Abstract
Purpose of review
Acute rejection is an immune process that begins with the recognition of the allograft as non-self and ends in graft destruction. Histological features of the allograft biopsy are currently used for the differential diagnosis of allograft dysfunction. In view of the safety and the opportunity for repetitive sampling, development of noninvasive biomarkers of allograft status is an important objective in transplantation. Herein we review some of the progress towards the development of noninvasive biomarkers of human allograft status.
Recent findings
Urinary cell and peripheral blood cell messenger RNA profiles have been associated with acute rejection of human renal allografts. Emerging data support the idea that development of noninvasive biomarkers predictive of antibody-mediated rejection is feasible. The demonstration that intragraft micro RNA expression predicts renal allograft status suggests that noninvasively ascertained miRNA profiles may be of value.
Summary
We are pleased with the progress to date, and anticipate clinical trials investigating the hypotheses that noninvasively ascertained mRNA profiles: (a) will minimize the need for invasive biopsy procedures; (b) predict the development of acute rejection and chronic allograft nephropathy; (c) facilitate preemptive therapy capable of preserving graft function; and (d) facilitate personalization of immunosuppressive therapy for the allograft recipient.
Keywords: Transplantation, Acute Rejection, mRNA Profiling
INTRODUCTION
Acute rejection is an allograft destructive immune response that may occur at any time during the life-span of an organ transplant. The mechanistic pathways of acute rejection are being resolved, and the consequences of immune rejection are evidenced by graft dysfunction and classified by histological features of the allograft biopsy specimen [1••]. Percutaneous needle biopsy of the renal allograft is currently used to diagnose acute rejection of renal allografts. Besides being an invasive procedure, sampling errors and inter-observer variations are additional concerns [2, 3].
We have hypothesized a time-line model to illustrate the development of acute rejection as a continuum, with initial events identified by molecular perturbations, and histological changes and clinical manifestations being relatively latter events ([4••], Figure 1). In this conceptualization, biomarkers may serve not only as diagnostic parameters but also as predictive biomarkers that anticipate the subsequent development of sub-clinical and clinical acute rejection. Towards these objectives, we and others have investigated the hypotheses that urine and peripheral blood cell profiles offer a noninvasive means of predicting the development of acute rejection and are diagnostic of biopsy confirmed acute rejection [reviewed in (4)]. In our laboratory, we have also tested the postulate that urinary cell mRNA profiles that include measurement of mRNA for FOXP3 predicts the outcome of an episode of acute rejection [5]. Whereas our studies have been hypothesis driven and focused on mechanistically related genes, others have cast a wider net by the clinical application of “omics” technology and have developed biomarkers predictive of allograft status [6, 7•].
Figure 1.
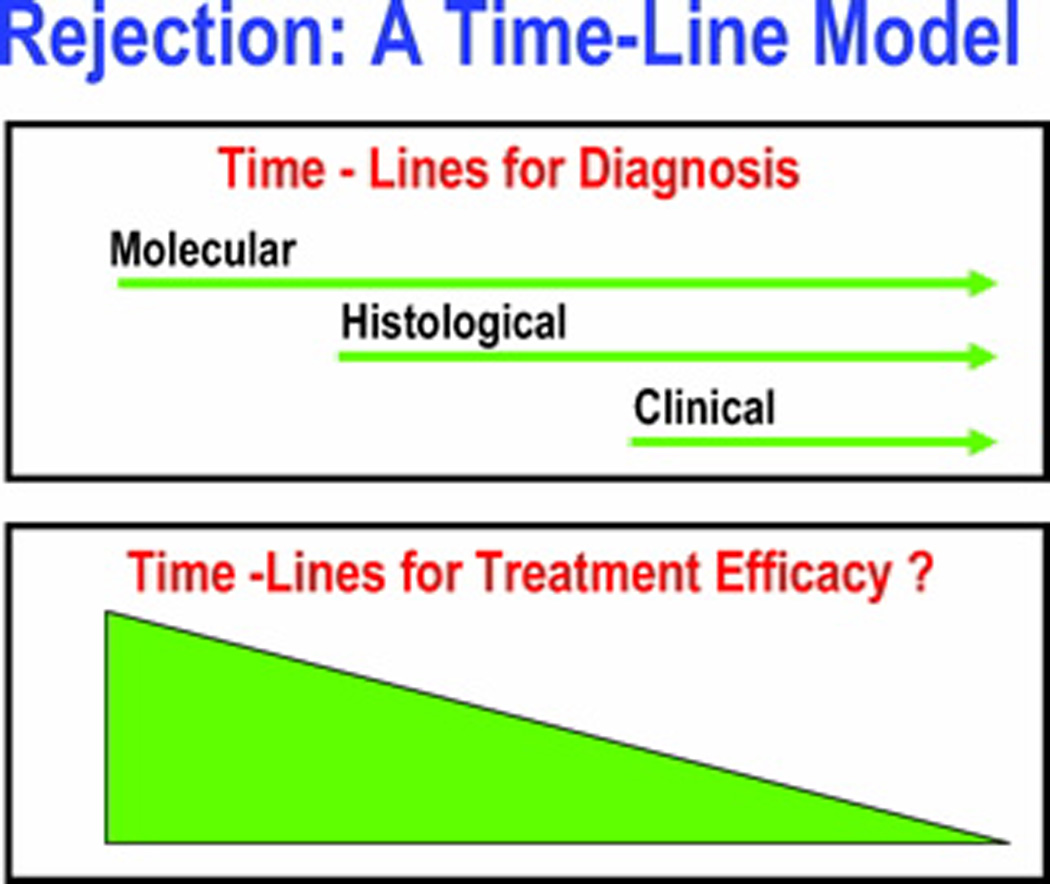
A time-line model for allograft rejection. In this formulation, rejection defined by molecular markers precedes histologically defined rejection and this precedes clinically defined rejection. The hypothesis that early intervention is efficacious is an important rationale for the development of molecular surveillance strategies to anticipate histologic and clinical rejection.
