lesion and how the different factors interact and communicate with each other to repair the damage and to guard the genome. In this review we will summarize the cellular response to DSBs. Hereby, we will particularly focus on signal transduction processes that occur during DNA damage recognition and repair.
Post-translational modifications of existing molecules play an important role in the cellular response to DSBs since they provide a means to alter the activity of a given protein without the necessity of de novo protein synthesis. Thus, the properties of a protein can be changed rapidly and without the need to transcribe damaged DNA which could be grossly hindered by the lesion or may result in non-functional protein products. While already addition or removal of small chemical groups modifies the characteristics of a target protein dramatically, small proteins of the family of ubiquitin-like proteins are also frequently attached to molecules involved in different cellular processes, to alter their activities or to extend their repertoire of protein-protein interactions. The most common post-translational modifications, also during the response to DSBs, are phosphorylation, acetylation, methylation, ubiquitination, SUMOylation and NED-Dylation. All these modifications are reversible and, accordingly, regulated by two types of enzymes. Phosphorylation is, for example, performed by kinases that covalently link a phosphate group onto a serine, threonine or tyrosine of the target protein [reviewed in: 8]. Phosphatases reverse this alteration by removing the phosphate group [reviewed in: 9]. Phosphorylation frequently results in a conformational change of the modified protein due to the introduction of a negative charge and may result in activation or inactivation of an enzyme. Alternatively, it can alter the capacity of a target protein to associate with other cellular factors [reviewed in: 8]. The property of a protein can also be changed by reversible addition of acetyl- or methyl-groups, a process that is similar to the attachment of phosphate groups [reviewed in: 10, 11]. Acetyl-groups are attached to lysine residues in the target protein by acetyltransferases and removed by deacetylases [reviewed in: 10]. Lysines as well as arginines can further be modified by addition of a methyl-group, which is catalyzed by methyltransferases [reviewed in: 11]. The functional outcome of acetylation and methylation depends on the target protein and the position of the acetylated/methylated lysine in the molecule. This type of modification affects frequently protein-protein interactions, protein-DNA interactions or protein degradation (due to competition with the ubiquitination machinery for the same lysine) [12, 13].
The function of certain proteins can further be influenced by covalent addition of members of the family of small ubiquitin-like proteins. The first protein of this group that was identified was ubiquitin itself, a protein of 76-amino acids that is ubiquitously expressed from yeast to man [reviewed in: 14]. Ubiquitin is implicated in a multitude of cellular processes like regulation of the cell cycle, DNA replication, DNA repair, stress response, apoptosis, signal transduction or in the biogenesis of ribosomes, nucleosomes and peroxisomes [reviewed in: 15-17]. The covalent ligation of ubiquitin is an extremely conserved process and involves several enzymes and accessory proteins that are generally highly conserved across different species [reviewed in: 15]. Moreover, the attachment of ubiquitin onto a target protein ranges from the attachment of only one ubiquitin molecule to one lysine (monoubiquitination) over the addition of one ubiquitin to several lysine residues within the same protein (multiubiquitination) to the formation of chains of ubiquitin molecules on one individual lysine (polyubiquitination) [reviewed in: 18]. Since the ubiquitin protein itself possesses 7 lysine residues (lysine 6, lysine 11, lysine 29, lysine 31, lysine 33, lysine 48, lysine 63), which all can serve as an anchor for ubiquitination, a variety of chains can be formed that control different properties [reviewed in: 19, 20]. Polyubiquitination of lysine 48 is the prototype for chains that lead to proteosomal degradation, although more recent reports showed that polyubiquitination of lysine 11 and eventually also of lysine 63 can target substrate proteins for destruction [21, reviewed in: 19, 20]. Proteins that are polyubiquitinated at lysine 63 are frequently implicated in DNA repair, while chain formation at lysine 29 seems to label their targets for lysosomal degradation [22, 23]. In addition, polyubiquitin chains are not necessarily homotypic. Chains, containing two types of linkages (lysine 6/11, lysine 27/29, lysine 29/48; lysine 29/33) have also been observed [24]. Like phosphorylation or acetylation, the modification of proteins with ubiquitin is reversible. Specific deubiquitinating enzymes (DUBs) hydrolyze the isopeptide bond between ubiquitin and its substrate and thus remove the ubiquitin molecule or chain from its target [reviewed in: 25].
Another member of the ubiquitin family is SUMO (small ubiquitin-like modifier), a small polypeptide of 10-11 kDa [reviewed in: 26, 27]. SUMOylation is frequently involved in the control of recombination, DNA repair, nucleocytoplasmatic transport, chromosomal dynamics, meiosis and mitosis and the maintenance of genomic stability [reviewed in: 26, 27]. In addition, it can protect proteins from proteasomal degradation, most likely through rivalry with the ubiquitination machinery for the same lysine residue [27]. The process of SUMOylation is mechanistically related to ubiquitination, as an enzyme cascade consisting of a SUMO-activating enzyme (E1), a SUMO-conjugating enzyme (E2) and a SUMO-ligase (E3) controls the process of SUMOylation of target proteins. Until today several SUMO-ligases have been identified including PIASxα, PIASxβ, PIAS1, PIAS3, PIASy, RanBP2 (Ran-binding protein 2), Pc2 and HDAC4 (histone deacetylase 4) [reviewed in: 26, 27]. In contrast to ubiquitination, a SUMO-ligase is not obligatory for SUMOylation but some mammalian SUMO-specific E3s stimulate the conjugation reaction, eventually even in a substrate independent manner [28]. The vast majority of SUMO substrates are directly recognized by the SUMO-conjugating enzyme, Ubc9, that binds to the consensus sequence ΨKxE/D (where Ψ is a large hydrophobic amino acid) [29, 30]. However, polymers of both, SUMO-1 and SUMO-2/3, have also been found at lysines that do not comply with the consensus motif [31, 32]. Since the SUMO-conjugating system utilizes only a single enzyme for the recognition and conjugation processes, a further level of regulation must be provided. This is frequently given by an extension of the recognition motif that may contain phosphorylated (PDSM; phosphorylation-dependent SUMOylation motif) or negatively charged (NDSM; negatively charged amino acid-dependent SUMOylation motif) amino acids in addition to the ΨKxE/D motif [33, 34]. Phosphorylation within the PDSM consensus sequence plays an important role in regulating SUMOylation of several substrates including p53, heat shock factors, GATA-1 and the myocyte enhancer factor-2 (MEF2A) [33-35]. Today, four isoforms of SUMO (SUMO-1 to SUMO-4) have been identified in mammals and transcripts of SUMO-1, -2 and -3 can be detected in all human and mouse tissues, whereas SUMO-4 expression seems to be restricted to immune tissues and to the kidney [36, 37]. SUMO-4 was initially identified as an intronless gene, encoding a protein that shares 86% amino acid homology with SUMO-2 [38]. SUMO-4 seems to be unable to form covalent isopeptide bonds with substrate proteins and may control protein functions via non-covalent interactions [39]. SUMO-2 and SUMO-3 are conjugated in a stress inducible manner [reviewed in: 26]. They are very similar on the amino acid level but share only 47% identity to SUMO-1. In contrast to ubiquitin, SUMOylation usually conjugates only a single SUMO molecule to a substrate. Nevertheless, recent reports also implicated the formation of SUMO-chains [31, 40]. Like ubiquitination, SUMOylation can be reversed. SUMO-hydrolyses of the SENP family (sentrin- or SUMO-specific protease) remove SUMO molecules from SUMOylated substrates [reviewed in: 27].
The 81 amino acid polypeptide NEDD8 (neural-precursor-cell-expressed developmentally down-regulated 8) is a relatively uncharacterized member of the family of ubiquitin-like proteins [reviewed in: 41]. Nevertheless, genetic approaches have exposed an obvious role for NEDD8 in cell proliferation, viability and development although the number of known substrates is rather small. Like ubiquitination and SUMOylation, the conjugation of NEDD8 to its substrate is initiated by an activating enzyme, followed by the activity of an E2 enzyme and an E3 ligase [reviewed in: 41]. In line with the other modifications, the process of NEDDylation is very dynamic and NEDD8 can be removed e.g. by the evolutionarily conserved COP9 signalosome [reviewed in: 42].
