Abstract
Although the rapid progress of NMR technology has significantly expanded the range of NMR-trackable systems, preparation of NMR-suitable samples that are highly soluble and stable remains a bottleneck for studies of many biological systems. The application of solubility-enhancement tags (SETs) has been highly effective in overcoming solubility and sample stability issues and has enabled structural studies of important biological systems previously deemed unapproachable by solution NMR techniques. In this review, we provide a brief survey of the development and successful applications of the SET strategy in biomolecular NMR. We also comment on the criteria for choosing optimal SETs, such as for differently charged target proteins, and recent new developments on NMR-invisible SETs.
Introduction
The advancement of NMR instrumentation and methodology has made solution NMR spectroscopy an increasingly powerful tool for the investigation of protein structure and dynamics under physiological conditions and for studies of ligand binding and reaction mechanisms in solution. However, the inherent sensitivity limitation of NMR requires protein samples to be stable at high concentrations (> 100 µM for structural studies) for an extended period (typically over a couple of days). Unfortunately, an estimated 75% of soluble proteins and many biologically important macromolecules are characterized by low solubility and instability (Christendat et al. 2000). Therefore, preparation of well-behaved, non-aggregated samples at sufficiently high protein concentrations remains a serious challenge for structural and dynamic studies by NMR.
Numerous efforts have been devoted to overcoming the solubility and sample stability issues. For example, extensive buffer screening (Bagby et al. 1997; Lepre and Moore 1998), addition of charged amino acids (Golovanov et al. 2004), or introduction of point mutants (Huang et al. 1996; Ito and Wagner 2004; Sun et al. 1999) have been successfully utilized to increase the solubility of the target proteins. However, these methods are often protein specific, largely based on trial and error, and may not be easily applicable to other systems. To overcome these issues and develop a generic approach, we introduced the concept of non-cleavable solubility-enhancement tags (SETs) for studies of poorly behaving proteins by solution NMR (Zhou et al. 2001b). Since then, this strategy has found wide applications in the NMR community, and has been used to improve the solubility and sample stability of ∼30 proteins. For many of these examples, this approach has enabled successful determination of high-resolution solution structures. Here, we give a brief overview of the initial development, the theory and the successful application of the SET strategy in biomolecular NMR studies, and we comment on recent improvements of the SET strategy. We refer readers to the excellent review by Waugh for applications of protein tags in a non-NMR setting (Waugh 2005).
Development and Application of SET
Protein tags such as GST and MBP have been widely used as affinity tags for purifying recombinant proteins (di Guan et al. 1988; Smith and Johnson 1988). It was frequently observed that these fusion proteins overexpress better and exhibit enhanced solubility and sample stability compared to their untagged counterparts. This observation has prompted the search of new fusion tags to improve the soluble expression of target proteins in E. coli ((Davis et al. 1999; DelProposto et al. 2009; Forrer and Jaussi 1998; Huth et al. 1997; LaVallie et al. 2000; Pilon et al. 1996; Samuelsson et al. 1994; Zou et al. 2008; Zuo et al. 2005); reviewed by Waugh (Waugh 2005)). Due to the size limit of NMR techniques (∼30 kDa), it is preferable to remove the protein tag before subsequent NMR studies. Unfortunately, once the fusion tag is cleaved by proteolytic digestion, the target protein often becomes unstable again and precipitates within hours, thereby prohibiting further NMR studies.
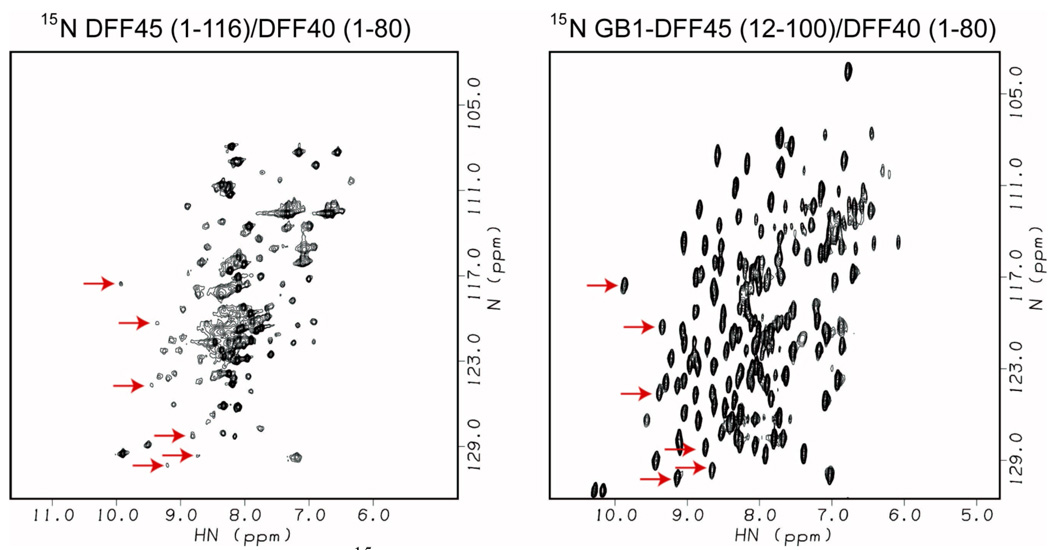
Because it is only the size limit that restricts the use of protein tags in solution NMR studies, we reasoned that a highly soluble and stable protein that is also sufficiently small can be used as a non-cleavable tag for NMR studies. Several small protein tags, such as protein G B1 domain (GB1, 56 residues) (Huth et al. 1997), protein D (110 residues) (Forrer and Jaussi 1998), the Z domain of Staphylococcal protein A (58 residues) (Samuelsson et al. 1994) and thioredoxin (109 residues) (LaVallie et al. 2000), have been shown to increase the yield of soluble proteins. We chose the smallest tag, GB1 as the solubility-enhancement tag for further evaluation. In our study of the DFF40/45 N-terminal CIDE domain complex, attachment of the non-cleavable GB1 tag to DFF45 not only increased the solubility of the DFF40/45 complex from 0.2 mM to 0.6 mM, but also increased the sample stability from 5 days to over a month at 23 °C (Zhou et al. 2001b). The use of the solubility-enhancement tag has resulted in a dramatic improvement of spectral quality (Figure 1) and has enabled subsequent structure determination of the DFF40/45 CIDE domain complex by NMR (Zhou et al. 2001a). To our knowledge, this is the first demonstration of using non-cleavable solubility-enhancement tags to overcome sample solubility and stability issues for structural studies by NMR.
