1. Introduction
DNA charge transport (CT) chemistry has received considerable attention by scientific researchers over the past 15 years since our first provocative publication on long range CT in a DNA assembly.1,2 This interest, shared by physicists, chemists and biologists, reflects the potential of DNA CT to provide a sensitive route for signaling, whether in the construction of nanoscale biosensors or as an enzymatic tool to detect damage in the genome. Research into DNA CT chemistry began as a quest to determine whether the DNA double helix, a macromolecular assembly in solution with π-stacked base pairs, might share conductive characteristics with π-stacked solids. Physicists carried out sophisticated experiments to measure the conductivity of DNA samples, but the means to connect discrete DNA assemblies into the devices to gauge conductivity varied, as did the conditions under which conductivities were determined. Chemists constructed DNA assemblies to measure hole and electron transport in solution using a variety of hole and electron donors. Here, too, DNA CT was seen to depend upon the connections, or coupling, between donors and the DNA base pair stack. Importantly, these experiments have resolved the debate over whether DNA CT is possible. Moreover these studies have shown that DNA CT, irrespective of the oxidant or reductant used to initiate the chemistry, can occur over long molecular distances but can be exquisitely sensitive to perturbations in the base pair stack.
Here we review some of the critical characteristics of DNA charge transport chemistry, taking examples from a range of systems, and consider these characteristics in the context of their mechanistic implications. This review is not intended to be exhaustive but instead to be illustrative. For instance, we describe studies involving measurements in solution using pendant photooxidants to inject holes, conductivity studies with covalently modified assemblies, and electrochemical studies on DNA-modified electrodes. We do not focus in detail on the differences amongst these constructs but instead on their similarities. It is the similarity among these various systems that allows us to consider different mechanisms to describe DNA CT. Thus we review also the various mechanisms for DNA CT that have been put forth and attempt to reconcile these mechanistic proposals with the many disparate measurements of DNA CT. Certainly the debate among researchers has shifted from "is DNA CT possible?" to "how does it work?". This review intends to explore this latter question in detail.
2. Properties of Long-Range Charge Transport in DNA
Among the most interesting characteristics of charge transport in DNA is the long distance over which it occurs (Figure 1).3–6 Nevertheless, there are some DNA systems that do not mediate charge over long distances. How DNA CT occurs depends upon coupling and the structure and dynamics of the DNA assembly. The chemistry and photophysics of the photoexcited acridine (Acr*+) containing systems, which mediate CT over only a few base pairs, have been particularly well-characterized in this regard.7,8 It is important to note that the same physical laws apply to all CT processes.9 The essential distinctions are with respect to the relative roles which different mechanisms play, and it is in this respect that long-range CT, with effective coupling to the base pair stack, differs from short-range CT, with poor coupling. Here we focus on long-range CT, where transport is through the base pair stack, and discuss short range systems only to the necessary extent to clarify the distinctions between the two regimes.
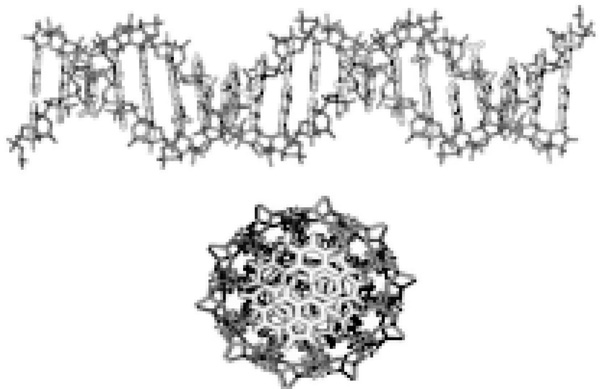
Figure 1.
Transverse and longitudinal perspectives of DNA. The sugar phosphate backbone envelops the hydrophobic base pairs. The planar base pairs form a one-dimensional π-stack down the center of the DNA, insulated by the backbone.
2.1. Coupling to the DNA
It is notable that initial measurements of DNA-mediated charge transport for both photooxidation experiments10 and device experiments11 found rates and conductivities spanning several orders of magnitude over comparable distances, depending on the experimental conditions. This foreshadowed the same observation in scanning tunneling microscopy (STM) measurements of conductivity through other molecular bridges,12 and, ultimately, was for the same reason. For short molecular bridges, it has been established both experimentally12 and theoretically13 that the coupling between the bridge and the donor (or acceptor) can dominate the observed conductivity. Similarly, when DNA is the bridge, the coupling can have a dramatic effect on both charge transport rates and yields (Figure 2).14–18 Characteristically, conductivity measurements that have not provided covalent contact between the DNA and the device yield a spectrum of behavior: from insulating to superconductive.11,19–23
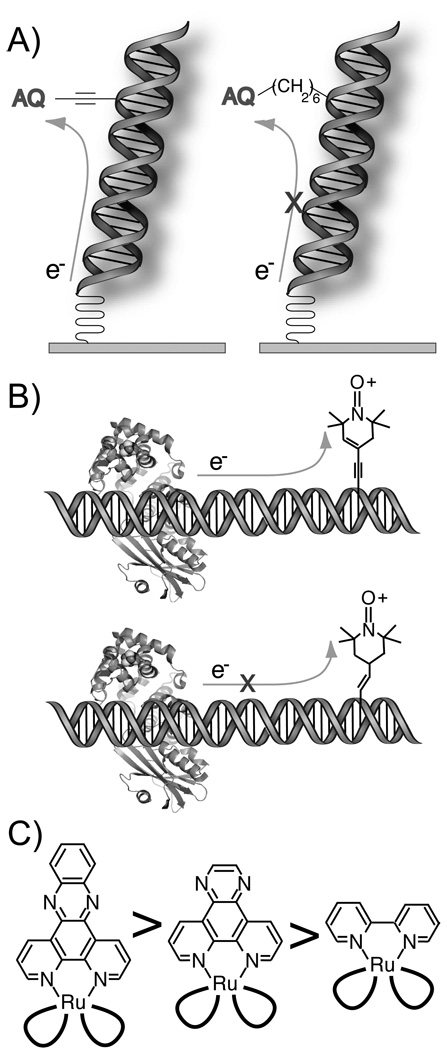
Figure 2.
DNA-mediated CT requires electronic coupling to the base pair stack. (A) Electrochemical reduction of an electronically well-coupled anthraquinone (AQ) is facile, while that of a poorly coupled AQ is suppressed.18 (B) MutY competently reduces an oxidized nitroxide spin label that is well coupled to thymidine, but not the nitroxide conjugated through the partially unsaturated linker.160 (C) For a series of polypyridyl RuIII(bpy)2L ground state oxidants, the yield of oxidative damage to DNA scales with the size and planarity of the intercalating ligand.15
In the case of DNA, the essential coupling is into the π-stack of the bases. This is a marked challenge, as DNA is essentially “insulated”, with sugars and phosphates flanking the periphery of the bases.24 This insulation, in part, explains why early experiments on dry DNA found insulating behavior, in contrast to that observed with conducting organic polymers. A series of well-conjugated charge donors and acceptors are now employed by various groups,25,26 including metallointercalators, organic intercalators, organic end-cappers, and modified bases. In several cases, direct comparison has been made between similar photooxidant pairs that differ primarily in their ability to couple well with the base stack. These examples include the adenine analogues ethenoadenine and 2-aminopurine (Ap),14 two different coupling strategies for ethidium bromide,27 and, most notably, a series of intercalating ruthenium analogues with decreasing planarity in the intercalating ligand.15 As an extreme case, for two ruthenium complexes that are unable to intercalate, and that are attached on opposite ends of a short DNA duplex via terminal phosphate modification, the CT rate was found to be ~10−6 s−1;28 this is what would be expected for the rate were the metal complexes connected solely through their σ tethers. Similarly, electrochemiluminescence studies find the same rates for DNA-mediated CT between a DNA-modified gold electrode and tethered Ru(bpy)32+ as are observed through solely the tether itself.29 In electrochemical studies of methylene blue covalently attached to a DNA duplex, effective transport is found only when the methylene blue is stacked in the helix, not under high salt conditions, where the dye, although still linked still by a σ-bonded tether, is unstacked.30 Indeed, electrochemical measurements on DNA films generally have been shown to be rate-limited by tether linking the DNA to the electrode surface31. In each case, it is clear that the coupling between the donor/acceptor pair and the bridge is dominating the measurement, and that the bridge is the π-stack of DNA.
2.2 Global Structural Integrity
The structure of DNA is central to its extraordinary effectiveness as the genetic template for the cell. This relationship between structure and function is underscored by the extent of the biological function that was first predicted in the landmark papers that reported the proper three-dimensional structure32,33. Hence, it is not surprising that DNA-mediated CT is also substantially affected by the global structure of a DNA sample. This is clear when considering the results of conductivity measurements on single or few DNA strands that have been performed in recent years. Various measurements from 1996 to the present have found DNA conductivities covering several orders of magnitude. Furthermore, conductivity has been found to be dependent on sequence, hydration, length, temperature, and hybridization in some experiments, while independent of each of those in others. Ultimately, the vast differences in observations can be largely reconciled by comparing the sample preparation methodologies of the individual studies.11 Conditions that cause global DNA conformational changes or damage can both increase or decrease the observed conductivity. In one extreme case, it was found that imaging conditions commonly used prior to conductance measurements lead to a morphological change in the structure of the DNA, that is itself correlated with increased conductivity.20
Among experiments that examine undamaged DNA, a profound difference is always observed between single-stranded and double-stranded DNA: single-stranded DNA does not mediate CT over long distances. This has been observed by direct conductivity studies,34 photooxidation,35 transient absorption,36 electrochemical AFM,37,38 STM,39 electrogenerated chemiluminescence,29 and electrochemical experiments in DNA monolayers.40 The caveat in interpreting studies on single stranded DNA is, however, that its structure and, importantly, stacking are heterogeneous and extremely dependent on sequence.
DNA, stabilized by a variety of hydrophobic and hydrophilic interactions and evolved for an aqueous environment, undergoes gross structural changes as a result of moving from a hydrated to a dehydrated environment.41 Critically, these changes are to the equilibrium conformation of DNA; the effects of dehydration on DNA dynamics are not well understood. Highly bound waters play a major role in the dynamics that gate molecular recognition and other biochemical interactions between macromolecules.42
Regrettably, the first and many recent measurements of DNA conductivity were performed under vacuum. Vacuum is ideal for conductivity measurements due to the suppression of voltage leak and the associated background current. Even experiments performed in the presence of water frequently deposit the DNA under vacuum conditions. Similar to the previous case of poorly coupled versus well-coupled systems, there is wide disparity between the conductivities observed under conditions of low humidity. Recently, progress has been made in understanding the role of humidity in many of the poorly coupled systems.43 Even without strong coupling into the DNA base stack, water adsorbed on the DNA and in DNA bundles can mediate ionic conduction. The amount of adsorbed water will depend strongly on humidity, and also on the adsorption environment of the DNA. This helps explain why many systems in which coupling to DNA was poor were still observed to conduct.19,23,44
Not surprisingly, experiments that have preserved the DNA in its native conformation, with leads covalently coupled to the bridge, have shown remarkably similar (and substantial) conductivities (Figure 3).34,37,44,45 The conductivity measured by Xu et al. across a dodecanucleotide with terminal propylthiol-Au contacts (> 40 Å) is comparable (6 × 10−4 G0, where G0 is the quantum unit of conductance) to that found across the much smaller benzenedimethanethiol (~10 Å) under the same experimental conditions,47 though this comparison is complicated by the possibility that DNA accommodates internal stretching during the measurement rather than extruding gold from the molecular junction, as is postulated for benzenedimethanethiol.
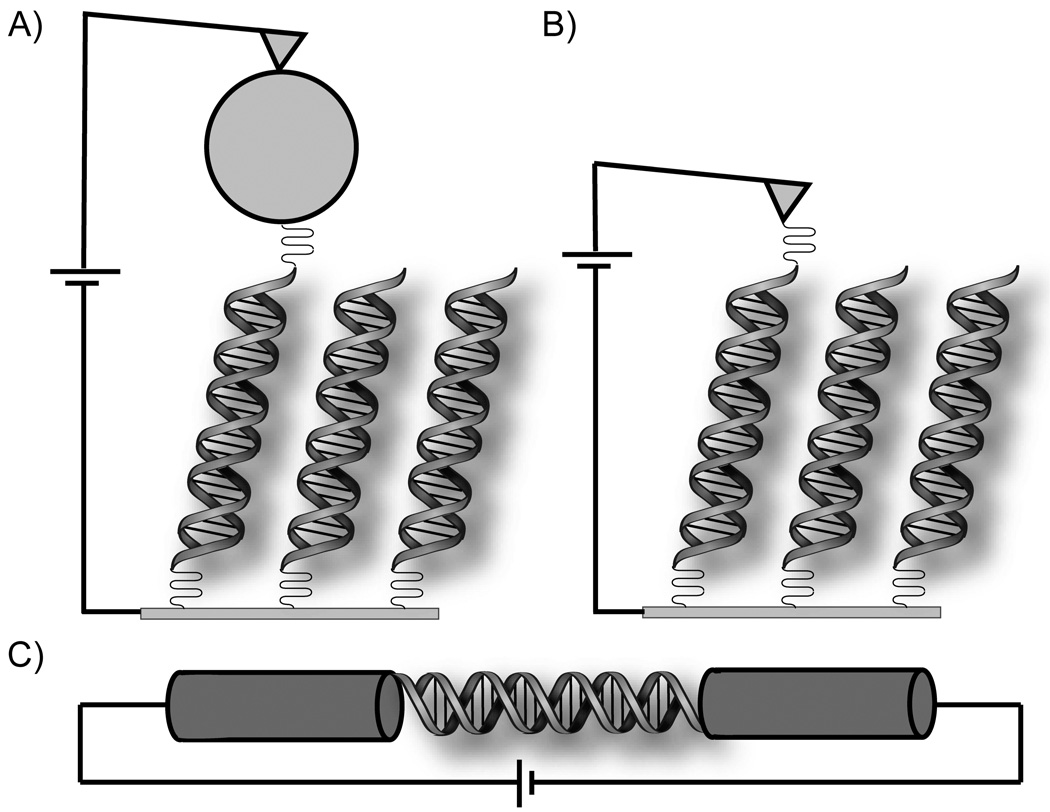
Figure 3.
Devices for measurement of single molecule DNA conductivity through molecular contacts. In each case, currents between 10 and 100 nA are obtained for modest source-drain and gating voltages. A) A gold nanoparticle allows strong coupling between the EC-AFM tip and an individual 26mer DNA molecule on a gold electrode.37 B) The gold STM tip is slowly brought in contact with thiol-modified DNA (8mer), allowing a histogram of conductance over many different orientations.45 C) A single 15mer DNA is covalently attached across a gap between single-walled carbon nanotubes.34
As is the case with water, ionic strength can dictate the conformation of DNA. High ionic strength drives the transition from B-form to the more extended Z-form of DNA. Poor base stacking, associated with this condensed structure, leads to less efficient DNA-mediated CT.48 Conversely, sufficiently low ionic strength leads to strand dehybridization, which also suppresses CT.