DNA DAMAGE RECOGNITION AND INITIAL SIGNALLING EVENTS
Once a DSB is generated, the information about this damage needs to be propagated to downstream effector machineries such as repair proteins, cell cycle checkpoints and cell death pathways. Accordingly, recognition of the lesion is the first and initial step in the DNA damage response. In a second step, the information about the lesion can then be transported across the damaged cell. Each specific lesion of the DNA is recognized by one or a set of individual DNA damage recognition factors that are associated with certain signaling components. Double strand breaks are generally recognized by the ataxia-telangiectasia-mutated (ATM) and by the DNA-damage-dependent protein kinase (DNA-PK).
ATM is a serine/threonine protein kinase with a multitude of activities. It signals the presence of DSBs to the cell cycle machinery where it mediates proliferation arrest at G1/S, intra-S and G2/M checkpoints (Fig. 1). The kinase is furthermore required for DNA repair and contributes to the initiation of apoptosis (Fig. 1) [reviewed in: 43]. Mutations in the ATM gene lead to the genetic disorder ataxia-telangiectasia (AT), which is characterized by cerebellar degeneration, immunodeficiency and an increased risk of cancer. Cells from AT patients are hypersensitive to ionizing radiation and display an increased frequency of chromosome breakage and defects in cell cycle control [reviewed in: 43]. ATM is primarily a nuclear protein where it is present as an inactive dimer or multimer (Fig. 2). The kinase is, furthermore, associated with protein phosphatase 2A (PP2A), a serine/threonine phosphatase that associates with ATM via its scaffolding A-subunit [44]. PP2A dephosphorylates ATM constitutively, which keeps the kinase in an inactive state [44]. In addition to PP2A, ATM was found associated with protein phosphatase 5 (PP5). This interaction of ATM with PP5 increases after exposure of cells to DNA damaging agents and appears to be important for ATM activity in response to DSBs [45]. How ATM becomes activated in the presence of DSBs is not entirely solved. A previous model suggested that upon sensing of a DSB, ATM becomes partially active, resulting in trans-autophosphorylation of ATM at serine 367, serine 1893 and serine 1981, and dissociation of ATM dimers/multimers into highly mobile and enzymatically active monomers that associate with damaged chromatin [46, 47]. More recently, this model of ATM activation has been questioned. Jean Gautier and co-workers showed that the MRN complex and DNA are sufficient to facilitate ATM monomerisation [48]. In addition, phosphorylation of serine 1981 appears to be optional for the dissociation of ATM or its function in general [49]. ATM even remained functional when additional autophosphorylation sites (serine 367 and/or serine 1893) were mutated [50]. Besides phosphorylation, ATM also becomes acetylated in response to DSBs. Tip60 (HIV-1 Tat-interacting protein 60 kDa) acetylates ATM on lysine 3016 and this modification is essential for ATM activation and dissociation into monomers [51]. It is, though, unclear by which mechanism Tip60 becomes activated in the presence of DSBs. Also Aven, an interaction partner of the anti-apoptotic protein Bcl-XL, has been reported to contribute to ATM activation [52]. Aven interacts with ATM in cellular extracts and overexpression of Aven promotes ATM phosphorylation on serine 1981 as well as phosphorylation of typical ATM substrates like p53 and Chk2 [52]. However, since Aven is less efficient than DSB in inducing ATM autophosphorylation it appears to act synergistically with DNA damage rather than being a sole activator of the kinase.
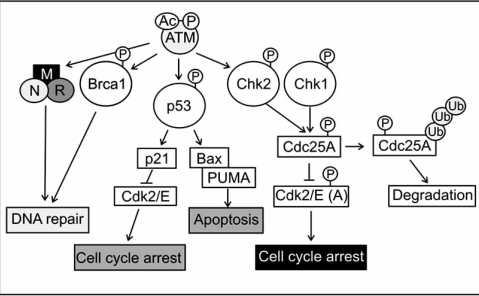
Fig. (1).
ATM and its target proteins. In the active state ATM phosphorylates target proteins that signal to cell cycle checkpoints, DNA repair proteins or to the cell death machinery. Phosphorylation of Brca1 or the activation of the MRN complex results in the initiation of DNA repair. Activation of cell cycle checkpoints that lead to cell cycle arrest are achieved by phosphorylation of the p53 tumor suppressor protein or the Chk2 kinase. In addition, by transcriptional induction of pro-apoptotic proteins like Bax and PUMA, p53 can also induce apoptosis.
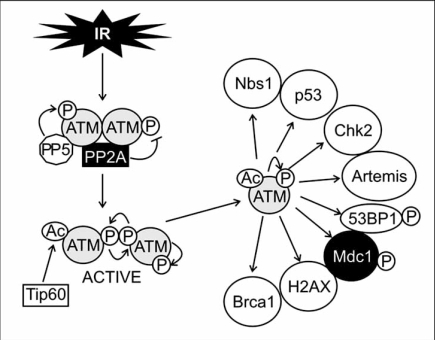
Fig. (2).
ATM activation and downstream signaling. (A) ATM is present in the nucleus as an inactive dimer/multimer that is associated with PP2A, a phosphatase that controls ATM phosphorylation. Upon ionizing radiation (IR), ATM is activated by trans-autophosphorylation and dissociation of the dimer/multimer into monomers as well as by Tip60-mediated acetylation, which enables ATM-mediated phosphorylation of mediator and DNA repair proteins.
Apart from ATM is DNA-PK (DNA-dependent protein kinase) able to recognize DSBs [53]. Like ATM, DNA-PK has a strong affinity to DNA. Its recruitment to sites of DNA damage is conducted by a heterodimer of the Ku70 and Ku80 proteins [54]. The Ku70/80 heterodimer forms a ring-shaped structure that can be threaded onto DNA ends [54]. Due to its high affinity for loose DNA ends and its capacity to bind DNA in a sequence-independent fashion, the Ku70/80 heterodimer is believed to serve as a DSB sensor. Once the Ku70/80 heterodimer is bound to damaged DNA, it serves as a docking site for the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs), which results in the formation of the DNA-PK holocomplex that displays serine/threonine kinase activity towards p53, H2AX, Artemis, XLF and others [55-58]. Formation of the heterodimeric holocomplex is mediated by the amino-terminal and the central region of both, the Ku70 and Ku80 protein. The Ku80 carboxy-terminus is, furthermore, required for DNA-PKcs autophosphorylation at threonine 2609 [59]. This phosphorylated form of DNA-PK co-localizes with γ-H2AX and 53PB1 at the DSB [59].
After recognition of the lesion, an operation platform for repair factors needs to be generated. Since eukaryotic DNA is organized in tightly packed nucleosomes, these structures need to be widened to allow recruitment of repair proteins and to enable their interaction with damaged DNA. For this purpose, different residues of certain histone proteins become post-translationally modified. Especially acetylation, methylation, phosphorylation and ubiquitination of histone proteins have been observed during the DNA damage response [60-65]. The main purpose of acetylation and phosphorylation seems to be to neutralize the positive charge of basic histone proteins. This would reduce the strength of the interaction of basic histone proteins with negatively charged DNA and may decondensate the chromatin. The earliest post-translational modification event at the chromatin as well as for the initiation of the DNA repair process is phosphorylation of the histone H2A variant H2AX at serine 139. The modified H2AX protein, which is called γ-H2AX, appears within a few minutes after exposure of cells to ionizing radiation at the break [64]. Beside H2AX is histone H3 post-translationally modified after a DSB. Within a few minutes after introduction of a DSB, H3 becomes methylated at lysine 79 [66]. Both histone modifications are required for recruitment of mediator proteins such as 53BP1 (p53-binding protein1), MDC1 (mediator of DNA damage checkpoint protein 1) or the MRN (Mre11, Rad50, Nbs1) complex to the DSB [66-69] (Fig. 3).
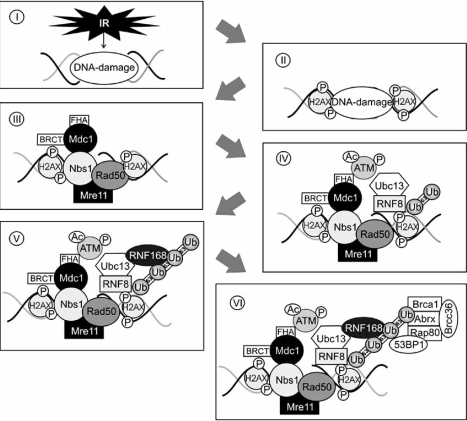
Fig. (3).