PLATFORMS FOR BIOMARKER DISCOVERY AND VALIDATION
Existing technologies to investigate gene expression patterns can be considered as those that are hypothesis testing (candidate-gene approach) or hypothesis generating (genome-wide sweep). Each approach has its own merits and limitations, and we believe that the two strategies are complementary rather than competitive. In addition to nucleic acid based assays, proteomics and metabolomics based approaches represent additional avenues for the development of biomarkers of allograft status.
Techniques have evolved to assess cell functions as well. Enzyme-linked immunosorbent spot assay (ELISPOT) testing detects the production of cytokines such as interferon-gamma (IFN-γ) in response to alloantigens, whereas the Immunknow® assay exploits ATP production in response to polyclonal mitogen phytohaemagglutinin as a measure of immune competence.
Flow cytometry and Luminex platform serve well to detect clinically relevant anti-allograft antibodies. Some of the tools used for biomarker discovery are summarized in Table 1, and reviewed by Hartono et al. [8].
Table 1.
Platforms for Biomarker Discovery and Validation
| TEST | PLATFORM | EXAMPLES OF POTENTIAL BIOMARKERS |
|
|---|---|---|---|
| Gene Transcripts |
Single gene | RT-PCR | mRNA: Granzyme B, Perforin, FoxP3; miRNA: miR155, miR223 |
| Multiple Genes | DNA Microarray | ||
| Proteins | Single Protein | Enzyme-linked immunosorbent assay (ELISA) |
Fractalkine, Amyloid A, β2 microglobulin |
| Multiple Proteins | Protein Microarray | ||
| Lymphocyte Function |
Cytokine Producing Cells |
Enzyme-linked immunosorbent spot assay (ELISPOT) |
IFN-γ |
| ATP levels in Activated T cells |
Immuknow® | ATP | |
| Alloantibodies | Single or Multiple Antibodies |
Luminex xMAP® | Anti-HLA Antibody, Anti-MICA Antibody |
NONINVASIVE BIOMARKERS OF ALLOGRAFT STATUS
A number of laboratories have investigated whether urinary cell and peripheral blood cell mRNA profiles are diagnostic of acute rejection in renal allografts [4]. A summary of data from the published studies is provided in Table 2.
Table 2.
mRNA Profiles Diagnostic of Acute Rejection of Renal Allografts.
| AUTHOR, YEAR [REF] | N* | URINARY CELL mRNA LEVELS DURING ACUTE REJECTION COMPARED TO NO ACUTE REJECTION |
|---|---|---|
| Li et al, 2001 [9] | 151/85 | Perforin and granzyme B higher |
| Muthukumar et al, 2003 [10] | 95/87 | PI-9, Granzyme B, and perforin mRNAs higher |
| Dadhania et al, 2003 [11] | 99/99 | Granzyme B higher; UTI did not increase granzyme B levels |
| Ding et al, 2003 [12] | 89/79 | CD103 higher |
| Kotsch et al, 2004 [13] | 221/26 | Granulysin higher. |
| Tatapudi et al, 2004 [14] | 63/58 | IP-10 and CXCR3 mRNAs higher |
| Muthukumar et al, 2005 [5] | 83/83 | FOXP3 mRNA higher |
| Matz et al, 2006 [15] | −/76 | IP-10 mRNA higher |
| Yannaraki et al, 2006 [16] | 162 /37 | Perforin, Granzyme B, and FasL mRNAs high during AR, UTI, CMV and DGF |
| Seiler et al, 2007 [17] | −/117 | NKG2D mRNA higher |
| Renesto et al, 2007 [18] | 72/72 | Tim-3 and IFN-γ mRNAs higher |
| Manfro et al, 2008 [19] | 165/115 | Tim-3 higher |
| Aquino-Dias et al, 2008 [20] | 48/35 | Perforin, granzyme B, FasL, Pi-9, FOXP3 higher |
| Ozbay et al, 2009 [21] | 64/64 | Perforin, granzyme B granulysin higher; granzyme B, granulysin but not perforin higher in AR compared to bacteriuria; perforin but not granzyme B and granulysin higher in AR compared to CMV |
| Vasconcellos et al, 1998 [22] | 31/25 | Granzyme B, perforin and FasL mRNAs increased |
| Dugre et al, 2000 [23] | −/21 | IL-4, IL-5, IL-6, IFN-γ, Granzyme B, and perforin mRNAs increased |
| Shoker et al, 2000 [24] | −/57 | CD40L mRNA increased in AR and/or CAN |
| Sabek et al, 2002 [25] | 27/27 | Granzyme B, perforin and HLA-DRA mRNAs increased |
| Netto et al, 2002 [26] | 206/29 | Granzyme B, perforin and FasL mRNAs increased |
| Simon et al, 2003 [27] | 364/67 | Granzyme B and perforin mRNAs increased |
| Shin et al, 2005 [28] | 88/15 | Perforin mRNA increased |
| Veale et al, 2006 [29] | 268/46 | Granzyme B and perforin mRNAs increased |
| Graziotto et al 2006 [30] | 64/− | Granzyme B, perforin, and FasL mRNAs increased |
| Manfro et al, 2008 [19] | 165/115 | Tim-3 higher |
| Aquino-Dias et al, 2008 [20] | 48/35 | Perforin, granzyme B, FasL, PI-9, FOXP3 higher |
number of samples/ number of patients
URINARY CELL MESSENGER RNA PROFILES DIAGNOSTIC OF ACUTE REJECTION OF RENAL ALLOGRAFTS