Beyond issues of ionic strength, there is conflicting evidence as to whether the identity of the counterion affects CT. Some calculations have shown that counterion identity does not affect the electric field inside the DNA,49 while others have found that movement of a single sodium has profound effects on base energies.50 Similarly contrasting results have been observed in experimental work.51–54 For solvent-exposed donors and acceptors, an ion-pair can form between dye and counter-ion that itself profoundly affects CT rate and yield.55
More chemically controlled experiments elucidate the structural basis of environmental effects. RNA/DNA hybrids and double stranded RNAs adopt the A-form while alternating purine-pyrimidine sequences under certain conditions adopt Z-form structures. Both conformations support DNA charge transport, though Z-form is an inferior bridge relative to A-form and B-form for electrochemical,48 but not photooxidation assays.56 Not surprisingly, the competence for mediating CT has been shown to follow the extent of base stacking, both in solution studies with Ap as the photooxidant57 and in electrochemical experiments monitoring the efficiency of reducing an intercalated redox probe.48 Again, different coupling of the redox probes into these different conformations means that they cannot be quantitatively compared.
Perhaps most interesting is the comparison of rates of intrastrand versus interstrand base-base CT in DNA assemblies modified with Ap.14,35,57 Here for the B-conformation, intrastrand CT is found to be three orders of magnitude faster than interstrand CT, consistent with the fact that stacking in the B-conformation is exclusively intrastrand; CT across strands requires CT across a hydrogen bond. However, in the A-form there is a mix of interstrand and intrastrand stacking down the helix, and here we observe that rates of intrastrand and interstrand CT are comparable.
2.3. Local Structural Integrity
A variety of studies have found similar effects of disrupting the base stack locally. The assays include electrochemical experiments in both films30,58,59 and devices,34 and solution experiments using time-resolved fluorescence,60 irreversible trapping of chemical product61 and transient absorption measurements.62 The presence of mismatches lowers both the rate and yield of DNA-mediated CT, and the extent of this attenuation scales with the base pair lifetime.63 Abasic sites64 and destabilizing lesions65 also interfere with CT through DNA films.
Regarding those experiments that utilize product trapping, however, it is important to note that the results are convoluted with two effective clocks. The first is the rate of back electron transfer (BET), if it occurs.66 The second is the rate of product trapping.66–69 A disruption of the π-stack will only be observable in product trapping experiments if it is sufficient to disrupt equilibration of the radical cation on the time-scale of BET and product trapping.70 Towards this end, guanine damage assays have recently been replaced by assays for fast decomposition of a radical trap. N-cyclopropylguanosine (CPG),71,72 N-cyclopropyladenosine (CPA),17,73,74 and N-cyclopropylcytidine (CPC)75,76 are synthetically accessible, cause minimal perturbation to the DNA-duplex, and undergo irreversible picosecond ring-opening upon oxidation or reduction.
Not all modifications of DNA suppress long-range CT. A dephosphorylation nick to the backbone, despite causing substantial change to local ion density, does not have a measurable effect.77,78 Furthermore, some modifications can enhance CT. Most notably, DNA with adenines replaced by the lower-potential base deazaadenine or the better-stacking benzodeazaadenine improves the rate and yield of DNA-mediated CT.79–81
In addition to global changes in structural integrity, subtle modulations to structure can also have profound effects on the rates and yields of DNA-mediated CT. DNA-mediated CT is attenuated by the presence of mismatches, even though mismatched base pairs cause only minor distortions to the structure of DNA.82,83 Nevertheless, mismatch discrimination has been observed in charge transport through DNA films,58,59,84 single molecule devices,34,85 and photooxidation systems.35,61 This attenuation is identical for oxidative and reductive CT,86 implying mechanistic similarity. Importantly, the extent of mismatch discrimination corresponds to the base-pair lifetime associated with the specific mismatch.63 In the extreme case, an abasic site completely suppresses CT.64,87,88 Subtle lesions, such as the oxidative guanine products O6-methyl-guanine and 8-oxoguanine, attenuate CT.65 It should be noted that although 8-oxoguanine terminates DNA-mediated CT as a thermodynamic and kinetic trap at high driving force,89,90 this and other damaged base products also attenuate CT even under driving forces incompetent for direct oxidation. This property of DNA-mediated CT has led to the development of a new class of DNA-detection devices,91,92 and might be relevant to damage detection in the cell.93
Local changes to structure can also be induced. Inside the cell, proteins can bend, twist, and dehybridize DNA, and some can extrude bases as well. Not surprisingly, many of these binding events have severe effects on DNA-mediated CT. Monitoring DNA-mediated electrochemistry to SoxR, a transcription factor that initiates the oxidative stress response in E. coli, is consistent with the prediction that the oxidized form twists DNA to initiate transcription,94 as has since been validated by a recent crystal structure.95 CT is also inhibited by the sequence-specific binding of TATA-binding protein (TBP),58,96 which bends DNA. One particularly informative experiment involved the methylase M.HhaI, which extrudes a cytidine within its recognition sequence, replacing it with glutamine. DNA with the HhaI recognition site shows attenuated CT in the presence of the protein. The Q237W mutant, in which the intercalating glutamate is replaced with the aromatic ligand tryptophan, however, barely affects CT compared to the absence of protein.58,97 Importantly, proteins that do not distort the DNA π-stack, such as antennapedia homeodomain protein or unactivated R.PvuII, do not attenuate DNA-mediated CT.97 Indeed, the rigidification associated with R.PvuII binding increased CT through the dynamically flexible TATA binding site. An interesting exception is the case of R.BamHI, a restriction endonuclease which does not bend the DNA π-stack, but contains an asparagine guanidinium that hydrogen bonds to a guanine in its cognate site. It has been shown that R.BamHI attenuates DNA CT to guanines both within and beyond its binding site, presumably due to electrostatic modulation of the intervening DNA via the hydrogen bonding interactions.98
Protein binding can also affect the fate of radicals in DNA.99–101 Guanine radical has been shown to crosslink to histones and short peptides. In one case,102 excitation of an anthraquinone (AQ)-DNA conjugate bound to a reconstituted nucleosome particle led to DNA-protein crosslinks; experiments with a different photooxidant with facile back electron transfer found no such effect.103 Since it is clear that migration can occur over long distances in both isolated nuclei104 and mitochondria,105 this might not be a fast pathway for radical quenching; the time-scale of protein-DNA crosslinking in the presence of guanine radical is similar to that for guanine radical decomposition to 8-oxoguanine.106 These crosslinks are reversible under the processing conditions used to convert base damage to strand cleavage, and hence are likely hidden in typical gel analysis experiments. Furthermore, protein cross-links can be formed as a secondary product after radical degradation in DNA to 8-oxoguanine.107 Recently, it has been shown that a protein that disrupts the π-stack, Hbb of Borrelia burgdorferi, affects the conversion of radical damage to interstrand crosslinks.108
DNA CT is sensitive to even more subtle deviations in stacking integrity. The strongest stacking interactions occur between consecutive purines, as has been shown both experimentally and computationally. Extended purine-pyrimidine runs correspond to the minimal extent of base-stacking, while purine-purine runs, particularly adenine tracts, correspond to the maximal extent for DNA containing natural nucleotides. This relationship is borne out in the sequence dependence of CT.51,109–111 Note that in photoactivated studies of electron transport, runs of pyrimidines, which are more easily reduced, are the preferred sequences.112 Likely here too, the decreased flexibility of homopurine-homopyrimidine sequences plays a role.
2.4. Conformational Gating
The rate and efficiency of charge transfer is centrally related to the structure of the individual pathway(s) that mediates CT between the donor and acceptor. Over long distances, however, it is inevitable that fluctuations will be induced at nonzero temperature, such that the equilibrium structure only reflects an average over the ensemble. If these fluctuations are sufficiently large, and slower than CT for the equilibrium conformation, then CT no longer is properly described by a unitary rate constant, but will instead proceed with a complicated time-dependence that convolutes the conformational dynamics with the CT rates of the different conformations.113 To avoid this, the best-performing molecular wires are chosen for, among other properties, rigid homogeneous conformations.114 For example, the conduction of certain oligophenyleneethynylenes can be substantially enhanced by the presence of bulky side-groups that limit the conjugation-breaking rotation around the acetylene bonds.115,116
It is not surprising that DNA, which even as a relatively short 15-mer contains several hundred atoms, exhibits substantial conformational flexibility. Interestingly, the effect of conformational gating in DNA is generally to increase CT rather than to decrease it. Duplexes frozen in glass show no attenuation in CT between the photooxidant Ap and neighboring guanine hole acceptor (Figure 4).117 The insertion of a single adenine, however, between donor and acceptor completely suppresses CT. The temperature-dependence of CT between Ap* and G for varying bridge length has been studied by both femtosecond transient absorption spectroscopy35 and steady-state fluorescence quenching measurements.54 The temperature-dependence has also been measured for CT between tethered, photoexcited [Rh(phi)2(bpy’)]3+ and CPG.68 For all bridges, CT efficiency increases with temperature, and the temperature dependence is greater with increasing bridge length.
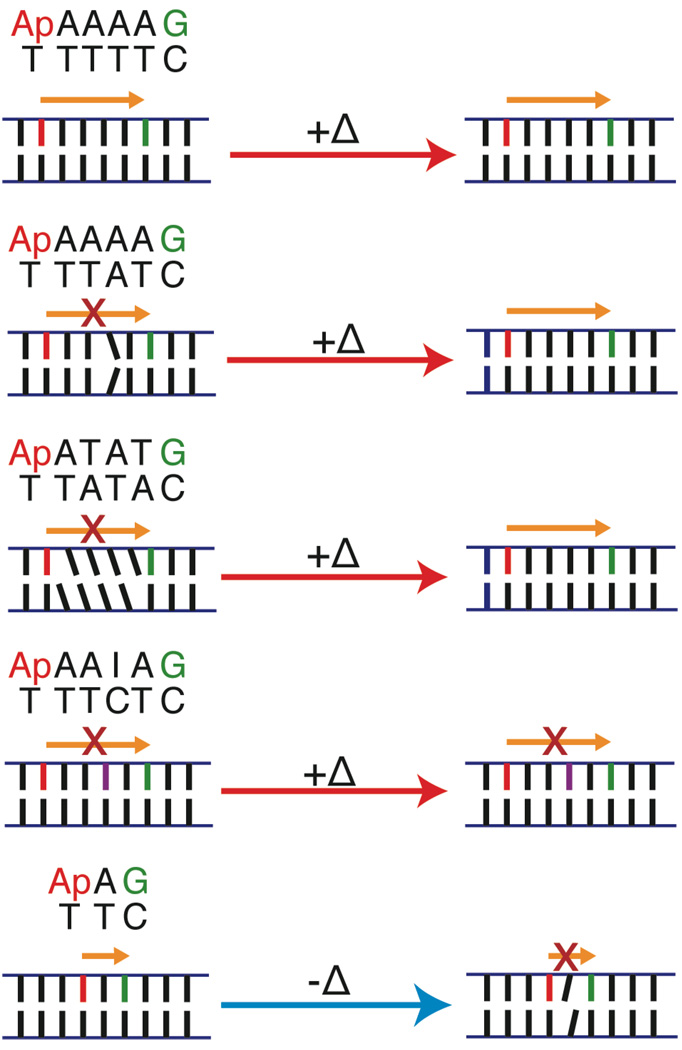
Figure 4.
The sequence and temperature dependence of single-step oxidation of guanine by photoexcited 2-aminopurine (Ap) in DNA.35,54,117 CT yield in well-matched Ap(A)4G increases with temperature (+Δ), up to duplex melting. Two perturbations that disturb CT due to poor stacking dynamics, an A–A mismatch and the sequence ATAT, attenuate CT at room temperature but are comparable to the A4 sequence at higher temperature, while CT through a perturbation that disturbs CT due to an electronic barrier, AAIA, is only partially recovered at high temperature. This argues that the CT activation is related to the flexibility of the bridge. At low temperature (77K), an intervening adenine eliminates CT from Ap to G, implying that the equilibrium conformation is not the CT active conformation.
Although thermal activation on its own does not imply a mechanism, the sequence dependence establishes a strong relationship between bridge structure and the activated process. The temperature dependence of the electronic contribution to the rate should be the same irrespective of increasing bridge length. The temperature dependence of the nuclear contribution, due to bridge-length dependences on driving force and reorganization energy (see below), can naturally be bridge-length dependent, but should disappear for distances above 10 to 15 Å,118 which are exceeded here. Conformational gating to reach a CT-active state, however, is expected to lead to increased CT with temperature, and for this increase to be greater when more bridge units are required to align, i.e. for longer bridges.
Interestingly, this increase in the rate with respect to increasing temperature is even more profound when the bridge is poorly stacked ATAT,54 or contains an AA mismatch,35 consistent with the model whereby these sequences disrupt DNA due to poor dynamic stacking (Figure 4). When the bridge is AAIA, which stacks well but attenuates CT from Ap* due to the inosine potential barrier, the increase in CT with respect to temperature is only modest. These experiments strongly suggest that the equilibrium conformation of DNA is not the active conformation for long-distance CT, but that conformational gating allows the formation of CT-active states. This is in direct contrast to the usual role dynamic disorder plays in molecular wires, where distortion from the equilibrium conformation decreases coupling and transport.119
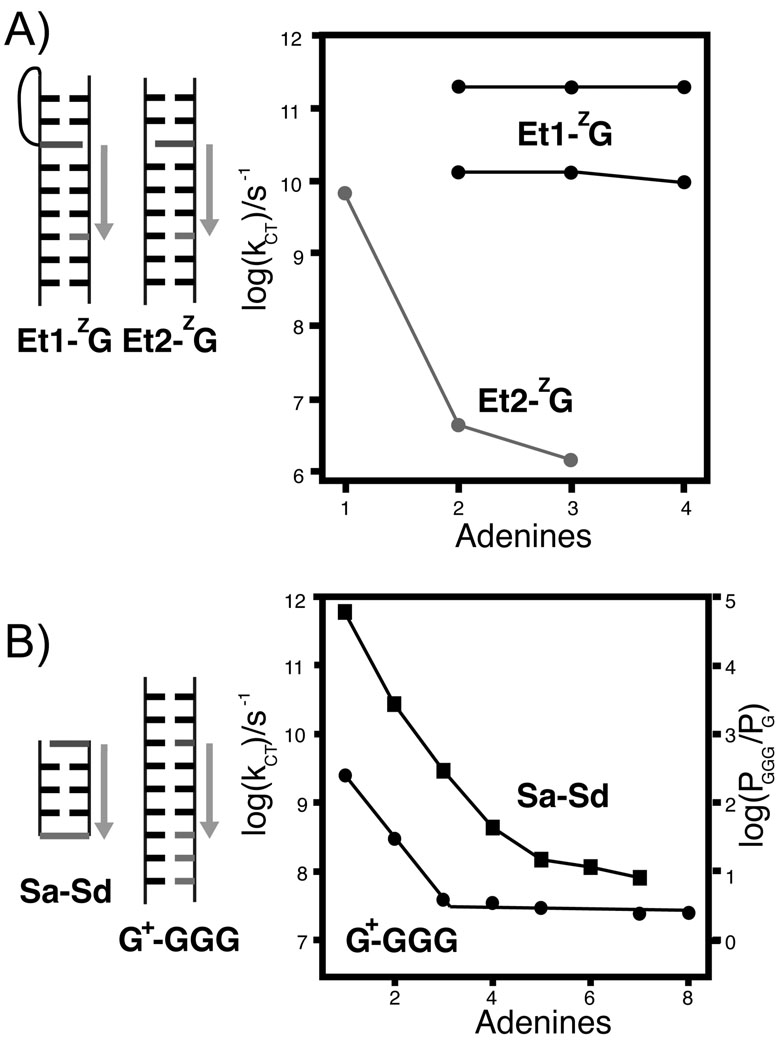
There are two different implications of conformational gating. In one sense, reorientation of the photooxidant with respect to the DNA to form a CT-active conformation can be rate-limiting. Alternatively, formation of a CT-active state in the DNA itself can be the rate-limiting step for CT. Both senses are represented by the case of ethidium bromide. Ethidium bromide (Et+) is competent for DNA-mediated oxidation and reduction of deazaguanine (ZG) and [Rh(phi)2(bpy’)]3+ respectively.27,60,120,121 Femtosecond transient absorption and fluorescence up-conversion spectroscopy of Et+ site-selectively intercalated in DNA found that the Et+ oxidized ZG with two rate constants.120 One (5 ps) corresponds to Et+ that is already present in a CT-active conformation. The other (75 ps) corresponds to the gating of Et+ to reach a CT-active conformation with respect to the DNA. This is the first sense of conformational gating. As the distance between Et+ and ZG increases, the rates of each component is unaffected, but the amplitudes monotonically decrease, suggesting that the increase in distance lowers the yields by virtue of changing the population of CT-active states, rather than by affecting the inherent rates of CT through the DNA. This represents the second sense of conformational gating.