DNA lesion recognition and recruitment of repair factors. After introduction of a DNA double strand break (I), the DNA damage response is initiated by phosphorylation of H2AX (II), which is required for the recruitment of mediator proteins such as MDC1 or the MRN complex. MDC1 attaches to phosphorylated H2AX via its BRCT domain and stabilizes the MRN complex (III). Additionally it is responsible for binding and accumulation of ATM at the DSB, which creates a docking site for RNF8/Ubc13 that also associates with phosphorylated H2AX and decorates histone proteins with lysine 63-linked ubiquitin molecules (IV). RNF168 recognizes the initial ubiquitin chains generated by RNF8 and, in association with Ubc13, extends and propagates lysine 63-linked ubiquitination (V) that is required for the accumulation of additional repair factors like Bcra1 or 53BP1 (VI).
The MRN complex, a highly conserved protein complex consisting of Mre11, Rad50 and Nbs1 (Nijmegen breakage syndrome 1), is implicated in DNA repair, cell cycle checkpoint control, DNA replication and telomere maintenance [70-72]. The three proteins, Mre11, Rad50 and Nbs1, form a tight complex which is homogenously distributed throughout the nuclei of mammalian cells but migrates rapidly to DSBs after ionizing radiation, independent of the phase of the cell cycle [73]. The Nbs1 protein appears to be responsible for nuclear localization and the proper assembly of the complex at DSB sites, probably by interacting with phosphorylated H2AX [68]. Mre11 displays endo- and exonuclease activity and is important for the processing of the broken DNA ends [74]. Rad50 also shows DNA binding capacity that possibly plays a role in tethering sister chromatids during HR [75, 76]. The MRN complex is composed of a single Nbs1 molecule that is associated with two dimers of Mre11 and Rad50. The Mre11 and Rad50 proteins themselves form a heterotetramer consisting of a dimer of Mre11 and Rad50. This heterotetramer contains two DNA-binding and processing domains that can bridge free DNA ends [76, 77]. The Mre11 dimer binds and holds the dsDNA ends in close proximity, near the Mre11 active site and it is thought that it aligns and bridges these loose DNA ends during DNA repair [78, 79]. Rad50 contains two long coiled-coil arms that promote intermolecular interactions through a terminal hook domain [76, 77]. Loss of Nbs1 or Mre11 is embryonic lethal [80, 81] and mutations in NBS1 and MRE11 lead to the chromosomal instability disorders Njimegen breakage syndrome (mutation in NBS) and ataxia telangiectasia-like disorder (ATLD; mutation in Mre11). Both of these syndromes are associated with enhanced sensitivity to ionizing radiation and chromosomal instability [reviewed in: 82, 83]. MRN binds to DSBs and leads to further activation of ATM [48, 70]. Accordingly, cells derived from patients with Nijmegen breakage syndrome or ataxia telangiectasia-like disorders exhibit decreased ATM kinase activity despite the presence of wild type ATM [70, 84-86]. The nuclease activity of Mre11 appears to be particularly important for this process [70, 87]. Despite the similar timing of the appearance of Ku70/80 and the MRN complex at DSBs, recruitment of MRN and Ku seems to be independent of each other [88]. Nbs1 and Mre11 are both targets for ATM and at least phosphorylation of Nbs1 is required for checkpoint signaling during S-phase [89]. Therefore the MRN complex acts on the one hand as a downstream mediator of ATM but is also important for the activation of ATM and phosphorylation of downstream substrates.
MDC1, also called NFBD1 (nuclear factor with BRCT domains protein 1), appears to be a master regulator of the microenvironment at damaged chromatin. The docking of MDC1 to DSBs allows retention of multiple checkpoint and adaptor proteins including Nbs1, 53BP1 or Brca1 (breast cancer 1) at the site of the lesion where they provide a molecular platform for efficient amplification of the DNA damage signal [68, 69, 90]. For this, MDC possesses two classes of phospho-binding motifs, a FHA (forkhead-associated) and a BRCT (Brca1 carboxy-terminal repeat) domain that serve as binding partners for phosphorylated proteins. BRCT and FHA modules are conserved throughout different species and are present in many proteins that are involved in the cellular response to DNA damage [reviewed in: 91, 92]. Two repeats of the BRCT domain are located at the carboxy-terminus of MDC1. These BRCT domains associate directly with phosphorylated H2AX and this interaction seems to be essential for the recruitment and docking of MDC1 to the site of the DNA lesion (Fig. 3). Accordingly, in H2AX-/- MEFs, MDC1 fails to accumulate at DSBs [90]. The FHA domain is positioned at the amino-terminus and binds to ATM which results in the accumulation of ATM at DSBs and enhanced phosphorylation of H2AX and of MDC1 itself [93]. In MDC1-deficient cells, recruitment of ATM to DSBs is impaired [93]. The central domain of MDC1 contains 14 repeats of a sequence that mediate its interaction with DNA-PKcs and the Ku heterodimer [94]. Once recruited to DSBs, MDC1 stabilizes the MRN complex that is bound to damaged DNA and acts as a molecular scaffold for the recruitment of 53BP1, BRCA1 and additional MRN molecules to nuclear foci [68, 90, 95]. Upon binding to MDC1, ATM phosphorylates the mediator protein and this phosphorylation creates a docking site for the recently identified E3-ligase RNF8 (ring finger 8) and for Ubc13 (ubiquitin-conjugating 13; Fig. 3) [22, 96]. These proteins associate with γ-H2AX and phosphorylated MDC1 via their FHA domains and decorate the histone protein H2A and its variant H2AX with lysine 63-linked polyubiquitin chains [22, 96]. Nevertheless, although RNF8 seems to be the first E3 ligase at the DSB, it appears to be insufficient for sustained ubiquitination of the chromatin. It probably rather primes the chromatin around the DSB, which then facilitates the recruitment of another E3 ligase, the RNF168 protein (Fig. 3) [97]. RNF168 recognizes the initial ubiquitin chains generated by RNF8 via its ubiquitin-binding domain and, in concert with Ubc13, propagates and extends the formation of lysine 63-linked ubiquitin chains of histone proteins. Eventually, a threshold of lysine 63-polyubiquitin chains may be required to recruit and hold additional repair factors at the DSB. Particularly recruitment of 53BP1 and Brca1 are assumed to depend on such an “interaction trap” made of polyubiquitinated lysine 63 chains (Fig. 3) [97]. After polyubiquitination of the chromatin, Brca1, a tumor suppressor protein that also possesses ubiquitin ligase activity, especially when it is complexed with Bard1, is recruited to the lesion together with Rap80, Abraxas, Brca1 and Brcc36, which form the BRCA1-A complex [98]. Brca1 can be found in three different complexes (Brca1 A, B and C), which depend on different adaptor proteins. The adaptor proteins for the complex A, B and C are Abraxas, Bach1/Brip1 (BRCA1-associated C-terminal helicase) and CtIP (CtBP-interacting protein), respectively. Each complex forms in a mutually exclusive manner [98-100]. Whereas complex A plays an important role in DNA damage repair, the Brca1/Bach1 and the Brca1/CtIP complex form at different stages of the cell cycle and are required for a prolonged G2 phase and for G2/M transition checkpoint control, respectively [99, 100]. Ionizing radiation leads to ATM-dependent phosphorylation of Rap80 (receptor-associated protein 80) at serine 140, serine 402 and serine 419 [22, 98]. RAP80, furthermore, possesses two ubiquitin interaction motifs (UIM) which recognize lysine 63-linked ubiquitin chains. These UIMs facilitate the association of the complexes with polyubiquitinated histone proteins [22, 97]. Brcc36 of the complex has ubiquitin hydrolyzing activity and plays an important role in the regulation of the E3 ligase activity of Brca1 in response to ionizing radiation [101, 102]. The Rap80/Brcc36 complex furthermore removes lysine 63-linked polyubiquitin chains from the chromatin and is thus also involved in the termination of RNF8-Ubc13-mediated polyubiquitination once the lesion has been repaired [103]. Although primarily implicated in HR, there is accumulating evidence that Brca1 may also be involved in NHEJ. The N-terminal region of Brca1 specifically associates with Ku80 and rapidly accumulates at DSBs in a Ku-dependent manner. Brca1 is furthermore assumed to control the fidelity of NHEJ [104, 105].