Cytotoxic T lymphocytes (CTL) have been implicated in allograft rejection, and data exist that both perforin and granzyme B contribute to the cytotoxic T cell machinery. Li et al. investigated the hypothesis that measurement of urinary cell levels of mRNA for perforin and granzyme B offers a noninvasive means of diagnosing acute rejection of renal allografts, and reported that acute rejection is predicted with a high degree of accuracy by urinary cell levels of perforin mRNA and granzyme B mRNA [9]. Muthukumar et al. and Dadhania et al. have confirmed that the levels of mRNAs encoding cytotoxic attack molecules are diagnostic of acute rejection of renal allografts [10,11], and showed further that mRNA for serine proteinase inhibitor-9 (PI-9), an endogenous antagonist of granzyme B, is increased in urine of patients with acute rejection [10], and bacterial urinary tract infection in renal allograft recipients is not associated with an increase in the levels of granzyme B in urine [11].
The cell surface protein CD103 is a natural ligand for E-cadherin, and Ding et al. reported that CD103 mRNA levels in urine are higher in patients with acute rejection compared to those without acute rejection [12]. Urinary cell levels of mRNA for granulysin, an effector molecule of expressed on cytotoxic T cells, were found to be diagnostic of acute rejection by Kotsch et al. [13]. Urinary cell levels of mRNA for chemokines and chemokine receptors have also been associated with acute rejection of renal allografts. Tatapudi et al. reported that levels of IP-10 mRNA and CXCR3 mRNA are diagnostic of acute rejection [14], and Matz et al. observed that levels of IP-10 mRNA as well as levels of IP-10 protein are diagnostic of acute rejection [15].
The role of FOXP3 expressing T regulatory cells in transplantation is being explored in a number of laboratories. Muthukumar et al. measured urinary cell levels of FOXP3 mRNA in renal allograft recipients and reported that levels of mRNA for FOXP3 in urine is increased during an episode of acute rejection, and the levels were inversely correlated with serum creatinine levels measured at the time of biopsy in the acute rejection group [5].
NKG2D, the activating cytotoxicity receptor, is expressed by all human NK cells and was reported by Seiler et al. that acute rejection of renal allografts is associated with an increase in urinary cell levels of NKG2D mRNA [17].
Urinary cell levels of mRNA for TIM-3, a type 1 membrane protein selectively expressed on the surface of terminally differentiated T-helper 1 cells, have been associated with acute rejection by Renesto et al. [18] and by Manfro et al. [19••]. Importantly, in the investigation by Manfro et al. urinary cell TIM-3 mRNA levels distinguished the 28 renal allograft recipients with delayed graft function (DGF) and biopsy diagnosis of acute rejection and acute tubular necrosis (ATN) from the 22 the recipients with DGF and biopsy diagnosis of ATN with a sensitivity of 100% and specificity of 100% [19]. Aqiuno-Dias et al. have extended the diagnostic accuracy of mRNA profiles in recipients with DGF, and reported that urinary cell levels of mRNA for perforin, granzyme B, FasL, PI-9 and FOXP3 predict acute rejection with a very high degree of accuracy [20•]. In this study, urinary cell FOXP3 mRNA levels were the most accurate and predicted acute rejection with a sensitivity of 100% and a specificity of 100%.
Immune response directed at infections could confound the diagnostic utility of inflammatory gene based signatures of acute rejection, and a number of laboratories have addressed this important concern. Whereas Dadhania et al. found that granzyme B mRNA levels are not increased in renal allograft recipients with bacterial urinary tract infection [11], Yannaraki et al. reported that mRNAs were not only increased during acute rejection but also in patients diagnosed with complications such as UTI, CMV infection, and DGF [16]. In accord with findings of Dadhania et al. that UTI is not associated with an increase in granzyme B mRNA in urine, Ozbay et al. found that granzyme B mRNA levels in urine are not increased in renal allograft recipients with bacteriuria and that both granzyme B and granulysin levels in urine distinguish acute rejection from bacteriuria [21]. It was also found in this study that perforin mRNA levels distinguish acute rejection from CMV infection.