Figure 1.
HSQC spectra of 15N-labeled DFF45 N-terminal CIDE Domain in complex with unlabeled DFF40 (1–80). Attachment of the GB1 tag significantly increased the solubility and stability of the DFF40/45 complex and generated superior NMR spectra. Arrows indicate distinct resonances from DFF45 in the DFF40/45 complex. (Reprinted with permission from Figure 1bc of (Zhou et al. 2001b), Journal of Biomolecular NMR)
Since the initial demonstration and application of the SET strategy to NMR structure determination (Zhou et al. 2001b; Zhou et al. 2001a), this fusion tag approach has found wide applications in the NMR community. Approximately 30 examples have now been reported in the literature, which show significant enhancement of protein solubility and/or sample stability using SETs (Table 1). Additionally, in many cases, the creation of SET-fusion proteins also significantly improved protein overexpression levels in E. coli and the final yields of the purified proteins. These target proteins cover a wide range of structural topologies and biological functions, which truly demonstrate the generality of the SET approach in biomolecular NMR studies.
Table 1.
Examples of NMR Studies Using the SET Approach
| Tag | Target Protein & Property | Reported Effect(s) | Notes and References |
|---|---|---|---|
| GB1 (6.2 kDa; pI=4.5) |
Mouse prion protein (mPrP) |
Increased the expression yield and solubility. |
GB1 was used to enhance the expression yield and the solubility of selected mPrP constructs. (Hornemann etal. 2009) |
| GB1basic (6.2 kDa; pI=8.0) |
HPV16 E6 constructs (1) E6 (17 kDa; pI=9.0) (2) E6N (8.9 kDa, pI=6.7) (3) E6C (7.5 kDa; pI=9.7) |
Improved solubility and sample stability of HPV-16 E6 protein. |
GB1basic is a GB1-mutant (D22N, D36R, and E42K) with a pI of 8.0. It was used to express basic target proteins to avoid aggregation. Use of the GB1basic tag allowed preparation of stable NMR samples of E6N and E6C at 2 mM, and E6 at 0.2 mM. The intrinsic solubility of E6N (after removing the GST-tag from GST-E6N) was in the range of hundreds of µM. (Liu etal. 2009) |
| GB1 (invisible C- terminal tag; 6.2 kDa; pI=4.5) |
Vav C-terminus SH3 (7.5 kDa; pI=6.4) |
Enhanced the solubility of VcSH3 by more than 10 fold. |
The target protein was initially expressed as an N- terminal GB1-fusion construct. A sortase-mediated protein ligation method was used to ligate a second, unlabeled GB1 to the C-terminus of the target protein. The N-terminal GB1 tag was subsequently removed by protease cleavage. The Vav C-terminus SH3 was almost insoluble at physiological pH. Using invisible C- terminal GB1 tag enabled preparation of stable NMR samples at 0.6 mM and subsequent structural determination (PDB: 2KBT). (Kobashigawa et al. 2009) |
| GB1 (6.2 kDa; pI=4.5) |
Borealin (1) Full length (31.3 kDa; pI=9.84) (2) residues13–92 (9.4 kDa; pI=5.48) |
Significantly improved the protein yield in the soluble fractions. |
(Zhou et al. 2009) |
| Calmodulin (CaM; invisible tag; 16.8 kDa, pI=4.1) |
Sterile alpha motif (SAM) from p63 (7.5 kDa; pI=5.8) |
Enhanced solubility by over 20-fold. |
The target protein was inserted between GST and the calmodulin binding peptide (CBP). The unlabeled calmodulin, which serves the role of a solubility- enhancement tag, was added to form a CBP-calmodulin complex. The N-terminal GST-tag was then removed by protease cleavage. (Durst et al. 2008) |
| GB1 (6.2 kDa; pI=4.5) |
17β-hydroxysteroid dehydrogenase type 1 (HSD17 β1). (homodimer with a molecular weight of 70 kDa) |
Increased sample stability. |
The fusion protein formed soluble aggregates at high concentrations, but maintained enzymatic activity to allow NMR-based inhibitor studies. (Ludwig et al.2008) |
| GB1 (6.2 kDa; pI=4.5) |
Potassium channel- interacting protein 4a (KChIP4a, residue 1–34; 3.7 kDa; pI=4.0) |
Enhanced solubility. | (Schwenk et al. 2008) |
| GB1 (c-terminal tag; 6.2 kDa; pI=4.5) |
CK2 substrate (XT111– 132; 2.4 kDa; pI=8.2) |
Enhanced solubility of fused peptide in live cells |
GB1 was used as a soluble carrier of a phosphorylation site and provided the solubility needed for recording spectra in live cells. (Selenko et al. 2008) |
| GB1 (6.2 kDa; pI=4.5) |
mRNA-decapping enzyme Dcp2 Nudix domain (17.3 kDa; pI=8.5) |
Enhanced solubility. | The untagged Nudix domain was only marginally soluble. The GB1-tagged protein (in the presence of Arg/Glu additives) was stable at 0.5 mM for several weeks. (PDB: 2JVB) (Deshmukh et al. 2008) |
| GB1 (6.2 kDa; pI=4.5) |
Eukaryotic translation initiation factor eIF5 (residues 241–405; 19.3 kDa; pI=5.2) |