53BP1 functions, together with MDC1 and Brca1, as an adaptor/mediator protein of the DNA damage response. As such, it contributes to the activation of downstream effector molecules that function in DNA repair and DNA damage signaling, although it has no enzymatic activity. 53BP1 was originally identified due to its ability to bind to p53 [106]. Upon ionizing radiation, it accumulates in foci at DSBs and becomes phosphorylated at several sites in an ATM-dependent manner. Among these is phosphorylation of serine 1219 particularly important as phosphorylation of this site assists in recruitment of MDC1 and γ-H2AX [107]. 53BP1 is furthermore required for p53 accumulation, G2/M and intra-S-phase arrest, ATM autophosphorylation at serine 1981 and for the formation of Brca1-containing foci in response to ionizing radiation [108, 109].
DOUBLE STRAND BREAK REPAIR AND SIGNAL TRANSDUCTION
After the generation of a protein interaction platform, repair factors are recruited to the DSB that ligate the broken DNA ends. In mammalian cells DSBs are usually repaired by non-homologous endjoining (NHEJ) and homologous recombination (HR) (Fig. 4). In dependence on the phase of the cell cycle the one or the other mechanism dominates. Nevertheless, while it was previously suggested that during the G1 and early S phase of the cell cycle, NHEJ is the pathway of choice and HR would only be operative in late S and G2, it becomes more and more evident that at least NHEJ is active in all phases of the cell cycle [110, 111]. This is, for example, evidenced by the appearance of Ku70/80, DNA-PKcs and ATP-dependent DNA ligase IV (LigIV) at DSBs not only in G1 but also in S and G2 phase. Nevertheless, although proteins of both repair pathways appear at the same DSB, assembly of the NHEJ machinery clearly precedes that of HR factors, even in the S and G2 phase of the cell cycle [88, 112, 113]. Moreover, the fact that factors of both repair machineries are present at DSBs at the same time suggests that NHEJ and HR are not two competing parallel pathways but rather that NHEJ may serve as an immediate early repair event whereas HR may repair persisting DNA lesions [88].
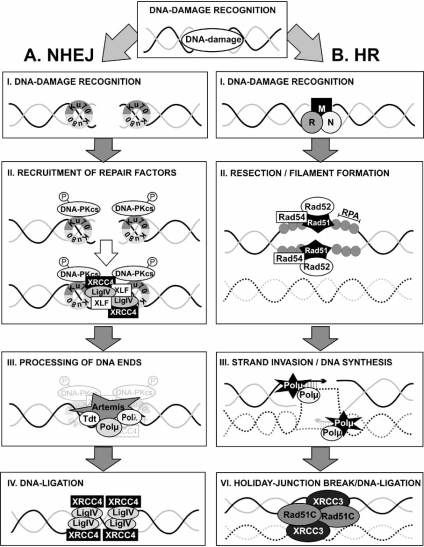
Fig. (4).
Mechanisms of DNA-repair by NHEJ and HR. Double strand breaks are repaired by two major pathways: non-homologous endjoining (A, NHEJ) and homologous recombination (B, HR). NHEJ: Loose DNA-ends are recognized by the Ku70/80 heterodimeric complex (A.I), which is needed for the recruitment of the additional repair factors DNA-PKcs, XRCC4, LigIV and XLF (A.II). Before broken DNA ends can be ligated, the ends are processed by Pol µ and λ, Artemis and Tdt (A.III). This step is followed by the ligation of the loose ends by LigIV and XRCC4 (A.IV). HR: HR is initiated by the MRN complex (B.I), followed by the recruitment of RPA that associates with single stranded DNA, and by Rad51/52 which are needed for filament formation and strand invasion (B.II). New DNA is synthesized while the homologous sister chromatid serves as a template (B.III). After branch migration, the holiday junction is resolved (B.IV).
Non-Homologous Endjoining (NHEJ)
NHEJ is the repair pathway of choice of non-dividing and of mitotic cells during G1 and early S-phase of the cell cycle. Since most cells of higher organisms are in a quiescent state, NHEJ is considered to be the major DSB repair pathway in mammals. The process of NHEJ is characterized by a “simple” ligation of broken ends without the requirement for larger sequence homologies. However, since the creation of ligatable DNA ends usually comes along with loss of genetic information, it is also an error-prone DNA repair mechanism.
The mechanism of NHEJ covers mainly four consecutive phases (i) recognition of the DNA lesion, (ii) sequential recruitment of repair factors, (iii) processing of DNA ends to yield ligatable termini and (iv) sealing of the break. To date seven proteins have been described to be required for NHEJ: the Ku70/Ku80 heterodimeric complex, DNA-PKcs, LigIV, XRCC4 (X-ray repair complementing group 4 protein), XLF (XRCC4-like factor) and Artemis (Fig. 4) [114-121]. If a cell is defective in one of these enzymes, neither DNA repair nor V(D)J recombination, a cut-and-paste mechanism that uses NHEJ to seal DSBs that arise during the generating of the diversity of antigen receptors in immune cells [reviewed in: 122], can be carried out. This results in enhanced sensitivity towards ionizing radiation and in immunodeficiency [114, 116, 120, 121, 123].
The Ku70/80 heterodimer serves as a recognition and first docking molecule at DSBs and it most likely also protects DNA ends from nucleolytic degradation (Fig. 4) [114, 124, 125]. In vivo studies with Ku-deficient yeast strains demonstrated that Ku is important for efficient NHEJ and for accurate rejoining of complementary and non-complementary DNA ends [114]. The recruitment of Ku70/80 is followed by the arrival of DNA-PKcs at the DSB and its autophosphorylation (Fig. 4). Mutation of the autophosphorylation site of DNA-PKcs impairs rejoining of broken DNA [59]. After the arrival of DNA-PK, the DNA at the break is processed. DSBs frequently lead to damaged bases or incompatible single strand overhangs at the break, which are unsuitable for direct ligation. Several enzymes with single-strand filling capacity or nucleolytic trimming potency have been suggested to play a role in the processing of the DNA, like polymerase λ, polymerase μ, Tdt (terminal deoxynucleotidyltransferase) and Artemis (Fig. 4). These enzymes add or remove nucleotides or extend the 3´-single stranded DNA tail in a template-independent manner. The DNA polymerases λ and μ are responsible for refilling the gaps that result from NHEJ intermediates while Tdt adds nucleotides to loose ends during V(D)J recombination [126-128]. Artemis, on the other hand, displays an intrinsic 5´-3´exonuclease activity in vitro and assists in the repair of a subset of DSBs [129]. Upon forming a complex with DNA-PKcs, Artemis becomes phosphorylated, which facilitates cleavage of 5´and 3´overhangs, nicks and hairpin structures at the loose end of the DNA [129]. Artemis is furthermore important for the opening of hairpin structures during V(D)J recombination [129].
LigIV is a core component of the NHEJ repair pathway and catalyzes the ligation of the DNA ends (Fig. 4). Structural studies have revealed that LigIV exists in a tight complex with XRCC4 that is accomplished by the central region of XRCC4 and that strongly enhances LigIV stability [117, 130, 131]. XRCC4 is a nuclear protein that is constitutively phosphorylated and its phosphorylation is further enhanced in response to ionizing radiation in a DNA-PK-dependent manner [132]. In addition to phosphorylation, a monoubiquitinated form of XRCC4 has been identified. The abundance of this modified form is enhanced when cells have been treated with etoposide, a topoisomerase inhibitor that generates DSBs [133]. The precise role of this ubiquitination of XRCC4 has, though, not been determined. In addition to phosphorylation and ubiquitination, XRCC4 is modified with SUMO at lysine 210, a modification that appears to be required for nuclear localization of the protein [134].
More recently XLF (XRCC4-like factor), also called Cernunnos, was identified as a new interaction partner of XRCC4 and an additional member of the NHEJ machinery [118, 119]. XLF forms homodimers with a XRCC4-like structure and modulates XRCC4/LigIV activity through direct interaction with XRCC4 [135, 136]. XLF is quickly recruited to DSBs in a Ku-dependent and XRCC4-independent manner [137]. Although XLF is phosphorylated by DNA-PK at serine 245 and by ATM at serine 251, no particular function could be attributed to these modifications [58].
Once repair is completed, the repair machinery needs to be disassembled, but how this is achieved has not yet been understood in detail. Only for the Ku70/80 proteins some insights into this process have been provided. Recently it was demonstrated that Ku80 bound to DSB is heavily modified with polyubiquitin chains. This modification appears to mediate efficient removal of Ku80 from repaired DNA [138]. Most interestingly, binding of Ku to DNA appears to initiate its polyubiquitination, which then allows its removal from the DNA once the lesion is fixed [138].