PERIPHERAL BLOOD CELL MESSENGER RNA PROFILES DIAGNOSTIC OF ACUTE REJECTION OF ALLOGRAFTS
Vasconcellos et al. investigated expression patterns of mRNA for perforin, granzyme B, and Fas ligand (FasL) in peripheral blood mononuclear cells (PBMCs) collected from renal transplant recipients and reported that perforin mRNA predicted acute rejection with a sensitivity of 82% and a specificity of 85%; granzyme B mRNA predicted with a sensitivity of 55% and a specificity of 85%, and FasL with a sensitivity of 100% and a specificity of 75% [22]. In this study, the up-regulated expression of 2 or more genes was diagnostic of acute rejection with a positive predictive value of 100%, and a lack of up-regulation of any gene ruled out rejection with a negative predictive value of 95%.
The expression patterns of mRNA for cytokines have also been found to be informative of allograft status. Dugre et al. reported that levels of mRNA for IL-4, IL-5, IL-6, IFN-γ, as well as perforin, and granzyme B mRNA levels were correlates of acute rejection of renal allografts [23]. Levels of mRNAs for proteins central to costimulation have been associated with acute rejection. Shoker et al. have reported that peripheral blood cell CD40L mRNA levels are higher in kidney allograft recipients with acute rejection and/or chronic allograft nephropathy, and their levels were also predictive of acute rejection severity [24].
The original observations of Vasconcellos et al. that peripheral blood cell levels of perforin and granzyme are informative of renal allograft status [22] have been confirmed and extended by Sabek et al. [25], Netto et al. [26], Simon et al. [27], Shin et al. [28] and Veale et al. [29]. In contrast, Graziotto et al. reported that peripheral blood cell levels of mRNA for perforin, granzyme-B, and FasL are not significantly higher during acute rejection compared to no acute rejection and that the mRNA levels are not informative of allograft biopsy diagnosis [30].
A significant percentage of recipients of deceased donor grafts suffer from DGF and noninvasive diagnosis of acute rejection in this setting is of considerable significance. Manfro et al. have reported that peripheral blood cell levels of TIM-3 mRNA predict acute rejection in renal allograft recipients with DGF with a sensitivity of 100% and a specificity of 100% [19]. Aquino-Dias et al. have reported that peripheral blood cell levels of perforin, granzyme B, FasL, PI-9 and FOXP3 predict acute rejection in renal allograft recipients with DGF with a high degree of precision [20].
BIOMARKERS PREDICTIVE OF SUBSEQUENT DEVELOPMENT OF ACUTE REJECTION
Table 3 provides a summary of published biomarker studies that have been reported to predict the development of an episode of acute rejection allograft status.
Table 3.
Biomarkers Predictive of Subsequent Development of Acute Rejection of Renal Allografts
| SAMPLE | BIOMARKERS DETECTED | N* | END-POINT | AUTHOR, YEAR [REF] |
|---|---|---|---|---|
| COMPETITIVE QUANTITATIVE PCR | ||||
| Urine | Granzyme B mRNA and Perforin mRNA, 1–9 days post-transplant |
37 | Development of AR within 10 days of transplant |
Li et al, 2001 [9] |
| REAL TIME QUANTITATIVE PCR | ||||
| Urine | Serine Proteinase Inhibitor -9 mRNA during acute rejection |
29 | Serum creatinine at 6 months following AR |
Muthukumar et al, 2003 [10] |
| Urine | Granulysin mRNA 1–90 days post-transplant |
26 | Development of AR | Kotsch et al, 2004 [13] |
| Urine | FOXP3 mRNA during AR | 36 | Graft loss within 6 months following AR |
Muthukumar et al, 2005 [5] |
| Urine | IP-10 mRNA and protein post- transplant |
mRNA: 58 Protein: 70 |
Development of AR | Matz et al, 2006 [15] |
| Urine | IP-10 mRNA and protein post- transplant |
mRNA: 58 Protein: 70 |
Development of AR | Matz et al, 2006 [15] |
| Urine | NKG2D mRNA post-transplant | 94 | Development of AR | Seiler et al, 2007 [17] |
| Blood | Granzyme B mRNA and Perforin mRNA 5–29 days post-transplant |
67 | Development of AR | Simon T et al, 2003 [27] |
| Blood | Granzyme B and Perforin mRNA 1–65 weeks post-transplant |
46 | Development of AR | Veale et al, 2006 [29] |
| Blood | Perforin mRNA and IL-18 mRNA 1–16 days post-transplant |
54 | Development of AR | Simon et al, 2004, [31] |
| ELISPOT | ||||
| Blood | Pre-transplant donor-specific IFN-γ producing cells |
19 | Development of AR | Heeger et al, 1999 [32] |
| Blood | Pre-transplant donor-specific IFN-γ producing cells |
42 | Development of AR | Nickel et al, 2004 [33] |
| Blood | Pre-transplant donor-specific IFN-γ producing cells |
37 | Development of AR | Augustine et al, 2005 [34] |
| Blood | Pre-transplant donor-specific IFN-γ producing cells |
22 | Development of AR | Nather et al, 2006 [35] |
| ELISA | ||||
| Urine | MIG protein 5–90 days post- transplant |
69 | Development of AR | Hauser et al, 2005 [36] |
| Serum | sCD30 protein pre and day15-post transplant |
50 | Development of AR | Sengul et al, 2006 [37] |
| ATP Release Assay | ||||
| Blood | ATP levels pre and day 14 post- transplant |
58 | Development of AR | Cadillo-Chávez et al, 2006 [38] |
| Flow Cytometry | ||||
| Blood | Anti-endothelial cell (anti-Tie-2) antibodies pre-transplant |
147 | Development of AR ≥3 months after transplant. |
Breimer et al, 2009 [39•] |
number of patients
EMERGING BIOMARKERS OF RENAL ALLOGRAFT REJECTION
The association between the presence of donor specific antibodies (DSA) and acute rejection has been noted in the late 1970s [40]. With the effective control of T cell mediated acute rejection with the current immunosuppressive regimens, antibody-mediated rejection (AMR) of renal allograft has re-emerged as an important post-transplant complication.