Enhanced solubility. | (Reibarkh et al. 2008) |
| GB1 (6.2 kDa; pI=4.5) |
Parkin ubiquitin like domain mutant (UbldR42P) (8.8 kDa; pI=6.7) |
The GB1 tag was used to overcome the poor expression and degradation of the UbldR42P mutant; without the GB1 tag, the UbldR42P could not be isolated. (Safadi and Shaw 2007) |
|
| GB1 (6.2 kDa; pI=4.5) |
A ubiquitin variant found at the N-terminus of S27a in Giardia lamblia (GlUbS27A; 7.0 kDa; pI=4.7) |
Enhanced solubility/sample stability. |
No protein expression was observed with the His- or HA-tagged constructs. The GB1-tagged GlUbS27A was stable at 1 mM for about a week at 25 °C. (Catic et al. 2007) |
| GB1 (6.2 kDa; pI=4.5) |
Fas Death Domain (Fas-DD; 9.9 kDa; pI=8.7) |
Increased sample stability/solubility. |
The untagged Fas-DD had an intrinsic tendency to form soluble aggregates at physiological pH. (Ferguson et al. 2007) |
| GB1 (6.2 kDa; pI=4.5) |
Inositol 1,4,5- trisphosphate receptor (IP3R) intraluminal loop L3-2 (2.3 kDa; pI=6.3) |
Increased sample stability/solubility. |
No protein expression was observed with a His-tagged construct. (Kang et al. 2007) |
| GB1 (6.2 kDa; pI=4.5) |
Murine eIF4E (25 kDa; pI=5.8) |
Greatly enhanced solubility. |
(Untagged) mammalian eIF4E behaved poorly in solution. (Moerke et al. 2007) |
| Poly Arg or Lys peptide tags |
BPTI-22 (a BPTI variant containing 22 alanines) |
Enhanced solubility by 4–6 folds. |
(Kato et al. 2007) |
| GB1 (6.2 kDa; pI=4.5) |
SRp20 RNA recognition motif (RRM; 9.6 kDa; pI=6.6) |
Enhanced solubility. | Poor solubility of the untagged protein prevented NMR studies. The GB1-SRp20 RRM was stable at 1 mM, which enabled structural studies. (PDB: 2I38 & 2I2Y) (Hargous et al. 2006) |
| GB1 (6.2 kDa; pI=4.5) |
9G8 RNA recognition motif (9G8 RRM; 11.3 kDa; pI=9.6) |
Enhanced solubility. | Poor solubility of the untagged protein prevented NMR studies. The GB1–9G8 RRM (in the presence of Arg/Glu additives) was stable at 1 mM. Hargous et al. 2006) |
| GB1 (6.2 kDa; pI=4.5) |
UBA domain of human bone marrow stromal cells ubiquitin-like protein (BMSC-UbP; 4.8 kDa; pI=4.0) |
Dramatically enhanced the solubility. |
The untagged UBA domain readily precipitated in solution. The GB1-UBA was stable at 1 mM. (PDB: 2CWB) (Chang et al. 2006) |
| GB1 (6.2 kDa; pI=4.5) |
Rat ADAR2 double- stranded RNA binding domain (dsRBD; 24.3 kDa; pI=6.2) |
Improved protein expression and solubility. |
The untagged rat ADAR2 dsRBD12 (74–-301) had low solubility in common NMR buffers. The GB1-fusion protein was stable at 0.8 mM. (Stefl et al. 2005; Stefl et al. 2006) |
| GB1 (invisible tag) |
Chitin-binding domain | Not reported | Used an intein-based strategy to incorporate the unlabeled GB1 tag into isotopically labeled proteins. (Züger and Iwai 2005) |
| GB1 (6.2 kDa; pI=4.5) |
eukaryotic translation initiation factor 2 gamma (eIF2γ; 51 kDa; pI=8.7) |
Enhanced solubility | GB1 was used to enhance the solubility of eIF2γ to enable studies of its interaction with eIF2α (Ito et al. 2004) |
| GB1 (6.2 kDa; pI=4.5) |
Mutant myotoxin a (MyoP20G; 4.7 kDa; pI=9.5) |
Increased the expression yield and enhanced the refolding efficiency. |
Untagged protein refolded poorly. The GB1 tag was removed after refolding. (Cheng and Patel 2004) |
| GB1 (6.2 kDa; pI=4.5) |
Human Ki67 FHA domain (hNIFK; 5 kDa; pI=4.5) |
Increased the protein yield and sample stability. |
(Li et al. 2004) |
| GB1 (6.2 kDa; pI=4.5) |
NALP1 Pyrin domain (10 kDa; pI=5.9) |
Enhanced solubility by ∼100-fold. |
Untagged protein aggregated at concentrations above ∼10 µM. The GB1-tagged protein was stable at 1 mM (PDB: 1PN5). (Hiller et al. 2003) |
| GB1 (6.2 kDa; pI=4.5) |
Human T-cell leukemia virus 1 (HTLV-1) Tax40N (4.3 kDa; pI=6.0) |
Not reported | (Li et al. 2003) |
| GB1 (6.2 kDa; pI=4.5) |
eIF5B–CTD (16.7 kDa; pI=8.7) |
Enhanced solubility | (Marintchev et al. 2003) |
| MBP (40.7 kDa; pI=5.2) |
Integrin αIIbβ3 (MW of β3 is 5.5 kDa; pI=9.2) |
Enhanced solubility | (PDB: 1M8O). (Vinogradova et al. 2002) |
Choice of SETs
Although GB1 has been a highly successful solubility-enhancement tag, other highly soluble and stable small protein domains can also serve similar functions. Unfortunately, how the SET enhances the solubility of a target protein remains poorly understood, and comparative proteomic studies have not revealed a universally good tag for all protein targets (Hammarström et al. 2002; Hammarström et al. 2006). Based on a thermodynamic analysis, we suggest here the following criteria for choosing a solubility-enhancement tag.