To further test this model, a new method of conjugating Et+ to DNA as a rigid base-pair surrogate was developed (Figure 5).27 For the Et+ separated from the hole acceptor, ZG, by a single base pair, the rate is similar to that for CT from the intercalated Et+. This rate drops four orders of magnitude if another adenine is inserted between the Et+ and ZG. Beyond a distance of two base pairs, the rate is constant. The authors interpreted this system as one that held the Et+ rigidly in a CT-inactive state. The rate-limiting step is injection, which for a poorly coupled donor exhibits a steep distance dependence. At sufficient donor-acceptor separation, re-orientation of the donor is competitive with the slow CT from poorly coupled Et+. Now the apparent distance-dependence is flat, for the same reason as for the intercalating Et+.
Figure 5.
(A) The rate of coherent deazaguanine (ZG) oxidation by ethidium bromide is the same over short distances for both the flexible linkage (Et1, black, both CT rates shown) and the rigid linkage (Et2, gray).27 For two intervening nucleotides, a sharp drop in rate is observed for the rigid Et+, but the rate is unaffected for the flexible Et+. (B) This steep drop in rate over short distances is consistent with that observed for oxidation of guanine by a photoinduced sugar radical (circles),207 and CT between hairpin capping stilbenes (squares, photooxidation of Sd by Sa),125 and has been attributed to a crossover between coherent superexchange and incoherent hopping. In the latter case, comparison of injection and hole arrival rates supports superexchange for one or two intervening base pairs, and hopping for three or more base pairs between the stilbenes.
A similar explanation might serve for the slow rate of charge injection into stilbene-capped hairpins where several AT base pairs separate the photoexcited stilbene hole donor from a guanine hole acceptor. Stilbene-4,4’-dicarboxamide incorporated as a bridging and capping element in short AT containing hairpins (Sa-AT) has an extended nanosecond lifetime and hence higher fluorescence quantum yield versus the free dye, but a single GC pair close to the stilbene dramatically decreases the fluorescence intensity, via CT quenching.122,123 The robust fluorescence in the absence of guanine was taken as evidence for a lack of CT between stilbene and adenine, despite the presence of several low-yield picosecond decay components. Eventually, these components were assigned to CT between the excited stilbene and adenine.124,125 The charge-separated state can either undergo recombination, or, in the presence of distal guanine or a lower energy stilbene-4,4’-diether (Sd), the hole migrates to the acceptor leading to CT quenching.126 It was argued that the recombination recovers the excited state, and that hence charge injection in the Sa-AT constructs only minimally quenches fluorescence.124 This is distinct from exciplex emission, as the emission spectrum is similar to the unconjugated fluorophore. The stilbene radical anion is solvent exposed, and the motions of DNA, associated counterions, and bound water allow rapid relaxation of dyes,127 so it is unlikely that recombination-induced emission would be of the same energy as radiation from the initial excited state. An alternate explanation is that this fast injection is limited to a small population that is in a CT-active conformation. The remaining population fluoresces normally in the absence of nearby guanine. In that context, the slow, strongly distance-dependent direct charge transfer from excited stilbene to guanine can be interpreted in the context of a donor that is not in a CT-active state with respect to the π-stack.122
2.5. Back Electron Transfer
Inevitably, photo-induced charge separation events are followed by charge recombination, also termed back electron transfer (BET). After all, if charge recombination is thermodynamically unfavorable, then charge separation is thermodynamically favorable and will not require photoexcitation. The effect of BET varies by the nature of the assay. Assays for the presence of the charge separated state, such as the slow oxidation of guanine cation radical, will generate yields that are convoluted with BET processes. In two extreme cases, thionine128 and Ap,73 which are competent for efficient charge separation, are not competent for the formation of permanent guanine oxidation products.
The case of the two excited electronic states of AQ offers a nice comparison of photooxidation with fast and slow BET. Irradiation of DNA-conjugated AQ at 350 nm promotes it to the singlet excited state, which relaxes to the triplet state. Both states are competent for direct oxidation of all four bases in DNA, but only the triplet radical anion reduces oxygen to superoxide. The singlet radical anion undergoes rapid BET, regenerating the initial state, while the charge injected by the triplet radical anion is persistent, and can equilibrate along the DNA on a longer timescale.129,130 This scheme explains the incompetence of AQ to oxidatively repair cyclobutane thymine dimer;131,132 repair can only proceed from the singlet state.
Experiments that rely on slow product trapping at guanine need BET to provide the fundamental clock that allows discrimination of CT attenuation.66 Hence, although the results will be qualitatively diagnostic, the quantitative accuracy will only hold relative to the BET rate for that system. For example, a single negatively charged phosphate group near the intercalation site of a tethered rhodium photooxidant changes the observed ratio of damage between a distal and proximal GG site by an order of magnitude, indicating that the distal/proximal damage ratio is not solely determined by the intervening bridge.52
Short-range CT is particularly subject to BET, as the recombination has a steeper distance dependence than separation, most likely due to greater separation in donor-bridge-acceptor energies, as discussed in section 3.2.122 This was first exploited in guanine damage systems using AQ as the donor, but has since been systematically studied in Acr+-phenothiazine (Ptz) and napthalimide (NI)-Ptz systems.133–136 In the former, suppressing BET allowed the extension of a canonical short-range CT system into one that exhibited persistent CT separation over a long range!136
2.6 Injection and Migration Effects
Even among well-coupled donors and acceptors, substantial variations in CT yields and rates have been observed. The fastest observed rates (subnanosecond) over long-distances are for the RuII*/III/RhIII/II pair1,137 and for the oxidation of ZG by Et+* 120 and the reduction of RhIII by Et+*.121 Oxidation of guanine by Ap*,138 excited stilbene,122 or even by guanine radical after initial oxidation by photoexcited stilbene,139,140 is substantially slower. To first order, this trend reflects the relative stacking of donors and acceptors with the DNA duplex.
In addition, some of these results may be reconciled in the context of considering the effect of electrostatics on hole migration.113 This effect was directly demonstrated by a study with RhIII* as the photooxidant wherein the position of a terminal phosphate was varied.52 In this experiment, comparative damage at GG sites proximal and distal to the Rh intercalation site was determined. Since the decomposition of the guanine cation radical is slow, and the DNA between the GG sites was short and undamaged, the final yield reflects both the relative extent of BET and the potentials at the GG sites. When a phosphate anion is added to the end opposite to the rhodium, there is a small increase in damage distribution towards the distal site. An extra phosphate anion on the same end as the rhodium, however, leads to a several-fold change in relative damage. For one sequence, relative damage at the proximal site increases from 16% to 56%. This argues that local charge can have a strong effect, both on the rate of BET and on the rate of migration. For AQ, which irreversibly injects a cation due to rapid oxygen quenching of the triplet AQ radical anion, this effect is not present,141 consistent with a lack of BET,66 although given the low amount of distal damage in these constructs, it is not clear whether a subtle change would be detectable.142 Importantly, in biological systems the initial oxidation product is generally a guanine cation radical, without an anion radical also being localized on the DNA. Hence, coulombic attraction will not inhibit transport away from the injection site, as is the case for Ap* and Ap(-H)•, stilbene, AQ, and other neutral photosensitizers.
2.7 Energetics
The natural nucleosides of DNA are resistant to mild oxidation and reduction and the radical cations and anions undergo secondary chemical reactions on the microsecond time-scale. Hence, the reversible potentials are not trivial to acquire.143 Approaches for determining the nucleoside potentials fall into four categories: computational, electrochemical, pulse radiolysis, and photooxidation studies (Table 1). A common conclusion from all of these studies is that the oxidation potentials increase in the order G<A<C~T.
Table 1.
Experimental and Calculated Oxidation Potentials of Nucleotides
| Method | Solvent | Nucleotide Oxidation Potential (vs.NHE) |
Reference | |||
|---|---|---|---|---|---|---|
| dG | dA | dC | dT | |||
| pulse radiolysis | aqueous | 1.29 V | 1.42 V | 1.6V | 1.7V | 272 |
| electrochemistry | MeCN | 1.49 V | 1.96 V | 2.14V | 2.11V | 144 |
| electrochemistry | DMSO | 1.44 V | 145 | |||
| CHCI3 | 1.62 V | |||||
| CHCI3 + dC | 1.28 V | |||||
| time-resolved quenching |
aqueous | 0.97 V | 1.2V | 174 | ||
| electrochemistry | DMF | 1.52 V | 146 | |||
| DFT | organic | 1.88 V | 2.01V | 2.18V | 2.25 V | 273 |
| organic + dC | 1.48 V | |||||
| organic + dT | 1.81V | |||||
| Pulse radiolysis | aqueous, DNA | 1.22 V | 149 | |||
Electrochemical measurements of base potentials are limited to organic solvents, generally acetonitrile, DMSO, or DMF, due to the relative facile oxidation of water versus the four bases. Considering the hydrophobic interior of DNA, potentials determined under these conditions may be more relevant to DNA than potentials determined in aqueous solution. To date, reversible electrochemistry has not been achieved. Irreversible oxidation potentials of all four nucleosides have been measured, which should be close to the standard potential.144 The values for dG are similar in acetonitrile and DMSO,145 but substantially more positive and more negative values have been found in chloroform145 and DMF,146 respectively. In chloroform, it was found that the presence of dC, which allows the possibility of proton-coupled electron transfer (PCET), lowers the oxidation peak of guanine by 340 mV.145
The closest that electrochemical experiments have come to measuring the oxidation of guanine in its natural context in DNA are the electrocatalytic experiments using mediators on indium tin oxide.147 That [Ru(bpy)3]3+ (E1/2 ~ 1.3 V; all potentials herein are vs. NHE) is a facile mediator for electrocatalytic oxidation of DNA indicates a comparable potential for guanine.148 Analysis of oxidation rate using a variety of metal complex mediators of different potentials supports this value. Notably, sufficiently high ionic strength was used in these experiments to deconvolute the potential from the affinity of the metal complex. This result was later validated by pulse radiolysis experiments in DNA,149 where a potential of 1.22 ± 0.02 eV was found for guanine in multiple sequence contexts. Although the absolute potentials from electrocatalysis are approximate, they provide strong evidence for 5'-GG-3' being about 0.15 V lower in potential than G with a 5'- pyrimidine.150
The latter result, that the 5'-G of GG doublets are lower in oxidation potential versus G, has been extensively exploited in studies of the migration of charge in DNA, where GG sites are used as shallow hole traps. Calculation and oxidative yield experiments indicate that GGG acts as an even deeper well, although smaller differences in potential between these sequences are found experimentally than by calculations,151 probably due to solvent interactions.152 Both calculation and experimental work support preferential hole density on the 5'-G.99,153–156 More generally, there is strong correlation between the calculated ab initio ionization potential of guanine in the different stacking environments and the relative oxidative damage found between different guanines under conditions that allow full equilibration of the injected charge.157 Stacking affects the energies of all the bases, with the strongest perturbation due to the 5'-base.150,158
Vitally, all of these experiments probe the equilibrium potentials of the bases. Random sequences of DNA have rugged potential landscapes, corresponding to extensive static disorder. There are many conformational modes in DNA and its hydration layer with timescales from picoseconds to seconds.127 As discussed in more detail below, the energetics of the bases are coupled to these modes, introducing both static and dynamic disorder to the system.