Ashton-Chess et al. investigated mRNA for Tribbles-1 (TRIB1) as a biomarker for chronic AMR [41••]. TRIB1, an intracellular human homolog of Drosophila tribbles is involved in toll-like receptor-mediated response and in the regulation of nuclear factor κB and mitogen-activated protein kinases. Intragraft expression of TRIB1 mRNA was higher in biopsies with chronic AMR compared to normal one. PBMC levels of TRIB1 mRNA were also higher during chronic AMR. Levels of TRIB1 mRNA in biopsy samples and PBMC distinguished transplant glomerulopathy with positive C4d staining and anti-HLA antibodies from transplant glomerulopathy without positive C4d staining and anti-HLA antibodies. The authors did not find urinary cell levels TRIB1 mRNA to be informative of chronic AMR. Further studies examining the clinical utility of TRIB1 as a biomarker for antibody mediated rejection are worthy of pursuit.
Sis et al. investigated 173 renal allograft biopsies for-cause for intragraft expression of endothelial-associated transcripts (ENDATs) with the use of Affymetrix microarrays [42••]. The mean ENDAT scores were higher in biopsies showing rejection, and was also higher in AMR compared to T cell-mediated rejection. Death censored graft survival rates were inferior in those with antibodies, C4d and a high ENDAT score (ACE group) compared to no ACE group. However, a high ENDAT score alone or the presence of antibodies alone did not impact graft survival.
MICRO RNAs AS BIOMARKERS OF ALLOGRAFT STATUS
Micro RNAs (miRNAs) are small non-coding RNAs approximately 22 nucleotides long that regulate gene expression by inducing translational repression, mRNA degradation, and/or transcriptional inhibition [43]. A single miRNA has the ability to regulate the expression of hundreds of mRNAs. miRNAs have been shown to control processes such as cellular survival, development, differentiation, proliferation [44] as well as modulate both innate and adaptive immunity [45]. Anglicheau et al. recently identified several miRNAs predictive of acute rejection of human renal allografts [46••]. The hypothesis that urinary cell and/or peripheral blood cell miRNA expression profiles are predictive, diagnostic and/or prognostic biomarkers of allografts is worthy of investigation.
POTENTIAL CONFOUNDING FACTORS
Issues related to bacterial infections confounding the diagnostic utility of inflammatory gene based signatures of acute rejection have been addressed in the earlier section. Herein we discuss the issues related to polyomavirus BK- associated nephropathy (PVAN) [47]. An existing challenge is to distinguish allograft dysfunction due to PVAN from acute rejection. Rogers et al. investigated whether immunophenotyping of renal allograft infiltrates help in the differential diagnosis of PVAN versus acute rejection [48]. The investigators performed immunohistochemical analysis of 10 biopsy samples from 10 renal allograft recipients with PVAN and 20 biopsy samples from 20 patients with acute rejection. They found that the percentage of perforin positive cells were significantly different between the PVAN and acute rejection biopsies; they also found that the percentages of CD20-stained cells and granzyme B positive cells were not different between the PVAN and acute rejection biopsies [48]. Mannon et al. investigated mRNA profiles of renal allograft biopsies from 10 patients with PVAN and from 17 patients with acute rejection [49]. Banff inflammation and tubulitis scores were not different between the two groups whereas transcripts for CD8, CXCR3, Perforin, HLA-DR, and IFN-γ were significantly higher in the PVAN biopsies compared to the acute rejection group. Moreover, mRNA for TGFβ, MMP2 and 9, collagen I and IV, fibronectin were higher in the PVAN group compared to the acute rejection group [49]. An important goal in this area is to investigate whether acute rejection could be distinguished from PVAN noninvasively. In this regard, a major challenge is to exclude the possibility that the inflammatory signal associated with PVAN is due to PVAN alone and not due to co-existing acute rejection.
Technical issues may also confound biomarker discovery. Excessive globin mRNA in red blood cells has been reported to influence expression profiling of whole blood specimens. Field et al. reported that globin reduction resulted in the detection of additional 2652±395 genes when the Affymetrix HU133A 2.0 arrays was used to profile whole blood collected using the PAXgene blood RNA system [50]. Tian et al. found a lower than expected present call rates and high degree of sample-to-sample variability when whole blood samples were profiled using Affymetrix microarrays for biomarker discovery, and recommended both globin gene reduction and hybridization on Illuminia BeadChips [51]. Li et al. recommended the use of isolated leukocytes instead of whole blood, globin reduction, and mathematical depletion to alleviate the confounding globin molecular signatures [52].