While the classical NHEJ pathway uses the XRCC4/LigIV complex to ligate broken DNA ends, the immune system can make use of an alternative NHEJ pathway that is XRCC4-independent [139]. This alternative NHEJ mechanism deletes sequences at the loose ends until short stretches of homology appear at the repair junction. These areas of microhomology assist in ligation in a process called microhomology-mediated endjoining (MMEJ). It is most likely that the enzymatic and scaffolding function of Mre11 participates in this process [140]. Whether this alternative route of NHEJ also contributes to the sealing of DSBs after ionizing radiation, e.g. when XRCC4 is absent, remains to be determined.
Homologous Recombination (HR)
HR is particularly activated during late S- and G2-phases of the cell cycle as this repair pathway requires sequence homologies of about 100 base pairs and more in close proximity to the damaged site. These conditions are normally only provided between DNA replication and cell division. Since the undamaged sister chromatid is used as a template, this type of repair is generally accurate. As HR requires the alignment of broken DNA ends with a homologous region of the genome, homology search and DNA strand invasion is a central component of HR. After introduction of a DSB, HR is generally initiated by nucleolytic resection of the broken DNA which results in long single stranded tails with 3´-hydroxyl overhangs. It is most likely that the Rad50/Mre11 complex contributes to this activity [78]. The resulting single stranded DNA (ssDNA) is bound by the replication protein A (RPA) (Fig. 4) [141]. RPA is a heterotrimeric, eukaryotic protein that binds to ssDNA in a sequence independent manner thus forming a nucleoprotein filament [reviewed in: 142]. Apart from protecting the ssDNA from nucleolytic attacks, RPA eliminates secondary structures, which allows more Rad51 molecules to access and bind ssDNA, thus stimulating the formation of a presynaptic complex of DNA, RPA and Rad51. RPA can have positive and negative effects on Rad51-mediated strand exchange. Pre-incubation of ssDNA with RPA protein prevents binding of Rad51 to the DNA but this inhibitory effect can be diminished by Rad52 or Rad55/57. The Rad55/57 complex interacts transiently with Rad51 which results in the abolishment of the RPA inhibitory effect and support of the formation of the Rad51 filament, similar to Rad52 [143]. In contrast, when RPA is present after the Rad51/DNA complex has formed, it is able to stimulate strand exchange via Rad51, possibly by removing secondary structures of the ssDNA, and to promote appropriate Rad51 filament formation [141, 144].
The main activity of Rad51 is to promote ATP-dependent strand exchange, catalyzed by an intrinsic ssDNA-dependent ATPase activity [141, 145]. Accordingly, yeast cells lacking Rad51 are hypersensitive to ionizing radiation and alkylating agents [141, 146]. In humans, five Rad51 paralogs exist named Rad51B, Rad51C, Rad51D, XRCC2 and XRCC3 [reviewed in: 147]. The mammalian counterpart of Rad52 is Brca2, a protein frequently mutated in breast and ovarian cancer. Similar to the yeast counterpart, Brca2 controls the activity of the recombinase, Rad51, and stimulates the loading of Rad51 onto ssDNA [148-150]. Once Rad51 has been recruited to the nucleoproteinfilament, it replaces RPA. The 3´-overhangs of the DNA associate with Rad52 and subsequently with polymerized Rad51 and form the so-called recombinase filament where Rad51 and Rad52 are wrapped around the ssDNA overhang. This interaction is stabilized by Rad54 that associates with the recombinase filament and enhances their catalytic activity [151]. Once this complex is complete, the tail of Rad52 starts to search for a homologous sequence in the sister chromatid and facilitates base pairing with the single-stranded overhang to form a D-loop intermediate. This homology search is furthermore assisted by Rad51 which, together with the ssDNA, invades into the intact homologous chromosome to form a heteroduplex. Rad54 appears to help in this process, although its precise function remains to be determined [152]. After formation of the heteroduplex, DNA polymerase µ and λ extend the 3´end of the invading strand in a process called branch migration (Fig. 4) [153]. Due to strand invasion, a joint molecule with crossed-over DNA strands is generated. To finish the repair process, these holiday junctions need to be resolved in order to reconstruct the two individual DNA double helices. The resolution of the holiday junction is performed by the resolvases Rad51C and XRCC3 (Fig. 4) [153-156].
CELL CYCLE ARREST AND APOPTOSIS
In mammalian cells, evolutionary conserved pathways exist that signal information about the state of the genome to a series of checkpoints, which can delay the progression into the next phase of the cell cycle. All these checkpoints are tightly regulated. In response to DNA damage, errors in DNA replication or chromosome segregation, cells can activate these checkpoints to allow time to fix the lesion before the cell reinitiates replicative DNA synthesis or begins mitosis. The most important checkpoints within the cellular response to DNA damage are the G1/S and the G2/M checkpoints. In addition, an intra-S-phase checkpoint exists [reviewed in: 157].
After sensing of the DSB, the information about the lesion is rapidly transduced to the checkpoints of the cell cycle. ATM and Chk2 (cell cycle checkpoint kinase 2) kinases play an important role in this process [158-160]. Chk2 is a stable protein which is expressed throughout the cell cycle. In the absence of DNA damage Chk2 is largely inactive. When DNA damage occurs, the kinase becomes rapidly phosphorylated at threonine 68 by ATM [158]. This phosphorylation stimulates Chk2 homodimerisation and activates the kinase, which results in autophosphorylation at threonine 383, threonine 387 and serine 516, and phosphorylation of Chk2 target proteins [159-161]. One of the targets of Chk2 is Cdc25A, a phosphatase that dephosphorylates and activates cyclin-dependent kinases (Cdks) (Fig. 1) [162]. These kinases drive the cell trough the cell cycle. Phosphorylation of Cdc25 by Chk2 in response to DSBs leads to rapid proteosomal degradation of the phosphatase and thus to inactivation of cdk2/cyclin E/A complexes as well as of cdk2/cyclin B/A complexes, which result in cell cycle arrest at the G1/S and G2M checkpoint, respectively [162, 163].
An additional important component of checkpoint activation in response to DNA damage is the p53 tumor suppressor protein (Fig. 1). Most characteristic for p53 are its proliferation-inhibiting and pro-apoptotic activities. Many, but not all of these activities are caused by its function as a transcription factor. Under normal growth conditions, the amount of the p53 protein is maintained low and the protein is transcriptionally inactive. Both parameters are mainly controlled by the E3 ubiquitin ligase Mdm2, although other proteins may also contribute to this process [reviewed in: Boehme and Blattner, in press]. Mdm2 binds to the N-terminal transactivation domain of p53 and mediates its ubiquitination and proteosomal degradation [164]. Upon DNA damage, p53 is protected from degradation and post-translationally modified at several sites in its amino- and carboxy-terminal part, which enhances its sequence-specific binding to promoters of target genes and activation of gene transcription [12, 165-168]. Among the target genes of p53 are bax, puma and p21, while transcription of other genes, such as bcl-2 are repressed [169-171, reviewed in: 172]. p21, also known as p21WAF1/Cip1 is a member of the Cip/Kip family of cyclin-dependent kinase inhibitors [reviewed in: 173]. Cyclin dependent kinases 2, 4 and 6 (Cdk2, Cdk4, Cdk6) and the associated cyclins, control progression through the cell cycle by phosphorylation of their target proteins [174]. An important substrate of theses cyclin/Cdk complexes is the retinoblastoma protein (Rb), a transcriptional repressor of the E2F family of transcription factors. E2F proteins control entry into and progression through the S-phase of the cell cycle. In its hyperphosphorylated state, Rb binds to E2F family members and prevents them from activating their target genes, which, at the same time, precludes entry into the S-phase of the cell cycle [175, 176]. In unstressed cells, Rb becomes phosphorylated at the end of S-phase by cyclin D1/Cdk4,6, which leads to the dissociation of E2F and Rb and transcription of E2F target genes [177]. Upon exposure to genotoxic agents, the Cdk-inhibitor p21 is induced and abolishes Cdk-mediated phosphorylation of Rb, which results in hypophosphorylation of Rb, sequestration of the E2F protein and cell cycle arrest at the G1/S boundary of the cell cycle [171].