1. The SET should not interact with the target protein or protein complex
Ideally, a solubility-enhancement tag should be “transparent” to the target protein, i.e., it should not perturb the structure or function of the target protein. In the absence of such prior knowledge, proper control experiments must be included to demonstrate the “inertness” of the solubility-enhancement tag for functional assays. Likewise, the lack of perturbations of tag resonances in the fusion protein provides a compelling argument that the solubility-enhancement tag does not interact with the target protein and is unlikely to alter its structure.
In this regard, GB1 appears to be remarkably “transparent” as demonstrated in a variety of GB1-fusion proteins in NMR studies (Table 1). Interestingly, many examples of the GB1-fusion proteins in NMR studies also display better sample stability at high concentrations (µM-mM). Because the “passive” GB1 tag is unlikely to alter the thermal stability of the target protein, the improved sample stability presumably results from the enhanced solubility and reduced aggregation of the fusion protein.
Because GB1 is slightly acidic (pI=4.5), it may cause non-specific electrostatic interactions when fused to proteins with basic pI values. To avoid these non-specific interactions, we created a GB1 mutant (GB1basic) by mutating D22N, D36R, and E42K, which increased the pI of GB1 to 8.0 (Zhou and Wagner, unpublished). This basic GB1 tag was successfully utilized to prepare highly soluble HPV16 E6 samples and prevent non-specific electrostatic interactions between the tag and the target protein (Liu et al. 2009). Without the tag, the solubility of the E6 constructs was too low to record spectra (J. Baleja, private communication). Consistent with this notion of choosing a SET based on matching its charge state with that of the target protein, Harrison and co-workers showed in their statistical model that avoidance of charge neutralization increases the probability of producing soluble proteins in E. coli (Davis et al. 1999; Wilkinson and Harrison 1991).
It should be noted that an “active” fusion tag can also be highly effective. For example, Ikura and co-workers fused the TAF N-terminal Domain 1 and 2 (TAND12) with its binding partner TATA-binding protein (TBP) to form a stable protein complex, which displayed enhanced solubility and sample stability (Mal et al. 2007). However, such an “active” fusion tag is target specific and cannot be easily applied to other proteins.
2. The SET should be highly soluble
Assuming that (1) there is no interaction between the tag and the target protein, (2) there is no structural change of either the tag or the target in the fusion protein, and (3) the contribution of the linker can be neglected, we give an estimation of the solubility-enhancement effect based on a simple thermodynamic model. Although the analysis below focuses on fusion proteins containing a single tag, it is straightforward to extend such an analysis to fusion proteins with multiple tags.
The free energies of individually transferring A (the tag) and B (the target protein) from the solid state to the solution state are given by:
| Eq.[1] |
At equilibrium (i.e. at saturation), the free energy of transferring the A and B from the solid state to the solution state is zero. Therefore one has:
| Eq.[2] |
, which can be re-arranged to give
| Eq.[3] |
With Eq. [3], one can rewrite Eq. [1] as
| Eq.[4] |
If there is no interaction between A and B, we can conceptually describe the transfer of the fusion protein A-B from the solid state to the solution state as two separate processes: transferring ASolid to ASolution and transferring BSolid to BSolution. The free energy of such a combined transfer is zero at equilibrium.
| Eq.[5] |
Because the covalent linker requires
| Eq.[6] |
, by substituting and with , we can rewrite Eq. [5] as
| Eq.[7] |
, which requires
| Eq.[8] |
Therefore, we have the saturation concentration of the fusion protein as:
| Eq.[9] |
We note that the above analysis does not account for changes of solid or solution state compositions, nor does it take into consideration of intermediate species (such as ASolid – BSolution and ASolution – BSolid ) of the solvation process. The latter approximation, in particular, can introduce a very large error in the solubility estimation of the fusion protein. Finally, strictly speaking, the concentration terms of Eq. [9] should be effective concentrations (i.e. activities), which may deviate from the apparent protein concentrations. This effect is expected to be larger at higher concentrations, which can result in an overestimation of the effective tag concentration at saturation. Because of these limitations, Eq. [9] can only be used in a qualitative way. It nevertheless gives a useful evaluation of the beneficial effect brought by a solubility-enhancement tag.