CONCLUSIONS
Noninvasive strategies by allowing repetitive sampling should facilitate detection of immune rejection prior to fixed tissue injury. Importantly, noninvasive ascertained parameters may inform the timing of the biopsy procedure, and the information gleaned from both strategies may complement one another. We are optimistic of the progress to date, and anticipate clinical trials investigating the hypotheses that noninvasively ascertained mRNA profiles will minimize the need for an invasive allograft biopsy procedure; predict the development of acute rejection and chronic allograft nephropathy and facilitate preemptive therapy capable of preserving graft function; and bring about personalization of immunosuppressive therapy for the organ graft recipient.
ACKNOWLEDGEMENT
The authors gratefully acknowledge the research contributions of our colleagues Drs. Baogui Li, Ruchuang Ding, Vijay Sharma and Darshana Dadhania. The research studies in the author’s laboratory were supported by the awards R37 AI51652, R01 AI60706 and R01 AI072790 from the NIAID/NIH, and by the Clinical and Translational Science Award (UL1-RR024996) to Weill Cornell Medical College from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as: • of special interest •• of outstanding interest
- 1. Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 2008;8:753. doi: 10.1111/j.1600-6143.2008.02159.x. An update on the collaborative international effort to refine the classification and interpretation of allograft biopsies.
- 2.Sorof JM, Vartanian RK, Olson JL, et al. Histopathological concordance of paired renal allograft biopsy cores. Effect on the diagnosis and management of acute rejection. Transplantation. 1995;60:1215. [PubMed] [Google Scholar]
- 3.Furness PN, Taub N. International variation in the interpretation of renal transplant biopsies: report of the CERTRAP project. Kidney Int. 2001;60:1998. doi: 10.1046/j.1523-1755.2001.00030.x. [DOI] [PubMed]
- 4. Anglicheau D, Suthanthiran M. Noninvasive prediction of organ graft rejection and outcome using gene expression patterns. Transplantation. 2008;86:192. doi: 10.1097/TP.0b013e31817eef7b. A comprehensive review of data regarding mRNA profiles in organ graft recipients
- 5.Muthukumar T, Dadhania D, Ding RC, et al. Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med. 2005;353:2342. doi: 10.1056/NEJMoa051907. [DOI] [PubMed] [Google Scholar]
- 6.Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4:1475. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khatri P, Sarwal MM. Using gene arrays in diagnosis of rejection. Curr Opin Organ Transplant. 2009;14:34. doi: 10.1097/MOT.0b013e32831e13d0. An update on the use of microarray technology in organ transplantation.
- 8.Hartono C, Dadhania D, Suthanthiran M. Noninvasive diagnosis of acute rejection of solid organ transplants. Frontiers in Bioscience. 2004;9:145. doi: 10.2741/1218. [DOI] [PubMed] [Google Scholar]
- 9.Li B, Hartono C, Ding RC, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001;344:947. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 10.Muthukumar T, Ding RC, Dadhania D, et al. Serine proteinase inhibitor-9, an endogenous blocker of granzyme B/perforin lytic pathway, is hyperexpressed during acute rejection of renal allografts. Transplantation. 2003;75:1565. doi: 10.1097/01.TP.0000058230.91518.2F. [DOI] [PubMed] [Google Scholar]
- 11.Dadhania D, Muthukumar T, Ding R, et al. Molecular signatures of urinary cells distinguish acute rejection of renal allografts from urinary tract infection. Transplantation. 2003;75:1752. doi: 10.1097/01.TP.0000063931.08861.56. [DOI] [PubMed] [Google Scholar]
- 12.Ding R, Li B, Muthukumar T, et al. CD103 mRNA levels in urinary cells predict acute rejection of renal allografts. Transplantation. 2003;75:1307. doi: 10.1097/01.TP.0000064210.92444.B5. [DOI] [PubMed] [Google Scholar]
- 13.Kotsch K, Mashreghi MF, Bold G, et al. Enhanced granulysin mRNA expression in urinary sediment in early and delayed acute renal allograft rejection. Transplantation. 2004;77:1866. doi: 10.1097/01.tp.0000131157.19937.3f. [DOI] [PubMed] [Google Scholar]
- 14.Tatapudi RR, Muthukumar T, Dadhania D, et al. Noninvasive detection of renal allograft inflammation by measurements of mRNA for IP-10 and CXCR3 in urine. Kidney Int. 2004;65:2390. doi: 10.1111/j.1523-1755.2004.00663.x. [DOI] [PubMed] [Google Scholar]
- 15.Matz M, Beyer J, Wunsch D, et al. Early post-transplant urinary IP-10 expression after kidney transplantation is predictive of short- and long-term graft function. Kidney Int. 2006;69:1683. doi: 10.1038/sj.ki.5000343. [DOI] [PubMed] [Google Scholar]
- 16.Yannaraki M, Rebibou J-M, Ducloux D, et al. Urinary cytotoxic molecular markers for a noninvasive diagnosis in acute renal transplant rejection. Transplant International. 2006;19:759. doi: 10.1111/j.1432-2277.2006.00351.x. [DOI] [PubMed] [Google Scholar]
- 17.Seiler M, Brabcova I, Viklicky O, et al. Heightened expression of the cytotoxicity receptor NKG2D correlates with acute and chronic nephropathy after kidney transplantation. Am J Transplant. 2007;7:423. doi: 10.1111/j.1600-6143.2006.01625.x. [DOI] [PubMed] [Google Scholar]
- 18.Renesto PG, Ponciano VC, Cenedeze MA, et al. High expression of Tim-3 mRNA in urinary cells from kidney transplant recipients with acute rejection. Am J Transplant. 2007;7:1661. doi: 10.1111/j.1600-6143.2007.01795.x. [DOI] [PubMed] [Google Scholar]
- 19. Manfro RC, Aquino-Dias EC, Joelsons G, et al. Noninvasive Tim-3 messenger RNA evaluation in renal transplant recipients with graft dysfunction. Transplantation. 2008;86:1869. doi: 10.1097/TP.0b013e3181914246. The clinically important issue of noninvasively diagnosing acute rejection in patients with DGF is reported in this manuscript.