If the damage of the DNA is too severe, programmed cell death (apoptosis) can be initiated to eliminate potential hazardous cells, which bear the risk of uncontrollable behavior or transmission of mutations to daughter cells. Apoptosis is a form of cell death that is characterized by specific morphological changes of the cell like overall shrinkage, chromatin condensation and fragmentation of genomic DNA [178]. Apoptosis can be initiated by different extracellular and intracellular factors like UV-light or ionizing radiation, chemotherapeutic drugs, by activation of death receptors or by reactive oxygen species. The apoptotic program can be started by two main pathways, an extrinsic one that is initiated by binding of ligands to membrane-bound death receptors and activation of caspase 8, and by an intrinsic one, that is initiated by mitochondrial membrane permeabilisation, release of cytochrome c from the intermembraneous space and activation of procaspase 9. Both pathways lead to activation of caspase 3, which initiates controlled degradation of the cell (Fig. 5) [reviewed in: 179].
Fig. (5).
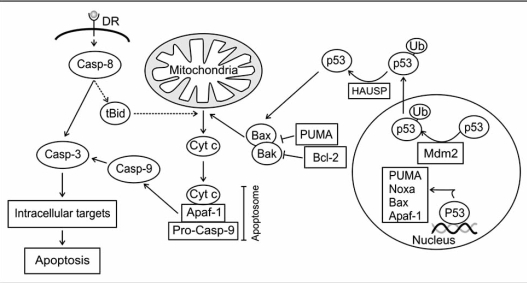
Apoptosis signaling pathways. Apoptosis can be induced upon activation of death receptors (DR) at the membrane by binding of cognate ligands or after release of pro-apoptotic factors from mitochondria. Upon DR activation, the initiator-caspase-8 becomes activated, which activates a caspase-cascade, and cleaves the pro-apoptotic proteins Bid. The cleavage of Bid results in the formation of active tBid, which, in cooperation with Bax and Bak, forms pores in the outer mitochondrial membrane resulting in the release of cytochrome c, formation of the apoptosome and activation of caspase 9. Caspases 9 activates caspase 3 leading to the degradation of intracellular targets. The proapoptotic proteins Bax and Bak can be inhibited by Bcl-2 and Puma. Besides its nuclear function p53 can become monoubiquitinated in a Mdm2-dependent manner, which leads to the translocation of p53 to mitochondria where it participates in mitochondrial membrane permeabilization and cytochrome c release.
Mitochondrial membrane permeabilisation is controlled by a number of pro- and anti-apoptotic proteins, most of which belong to the Bcl-2 superfamily. Members of this family fall into three groups and all either stimulate or inhibit programmed cell death. Bcl-2, Bcl-XL and Mcl1 belong to the group of anti-apoptotic proteins while Bax and Bak are pro-apoptotic proteins. The third group is composed of the BH3-only proteins, which can be subdivided into “activators” and “de-repressors”. The activators (e.g. tBid) can directly interact with the pro-apoptotic effector proteins Bax and Bak and induce their oligomerization which leads to the formation of pores in the mitochondrial membrane. The de-repressors (e.g. PUMA) act in a more indirect way on the activation of Bax and Bak as they bind to anti-apoptotic proteins that guard Bax and Bak and force their release from the inactivating complex [reviewed in: 180].
p53 initiates apoptosis in response to DNA damage primarily through transcriptional activation of genes encoding apoptotic factors such as Bax, PUMA or Noxa, which neutralize anti-apoptotic proteins from the IAP family [169, 170, 181]. However, a significant portion of the tumor suppressor has also been found in the cytoplasm, where it is also prevented from degradation and contributes to the initiation of cell death. In response to DNA damage, cytoplasmic p53 induces the oligomerization of Bax, Bak, VDAC and cyclophilin D, which triggers the permeabilisation of the mitochondrial membrane, resulting in the release of the pro-apoptotic molecules cytochrome c, smac and AIF [182, 183]. The mechanism how p53 translocates to the mitochondria is not fully understood. Neither phosphorylation nor acetylation appears to contribute to this process. However, it has been suggested that this process may be triggered by Mdm2-mediated monoubiquitination of p53 [184]. Once p53 has arrived at the mitochondrial membrane it is rapidly deubiquitinated by the local mitochondrial HAUSP protein via a stress-induced mitochondrial p53-HAUSP complex, creating the apoptotically active, non-ubiquitinated p53 protein [184].
CONCLUSIONS
The whole process of DNA repair, starting with the sensing and recognition of a DNA strand break and finally resulting in the ligation of the loose ends, is accomplished by a very complex network of interacting proteins that is only incompletely understood. Efforts of more recent years show that a bunch of different players are involved in the DNA damage response. Moreover, there is more and more evidence that many of the sensing, signaling and repair factors interact with each other. Other repair factors even contribute to several different signaling and repair pathways. Examples of such multitalented factors are the MRN complex or ATM, but also Brca1, H2AX, PARP-1, RAD18 or DNA-PKcs. But even though this complex network began to become clearer in more recent years, a lot of open questions are left, including the exact sensing mechanism of DNA damage and the identity of all the proteins involved in this process, the communication of the factors with each other and the basics of the choice between different repair pathways.
Defects in the key players of the DNA damage response can result in a very severe outcome like genomic instability and predisposition for cancers. However, while DNA repair is of utmost importance for healthy individuals, it can be counterproductive during cancer therapy, which frequently uses the implementation of DNA damage to kill tumors by apoptosis. Therefore, attempts are currently under way to inhibit DNA repair during cancer therapy by specifically targeting key proteins of the repair machinery. Along this line, several inhibitors targeting DNA-PK or ATM have been synthesized, which are currently tested for their application in humans. A detailed understanding of the DNA repair networks would help to identify further targets and greatly support the development of new drugs for the treatment of cancer.
ACKNOWLEDGEMENTS
We apologize for having omitted several references due to space limitations. This work was supported by the BMBF, grant 02S8477.
REFERENCES
- 1.Hoeijmakers JH. DNA repair mechanisms. Maturitas. 2001;38:17–22. doi: 10.1016/s0378-5122(00)00188-2. [DOI] [PubMed] [Google Scholar]
- 2.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 3.Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem. Soc. Trans. 2006;34:523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- 4.Jolly CJ, Cook AJ, Manis JP. Fixing DNA breaks during class switch recombination. J. Exp. Med. 2008;205:509–513. doi: 10.1084/jem.20080356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fortini P, Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Nouspikel T. DNA repair in mammalian cells: Nucleotide excision repair: variations on versatility. Cell Mol. Life Sci. 2009;66:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech. Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salazar C, Hofer T. Multisite protein phosphorylation--from molecular mechanisms to kinetic models. FEBS J. 2009;276:3177–3198. doi: 10.1111/j.1742-4658.2009.07027.x. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paik WK, Paik DC, Kim S. Historical review: the field of protein methylation. Trends Biochem. Sci. 2007;32:146–152. doi: 10.1016/j.tibs.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 13.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 14.Hershko A. Ubiquitin-mediated protein degradation. J. Biol. Chem. 1988;263:15237–15240. [PubMed] [Google Scholar]
- 15.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 16.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ande SR, Chen J, Maddika S. The ubiquitin pathway: An emerging drug target in cancer therapy. Eur. J. Pharmacol. 2009;625(1-3):199–205. doi: 10.1016/j.ejphar.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 18.Haglund K, Stenmark H. Working out coupled monoubiquitination. Nat. Cell Biol. 2006;8:1218–1219. doi: 10.1038/ncb1106-1218. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrmann J, Lerman LO, Lerman A. Ubiquitin and ubiquitin-like proteins in protein regulation. Circ. Res. 2007;100:1276–1291. doi: 10.1161/01.RES.0000264500.11888.f0. [DOI] [PubMed] [Google Scholar]
- 21.Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-e A, Tanaka K. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009;28:359–371. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. USA. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chastagner P, Israel A, Brou C. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 2006;7:1147–1153. doi: 10.1038/sj.embor.7400822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 25.Wickliffe K, Williamson A, Jin L, Rape M. The multiple layers of ubiquitin-dependent cell cycle control. Chem. Rev. 2009;109:1537–1548. doi: 10.1021/cr800414e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hay RT. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Guo B, Yang SH, Witty J, Sharrocks AD. Signalling pathways and the regulation of SUMO modification. Biochem. Soc. Trans. 2007;35:1414–1418. doi: 10.1042/BST0351414. [DOI] [PubMed] [Google Scholar]
- 28.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and Ran-GAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 31.Pedrioli PG, Raught B, Zhang XD, Rogers R, Aitchison J, Matunis M, Aebersold R. Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software. Nat. Methods. 2006;3:533–539. doi: 10.1038/nmeth891. [DOI] [PubMed] [Google Scholar]