To give an example, we were able to make 15–20 mM GB1 solutions routinely without any noticeable precipitations. Using these numbers as the solubility of GB1, we estimate that the SET approach yields a saturation concentration of 1.2–1.4 mM or 0.38–0.44 mM for a target protein with inherent solubility of 0.1 mM or 0.01 mM respectively, corresponding to a ∼10–40 fold enhancement of the solubility over the untagged protein! Experimentally, approximately 3–100 fold enhancements of solubility have been reported for GB1-fusion proteins (Hiller et al. 2003; Kobashigawa et al. 2009; Zhou et al. 2001b). The largest effect was reported for the pyrin domain of NALP1, which saw its solubility increased from ∼10 µM to 1 mM (Hiller et al. 2003).
Eq. [9] argues that proteins with higher intrinsic solubility, but not with larger molecular weights, function as better tags. Although this conclusion may seem counterintuitive, several large scale solubility studies have consistently categorized the small GB1 tag (5.6 kDa) as one of the most effective tags to use (Hammarström et al. 2002; Hammarström et al. 2006). For example, Hammarström compared the effect of different tags on the solubility of 27 small- to medium-sized human proteins, and ranked GB1, MBP and thioredoxin as the best tags (Hammarström et al. 2002). The authors concluded that the there was no statistical difference of GB1, MBP and thioredoxin in their ability to enhance the solubility of a target protein. It is important to note that in most of the studies, the solubility (often reported as gel intensity) reflects the mass yield of the fusion proteins, but not the untagged target proteins. This could lead to an overestimation of the solubility-enhancement effect for large tags such as MBP or NusA. After correcting for the molecular weight contributions from different tags, Hammarstrom et al. concluded that GB1 gave a significantly larger amount of soluble target proteins for the 45 human proteins tested (Hammarström et al. 2006).
Finally, we would like to emphasize that Eq. [9] is based on a thermodynamic analysis. It assumes no interaction between the tag and the target protein and requires the solvation process to be fully reversible. Several protein tags have been shown to facilitate protein folding in E. coli by promoting disulfide bond formation (Stewart et al. 1998), by serving as a molecular chaperone (Bach et al. 2001; Kapust and Waugh 1999) or by enhancing transcription pausing (Davis et al. 1999). In these scenarios, the significantly better “solubilizing” effect of the “active” tags over “passive” tags may reflect the benefit of folding kinetics, but not thermodynamics.
3. The SET should be highly stable
Because NMR experiments are performed under a variety of pH, temperature and buffer conditions, a good solubility-enhancement tag should be stable under these conditions. The rapid two-state refolding property of a tag can also be highly beneficial. For example, in the study of mutant myotoxin a (MyoP20G), Cheng and Patel reported that GB1 appears to increase protein (re)folding efficiency (Cheng and Patel 2004), which likely comes from the enhanced solubility (and reduced aggregation) of the denatured fusion protein.
4. The SET can increase the overexpression level and yield of the target protein
As reported in early literature, a successful solubility-enhancement tag often enhances protein overexpression levels and increases the yields of the purified proteins. Some tags, such as MBP and thioredoxin, have been suggested to serve as chaperones to promote proper folding of target proteins (Bach et al. 2001; Kapust and Waugh 1999; Kern et al. 2003). Although similar benefits in protein expression levels and yields have been observed for GB1-fusion proteins (Table 1; also see studies by Hammarström et al. (Hammarström et al. 2002; Hammarström et al. 2006)), the experimental evidence for the chaperone activity of GB1 is lacking. It should be noted that such effects do not have to derive from the chaperone activity. The enhanced solubility of the fusion protein itself is expected to facilitate protein folding and overexpression in vivo and increase the yield of protein purification in vitro by reducing protein aggregation and precipitation.
Several studies reported diminished effects of SETs on the E. coli expression of large proteins (>25–30 kDa) in soluble fractions (Hammarström et al. 2002; Hammarström et al. 2006). Because large proteins frequently require chaperones or binding partners to fold properly, it is likely that these observations reflect an intrinsic folding (kinetic) problem of the large proteins, rather than the ineffectiveness of SETs.
Invisible SETs
Despite the success of the SET approach, it still brings a sizeable amount of extra signals from the protein tag. For a target protein of 10–20 kDa, inclusion of a small GB1 tag (56 residues) easily adds about a quarter to a half of “extra” signals to those from the untagged protein. Although the excellent signal dispersion and the lack of resonance perturbation make the tag signals easy to identify, they nevertheless bring extra burden and complexity for resonance assignment.