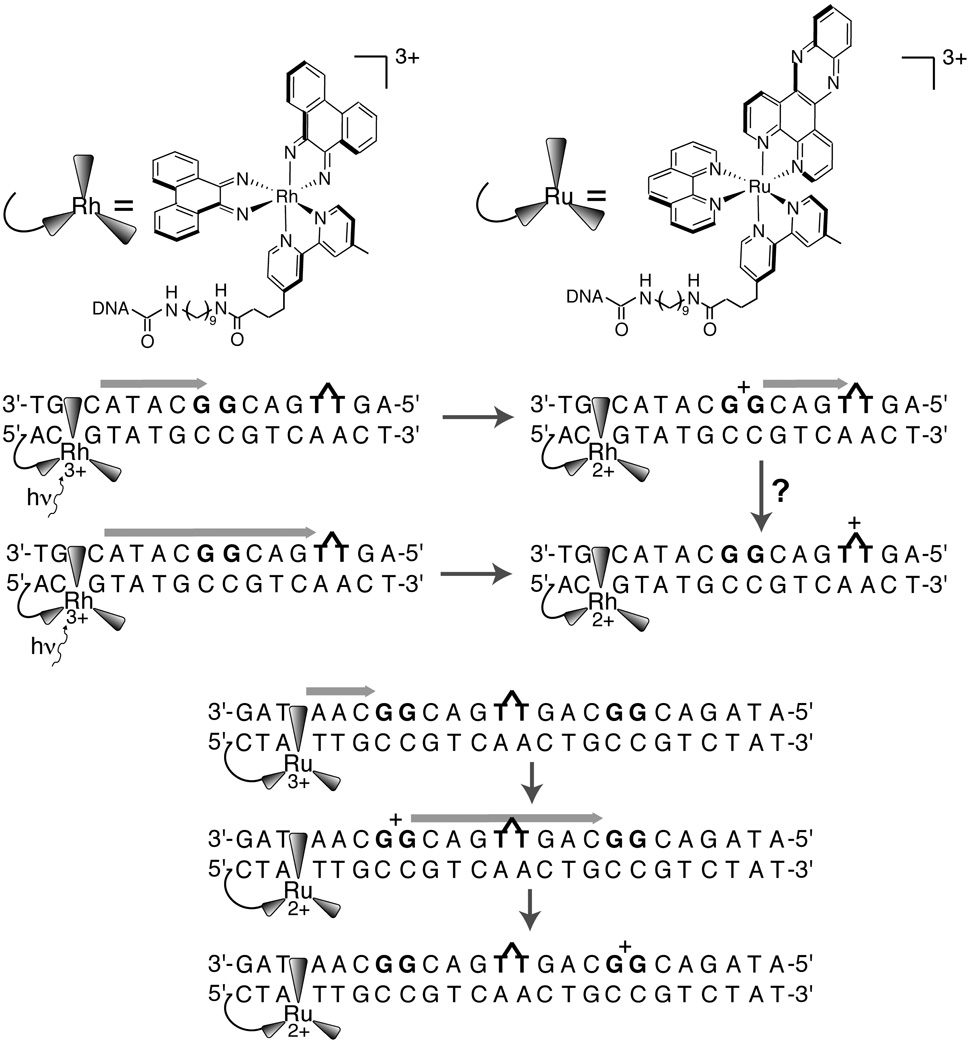
Given the challenge in properly coupling an individual photooxidant to DNA, it is not surprising that few studies have attempted to determine rationally the driving force dependence of CT. It has been established from several studies that CT does not occur from an excited photooxidant hole donor to a higher energy hole acceptor. For example, the metallointercalator [Rh(phi)2(bpy’)]3+, tethered to DNA, can oxidize A, G, or C from its excited state (E3+*/E2+ = 2.0 eV), but the metallointercalator [Ru(phen)(dppz)(bpy’)]3+ (E3+/E2+ = 1.6 eV) oxidizes G, but not C.76 Similarly, [Ru(phen)(dppz)(bpy’)]3+ and ethidium bromide (E*+/E0 = 1.2V)121 are not competent to repair thymine dimers, but photoexcited [Rh(phi)2(bpy’)]3+ performs this repair, as does napthaldiimide (NDI) (E*/E1− > 1.9 V).131,159 These studies include measurements with the hole donor tethered far from the acceptor, with intervening low-potential double guanine sites, indicating that even after charge is injected into DNA, there is some memory of the energy of its initial state (Figure 6).
Figure 6.
Oxidation and repair of thymine dimer (~1.8 eV) by tethered photoexcited [Rh(phi)2(bpy’)]3+ (2.0 V) is unaffected by the intervening double guanine site (1.2 eV). Oxidation of double guanine sites by [Ru(phen)(bpy’)(dppz)]3+ (1.5 eV) is unaffected by the presence of thymine dimer, which this oxidant lacks sufficient driving force to repair. The latter result implies that guanine radical is not competent to repair thymine dimers, in accordance with the known potentials. Hence either the guanine radical oxidized by [Rh(phi)2(bpy’)]3+ does not relax prior to migration to the thymine dimer, or the guanine radical is not an intermediate in DNA-mediated oxidation of thymine dimer by [Rh(phi)2(bpy’)]3+.
In support of this interpretation, CT can still occur far below the potential of the DNA. In one example,160 an oxidized nitroxide (NO+/NO• ~ 1 V) was incorporated into a duplex by covalent attachment to thymine. The [4Fe-4S] protein MutY (E3+/E2+ = 0.1 V) was added, and the generation of the reduced nitroxide spin radical was observed by EPR (yield ~ 50%). This chemistry was demonstrated to be DNA-mediated by two controls. A noncleavable substrate analogue for MutY incorporated into DNA far from the label increased the reduction yield, and partially saturating the electronic conjugation between the label and the DNA substantially decreased the reduction yield.
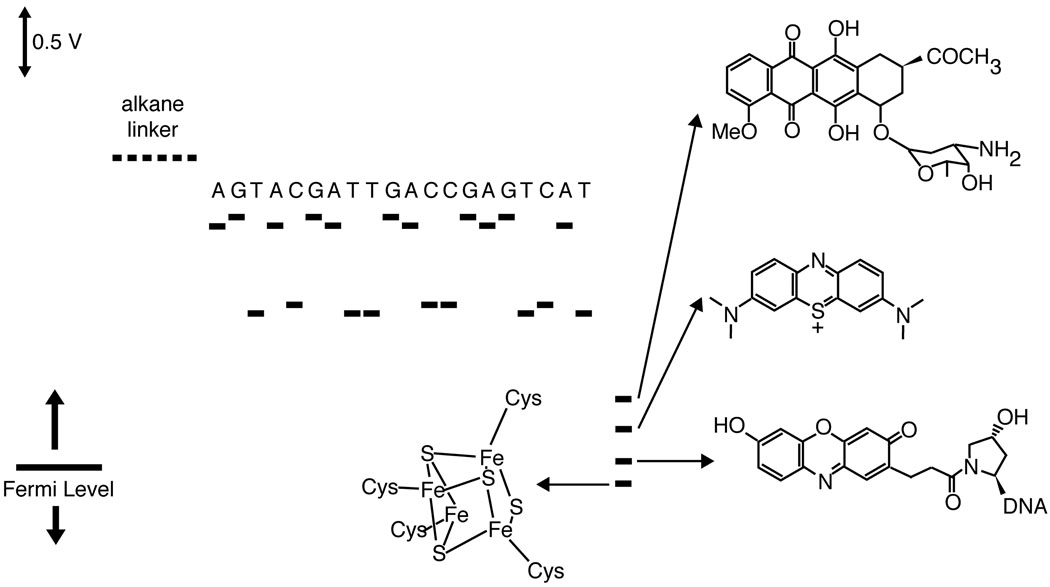
A similar conundrum is involved in DNA-mediated CT in self-assembled monolayers on gold (Figure 7).161 In these systems, a DNA monolayer is covalently self-assembled on an electrode, and CT through the DNA is measured with an electrochemical probe. At an applied potential greater than 0.4 V, the DNA adopts an upright conformation. Although elastic motions can bring the DNA in contact with the surface,162 these conditions require high salt and fairly positive potentials. DNA mediation has been established for our system by the methods discussed above, i.e. mismatch and binding event discrimination, probe conjugation, linker-length dependence, and by differential redox potential between direct contact and DNA-mediated reduction.31,163–165 The window for these experiments on Au is limited by the potential of Au-S reduction, about −0.5 V. DNA-mediated CT has been observed for a dozen different probes spanning a full volt below this value.91,163–166 Importantly, the reductive limit is 600 mV below the reduction potential of cytosine.
Figure 7.
Scale diagram describing the relevant potentials for DNA-mediated CT through DNA self-assembled monolayers on gold. The potentials of the individual nucleotides are not accessible within the window of electrochemistry of DNA monolayers on Au. Nevertheless, facile DNA-mediated electrochemistry is observed for redox probes over DNA bridges. For all probes and sequences of well-matched duplexes, the tunneling through the alkane linker is rate limiting (~30 s−1). Shown, in order from top, are daunomycin, methylene blue, Redmond Red, and a [4Fe-4S] cluster similar to those in the redox-active repair proteins EndoIII and MutY.
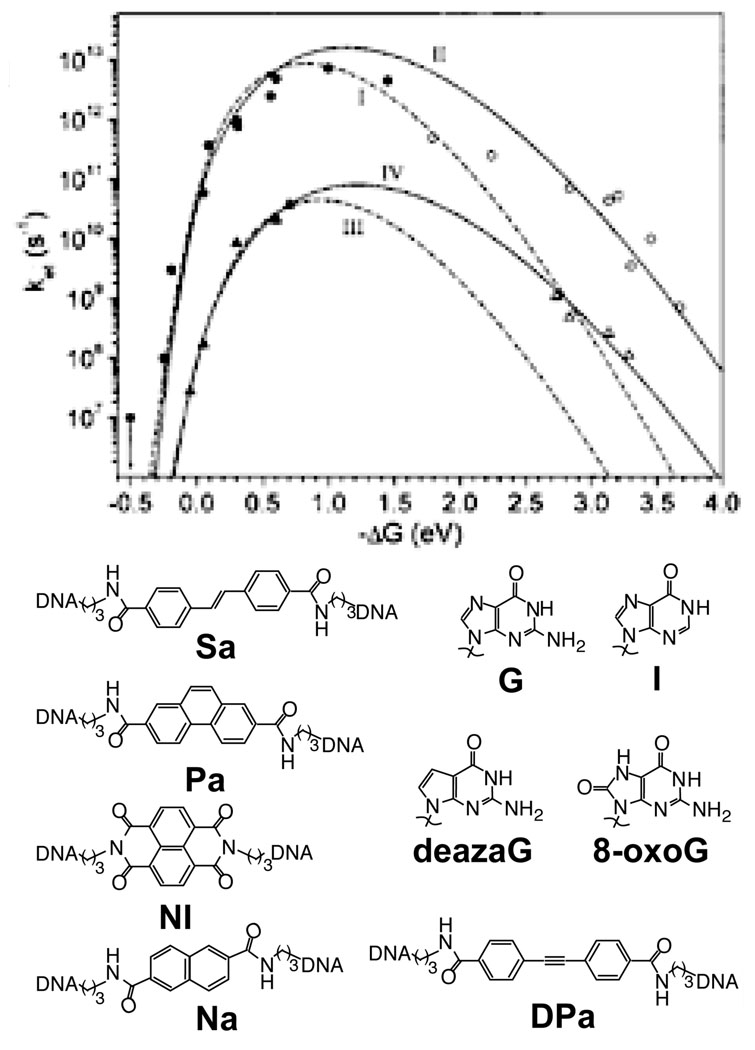
Photooxidant-bridged DNA hairpins were employed to measure systematically the dependence of CT rate on driving force.167 In these experiments, five photooxidants, stilbene-4,4’-dicarboxamide (Sa, E*/− = 1.68 V), naphthalene-2,6-dicarboxamide (E*/− = 1.74 V), diphenylacetylene-4,4’-dicarboxamide (PA, E*/− = 2.02 V), NDI (E*/− = 2.93 V), and phenanthrene-2,7-dicarboxamide (E*/− = 1.43 V), were employed as hole donors. Guanosine, inosine, deazaguanosine, and 8-oxoguanosine were used as the hole acceptors, either immediately adjacent to the bridging dye, or separated by a bridge of two A–T base pairs. The measured charge separation and recombination rates fit well to the Marcus-Levich-Jortner equation, with reorganization energies of 1.2 eV and 1.3 eV respectively (Figure 8). The agreement is improved if a molecular based model for the solvent is used and the Q-model is employed for the energy surfaces; in this case higher reorganization energies are found.168 Further experiments varied the bridge length for the St and PA duplexes, and found that the distance dependence increased for greater donor-bridge energy separation.169
Figure 8.
The driving force dependence for CT in photooxidant-bridged DNA hairpins is determined from time-resolved transient absorption studies of a series of five stilbene-derived photooxidants and four hole acceptor bases, following both charge separation (filled) and charge recombination (empty). Case I,II (circles): donor and acceptor are in contact. Case III,IV (triangles): donor and acceptor are separated by two TA base pairs. Case I,III are fit only to charge separation rates (dotted), while Case II,IV are fit to both charge separation and charge recombination rates (solid). Similar reorganization energies, about 1 eV for the nuclear reorganization energy and ~0.2 V for the solvent reorganization energy, are found for both Case II and Case IV. Reprinted with permission from Reference 167. Copyright American Chemical Society.
2.8. Proton-Coupled Electron Transfer
If a participant in electron transfer is also an acid or base, the reduced and oxidized species will often have different pKA values as well. In this case, it is likely that CT will be coupled to proton transfer.170 Proton-coupled electron transfer (PCET) may be more the rule than the exception in biological systems. Most redox-active groups in biology are also subject to protonation or deprotonation, with the pKA dependent on the redox state. Since complete proton transfer is unnecessary for substantial effects on the coupled electron transfer rate, the question is not whether the electron transfer is proton-coupled, but whether the coupling is significant so that proton transfer becomes rate-limiting. In the case of Class I E. coli ribonucleotide reductase, PCET has been found to occur over multiple steps.171–173
Each nucleotide in the double strand participates in stable hydrogen bonding with its complement in a base pair. Hence, it is not surprising that CT between nucleotides should be proton-coupled, although likely in a way that cannot be probed by the usual assay of pH dependence, given that the base-pair protons are excluded from solvent. It has been shown that oxidation of the aqueous, isolated nucleosides by Ap* is not proton-coupled,174 but that does not exclude the possibility of PCET in the context of protons in the base pair. Theoretical work predicts that double proton transfer between guanine and cytosine lowers the guanine potential.175 Indeed, the cytosine radical has been directly observed by transient absorption spectroscopy, after oxidation of DNA by SeO3•− and SO4•− ions generated by pulse radiolysis176. This might not be general, though, as the mechanism of G-C oxidation in pulse radiolysis is strongly dependent on the chemical interaction with the oxidizing radical.149,177 Similar evidence for PCET reduction of thymidine base-paired to adenine has also been found.178
Furthermore, the pKA of the guanine cation radical has been measured to be 4, near that of cytosine (4.5),179 and the neutral guanine radical is observed by nanosecond transient absorption and EPR after oxidation by intercalated Δ-[Ru(phen)2(dppz)]3+,180,181 supporting deprotonation of the guanine cation radical, presumably to the paired cytosine, on a faster timescale. For DNA ionized by γ-irradiation at 77 K, ESR measurements find that the equilibrium strongly favors the neutral guanine radical over the cation radical; this held for guanine stacked between cytidine and for each guanine of the GGG triplet.155 Although this evidence strongly supports proton transfer, it does not establish coherent PCET, as proton transfer consequent to oxidation has been shown to be favorable.182
Isotope experiments, which would establish whether proton transfer is rate-limiting, have not been straightforward. Certainly, for oxidation by SO4•− ions, the charge injection yield is decreased in D2O.177. In one experiment, deuterium replacement of acid protons led to a three-fold decrease in the relative yield of damage of distal GGG to proximal G sites.87 In another, CT between Ap(-H)• and G was found to exhibit a small differential between D2O and H2O, consistent with PCET.183 A similar small differential was observed in some sequences, but not in others, for CT between photoexcited NI and Ptz.184 However, in the fluorescence experiments, the substitution of D2O for H2O also affects excited state lifetimes.
At first, it might seem that the facile oxidation of CPC in competition with CPG supports a PCET model73. However, CPC oxidation is increased by base-pairing with inosine, a high potential guanine analogue, indicating that PCET is not the mechanism of CPC oxidation. Instead, this mechanism is enticingly similar to the proposed mechanism for excited state relaxation in GC base pairs,185,186 which involves proton-coupled exciplex formation and has also received experimental support,187,188 although it appears that guanine-guanine stacking might prevent this relaxation.189 Based on this accumulated evidence, it seems likely that PCET is involved in at least some charge injections to guanine, and that neutral guanine radical is the persistent form of injected radical.