- 20. Aquino-Dias EC, Joelsons G, da Silva DM, et al. Non-invasive diagnosis of acute rejection in kidney transplants with delayed graft function. Kidney Int. 2008;73:877. doi: 10.1038/sj.ki.5002795. The report extends the findings in the report of Manfro et al. that noninvasive diagnosis acute rejection is feasible in patients with DGF.
- 21.Ǿzbay A, Torring C, Olsen R, et al. Transcriptional profiles in urine during acute rejection, bacteriuria, CMV infection and stable graft function after renal transplantation. Scandinavian Journal of Immunology. 2009;69:357. doi: 10.1111/j.1365-3083.2009.02226.x. [DOI] [PubMed] [Google Scholar]
- 22.Vasconcellos LM, Schachter AD, Zheng XX, et al. Cytotoxic lymphocyte gene expression in peripheral blood leukocytes correlates with rejecting renal allografts. Transplantation. 1998;66:562. doi: 10.1097/00007890-199809150-00002. Erratum in: Transplantation 1998;66:1264. [DOI] [PubMed] [Google Scholar]
- 23.Dugre FJ, Gaudreau S, Belles-Isles M, et al. Cytokine and cytotoxic molecule gene expression determined in peripheral blood mononuclear cells in the diagnosis of acute renal rejection. Transplantation. 2000;70:1074. doi: 10.1097/00007890-200010150-00014. [DOI] [PubMed] [Google Scholar]
- 24.Shoker A, George D, Yang H, et al. Heightened CD40 ligand gene expression in peripheral CD4+ T cells from patients with kidney allograft rejection. Transplantation. 2000;70:497. doi: 10.1097/00007890-200008150-00018. [DOI] [PubMed] [Google Scholar]
- 25.Sabek O, Dorak MT, Kotb M, et al. Quantitative detection of T-cell activation markers by real-time PCR in renal transplant rejection and correlation with histopathologic evaluation. Transplantation. 2002;74:701. doi: 10.1097/00007890-200209150-00019. [DOI] [PubMed] [Google Scholar]
- 26.Netto MV, Fonseca BA, Dantas M, et al. Granzyme B, FAS-ligand and perforin expression during acute cellular rejection episodes after kidney transplantation: comparison between blood and renal aspirates. Transplant Proc. 2002;34:476. doi: 10.1016/s0041-1345(02)02601-5. [DOI] [PubMed] [Google Scholar]
- 27.Simon T, Opelz G, Wiesel M, et al. Serial peripheral blood perforin and granzyme B gene expression measurements for prediction of acute rejection in kidney graft recipients. Am J Transplant. 2003;3:1121. doi: 10.1034/j.1600-6143.2003.00187.x. [DOI] [PubMed] [Google Scholar]
- 28.Shin GT, Kim SJ, Lee TS, et al. Gene expression of perforin by peripheral blood lymphocytes as a marker of acute rejection. Nephron Clin Pract. 2005;100:c63. doi: 10.1159/000085050. [DOI] [PubMed] [Google Scholar]
- 29.Veale JL, Liang LW, Zhang Q, et al. Noninvasive diagnosis of cellular and antibody-mediated rejection by perforin and granzyme B in renal allografts. Human Immunol. 2006;67:777. doi: 10.1016/j.humimm.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Graziotto R, Del Prete D, Rigotti P, et al. Perforin, granzyme B, and Fas ligand for molecular diagnosis of acute renal-allograft rejection: analyses on serial biopsies suggest methodological issues. Transplantation. 2006;81:1125. doi: 10.1097/01.tp.0000208573.16839.67. [DOI] [PubMed] [Google Scholar]
- 31.Simon T, Opelz G, Wiesel M, et al. Serial peripheral blood interleukin-18 and perforin gene expression measurements for prediction of acute kidney graft rejection. Transplantation. 2004;77:1589. doi: 10.1097/01.tp.0000121764.92730.fb. [DOI] [PubMed] [Google Scholar]
- 32.Heeger PS, Greenspan NS, Kuhlenschmidt S, et al. Pretransplant frequency of donor-specific IFN-γ-producing lymphocytes is a manifestation of immunologic memory and correlates with the risk of posttransplant rejection episodes. J Immunol. 1999;163:2267. [PubMed] [Google Scholar]
- 33.Nickel P, Presber F, Bold G, et al. Enzyme-linked immunosorbent spot assay for donor-reactive interferon-gamma-producing cells identifies T-cell presensitization and correlates with graft function at 6 and 12 months in renal-transplant recipients. Transplantation. 2004;78:1640. doi: 10.1097/01.tp.0000144057.31799.6a. [DOI] [PubMed] [Google Scholar]
- 34.Augustine JJ, Siu DS, Clemente MJ, et al. Pre-transplant IFN-gamma ELISPOTs are associated with post-transplant renal function in African-American renal transplant recipients. Am J Transplant. 2005;8:1971. doi: 10.1111/j.1600-6143.2005.00958.x. [DOI] [PubMed] [Google Scholar]