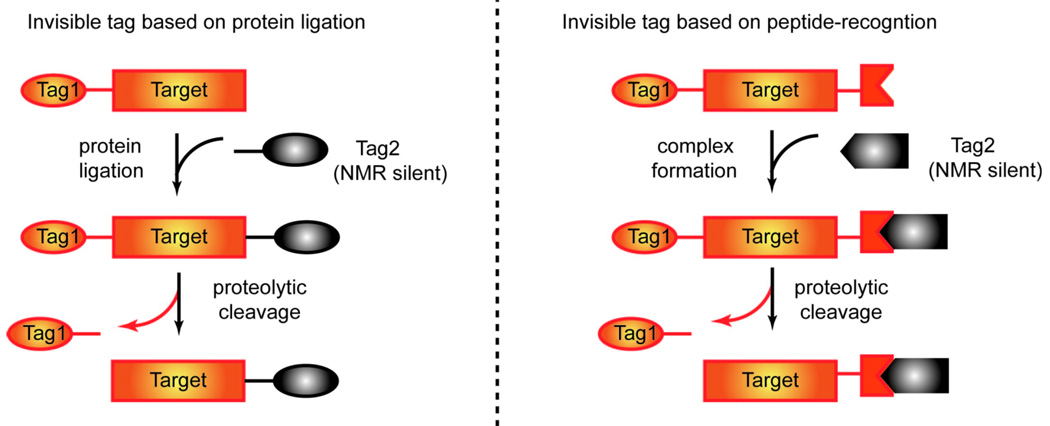
Recently, two types of NMR-invisible tags have used to overcome this issue (Figure 2) (Durst et al. 2008; Kobashigawa et al. 2009; Züger and Iwai 2005). Both approaches start from an isotopically enriched fusion protein containing a cleavable solubility tag. A second and unlabeled solubility tag—which is invisible by NMR—is then introduced to maintain solubility. The isotopically labeled tag is subsequently removed to generate the final form of the NMR sample.
Figure 2.
NMR-invisible solubility-enhancement tags.
The two approaches differ in how the NMR-invisible tag was introduced. In the first approach, the unlabeled GB1 tag was attached to the isotopically labeled chitin-binding domain or the Vav C-terminus SH3 domain using either an intein-based or a sortase-mediated protein ligation strategy (Kobashigawa et al. 2009; Züger and Iwai 2005). Because the yield of the final fusion protein depends on the ligation efficiency, optimization of the ligation condition is critical for the general application of this approach. In the second approach, a calmodulin-binding peptide (CBP, 23 residues) was included in the construct of the GST-tagged target protein (Durst et al. 2008). The unlabeled calmodulin, which binds the CBP, was added to the solution. After formation of the calmodulin/CBP complex, the isopotically labeled GST-tag was removed by proteolytic cleavage, and the unlabeled calmodulin served as the NMR-invisible solubility-enhancement tag. Because the latter approach bypasses the protein ligation step completely, it is more convenient to use. However, there is no reason why one should be restricted to the CBP tag of 23 residues; systems using shorter peptides and the corresponding high-affinity binding partners are likely to emerge in the future.
Conclusion
The preparation of highly soluble and stable samples represents a significant challenge for solution NMR studies of proteins with inherent poor solubility and stability. The use of solubility-enhancement tags has been demonstrated to overcome sample solubility and stability barriers and has enabled detailed structural analyses of many poorly-behaving proteins. The recent development of NMR-invisible tags promises to further expand the application of the SET strategy in biomolecular NMR.
References
- Bach H, Mazor Y, Shaky S, Shoham-Lev A, Berdichevsky Y, Gutnick DL, Benhar I. Escherichia coli maltose-binding protein as a molecular chaperone for recombinant intracellular cytoplasmic single-chain antibodies. Journal of molecular biology. 2001;312:79–93. doi: 10.1006/jmbi.2001.4914. [DOI] [PubMed] [Google Scholar]
- Bagby S, Tong KI, Liu D, Alattia JR, Ikura M. The button test: a small scale method using microdialysis cells for assessing protein solubility at concentrations suitable for NMR. Journal of biomolecular NMR. 1997;10:279–282. doi: 10.1023/a:1018359305544. [DOI] [PubMed] [Google Scholar]
- Catic A, Sun ZY, Ratner DM, Misaghi S, Spooner E, Samuelson J, Wagner G, Ploegh HL. Sequence and structure evolved separately in a ribosomal ubiquitin variant. The EMBO journal. 2007;26:3474–3483. doi: 10.1038/sj.emboj.7601772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YG, Song AX, Gao YG, Shi YH, Lin XJ, Cao XT, Lin DH, Hu HY. Solution structure of the ubiquitin-associated domain of human BMSC-UbP and its complex with ubiquitin. Protein Sci. 2006;15:1248–1259. doi: 10.1110/ps.051995006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Patel DJ. An efficient system for small protein expression and refolding. Biochemical and biophysical research communications. 2004;317:401–405. doi: 10.1016/j.bbrc.2004.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christendat D, Yee A, Dharamsi A, Kluger Y, Savchenko A, Cort JR, Booth V, Mackereth CD, Saridakis V, Ekiel I, et al. Structural proteomics of an archaeon. Nature structural biology. 2000;7:903–909. doi: 10.1038/82823. [DOI] [PubMed] [Google Scholar]
- Davis GD, Elisee C, Newham DM, Harrison RG. New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnology and bioengineering. 1999;65:382–388. [PubMed] [Google Scholar]
- DelProposto J, Majmudar CY, Smith JL, Brown WC. Mocr: a novel fusion tag for enhancing solubility that is compatible with structural biology applications. Protein expression and purification. 2009;63:40–49. doi: 10.1016/j.pep.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh MV, Jones BN, Quang-Dang DU, Flinders J, Floor SN, Kim C, Jemielity J, Kalek M, Darzynkiewicz E, Gross JD. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Molecular cell. 2008;29:324–336. doi: 10.1016/j.molcel.2007.11.027. [DOI] [PubMed] [Google Scholar]
- di Guan C, Li P, Riggs PD, Inouye H. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene. 1988;67:21–30. doi: 10.1016/0378-1119(88)90004-2. [DOI] [PubMed] [Google Scholar]
- Durst FG, Ou HD, Lohr F, Dotsch V, Straub WE. The better tag remains unseen. Journal of the American Chemical Society. 2008;130:14932–14933. doi: 10.1021/ja806212j. [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Esposito D, Jovanovic J, Sankar A, Driscoll PC, Mehmet H. Biophysical and cell-based evidence for differential interactions between the death domains of CD95/Fas and FADD. Cell death and differentiation. 2007;14:1717–1719. doi: 10.1038/sj.cdd.4402191. [DOI] [PubMed] [Google Scholar]