2.9. Characteristics of DNA CT
It is apparent that DNA mediates CT over long distances, and that the rate and yield are sensitive to both the donor and acceptor identities and to the integrity of the intervening π-stack. Structural distortion of the DNA, or poor coupling of the donor or acceptor to the DNA, sharply attenuate long-range CT. Furthermore, rapid CT is conformationally gated, and the equilibrium conformation is not necessarily the CT-active conformation.
3. DNA-mediated CT Mechanisms
Tautologically, all mechanisms of charge transport incorporate an electron moving from a donor orbital to an acceptor orbital. The variation consists in the identification of orbitals that mediate this transition, and the pathways that are coupled to it. In a large biomolecule, such as DNA, complexity arises from the sheer number of atoms involved. In this section, we will evaluate postulated mechanisms of long-range CT in DNA in the context of the properties discussed above.
3.1. Transport through Water, Ions, Phosphates
An obvious source of conductivity in DNA is the highly charged phosphate backbone. Indeed, one of the earliest models of CT through DNA involved transport through the phosphates.190 A recent measurement of delocalization of a hole produced on a single phosphate lends some credence to this model,191 although it is unclear whether this delocalization can be transduced into conduction, and comparable measurements have not observed this delocalization.192 In the phosphate conduction model, phosphates on the edge of the DNA are directly ionized, and the hole rapidly hops through isoenergetic phosphates. For this to occur, coupling between the phosphates must be substantial. Even more importantly, oxidative damage must preferentially occur at phosphates versus the base stack. Some calculations found that this was the case,193 but later work demonstrated that this was due to neglecting the presence of water and counterions that can shield the phosphate group’s negative charge.194 Theoretical and experimental work suggests that photoionization is initiated at bases and not at phosphates195 and that the energies of the ions, phosphates and sugar states are far from the Fermi energy.196
Alternatively, the motions of water and ions can lead to apparent conduction. DNA adsorbs several layers of high dielectric water,42 a primary condensation layer of cations, and a secondary layer of condensed anions. Even under relatively dry conditions, water and cations are still adsorbed. This layer plays a major role in the conformational dynamics of DNA and mediates molecular recognition events with other biomolecules. In particular, it seems certain that early conduction measurements were-measuring ionic conduction along the DNA, rather than properties of the DNA molecule itself.43
Ultimately, however, it is difficult to rationalize these models with the marked sensitivity of long-range DNA-mediated CT to the integrity of stacking, as described in section 2. In contrast, changing the pH, ionic strength, or the identity of the salt has at best a minor effect on CT, as long as well-coupled donors and acceptors are employed. Even the removal of a phosphate along the bridge does not cause a measurable difference in CT yield.77,78 Adding extra intervening phosphates, via the construction of triplex DNA, actually lowers the competence for CT.197 Hence, it is apparent that DNA CT must proceed through the base pairs, in the interior of the duplex.
3.2. Superexchange
Any medium is a superior pathway for charge transport in comparison to vacuum. Superexchange is coherent orbital-mediated tunneling, where, for electron (hole) transport, the high-energy LUMOs (HOMOs) on the pathway are virtually occupied, allowing a probability and corresponding rate of transmission from the donor to the acceptor. Following the Born-Oppenheimer approximation, the rate of superexchange can be separated into the nuclear factor, υn, and an electronic factor, υe:
where
and
and ΔG is the driving force, H0DA is the donor-acceptor coupling extrapolated to zero bridge length, β is a decay parameter characteristic of the bridge, and d is the bridge length. The nuclear factor is a function solely of the identities and the environment of the donor and acceptor. The electronic factor represents the electronic coupling between the donor and acceptor, mediated by the bridge states. In the adiabatic limit, electronic coupling is sufficiently strong such that the nuclear motion will determine the rate of charge transfer. In the non-adiabatic limit, the electronic coupling is sufficiently weak such that the electronic transition probability is less than unity at the transition state.9 Hence, long-range (greater than 1 nm) CT systems are generally treated as non-adiabatic, though it has increasingly been recognized that changing the structure of even long bridges can have a non-trivial affect on both ΔG and λ.7,168,185,198 Ignoring this effect, the only dependence of the rate on donor-acceptor distance is the exponential decay of the donor-acceptor coupling with d, the length of the bridge, characterized by the parameter β. It is important to note that β is generally not what is directly measured in experimental systems. For systems that measure the yield of irreversible chemical products, competing processes such as BET or equilibration will inevitably convolute with the inherent rate of charge separation. Even for very fast charge traps, or spectroscopic based measurements that can directly measure kCT, the exponential drop-off will not necessarily correspond to β if the nuclear factor is itself distance dependent.199 This restriction can be mitigated for long-range CT, where the iterative changes in the bridge length are unlikely to affect ΔG and λ, but is significant for short-range CT.7
Furthermore, it is important to note that calculation of the CT rate requires precise knowledge of the intervening electronic structure, which in turn is dependent on molecular structure. If the mechanism or pathway changes with an increase in bridge length, then the distance dependence will not be well represented by the electronic factor β. Also, conformational dynamics can lead to a time-dependent rate. If the equilibrium structure is the best-coupled structure, dynamics will decrease the apparent CT rate.
Another important consideration is that β is not independent of the bridge and donor energies. For increasing difference between the donor and bridge energies, β increases according to:
where a is the intersite separation, V is the intersite coupling and Δε is the donor-bridge energy separation. As this separation decreases to below V, direct injection will successfully compete with tunneling. Hence, if tunneling is occurring, β is limited to about 0.3 Å−1 (numerical calculations that properly treat the bridge as finite find about 0.2 Å−1).200 This supports the assignment of extremely shallow distance dependences to incoherent processes; at least it excludes superexchange mediated by orbitals on the individual bases of DNA. This model was supported by experiments in photooxidant capped adenine tract hairpins.169 The oxidations of guanine by photoexcited Sa and of ZG by photoexcited phenanthrene-2,7-dicarboxamide are of similar driving force, but the latter pair are 0.25 eV lower in potential than the former pair. For each pair, the rate constants were measured for varying lengths of an intervening adenine tract, and the distance dependence was greater for the PA-ZG pair.
Superexchange has been most thoroughly characterized as a mechanism for CT within and between redox-active proteins; charge transfer reactions among proteins are essential to all organisms. To a rough approximation, proteins can be treated as a homogeneous medium with a single characteristic β of 0.9 Å−1.201 The scatter for individual proteins, however, spans several orders of magnitude, indicating that the electronic structure and pathways vary strongly with the identity of the protein, and the location of the donor and acceptor.202 For some pathways in proteins, conformational dynamics have been shown to play an important role in dictating which pathways are available.203 It is clear that proteins optimize charge transport not only by controlling donor-acceptor distance and driving force, but by allowing a specific pathway, or combination of pathways, to be available for superexchange. An essential lesson from superexchange in proteins is that the most facile pathways determine the overall rate and yield. Although DNA might appear to be a simple one-dimensional system, owing to the extensive π-stacking, experiments suggest a more complicated system.
3.2.1. Coupling Constants in DNA
No model of superexchange can be properly constructed without first considering appropriate values for the coupling constants along the bridge.204 Given the structural complexity, non-trivial assumptions are necessary to allow tractable calculations, each of which have certain disadvantages. Furthermore, the stacking interaction of bases is a particularly challenging one to computationally describe.205 It is important to consider these couplings when developing a theoretical model. For example, a two-stranded model206 for coherent DNA-mediated CT was recently published to fit the results of work207 by Giese and coworkers. The model was only able to fit the data by taking intrastrand AA coupling to be 0.52 eV, nearly an order of magnitude greater than the time-averaged value found in typical calculations.208,209 The average couplings appear to be about 80 meV for intrastrand GG, with somewhat lower instrastrand coupling between AA, and smaller values for interstrand purine-purine coupings.
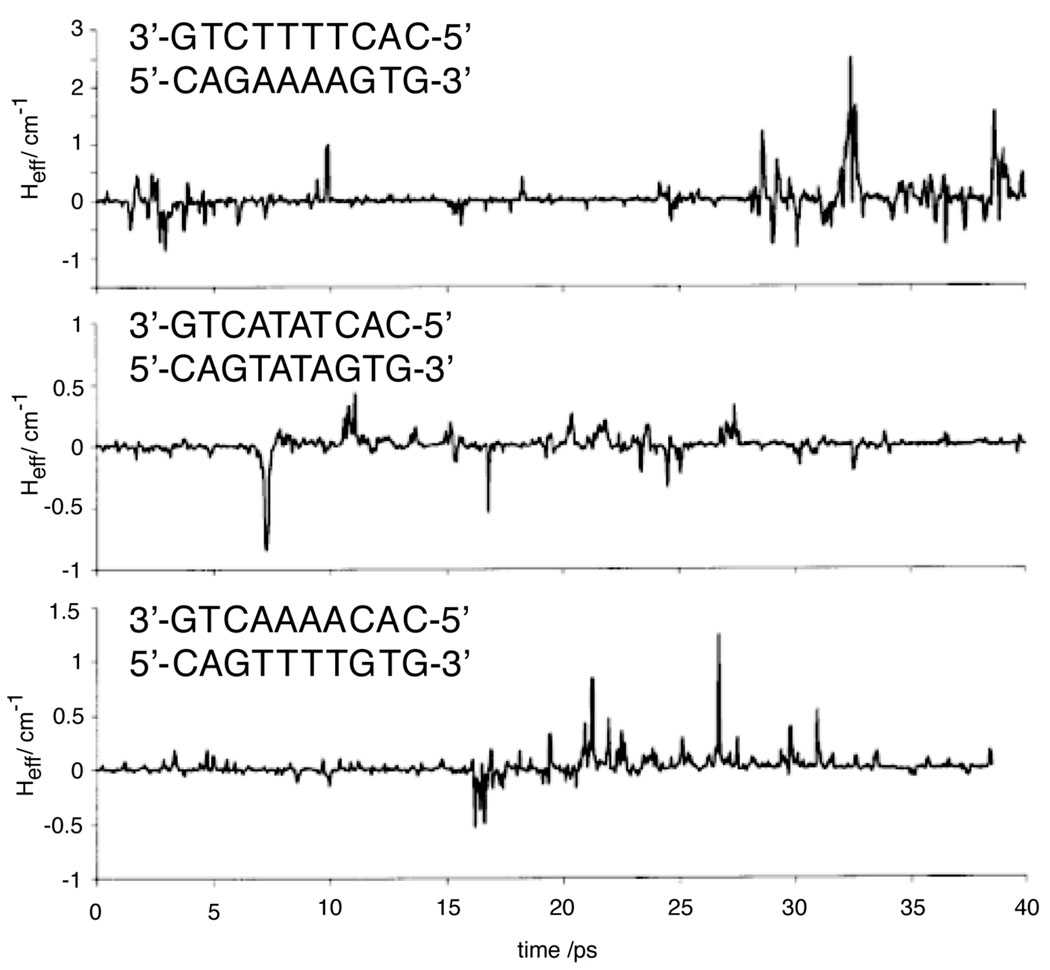
Increasingly, it has been clear that couplings are highly dependent upon the geometry of the stacked bases. An early demonstration of this concept was the calculation of coupling constants between base pairs for coordinates drawn for a large family of crystallized duplexes.210 Even though this measurement was only for coordinates from a set of crystals, each of which presumably corresponds to an equilibrium conformation, variations in couplings were on the order of half of the values. In addition to this static disorder, DNA is subject to extensive dynamic disorder on a full spectrum of time-scales. A later study considered fluctuations from equilibrium conformations, using MD simulations to access the transient structures (Figure 10).211 This study found even larger variations in coupling, and found that HDA for GAAAAG varied by more than an order of magnitude over the course of the 40 ps simulation. Interestingly, they also found that transverse base motions, which affect stacking, are more significant than longitudinal motions; this is consistent with recent work that found that shear, twist, and stretching within base-pairs also affects coupling constants.212 They also found that peaks in coupling over the bridge were more significant for GAAAAG than GTTTTG and nearly absent in GATATG, in accordance with measured CT yields. Since then, similarly large fluctuation-dependent variations have been found using a variety of computational approaches,49,208,213,214 with fluctuations being most significant on the picosecond time-scale.213 These studies have demonstrated that conformation also has a profound effect on the transient nucleobase energies, as does solvent polarization.215 Interestingly, calculations indicated that base energies tend to be correlated in the duplex,182 although the relative ordering of base energies are preserved.208 These results offer a natural explanation for the conformational gating that has been observed in long-range systems.
Figure 10.
The time-dependent couplings between guanines separated by three different four-base sequence contexts, based on conformations generated through molecular dynamics. It is clear that the average value of coupling can be several orders of magnitude lower than the maximum coupling. For the poorly stacked, flexible ATAT sequence, strong coupling between the guanines is not achieved over the time-scale of the simulation. Reprinted with permission from Reference 211. Copyright American Chemical Society.
3.2.2 Reorganization energy
Given the excellent correlation of theory and experiment for the driving force dependence of CT in stilbene-capped hairpins,167 the reorganization energy for those systems is certainly close to 1 eV. For systems where the donor and acceptor are internal to the π-stack, as with intercalators, it is less clear, as these sites are far less solvent exposed than the end-capped agents, such as Sa, Ptz, AQ, and napthalimide (NI).
Even for transfers between sites in DNA, the reorganization energy can vary substantially. Sequence context, which affects couplings and site energies, is expected to affect reorganization energy as well. Delocalization among multiple bases, which decreases the effective amount of charge that must be transferred, lowers the reorganization energy.216 One study found, by sampling many molecular dynamics configurations of oligopurine•oligopyrimidine DNA, reorganization energy for nearest neighbor hops to be about 1.1 eV for both adenine to adenine and for guanine to guanine.49 Another study estimated 0.5 eV for adenine to adenine based on the spectral density of intercalated ethidium bromide.217
It has been found that for short range CT in DNA, changes in bridge length can induce substantial changes in the reorganization energies.7,118 This finding is consistent with Marcus’ classical description of the solvent reorganization energy:
where Δq is the change in charge, aD and aA are the donor and acceptor radii respectively, and εop and εst are the optical and static dielectric constants, λ explicitly depends on the donor-acceptor separation.9 An indirect length dependence of λ will also be incorporated through the effect of molecular structure on the dielectric.