- 35.Nather BJ, Nickel P, Bold G, et al. Modified ELISPOT technique – highly significant inverse correlation of post-Tx donor-reactive IFNγ-producing cell frequencies with 6 and 12 months graft function in kidney transplant recipients. Transpl Immunol. 2006;16:232. doi: 10.1016/j.trim.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 36.Hauser IA, Spiegler S, Kiss E, et al. Prediction of acute renal allograft rejection by urinary monokine induced by IFN-γ (MIG) J Am Soc Nephrol. 2005;16:1849. doi: 10.1681/ASN.2004100836. [DOI] [PubMed] [Google Scholar]
- 37.Sengul S, Keven K, Gormez U, et al. Identification of patients at risk of acute rejection by pretransplantation and posttransplantation monitoring of soluble CD30 levels in kidney transplantation. Transplantation. 2006;81:1216. doi: 10.1097/01.tp.0000203324.49969.30. [DOI] [PubMed] [Google Scholar]
- 38.Cadillo-Chavez R, de Echegaray S, Santiago-Delpin EA, et al. Assessing the risk of infection and rejection in Hispanic transplant recipients by means of an adenosine triphosphate release assay. Transplant Proc. 2006;38:918. doi: 10.1016/j.transproceed.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 39. Breimer ME, Rydberg L, Jackson AM, et al. Multicenter evaluation of a novel endothelial cell crossmatch test in kidney transplantation. Transplantation. 2009;87:549. doi: 10.1097/TP.0b013e3181949d4e. This article introduced a novel flow-cytometry crossmatch technique to predict acute rejection.
- 40.Suthanthiran M, Garovoy MR. Immunologic monitoring of the renal transplant recipient. Urol Clin North Am. 1983;10:315. [PubMed] [Google Scholar]
- 41. Ashton-Chess J, Giral M, Mengel M, et al. Tribbles-1 as a novel biomarker of chronic antibody-mediated rejection. J Am Soc of Nephrol. 2008;19:1116. doi: 10.1681/ASN.2007101056. This article identified a potential biomarker for antibody-mediated rejection in kidney transplant.
- 42. Sis B, Jhangri GS, Bunnag S, et al. Endothelial gene expression in kidney transplants with alloantibody indicates antibody-mediated damage despite lack of C4d staining. Am J Transplant. 2009;9:1. doi: 10.1111/j.1600-6143.2009.02761.x. This article identified novel endothelial related molecular biomarkers for antibody-mediated rejection in kidney transplant.
- 43.Hoefig KP, Heissmeyer MicroRNAs grow up in the immune system. Curr Opin in Immunology. 2008;20:281. doi: 10.1016/j.coi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 44.Lodish HF, Zhou B, Liu G, et al. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 45.Valencia-Sanchez MA, Liu J, Hannon GJ, et al. Control of translational and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 46. Anglicheau D, Sharma VK, Ding RC, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci USA. 2009;106:5330. doi: 10.1073/pnas.0813121106. This article demonstrated miRNA expression patterns to be biomarkers of acute rejection of renal allografts.
- 47.Ramos E, Drachenberg CB, Wali R, et al. The decade of polyomavirus BK-associated nephropathy: state of affairs. Transplantation. 2009;87:621. doi: 10.1097/TP.0b013e318197c17d. [DOI] [PubMed] [Google Scholar]
- 48.Rogers NM, Russ GR, Cooper J, et al. Immunophenotyping of interstitial infiltrate does not distinguish between BK virus nephropathy and acute cellular rejection. Nephrology. 2009;14:118. doi: 10.1111/j.1440-1797.2008.01050.x. [DOI] [PubMed] [Google Scholar]
- 49.Mannon RB, Hoffmann SC, Kampen RL, et al. Molecular evaluation of BK polyomavirus nephropathy. Am J Transplant. 2005;5:2883. doi: 10.1111/j.1600-6143.2005.01096.x. [DOI] [PubMed] [Google Scholar]
- 50.Field LA, Jordan RM, Hadix JA, et al. Functional identity of genes detectable in expression profiling assays following globin mRNA reduction of peripheral blood samples. Clin Biochem. 2007;40:499. doi: 10.1016/j.clinbiochem.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Tian Z, Palmer N, Schmid P, et al. A practical platform for blood biomarker study by using global gene expression profiling of peripheral whole blood. PLoS One. 2009;4:5157. doi: 10.1371/journal.pone.0005157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L, Ying L, Naesens M, et al. Interference of globin genes with biomarker discovery for allograft rejection in peripheral blood samples. Physiol Genomics. 2008;32:190. doi: 10.1152/physiolgenomics.00216.2007. [DOI] [PubMed] [Google Scholar]