- 32.Cooper HJ, Tatham MH, Jaffray E, Heath JK, Lam TT, Marshall AG, Hay RT. Fourier transform ion cyclotron resonance mass spectrometry for the analysis of small ubiquitin-like modifier (SUMO) modification: identification of lysines in RanBP2 and SUMO targeted for modification during the E3 auto-SUMOylation reaction. Anal. Chem. 2005;77:6310–6319. doi: 10.1021/ac058019d. [DOI] [PubMed] [Google Scholar]
- 33.Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin JY, Ohshima T, Shimotohno K. Association of Ubc9, an E2 ligase for SUMO conjugation, with p53 is regulated by phosphorylation of p53. FEBS Lett. 2004;573:15–18. doi: 10.1016/j.febslet.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 36.Melchior F. SUMO--nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 37.Guo D, Li M, Zhang Y, Yang P, Eckenrode S, Hopkins D, Zheng W, Purohit S, Podolsky RH, Muir A, Wang J, Dong Z, Brusko T, Atkinson M, Pozzilli P, Zeidler A, Raffel LJ, Jacob CO, Park Y, Serrano-Rios M, Larrad MT, Zhang Z, Garchon HJ, Bach JF, Rotter JI, She JX, Wang CY. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 2004;36:837–841. doi: 10.1038/ng1391. [DOI] [PubMed] [Google Scholar]
- 38.Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 2004;279:27233–27238. doi: 10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- 39.Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 2005;337:517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 40.Yang M, Hsu CT, Ting CY, Liu LF, Hwang J. Assembly of a polymeric chain of SUMO1 on human topoisomerase I in vitro. J. Biol. Chem. 2006;281:8264–8274. doi: 10.1074/jbc.M510364200. [DOI] [PubMed] [Google Scholar]
- 41.Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem. Soc. Trans. 2008;36:802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- 42.Chamovitz DA. Revisiting the COP9 signalosome as a transcriptional regulator. EMBO Rep. 2009;10:352–358. doi: 10.1038/embor.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 44.Goodarzi AA, Jonnalagadda JC, Douglas P, Young D, Ye R, Moorhead GB, Lees-Miller SP, Khanna KK. Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. EMBO J. 2004;23:4451–4461. doi: 10.1038/sj.emboj.7600455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali A, Zhang J, Bao S, Liu I, Otterness D, Dean NM, Abraham RT, Wang XF. Requirement of protein phosphatase 5 in DNA-damage-induced ATM activation. Genes Dev. 2004;18:249–254. doi: 10.1101/gad.1176004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 47.Kozlov SV, Graham ME, Peng C, Chen P, Robinson PJ, Lavin MF. Involvement of novel autophosphorylation sites in ATM activation. EMBO J. 2006;25:3504–3514. doi: 10.1038/sj.emboj.7601231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dupre A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat. Struct. Mol. Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- 49.Pellegrini M, Celeste A, Difilippantonio S, Guo R, Wang W, Feigenbaum L, Nussenzweig A. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature. 2006;443:222–225. doi: 10.1038/nature05112. [DOI] [PubMed] [Google Scholar]
- 50.Daniel JA, Pellegrini M, Lee JH, Paull TT, Feigenbaum L, Nussenzweig A. Multiple autophosphorylation sites are dispensable for murine ATM activation in vivo. J. Cell Biol. 2008;183:777–783. doi: 10.1083/jcb.200805154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol. Cell Biol. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo JY, Yamada A, Kajino T, Wu JQ, Tang W, Freel CD, Feng J, Chau BN, Wang MZ, Margolis SS, Yoo HY, Wang XF, Dunphy WG, Irusta PM, Hardwick JM, Kornbluth S. Aven-dependent activation of ATM following DNA damage. Curr. Biol. 2008;18:933–942. doi: 10.1016/j.cub.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 54.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 55.Sheppard HM, Liu X. Phosphorylation by DNAPK inhibits the DNA-binding function of p53/T antigen complex in vitro. Anticancer Res. 1999;19:2079–2083. [PubMed] [Google Scholar]
- 56.Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 57.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, Lees-Miller SP. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu Y, Mahaney BL, Yano K, Ye R, Fang S, Douglas P, Chen DJ, Lees-Miller SP. DNA-PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks. DNA Repair (Amst) 2008;7:1680–1692. doi: 10.1016/j.dnarep.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maas NL, Miller KM, DeFazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell. 2006;23:109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 61.Qin S, Parthun MR. Recruitment of the type B histone acetyl-transferase Hat1p to chromatin is linked to DNA double-strand breaks. Mol. Cell Biol. 2006;26:3649–3658. doi: 10.1128/MCB.26.9.3649-3658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 64.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 65.Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 66.Huyen Y, Zgheib O, Ditullio RA, Jr., Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 67.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 68.Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 70.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bressan DA, Baxter BK, Petrini JH. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seno JD, Dynlacht JR. Intracellular redistribution and modification of proteins of the Mre11/Rad50/Nbs1 DNA repair complex following irradiation and heat-shock. J. Cell Physiol. 2004;199:157–170. doi: 10.1002/jcp.10475. [DOI] [PubMed] [Google Scholar]
- 74.Paull TT, Gellert M. The 3' to 5' exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 75.de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 76.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JH, Tainer JA. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 77.Wiltzius JJ, Hohl M, Fleming JC, Petrini JH. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 2005;12:403–407. doi: 10.1038/nsmb928. [DOI] [PubMed] [Google Scholar]
- 78.Helmink BA, Bredemeyer AL, Lee BS, Huang CY, Sharma GG, Walker LM, Bednarski JJ, Lee WL, Pandita TK, Bassing CH, Sleckman BP. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J. Exp. Med. 2009;206:669–679. doi: 10.1084/jem.20081326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- 81.Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Antoccia A, Kobayashi J, Tauchi H, Matsuura S, Komatsu K. Nijmegen breakage syndrome and functions of the responsible protein, NBS1. Genome Dyn. 2006;1:191–205. doi: 10.1159/000092508. [DOI] [PubMed] [Google Scholar]
- 83.Taylor AM, Groom A, Byrd PJ. Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis. DNA Repair (Amst) 2004;3:1219–1225. doi: 10.1016/j.dnarep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 84.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 85.Girard PM, Riballo E, Begg AC, Waugh A, Jeggo PA. Nbs1 promotes ATM dependent phosphorylation events including those required for G1/S arrest. Oncogene. 2002;21:4191–4199. doi: 10.1038/sj.onc.1205596. [DOI] [PubMed] [Google Scholar]
- 86.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 2008;27:1953–1962. doi: 10.1038/emboj.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 90.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 91.Durocher D, Jackson SP. The FHA domain. FEBS Lett. 2002;513:58–66. doi: 10.1016/s0014-5793(01)03294-x. [DOI] [PubMed] [Google Scholar]
- 92.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 93.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, Alt FW, Chen J. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 94.Lou Z, Chen BP, Asaithamby A, Minter-Dykhouse K, Chen DJ, Chen J. MDC1 regulates DNA-PK autophosphorylation in response to DNA damage. J. Biol. Chem. 2004;279:46359–46362. doi: 10.1074/jbc.C400375200. [DOI] [PubMed] [Google Scholar]
- 95.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 97.Stewart GS, Panier S, Townsend K, Al Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 98.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 100.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, Shiekhattar R. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a sig-nalosome-like subunit and its role in DNA repair. Mol. Cell. 2003;12:1087–1099. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 102.Chen X, Arciero CA, Wang C, Broccoli D, Godwin AK. BRCC36 is essential for ionizing radiation-induced BRCA1 phosphorylation and nuclear foci formation. Cancer Res. 2006;66:5039–5046. doi: 10.1158/0008-5472.CAN-05-4194. [DOI] [PubMed] [Google Scholar]
- 103.Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morris-sey DE, Greenberg RA. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc. Natl. Acad. Sci. USA. 2009;106:3166–3171. doi: 10.1073/pnas.0807485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhuang J, Zhang J, Willers H, Wang H, Chung JH, van Gent DC, Hallahan DE, Powell SN, Xia F. Checkpoint kinase 2-mediated phosphorylation of BRCA1 regulates the fidelity of nonhomologous end-joining. Cancer Res. 2006;66:1401–1408. doi: 10.1158/0008-5472.CAN-05-3278. [DOI] [PubMed] [Google Scholar]
- 105.Wei L, Lan L, Hong Z, Yasui A, Ishioka C, Chiba N. Rapid recruitment of BRCA1 to DNA double-strand breaks is dependent on its association with Ku80. Mol. Cell Biol. 2008;28:7380–7393. doi: 10.1128/MCB.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S. Two cellular proteins that bind to wild-type but not mutant p53. Proc. Natl. Acad. Sci. USA. 1994;91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee H, Kwak HJ, Cho IT, Park SH, Lee CH. S1219 residue of 53BP1 is phosphorylated by ATM kinase upon DNA damage and required for proper execution of DNA damage response. Biochem. Biophys. Res. Commun. 2009;378:32–36. doi: 10.1016/j.bbrc.2008.10.150. [DOI] [PubMed] [Google Scholar]