3.2.3. Superexchange in DNA
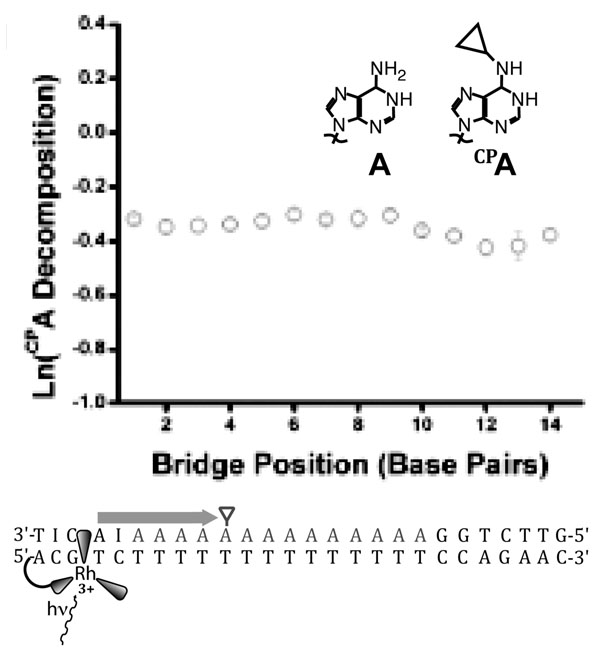
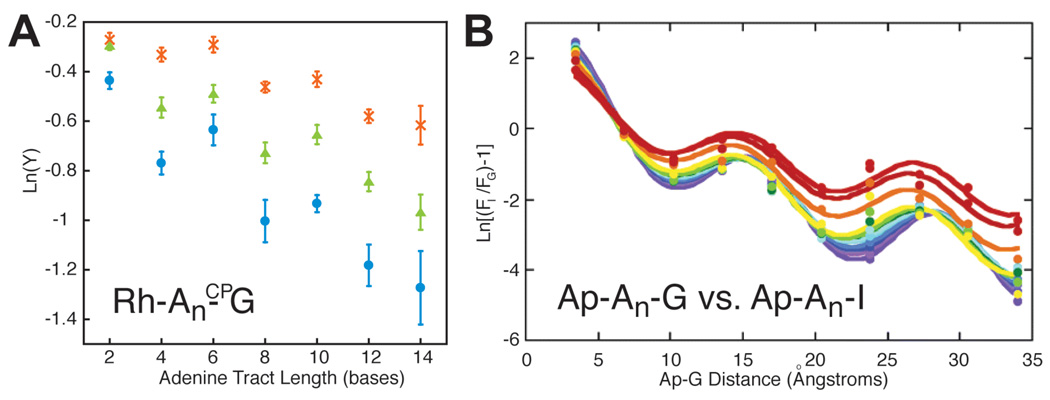
Long range charge transport over more than 50 Å seems incompatible with superexchange, given its inherently strong distance dependence. Even a β of 0.1 Å−1 implies a loss of over eight orders of magnitude in rate over 200 Å. However, most long range measurements either neglect yield122,159 or measure products formed on long timescales.3 In the former case, long range transport can reflect a small yield, while in the latter case, products might be formed only on the timescale of milliseconds to seconds. Fluorescence quenching of photoexcited 2-aminopurine by guanine 35 Å away54 has shown that long-range CT can occur on a timescale that is defined by the nanosecond lifetime of the 2-aminopurine excited state. Furthermore, the distance-independent decomposition ofCP A by photoexcited [Rh(phi)2(bpy’)]3+ over 40 Å17 (Figure 9) demonstrates that CT occurs at high yield at least as fast as BET between oxidized adenine and [Rh(phi)2(bpy’)]2+; the energetically similar reduction of [Rh(phi)2(phen’)]3+ by [Ru(phen’)2dppz]2+ over 41 Å is faster than 3 nanoseconds.1 Hence, it is clear that DNA CT can occur over long distances on relatively short timescales, and any model must account for this. For these reasons, superexchange models are not satisfactory for DNA-mediated CT over long distances.
Figure 9.
CT from photoexcited [Rh(phi)2(bpy’)]3+ to N-cyclopropyladenosine (CPA) across an adenine tract is distance-independent over 14 adenines.17 The rate of CT across the adenine tract, then, must be much faster than BET from the first adenine to the reduced rhodium. The driving force for recombination is only about 1.7 V, implying that BET should not be in the inverted region, consistent with evidence that BET from adenine to this rhodium complex is facile.68 The lack of distance-dependence, in a system with a rapid competing process in BET and a charge trap that samples pre-equilibrium CT dynamics, implies extensive delocalization across the bridge.
3.3. Localized Hopping
The apparent contrast between theory and experiment led to extraordinary efforts to challenge the validity of the measured CT rates and yields. Hopping models offer an alternative that does not require exceptional coupling between bridge sites. Hopping, a type of diffusive, incoherent transport, is the concatenation of multiple superexchange steps, or “hops”, in which charge occupies the bridge between each hop. Hopping has been proven as a mechanism in both natural171 and synthetic218 protein models. The distance dependence of hopping is geometric, and hence shallower than the distance dependence of superexchange.202,219 The reason for the shallower distance dependence is intuitive; long, slow, superexchange steps are avoided.
3.3.1. Nearest-Neighbor Models
Although formal ballistic models do not distinguish superexchange from hopping, it is most straightforward to treat hopping as a multi-step process. In this case, an injected charge resides on the lowest potential base, guanine. This charge can diffusively migrate along the DNA, mediated by short single-step superexchange with neighboring guanines.219 The rate of a hop will depend on the distance and sequence context. The fastest hop will be to neighboring guanine; hops through other nucleotides (i.e. GAG, GCG, or GTG) will be slower. For the case of GCG, hopping is also allowed to the guanine on the complementary strand (G+CG/CGC➔ GCG/CG+C➔ GCG+/CGC).
The most impressive evidence in support of this model are a series of photooxidation experiments by the Majima group, and a series of STM conduction experiments performed by the Tao group.45 Using an elegant experimental setup,220 they form unambiguously covalent contacts to single DNA molecules under aqueous conditions. They found a geometric dependence of the conductance on length for (GC)n sequences, but insertion of an (AT)m sequence into the (GC)n sequence led to an exponential decrease of the conductance with the length of the (AT)m sequence, as predicted by the hopping model. However, they did not investigate sequences that allowed purine-purine stacking, and were unable to find evidence for thermal activation,221 although this was possibly due to the limited window of temperatures and potentials that allowed device stability. Calculations on averaged structures confirm that the alternating purine-pyrimidine sequence attenuates delocalization for this system and reproduce the data well.222 Furthermore, more recent experiments using the same system223 and a similar approach using a mechanical break junction224 found a much smaller effect on conduction from increasing AT content. This was ascribed to the latter experiments being performed with DNA that was more likely to form the B-conformation versus the (GC)n sequences. For DNA covalently bridging a carbon nanotube gap, there appears to be no sequence dependence when the GC content of random sequence DNA is varied.225
Majima and his colleagues have used transient absorption to study the oxidation of Ptz by photoexcited NI, linked by varying sequences of DNA.4 The sequences near the hole acceptors and donors were kept constant, and a central region varied with (GA)n or (GT)n; complementary studies separated the donor and acceptor by only adenines.226,227 Each system fits well to a geometric dependence of rate on bridge length. They found the rate of hopping to be 2×1010 s−1 for adenine to adenine hopping, 7.6×107 s−1 for G+-A-G ➔ G-A-G+ hopping, and about 2×105 s−1 for G+-T-G ➔ G-T-G+ hopping. Although this is strong evidence for multistep hopping in these systems, they have noted that it does not necessarily require that the intermediate states be completely localized on individual nucleotides.227 More generally, the researchers were able to distinguish between hole injection and hole arrival, showing that the two are not coincident over long distances. Similar results have been observed for CT between DNA-capping stilbenes, with the transition between single and multistep CT at about two intervening AT base pairs.125,126
3.3.2. Thermally Induced Hopping
Although hopping between guanine sites can explain many features of the propagation of holes in mixed sequences, it is not sufficient to explain facile charge transport through adenine tracts.228 Occupation of adenines during CT has been demonstrated both by a direct chemical probe73 and by the observation of facile and weakly distance-dependent transport of holes across long adenine bridges,17,31,207,229–231 even when BET competes with equilibration.31 This can be explained by a reasonable modification of the hopping model, whereby a hole on guanine can be thermally excited to occupy an adenine tract.228,232,233 Although this will be disfavored in the case of mixed-sequence DNA in preference to guanine hopping, it can be much more favorable than the slow hop through a long AT sequence. For isoenergetic adenines, this model does not sufficiently explain the distance independence,234 but if the adenines on the edge of the tract are higher in potential than the interior adenines, then the apparent yield of CT will be distance-independent.235 This explanation, although reasonable for (A)n bridges on the strand complementary to the guanine sites, does not support the shallow distance dependence observed when the (A)n bridge is in the same strand,229,236 for which the edge adenines should be of lower energy versus internal adenines. It would be interesting to determine what effect incorporating the stacking-dependent adenine energetics would have on the theoretical predictions of the thermally induced hopping model.228
The most compelling evidence for thermal activation comes from a biochemical trapping assay of G+/An/GGG, where the yield of GGG versus G damage was quantified after hole injection from a sugar radical near the single G site.207 A steep distance dependence for n ≤ 3 was followed by a flat distance dependence for n ≥ 3. This is consistent with two mechanisms at play, where the steep distance dependence corresponds to CT through superexchange across the AT bridge, and the flat regime is where superexchange is sufficiently slow for thermally induced hopping across the adenine tract to become the dominant mechanism. It is important to note, however, that this dependence looks identical to that found for the rigid Et+ base-pair surrogate.27 In the latter case, the dependence was caused not by a fundamental shift in mechanism, but rather by the rate-limiting injection from the hole-donor. Stilbene-capped hairpin systems125 and AQ-capped duplexes236 show a similar, though much more gradual, positive second-order change in the slope with distance. As shall be discussed in greater detail below, delocalized mechanisms can also explain facile transport through adenine tracts. Ultimately, a change in slope on its own is not sufficient to justify a crossover in mechanism.237
3.3.3. Variable Range Models
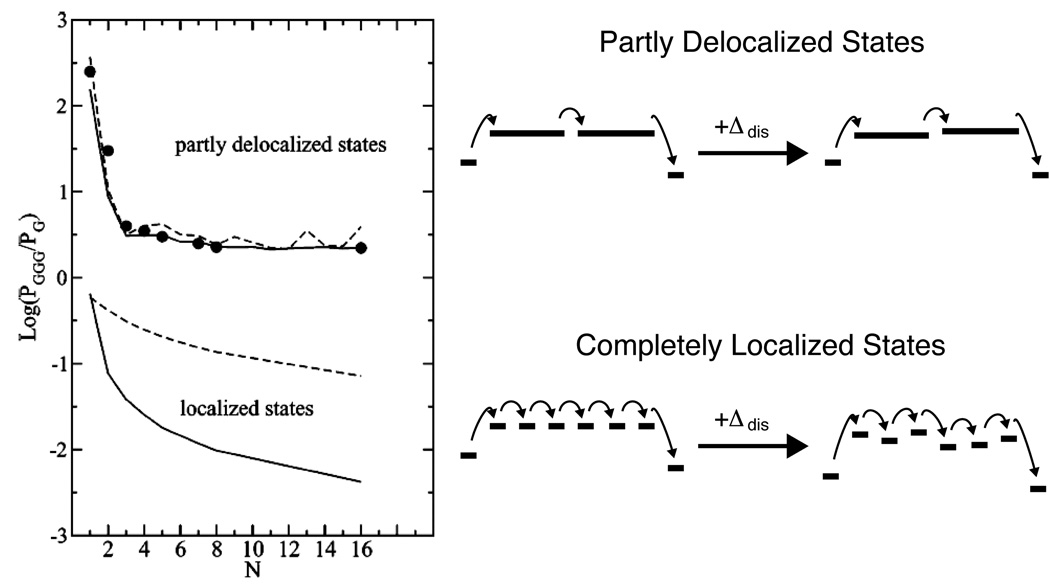
All the mechanisms listed above can be considered together, as components of a variable range hopping model. Here, a hole is allowed to migrate by superexchange to any other site, rather than being limited to nearest neighbors.238 The most probable sites will be the closest low-potential sites, i.e. guanines. Hence, the hole will hop from guanine to guanine through the DNA, preferring intrastrand transfer, but able to exploit interstrand transfer or thermally induced hopping onto adenine tracts where the sequence does not allow more favorable pathways. Even unfavorable pathways are possible, although slow. Theoretical treatments using this model have been successful in modeling some biochemical experiments,239 although it was demonstrated that introducing static disorder substantially degrades the success of variable range hopping models that rely on localized states (Figure 11).217 In turn, dynamic disorder, analogous to the conformational gating discussed above, can assist hopping in a rugged landscape.113
Figure 11.
The variable range hopping model predicts a shallow distance dependence for the rate of CT between G and GGG across an adenine tract of the opposing strand. Delocalized states, even in the absence of disorder (dashed lines), yield larger and shallower CT rates due to the smaller reorganization energy, and a shorter effective bridge length. In the presence of static disorder (solid lines), localized hopping is substantially attenuated due to the rugged energy landscape. Delocalized hopping, however, is relatively unaffected by static disorder, as the coupling is strong enough to allow tunneling through local barriers. Reprinted from Reference 217 with permission. Copyright American Chemical Society.
One challenge that is common to all localized hopping models is the explanation of the mismatch discrimination that has been observed in nearly every system studied. One proposal was that mismatches allow water access to preferentially quench CT at guanine through proton abstraction or other chemical reaction.232 This model was supported by the observation that GT mismatches affected distal yield more than AA mismatches, and that methylation of the most acidic residue of a guanine opposite an abasic site restores CT.87 This model has not stood up to more extensive measurements, however, as AC mismatches attenuate CT more than GT mismatches. It has also been shown that GT mismatches lower the yield of CT by lowering the rate,4 and GT mismatches attenuate CT even under applied potentials insufficient for guanine oxidation.30 Alternatively, mismatched base pairs might have lower couplings to the neighboring bases than matched pairs.62 Particularly, they are less stacked, and sample more unstacked configurations. A similar argument can then be made to explain how DNA-binding proteins that bend the π-stack also attenuate CT.
A more profound problem with localized hopping models is the apparent “memory” that a charge has of the energy of the state from which is was injected. The intermediate in a localized hopping model is the cation or neutral radical on guanine or adenine. Oxidation of cytidine or thymidine by these species is taken to be highly unfavorable. Hence, the energy of injected charge should not affect the nature of the intermediate over long distances. This is not consistent with the evidence from thymine dimer159 or CPC75,76 oxidation, where oxidants competent for guanine oxidation, but not pyrimidine oxidation, were unable to decompose these species over long distance. Oxidants that are competent for thymine dimer repair or CPC decomposition, however, remain competent to decompose these oxidation reporters even with intervening low energy guanines. For an extended (A)20 bridge separating Ptz and photoexcited NI, central double guanine does attenuate CT yield by about half, indicating that over a very long piece of DNA relaxation of the cation does occur.227
Localized hopping is also inconsistent with electrochemical measurements, where the Fermi level is maintained up to a volt below the potentials of the bases.58,88,164,240,241 Consider as an example where the electrode is at the potential of an [4Fe-4S] containing protein (0.1 V), and injection is into cytidine (certainly < −0.9 V), the most readily reduced base, for an unfavorable driving force of at least 1.0 V. Although the coupling between cytidine and the metal is mediated by a long saturated linker, and hence will be small, let the coupling correspond to a generous value for two stacked bases, HDA ~ 0.2 eV, and take the reorganization energy as being 0.5 V. According to a simple nonadiabatic Marcus-derived expression,116,242 the injection rate is no greater than 0.007 s−1; for realistic values for the molecule-metal coupling this injection rate would necessarily be far lower. This is slower than the linker-limited rate found through DNA of about 30 s−1.18,240 Effectively, this discrepancy reflects the inherent unfavorability of thermal activation far from the bridge potential.