- Forrer P, Jaussi R. High-level expression of soluble heterologous proteins in the cytoplasm of Escherichia coli by fusion to the bacteriophage lambda head protein D. Gene. 1998;224:45–52. doi: 10.1016/s0378-1119(98)00538-1. [DOI] [PubMed] [Google Scholar]
- Golovanov AP, Hautbergue GM, Wilson SA, Lian LY. A simple method for improving protein solubility and long-term stability. Journal of the American Chemical Society. 2004;126:8933–8939. doi: 10.1021/ja049297h. [DOI] [PubMed] [Google Scholar]
- Hammarström M, Hellgren N, van Den Berg S, Berglund H, Hard T. Rapid screening for improved solubility of small human proteins produced as fusion proteins in Escherichia coli. Protein Sci. 2002;11:313–321. doi: 10.1110/ps.22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarström M, Woestenenk EA, Hellgren N, Hard T, Berglund H. Effect of N-terminal solubility enhancing fusion proteins on yield of purified target protein. Journal of structural and functional genomics. 2006;7:1–14. doi: 10.1007/s10969-005-9003-7. [DOI] [PubMed] [Google Scholar]
- Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. The EMBO journal. 2006;25:5126–5137. doi: 10.1038/sj.emboj.7601385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller S, Kohl A, Fiorito F, Herrmann T, Wider G, Tschopp J, Grutter MG, Wuthrich K. NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain. Structure. 2003;11:1199–1205. doi: 10.1016/j.str.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Hornemann S, Christen B, von Schroetter C, Perez DR, Wüthrich K. Prion protein library of recombinant constructs for structural biology. The FEBS journal. 2009;276:2359–2367. doi: 10.1111/j.1742-4658.2009.06968.x. [DOI] [PubMed] [Google Scholar]
- Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- Huth JR, Bewley CA, Jackson BM, Hinnebusch AG, Clore GM, Gronenborn AM. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 1997;6:2359–2364. doi: 10.1002/pro.5560061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Marintchev A, Wagner G. Solution structure of human initiation factor eIF2alpha reveals homology to the elongation factor eEF1B. Structure. 2004;12:1693–1704. doi: 10.1016/j.str.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Ito T, Wagner G. Using codon optimization, chaperone co-expression, and rational mutagenesis for production and NMR assignments of human eIF2 alpha. Journal of biomolecular NMR. 2004;28:357–367. doi: 10.1023/B:JNMR.0000015405.62261.cb. [DOI] [PubMed] [Google Scholar]
- Kang J, Kang S, Yoo SH, Park S. Identification of residues participating in the interaction between an intraluminal loop of inositol 1,4,5-trisphosphate receptor and a conserved N-terminal region of chromogranin B. Biochimica et biophysica acta. 2007;1774:502–509. doi: 10.1016/j.bbapap.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Maki K, Ebina T, Kuwajima K, Soda K, Kuroda Y. Mutational analysis of protein solubility enhancement using short peptide tags. Biopolymers. 2007;85:12–18. doi: 10.1002/bip.20596. [DOI] [PubMed] [Google Scholar]
- Kern R, Malki A, Holmgren A, Richarme G. Chaperone properties of Escherichia coli thioredoxin and thioredoxin reductase. The Biochemical journal. 2003;371:965–972. doi: 10.1042/BJ20030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobashigawa Y, Kumeta H, Ogura K, Inagaki F. Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method. Journal of biomolecular NMR. 2009;43:145–150. doi: 10.1007/s10858-008-9296-5. [DOI] [PubMed] [Google Scholar]
- LaVallie ER, Lu Z, Diblasio-Smith EA, Collins-Racie LA, McCoy JM. Thioredoxin as a fusion partner for production of soluble recombinant proteins in Escherichia coli. Methods in enzymology. 2000;326:322–340. doi: 10.1016/s0076-6879(00)26063-1. [DOI] [PubMed] [Google Scholar]
- Lepre CA, Moore JM. Microdrop screening: a rapid method to optimize solvent conditions for NMR spectroscopy of proteins. Journal of biomolecular NMR. 1998;12:493–499. doi: 10.1023/a:1008353000679. [DOI] [PubMed] [Google Scholar]
- Li H, Byeon IJ, Ju Y, Tsai MD. Structure of human Ki67 FHA domain and its binding to a phosphoprotein fragment from hNIFK reveal unique recognition sites and new views to the structural basis of FHA domain functions. Journal of molecular biology. 2004;335:371–381. doi: 10.1016/j.jmb.2003.10.032. [DOI] [PubMed] [Google Scholar]
- Li J, Li H, Tsai MD. Direct binding of the N-terminus of HTLV-1 tax oncoprotein to cyclin-dependent kinase 4 is a dominant path to stimulate the kinase activity. Biochemistry. 2003;42:6921–6928. doi: 10.1021/bi034369n. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cherry JJ, Dineen JV, Androphy EJ, Baleja JD. Determinants of stability for the E6 protein of papillomavirus type 16. Journal of molecular biology. 2009;386:1123–1137. doi: 10.1016/j.jmb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig C, Michiels PJ, Lodi A, Ride J, Bunce C, Gunther UL. Evaluation of solvent accessibility epitopes for different dehydrogenase inhibitors. ChemMedChem. 2008;3:1371–1376. doi: 10.1002/cmdc.200800110. [DOI] [PubMed] [Google Scholar]