- 108.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 109.Mochan TA, Venere M, Ditullio RA, Jr, Halazonetis TD. 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer Res. 2003;63:8586–8591. [PubMed] [Google Scholar]
- 110.Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 114.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Smith GC, Divecha N, Lakin ND, Jackson SP. DNA-dependent protein kinase and related proteins. Biochem. Soc. Symp. 1999;64:91–104. [PubMed] [Google Scholar]
- 116.Riballo E, Critchlow SE, Teo SH, Doherty AJ, Priestley A, Broughton B, Kysela B, Beamish H, Plowman N, Arlett CF, Lehmann AR, Jackson SP, Jeggo PA. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 117.Grawunder U, Zimmer D, Kulesza P, Lieber MR. Requirement for an interaction of XRCC4 with DNA ligase IV for wild-type V(D)J recombination and DNA double-strand break repair in vivo. J. Biol. Chem. 1998;273:24708–24714. doi: 10.1074/jbc.273.38.24708. [DOI] [PubMed] [Google Scholar]
- 118.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 119.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 120.Rooney S, Alt FW, Lombard D, Whitlow S, Eckersdorff M, Fleming J, Fugmann S, Ferguson DO, Schatz DG, Sekiguchi J. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 2003;197:553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, DeVido J, Foy D, Chaudhuri J, Lombard D, Alt FW. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol. Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 122.Weterings E, van Gent DC. The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair (Amst) 2004;3:1425–1435. doi: 10.1016/j.dnarep.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 123.Chang C, Biedermann KA, Mezzina M, Brown JM. Characterization of the DNA double strand break repair defect in scid mice. Cancer Res. 1993;53:1244–1248. [PubMed] [Google Scholar]
- 124.Pang D, Yoo S, Dynan WS, Jung M, Dritschilo A. Ku proteins join DNA fragments as shown by atomic force microscopy. Cancer Res. 1997;57:1412–1415. [PubMed] [Google Scholar]
- 125.Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 127.Mahajan KN, Gangi-Peterson L, Sorscher DH, Wang J, Gathy KN, Mahajan NP, Reeves WH, Mitchell BS. Association of terminal deoxynucleotidyl transferase with Ku. Proc. Natl. Acad. Sci. USA. 1999;96:13926–13931. doi: 10.1073/pnas.96.24.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 129.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 130.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 131.Bryans M, Valenzano MC, Stamato TD. Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutat. Res. 1999;433:53–58. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- 132.Matsumoto Y, Suzuki N, Namba N, Umeda N, Ma XJ, Morita A, Tomita M, Enomoto A, Serizawa S, Hirano K, Sakaia K, Yasuda H, Hosoi Y. Cleavage and phosphorylation of XRCC4 protein induced by X-irradiation. FEBS Lett. 2000;478:67–71. doi: 10.1016/s0014-5793(00)01800-7. [DOI] [PubMed] [Google Scholar]
- 133.Foster RE, Nnakwe C, Woo L, Frank KM. Monoubiquitination of the nonhomologous end joining protein XRCC4. Biochem. Biophys. Res. Commun. 2006;341:175–183. doi: 10.1016/j.bbrc.2005.12.166. [DOI] [PubMed] [Google Scholar]
- 134.Yurchenko V, Xue Z, Sadofsky MJ. SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol. Cell Biol. 2006;26:1786–1794. doi: 10.1128/MCB.26.5.1786-1794.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol. Cell. 2007;28:1093–1101. doi: 10.1016/j.molcel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 136.Hentges P, Ahnesorg P, Pitcher RS, Bruce CK, Kysela B, Green AJ, Bianchi J, Wilson TE, Jackson SP, Doherty AJ. Evolutionary and functional conservation of the DNA non-homologous end-joining protein, XLF/Cernunnos. J. Biol. Chem. 2006;281:37517–37526. doi: 10.1074/jbc.M608727200. [DOI] [PubMed] [Google Scholar]
- 137.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. Ku80 removal from DNA through double strand break-induced ubiquitylation. J. Cell Biol. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, Shah S, Brandt VL, Meek K, Roth DB. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 140.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 142.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 143.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 144.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 145.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 146.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 147.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 2001;477:131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 148.Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 149.Galkin VE, Esashi F, Yu X, Yang S, West SC, Egelman EH. BRCA2 BRC motifs bind RAD51-DNA filaments. Proc. Natl. Acad. Sci. USA. 2005;102:8537–8542. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J. Biol. Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wolner B, Van Komen S, Sung P, Peterson CL. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- 152.Rossi MJ, Mazin AV. Rad51 protein stimulates the branch migration activity of Rad54 protein. J. Biol. Chem. 2008;283:24698–24706. doi: 10.1074/jbc.M800839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Maloisel L, Fabre F, Gangloff S. DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol. Cell Biol. 2008;28:1373–1382. doi: 10.1128/MCB.01651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu Y, Masson JY, Shah R, O'Regan P, West SC. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- 155.Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, Na-rayana LS, Zhou ZQ, Adamson AW, Sorensen KJ, Chen DJ, Jones NJ, Thompson LH. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 156.Liu Y, Tarsounas M, O'Regan P, West SC. Role of RAD51C and XRCC3 in genetic recombination and DNA repair. J. Biol. Chem. 2007;282:1973–1979. doi: 10.1074/jbc.M609066200. [DOI] [PubMed] [Google Scholar]
- 157.Lukas J, Lukas C, Bartek J. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair (Amst) 2004;3:997–1007. doi: 10.1016/j.dnarep.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 158.Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 2000;60:5934–5936. [PubMed] [Google Scholar]
- 159.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 160.Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 161.Schwarz JK, Lovly CM, Piwnica-Worms H. Regulation of the Chk2 protein kinase by oligomerization-mediated cis- and transphosphorylation. Mol. Cancer Res. 2003;1:598–609. [PubMed] [Google Scholar]
- 162.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 163.Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, Lukas J. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 164.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 165.Knights CD, Catania J, Di Giovanni S, Muratoglu S, Perez R, Swartzbeck A, Quong AA, Zhang X, Beerman T, Pestell RG, Avantaggiati ML. Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J. Cell Biol. 2006;173:533–544. doi: 10.1083/jcb.200512059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Roy S, Packman K, Jeffrey R, Tenniswood M. Histone deacetylase inhibitors differentially stabilize acetylated p53 and induce cell cycle arrest or apoptosis in prostate cancer cells. Cell Death Differ. 2005;12:482–491. doi: 10.1038/sj.cdd.4401581. [DOI] [PubMed] [Google Scholar]
- 167.Unger T, Sionov RV, Moallem E, Yee CL, Howley PM, Oren M, Haupt Y. Mutations in serines 15 and 20 of human p53 impair its apoptotic activity. Oncogene. 1999;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- 168.Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 1998;273:33048–33053. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]
- 169.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 170.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 171.el Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 172.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 173.Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 174.Arellano M, Moreno S. Regulation of CDK/cyclin complexes during the cell cycle. Int. J. Biochem. Cell Biol. 1997;29:559–573. doi: 10.1016/s1357-2725(96)00178-1. [DOI] [PubMed] [Google Scholar]
- 175.Hiebert SW, Chellappan SP, Horowitz JM, Nevins JR. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 1992;6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- 176.Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 177.Hinds PW, Mittnacht S, Dulic V, Arnold A, Reed SI, Weinberg RA. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 178.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 180.Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 181.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yama-shita T, Tokino T, Taniguchi T, Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 182.Wolff S, Erster S, Palacios G, Moll UM. p53's mitochondrial translocation and MOMP action is independent of Puma and Bax and severely disrupts mitochondrial membrane integrity. Cell Res. 2008;18:733–744. doi: 10.1038/cr.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nemajerova A, Erster S, Moll UM. The post-translational phosphorylation and acetylation modification profile is not the determining factor in targeting endogenous stress-induced p53 to mitochondria. Cell Death Differ. 2005;12:197–200. doi: 10.1038/sj.cdd.4401526. [DOI] [PubMed] [Google Scholar]
- 184.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]