3.4. Delocalized Mechanisms
The models discussed so far each assume localization of a hole on a single base. Although the couplings between bases might be expected to allow delocalization, disorder in the bath should rapidly localize charges onto a single site as long as reorganization energy is greater than interbase coupling. However, there is some experimental evidence for delocalization of charges, such as the effect of stacking interactions on the pKA of the adenine cation radical,243 or the competition of CPC with CPG for oxidation.76 It has also been demonstrated that static disorder attenuates rapid hopping by creating low potential bottlenecks.238 This can be alleviated by allowing delocalization of the charge; in this case, static disorder is partially averaged.217,244 In conjuction with the known role of conformational gating, the obvious candidate for the delocalized state is the polaron.245,246
3.4.1. Polaron Hopping and Gating Mechanisms
Whenever charge is injected into a molecule, the environment will polarize in response, effectively partially delocalizing the charge and lowering the energy of the system.247 Since the energy of the polaron is different than that of the purely localized charge, the presence of a polaron will affect the CT behavior of the system, in a way largely dependent on the polaron size. Much like PCET is inevitable for any charge transfer participant with acidic protons, polaron formation is inevitable whenever CT proceeds with bridge occupation. The essential questions are whether the polarization occurs on a time-scale that can impact the CT process, which relaxation modes will be coupled to the polaron formation, and how much the polaron is stabilized relative to the completely localized state.
At first order, polarization of the environment in response to charge injection does not violate the tight-binding assumption. In this case, although DNA conformation, ion distribution, and water orientation all restructure as a result of charge migration, the affect on CT efficiency and rate will be via a change in the site energies on the bases, and gated by the time-scale of environmental polarization. It is important to note that small polaron formation slows charge migration, as the site energy is lowered, and hence the activation energy of each hop is increased; this leads to dynamic disorder, distinct from the static disorder discussed above. The exception is if the polaron can move by drift, where the orbitals of the donor and acceptor states overlap, so that CT occurs in the adiabatic limit. This results in transport that is faster than hopping, especially as it can be activationless.247 However, drift is most rapid between isoenergetic sites, so it is not a likely mechanism in the presence of static disorder, unless gated by conformational fluctuation of site energies.
Any description of polarons must take into account the structural rearrangement that provides the polarization. Lattice motion has been well treated in terms of deformations along the hydrogen bonds between the base pairs.248–250 This treatment is particularly instructive regarding the effect of increased coupling between the lattice motion and the charge; high coupling implies a higher activation energy for individual hops and a higher probability for trapping. Hence, thermal activation is taken as evidence for small polaron trapping. There have been contradictory results on the temperature dependence of CT in conductivity measurements,23,221 though photooxidation studies have unambiguously shown an increase in long-range CT rate184 and yield54,68 with temperature. Whether this temperature dependence is due to conformational gating, small polaron activation, or activation of localized hopping is not immediately obvious. Ultimately, the distinction between these cases is not sharp. Conformational dynamics influence bridge energy, and hence the activation energy for polaron drift. Calculations suggest that ion fluctuations, in particular, could sufficiently modulate the potential of a bridging sequence of DNA to permit polaron equilibration between two sites.50 Given the ambiguity in experiments where counterion identity and concentration have been varied,51–54 water re-orientation is more likely than ion motion to gate polaron formation.
Sufficient polarization of the environment will lead to formation of a large polaron. In this case, the polarization distortion extends far beyond the lattice site, i.e. the individual base (Figure 12). Polarization over a large range must involve the medium, water. Calculation has supported that these polarons form by water reorientation delocalized over 2 to 5 base pairs, depending on the sequence.251–252 Large polaron formation can have both positive and negative consequences for CT: self-trapping can decrease the rate of individual hopping steps, as is the case for small polarons, but delocalization decreases the distance between individual steps. Furthermore, for periodic sequences, drift can substantially increase the rate of individual hops, by lowering the activation energy for incoherent transport.
Figure 12.
Formation of a large polaron. Upon charge injection, a hole is initially localized on a single base (red). Reorientation of the environment, including neighboring bases and the hydration layer, lowers the energy of the hole. Delocalization occurs to the extent that the coupling between the bases balances the unfavorable decrease in the reorganization energy.
Critically, polaron drift can explain important features of DNA-mediated CT as discussed in the previous sections. As discussed above, the observed dependences of CT rates and yield on distance across adenine tracts is too shallow to be readily reconciled with thermal activation and localized hopping.17,125,207,236 Rapid polaron drift across adenine tracts, in concert with inhibition of BET from adenine to guanine due to polaron self-trapping, has been predicted to provide a shallow distance dependence.253 Furthermore, since the calculated polaron size is ~ 4 adenines, the steep distance dependence that has been found for tracts shorter than this length naturally corresponds to these sequences not supporting polaron formation.
This mechanism is not limited to adenine tracts, as polaron formation is predicted over mixed polypurine sites, with significant population of high-energy pyrimidines.252,254 This is consistent with the oxidative damage observed at the fast trap CPC despite the presence of guanine75 and the preferential damage at thymine in constructs containing only thymine or adenine.69 As described above, the resulting delocalization serves as a mechanism for dynamic motions, that allow polaron formation, to alleviate the barrier to CT generated by the static disorder of site energies in DNA, and is consistent with the observed long-range migration of CT through mixed DNA sequences.3,103 In this model, the effective hopping rates observed by the Majima group could correspond to hops between delocalized polaron sites.4,226,227
Physical identification of the polarization medium allows calculation of the polaron properties, particularly the speed limit on polaron migration imposed by the rate of repolarization. For drift along an adenine tract, water reorientation limits polaron mobility to about 3×10−3 cm2/(V s).255 This mobility can be related to the conductivity of a single DNA between two carbon nanotubes,34 where a mixed, aperiodic sequence should decrease the mobility of a polaron. Here, a resistance of about 3 MΩ was observed in a fifteen base pair duplex. Although the number of charge carriers was unknown, it certainly cannot be less than unity or greater than fifteen, the number of base pairs. Within that range, the mobility is constrained to between 3 × 10−2 cm2/(V sec) and 5 × 10−1 cm2/(V sec). It will be interesting if theoretical evaluation is able to rationalize these values, as they appear inconsistent with polaron drift.
3.4.2. Domain Delocalization
Evidence for delocalization in DNA has come from recent insights into long-lived excited states and exciplexes. It is well known that the individual nucleotides of DNA rapidly relax upon excitation, with a low fluorescence quantum yield.256 This property is essential for the molecule of life, as long-lived excited states would render the genetic material prone to photodamage. For mixed sequence DNA as well, relaxation is rapid (subpicosecond to picosecond), but in a manner highly dependent on the specific sequence context.257 Recently, however, femtosecond studies of purine repeats in both oligonucleotides and duplexes have found much longer lifetimes for ground state recovery,189 possibly due to exciton or exciplex states. Critically, for B-form DNA, it is base stacking rather than base-pairing interactions that are most critical in achieving long-lived states.258 The extent of delocalization is still under debate. Although some states might extend over 4 to 8 base pairs,259,260 others are delocalized over only two base pairs,261 particularly in single strands. There is some computational support for this delocalization, as well. DFT calculations find that eximers can delocalize over several adenines,262 and it has been found that fluctuations in the on-site energies of neighboring bases is highly correlated.214 Recent calculations that incorporate static and dynamic disorder and solvent effects have shown that such transient delocalization can occur over several base-pairs.263
Despite experimental and computational support for delocalization, as described above, there are profound theoretical arguments against delocalization models in DNA. A variety of computational studies have found that solvent and ion motions will strongly localize injected charge to a single or only a few nucleotides.152,264,265 Importantly, these studies have mostly been limited to considerations of equilibrated or averaged structures. It will be important to determine whether solvent-induced localization is maintained in the presence of dynamic disorder.
A recent computational study266 on the dynamics of electric fields produced by DNA, water, and ions was able to reproduce time-resolved Stokes shift data that directly measures those dynamics.127 This study found no evidence of subpopulations where the DNA had particular electric fields beyond the Gaussian distribution. However, the Gaussian tails could in principle be the very states that mediate CT. All the data suggest that delocalization must be highly transient, and over very long distances, a substantial amount of incoherent hopping will also occur.
Perhaps the most distinctive feature of DNA-mediated CT that has been observed in recent years is the periodic length dependence of CT yield across adenine tracts for some systems (Figure 13). This dependence was clear, with the same period of 3 to 4 base pairs, for coherent transport from Ap* to guanine54 and for total CT from [Rh(phi)2(bpy’)]*3+ to CPG.68 This periodicity was shown to be with respect to adenine tract length, rather than with respect to donor-acceptor distance; by measuring the decomposition of CPA moved serially along an adenine tract of constant length, no periodicity is observed.17 For CT from photoexcited AQ to guanine, the periodicity is less apparent, but this is likely due to the quenching of the radical anion of AQ by oxygen, which allows charge equilibration. Interestingly, Ap* oxidation of CPG across adenine tracts is smoothly monotonic, but separating the Ap* from the adenine tract with three inosines restores the periodicity. Clearly, the rapid BET associated with Ap* allows duplexes that are well suited to forward transport also to better mediate BET, suppressing the periodicity. It should be noted that the inosine tract is a high potential barrier to oxidation by Ap.120 It lowers both forward CT and BET, but since the former competes with the nanosecond Ap* fluorescence lifetime, and the latter competes with picosecond ring opening, BET should be comparatively more attenuated. With BET suppressed, periodicity is again apparent.
Figure 13.
Equivalent periodicities with the same period and temperature dependence are observed for (B) the single-step oxidation of guanine by Ap* and (A) the total oxidation of CPG by photoexcited [Rh(phi)2(bpy’)]3+. Temperature increases from purple to red. Errors are given in (A) as 90% SEM.54,68 Reprinted with permission from References 54 and 68. Copyright American Chemical Society.
A periodic A-tract dependence indicates that some adenine tract lengths mediate CT superior to others. Based on our experiments, this length is about three or four base pairs. In light of the extensive evidence for delocalization cited above, we characterize this CT-active tract as a delocalized domain. The role of conformational gating, then, is to generate this CT-active state. An adenine tract length that allows an integer number of these states allows facile CT; transport across other tracts requires dephasing processes, such as drift or hopping. Because these domains are, by their nature, transient, these effects will only be seen in experiments where the donor and acceptor are well-coupled to the bridge, and where injection and arrival can be observed on a fast timescale, decoupled from other pathways, such as BET. Critically, domain delocalization readily explains the facile competition between CPC and CPG,74 and the ability of DNA to mediate CT far below the base potentials.161,164
4. Summary
It is clear that DNA, when adequately coupled between the donor and acceptor, can competently mediate CT over long distances. This property is dependent on, and hence diagnostic of, the integrity of base stacking. Furthermore, long-range DNA-mediated CT is thermally activated in a manner dependent on the dynamical stacking of the bridge, indicating that conformational gating is convoluted with the CT rate. Theoretically, CT over long molecular distances cannot be assigned to superexchange. Incoherent transport must play a role, although evidence does support coherent transport over at least 30 Å in some systems. Assigning the intermediates as guanine cation radicals in the context of a variable range hopping model is sufficient to explain some gross features of DNA-mediated CT, but this model cannot explain long-range coherence. Transient delocalization plays an important role, at least with some sequences. Identifying the extent to which delocalization occurs, including via polaron formation, will be particularly important for understanding DNA CT mediated at potentials below those of the individual nucleotides.
Any model for DNA CT must consider the effects of static and dynamic disorder. For most models, static disorder attenuates long-range CT. Since DNA has many sources of static disorder in the site energies, inter-site couplings, and reorganization energies, it is unlikely that calculations performed on uniform ideal structures with a single repeating base-pair will be relevant to understanding experimental results. On the other hand, dynamic disorder has the potential to alleviate the challenge posed by static disorder, by allowing transient structures to form with less rugged energetic landscapes. As long as the equilibrium conformation is not the most CT-active conformation, this condition will hold for most pathways, whether incoherent or coherent. Computational studies have begun to appear that consider what CT-active states look like267; it will be a challenge to experimentalists to evaluate these exciting predictions.
CT between a donor and acceptor will always proceed through the fastest pathways available. In a dynamic, structurally complex molecule like DNA, multiple timescales describe the energetic and coupling landscapes, and hence there will be a time-dependent ensemble of pathways. This ensemble is even larger when delocalized states are allowed, whether they are transiently formed prior to, concurrently with, or after charge injection. For conditions that deplete available pathways, whether through rigidifying the duplex, disrupting donor and acceptor coupling to the bridge, or by introducing structural distortion, slower CT and conduction will inevitably result. In this context, correlating the distance dependence to the β value of the electronic factor of the CT rate equation requires a high level of experimental support. It is unlikely that any of the measured distance dependences correspond to the distance dependence of the purely electronic component of CT through DNA. Nevertheless, the effective distance dependence over long distances compares favorably with common molecular wires such as oligophenylenevinylene and oligophenyleneethynylene, indicating a promising role for DNA in molecular electronics.
5. Unanswered Questions
It should be clear from this review that DNA-mediated CT does not pose a challenge to the fundamental theories of electron and hole transport. Ultimately, charge transfer events only occur with the rates predicted by Marcus’ theory. For a molecule as large and complicated as DNA, however, the parameters for the Marcus equation are not trivial to determine. Each conformation of a given DNA offers many pathways, and the extent of dynamical disorder leads to the failure of the Condon approximation. Furthermore, in the context of hopping and drift, the nature of the states that mediate charge transport vary with sequence and sequence-dependent dynamics. What these states are: localized radical cations, localized neutral radicals, large polarons, delocalized domains, or a combination, will be different based on the properties of the specific donor, DNA bridge, and acceptor. Understanding what conditions lead to what mechanism of transport is important, as the physical nature of charge injection and migration in DNA undoubtedly influences CT between DNA and redox-active DNA-binding proteins,5,17,93,94 and the cellular defense against oxidizing radicals.105,268–271
Particular experiments that require more attention by theorists are the electrochemical experiments in DNA films. In these experiments, the Fermi level is held to potentials far from those of the bridge states, and yet many of the same properties are observed here as are observed in solution and device experiments that are at profoundly different energies. Insight into this process will undoubtedly also help elucidate DNA-mediated CT in general.
Ultimately, single-molecule conductivity experiments have the most potential for determining details of DNA-mediated CT, due to the strong control of driving force and online measurement of current. The main challenges for these experiments is maintaining the DNA in its native structure, and establishing that the observed current is, in fact, due to the DNA. These can be easily determined by the proper choice of controls.
If the past fifteen years of DNA-mediated CT are any indication, the synergy between the applications of DNA in devices and biology, and theoretical and experimental efforts to elucidate the mechanism, will continue to advance both areas of study. Certainly, bringing these different perspectives together offer both a challenge and an opportunity.