- Mal TK, Takahata S, Ki S, Zheng L, Kokubo T, Ikura M. Functional silencing of TATA-binding protein (TBP) by a covalent linkage of the N-terminal domain of TBP-associated factor 1. The Journal of biological chemistry. 2007;282:22228–22238. doi: 10.1074/jbc.M702988200. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Kolupaeva VG, Pestova TV, Wagner G. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1535–1540. doi: 10.1073/pnas.0437845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Pilon AL, Yost P, Chase TE, Lohnas GL, Bentley WE. High-level expression and efficient recovery of ubiquitin fusion proteins from Escherichia coli. Biotechnology progress. 1996;12:331–337. doi: 10.1021/bp9600187. [DOI] [PubMed] [Google Scholar]
- Reibarkh M, Yamamoto Y, Singh CR, del Rio F, Fahmy A, Lee B, Luna RE, Ii M, Wagner G, Asano K. Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. The Journal of biological chemistry. 2008;283:1094–1103. doi: 10.1074/jbc.M708155200. [DOI] [PubMed] [Google Scholar]
- Safadi SS, Shaw GS. A disease state mutation unfolds the parkin ubiquitin-like domain. Biochemistry. 2007;46:14162–14169. doi: 10.1021/bi7016969. [DOI] [PubMed] [Google Scholar]
- Samuelsson E, Moks T, Nilsson B, Uhlen M. Enhanced in vitro refolding of insulin-like growth factor I using a solubilizing fusion partner. Biochemistry. 1994;33:4207–4211. doi: 10.1021/bi00180a013. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Zolles G, Kandias NG, Neubauer I, Kalbacher H, Covarrubias M, Fakler B, Bentrop D. NMR analysis of KChIP4a reveals structural basis for control of surface expression of Kv4 channel complexes. The Journal of biological chemistry. 2008;283:18937–18946. doi: 10.1074/jbc.M800976200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P, Frueh DP, Elsaesser SJ, Haas W, Gygi SP, Wagner G. In situ observation of protein phosphorylation by high-resolution NMR spectroscopy. Nature structural & molecular biology. 2008;15:321–329. doi: 10.1038/nsmb.1395. [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Stefl R, Skrisovska L, Xu M, Emeson RB, Allain FH. Resonance assignments of the double-stranded RNA-binding domains of adenosine deaminase acting on RNA 2 (ADAR2) Journal of biomolecular NMR. 2005;31:71–72. doi: 10.1007/s10858-004-6058-x. [DOI] [PubMed] [Google Scholar]
- Stefl R, Xu M, Skrisovska L, Emeson RB, Allain FH. Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs. Structure. 2006;14:345–355. doi: 10.1016/j.str.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Stewart EJ, Aslund F, Beckwith J. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. The EMBO journal. 1998;17:5543–5550. doi: 10.1093/emboj/17.19.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZY, Dötsch V, Kim M, Li J, Reinherz EL, Wagner G. Functional glycan-free adhesion domain of human cell surface receptor CD58: design, production and NMR studies. The EMBO journal. 1999;18:2941–2949. doi: 10.1093/emboj/18.11.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas T, Plow E, Qin J. A structural mechanism of integrin alpha(IIb)beta(3) "inside-out" activation as regulated by its cytoplasmic face. Cell. 2002;110:587–597. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- Waugh DS. Making the most of affinity tags. Trends in biotechnology. 2005;23:316–320. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Wilkinson DL, Harrison RG. Predicting the solubility of recombinant proteins in Escherichia coli. Bio/technology (Nature Publishing Company) 1991;9:443–448. doi: 10.1038/nbt0591-443. [DOI] [PubMed] [Google Scholar]
- Zhou L, Li J, George R, Ruchaud S, Zhou HG, Ladbury JE, Earnshaw WC, Yuan X. Effects of Full-Length Borealin on the Composition and Protein-Protein Interaction Activity of a Binary Chromosomal Passenger Complex (dagger) Biochemistry. 2009 doi: 10.1021/bi801298j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Lugovskoy AA, McCarty JS, Li P, Wagner G. Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45. Proceedings of the National Academy of Sciences of the United States of America. 2001a;98:6051–6055. doi: 10.1073/pnas.111145098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Lugovskoy AA, Wagner G. A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. Journal of biomolecular NMR. 2001b;20:11–14. doi: 10.1023/a:1011258906244. [DOI] [PubMed] [Google Scholar]
- Zou Z, Cao L, Zhou P, Su Y, Sun Y, Li W. Hyper-acidic protein fusion partners improve solubility and assist correct folding of recombinant proteins expressed in Escherichia coli. Journal of biotechnology. 2008;135:333–339. doi: 10.1016/j.jbiotec.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Züger S, Iwai H. Intein-based biosynthetic incorporation of unlabeled protein tags into isotopically labeled proteins for NMR studies. Nature biotechnology. 2005;23:736–740. doi: 10.1038/nbt1097. [DOI] [PubMed] [Google Scholar]
- Zuo X, Mattern MR, Tan R, Li S, Hall J, Sterner DE, Shoo J, Tran H, Lim P, Sarafianos SG, et al. Expression and purification of SARS coronavirus proteins using SUMO-fusions. Protein expression and purification. 2005;42:100–110. doi: 10.1016/j.pep.2005.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]