Acknowledgments
We thank the NIH (GM49216) for their financial support of this work. We also thank the many researchers in the field of DNA CT chemistry that have contributed their many and varied perspectives.
Biographies

Joseph C. Genereux was born in Burbank, CA in 1981. He received his B.A. with honors in Chemistry from Swarthmore College in 2001, where he worked in the laboratory of Prof. Robert F. Pasternack. He received a B.S. in Physics from the University of California – Irvine in 2002. He is currently a graduate student in the research group of Professor Jacqueline K. Barton at Caltech, studying DNA-mediated charge transport.

Dr. Jacqueline K. Barton is the Arthur and Marian Hanisch Memorial Professor of Chemistry and Chair of the Division of Chemistry and Chemical Engineering at the California Institute of Technology. Barton was awarded the A.B. summa cum laude at Barnard College in 1974 and a Ph.D. at Columbia University in 1978. After a postdoctoral fellowship at Bell Laboratories and Yale University, she became an assistant professor at Hunter College, City University of New York. In 1983, she returned to Columbia University, becoming an associate professor of chemistry and biological sciences in 1985 and professor in 1986. In the fall of 1989, she joined the faculty at Caltech. In 2009, she began her term as Chair of the Division. Professor Barton has pioneered the application of transition metal complexes to probe recognition and reactions of double helical DNA, in particular studies to elucidate electron transfer chemistry mediated by the DNA double helix. Barton has received numerous awards. These include the NSF Waterman Award, the ACS Award in Pure Chemistry, the ACS Breslow Award in Biomimetic Chemistry and a MacArthur Foundation Fellowship. She is a member of the American Academy of Arts and Sciences, the American Philosophical Society, and the National Academy of Sciences.
References
- 1.Murphy CJ, Arkin MR, Jenkins Y, Ghatlia ND, Bossmann SH, Turro NJ, Barton JK. Science. 1993;262:1025. doi: 10.1126/science.7802858. [DOI] [PubMed] [Google Scholar]
- 2.Murphy CJ, Arkin MR, Ghatlia ND, Bossman SH, Turro NJ, Barton JK. Proc. Natl. Acad. Sci. USA. 1994;91:5315. doi: 10.1073/pnas.91.12.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nuñez ME, Hall DB, Barton JK. Chem. Biol. 1999;6:85. doi: 10.1016/S1074-5521(99)80005-2. [DOI] [PubMed] [Google Scholar]
- 4.Takada T, Kawai K, Fujitsuka M, Majima T. Proc. Natl. Acad. Sci. USA. 2004;101:14002. doi: 10.1073/pnas.0402756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee PE, Demple B, Barton JK. Proc. Natl. Acad. Sci. USA. 2009;106:13164. doi: 10.1073/pnas.0906429106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osakada Y, Kawai K, Fujitsuka M, Majima T. Proc. Natl. Acad. Sci. USA. 2006;103:18072. doi: 10.1073/pnas.0607148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis WB, Hess S, Naydenova I, Haselsberger R, Ogrodnik A, Newton MD, Michel-Beyerle M-E. J. Am. Chem. Soc. 2002;124:2422. doi: 10.1021/ja017304l. [DOI] [PubMed] [Google Scholar]
- 8.von Feilitszch T, Tuma J, Neubauer H, Verdier L, Haselsberger R, Feick R, Gurzadyan G, Voityuk AA, Griesinger C, Michel-Beyerle ME. J. Phys. Chem. B. 2008;112:973. doi: 10.1021/jp076405o. [DOI] [PubMed] [Google Scholar]
- 9.Marcus RA, Sutin N. Biochim. Biophys. Acta. 1985;811:265. [Google Scholar]
- 10.Turro NJ, Barton JK. J. Biol. Inorg. Chem. 1998;3:201. [Google Scholar]
- 11.Endres RG, Cox DL, Singh RRP. Rev. Mod. Phys. 2004;76:195. [Google Scholar]
- 12.Chen F, Hihath J, Huang Z, Li X, Tao NJ. Annu. Rev. Phys. Chem. 2007;58:535. doi: 10.1146/annurev.physchem.58.032806.104523. [DOI] [PubMed] [Google Scholar]
- 13.Müller K-H. Phys. Rev. B. 2006;73:045403. [Google Scholar]
- 14.Kelley SO, Barton JK. Science. 1999;283:375. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 15.Delaney S, Pascaly M, Bhattacharya PK, Han K, Barton JK. Inorg. Chem. 2002;41:1966. doi: 10.1021/ic0111738. [DOI] [PubMed] [Google Scholar]
- 16.Tada T, Kondo M, Yoshizawa K. ChemPhysChem. 2003;4:1256. doi: 10.1002/cphc.200300811. [DOI] [PubMed] [Google Scholar]
- 17.Augustyn KE, Genereux JC, Barton JK. Angew. Chem. Int. Ed. 2007;46:5731. doi: 10.1002/anie.200701522. [DOI] [PubMed] [Google Scholar]
- 18.Gorodetsky AA, Green O, Yavin E, Barton JK. Bioconj. Chem. 2007;18:1434. doi: 10.1021/bc0700483. [DOI] [PubMed] [Google Scholar]
- 19.Fink HW, Schoenberger C. Nature. 1999;398:407. doi: 10.1038/18855. [DOI] [PubMed] [Google Scholar]
- 20.de Pablo PJ, Moreno-Herrero F, Colchero J, Gómez Herrero J, Herrero P, Baró AM, Ordejón P, Soler JM, Artacho E. Phys. Rev. Lett. 2000;85:4992. doi: 10.1103/PhysRevLett.85.4992. [DOI] [PubMed] [Google Scholar]
- 21.Kasumov AYu, Kociak M, Guéron S, Reulet B, Volkov VT, Klinov DV, Bouchiat H. Science. 2001;291:280. doi: 10.1126/science.291.5502.280. [DOI] [PubMed] [Google Scholar]
- 22.Storm AJ, van Noort J, de Vries S, Dekker C. Appl. Phys. Lett. 2001;79:3881. [Google Scholar]
- 23.Yoo K-H, Ha DH, Lee J-O, Park JW, Kim J, Kim JJ, Lee H-Y, Kawai T, Choi HY. Phys. Rev. Lett. 2001;87:198102. doi: 10.1103/PhysRevLett.87.198102. [DOI] [PubMed] [Google Scholar]
- 24.Odom DT, Dill EA, Barton JK. Chem. Biol. 2000;7:475. doi: 10.1016/s1074-5521(00)00133-2. [DOI] [PubMed] [Google Scholar]
- 25.Schuster GB, editor. Long-Range Charge Transfer in DNA, I and II. Vols. 236, 237. New York: Springer; 2004. [Google Scholar]
- 26.Wagenknecht HA, editor. Charge Transfer in DNA. Weinheim: Wiley-VCH; 2005. [Google Scholar]
- 27.Valis L, Wang Q, Raytchev M, Buchvarov I, Wagenknecht H-A, Fiebig T. Proc. Natl. Acad. Sci. USA. 2006;103:10192. doi: 10.1073/pnas.0600957103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meade TJ, Kayyem JF. Angew. Chem. Int. Ed. 1995;34:352. [Google Scholar]
- 29.Lyndon Pittman T, Miao W. J. Phys. Chem. C. 2008;112:16999. [Google Scholar]
- 30.Boon EM, Jackson NM, Wrightman MD, Kelley SO, Hill MG, Barton JK. J. Phys. Chem. B. 2003;107:11805. [Google Scholar]
- 31.Drummond TG, Hill MG, Barton JK. J. Am. Chem. Soc. 2004;126:15010. doi: 10.1021/ja044910i. [DOI] [PubMed] [Google Scholar]
- 32.Watson JD, Crick FHC. Nature. 1953;171:737. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 33.Watson JD, Crick FHC. Nature. 1953;171:964. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 34.Guo X, Gorodetsky AA, Hone J, Barton JK, Nuckolls C. Nat. Nanotech. 2008;3:163. doi: 10.1038/nnano.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Neill MA, Becker HC, Wan C, Barton JK, Zewail AH. Angew. Chem. Int. Ed. 2003;42:5896. doi: 10.1002/anie.200352831. [DOI] [PubMed] [Google Scholar]
- 36.Melvin T, Botchway S, Parker AW, O’Neill P. J. Chem. Soc., Chem. Commun. 1995;6:653. [Google Scholar]
- 37.Cohen H, Nogues C, Naaman R, Porath D. Proc. Natl. Acad. Sci. USA. 2005;102:11589. doi: 10.1073/pnas.0505272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen H, Nogues C, Ullien D, Daube S, Naaman R, Porath D. Far. Disc. 2006;131:367. doi: 10.1039/b507706k. [DOI] [PubMed] [Google Scholar]
- 39.Ceres DM, Barton JK. J. Am. Chem. Soc. 2003;125:14964. doi: 10.1021/ja0384476. [DOI] [PubMed] [Google Scholar]
- 40.Boon EM, Ceres DM, Drummond TG, Hill MG, Barton JK. Nat. Biotech. 2000;18:1096. doi: 10.1038/80301. [DOI] [PubMed] [Google Scholar]
- 41.Calladine CR, Drew HR, Luisi BF, Travers AA. Understanding DNA: The Molecule and How it Works. San Diego: Elsevier Academic Press; 2004. p. 304. [Google Scholar]
- 42.Pal SK, Zewail AH. Chem. Rev. 2004;104:2099. doi: 10.1021/cr020689l. [DOI] [PubMed] [Google Scholar]
- 43.Yamahata C, Collard D, Takekawa T, Kumemura M, Hashiguchi G, Fujita H. Biophys. J. 2008;94:63. doi: 10.1529/biophysj.107.115980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han Ha D, Nham H, Yoo K-H, So H-m, Lee H-Y, Kawai T. Chem. Phys. Lett. 2002;355:405. [Google Scholar]
- 45.Xu B, Zhang P, Li X, Tao N. Nano Lett. 2004;4:1105. [Google Scholar]
- 46.Nogues C, Cohen SR, Daube S, Apter N, Naaman R. J. Phys. Chem. B. 2006;110:8910. doi: 10.1021/jp060870o. [DOI] [PubMed] [Google Scholar]
- 47.Xiao X, Xu B, Tao NJ. Nano Lett. 2004;4:267. [Google Scholar]
- 48.Boon EM, Barton JK. Bioconj. Chem. 2003;14:1140. doi: 10.1021/bc034139l. [DOI] [PubMed] [Google Scholar]
- 49.Steinbrecher T, Koslowski T, Case DA. J. Phys. Chem. B. 2008;112:16935. doi: 10.1021/jp8076134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barnett RN, Cleveland CL, Joy A, Landman U, Schuster GB. Science. 2001;294:567. doi: 10.1126/science.1062864. [DOI] [PubMed] [Google Scholar]
- 51.Williams TT, Odom DT, Barton JK. J. Am. Chem. Soc. 2000;122:9048. [Google Scholar]
- 52.Williams TT, Barton JK. J. Am. Chem. Soc. 2002;124:1841. doi: 10.1021/ja012217e. [DOI] [PubMed] [Google Scholar]
- 53.Douki T, Angelov D, Cadet J. J. Am. Chem. Soc. 2001;123:11360. doi: 10.1021/ja016426a. [DOI] [PubMed] [Google Scholar]
- 54.O’Neill MA, Barton JK. J. Am. Chem. Soc. 2004;126:11471. doi: 10.1021/ja048956n. [DOI] [PubMed] [Google Scholar]
- 55.Blaustein GS, Demas B, Lewis FD, Burin AL. J. Am. Chem. Soc. 2009;131:400. doi: 10.1021/ja805589r. [DOI] [PubMed] [Google Scholar]
- 56.Abdou IM, Sartor V, Cao H, Schuster GB. J. Am. Chem. Soc. 2001;123:6696. doi: 10.1021/ja004274y. [DOI] [PubMed] [Google Scholar]
- 57.O’Neill MA, Barton JK. J. Am. Chem. Soc. 2002;124:13053. doi: 10.1021/ja0208198. [DOI] [PubMed] [Google Scholar]
- 58.Boon EM, Salas JE, Barton JK. Nat. Biotech. 2002;20:282. doi: 10.1038/nbt0302-282. [DOI] [PubMed] [Google Scholar]
- 59.Okamoto A, Kamei T, Saito I. J. Am. Chem. Soc. 2006;128:658. doi: 10.1021/ja057040t. [DOI] [PubMed] [Google Scholar]
- 60.Kelley SO, Barton JK. Chem. Biol. 1998;5:413. doi: 10.1016/s1074-5521(98)90158-2. [DOI] [PubMed] [Google Scholar]
- 61.Bhattacharya PK, Barton JK. J. Am. Chem. Soc. 2001;123:8649. doi: 10.1021/ja010996t. [DOI] [PubMed] [Google Scholar]
- 62.Osakada Y, Kawai K, Fujitsuka M, Majima T. Nuc. Acids Res. 2008;36:5562. doi: 10.1093/nar/gkn505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bhattacharya PK, Cha J, Barton JK. Nuc. Acids Res. 2002;30:4740. doi: 10.1093/nar/gkf601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buzzeo MC, Barton JK. Bioconj. Chem. 2008;19:2110. doi: 10.1021/bc800339y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boal AK, Barton JK. Bioconj. Chem. 2005;16:312. doi: 10.1021/bc0497362. [DOI] [PubMed] [Google Scholar]
- 66.Williams TT, Dohno C, Stemp EDA, Barton JK. J. Am. Chem. Soc. 2004;126:8148. doi: 10.1021/ja049869y. [DOI] [PubMed] [Google Scholar]
- 67.Furrer E, Giese B. Helv. Chim. Acta. 2003;86:3623. [Google Scholar]
- 68.Genereux JC, Augustyn KE, Davis ML, Shao F, Barton JK. J. Am. Chem. Soc. 2008;130:15150. doi: 10.1021/ja8052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joy A, Ghosh AK, Schuster GB. J. Am. Chem. Soc. 2006;128:5346. doi: 10.1021/ja058758b. [DOI] [PubMed] [Google Scholar]
- 70.Delaney S, Yoo J, Stemp EDA, Barton JK. Proc. Natl. Acad. Sci. USA. 2004;101:10511. doi: 10.1073/pnas.0403791101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakatani K, Dohno C, Saito I. J. Am. Chem. Soc. 2001;123:9681. doi: 10.1021/ja010479a. [DOI] [PubMed] [Google Scholar]
- 72.O’Neill MA, Dohno C, Barton JK. J. Am. Chem. Soc. 2004;126:1316. doi: 10.1021/ja037802p. [DOI] [PubMed] [Google Scholar]
- 73.Dohno C, Ogawa A, Nakatani K, Saito I. J. Am. Chem. Soc. 2003;125:10154. doi: 10.1021/ja035887o. [DOI] [PubMed] [Google Scholar]
- 74.Elias B, Genereux JC, Barton JK. Angew. Chem. Int. Ed. 2008;47:9067. doi: 10.1002/anie.200803556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shao F, O’Neill MA, Barton JK. Proc. Natl. Acad. Sci. USA. 2004;101:17914. doi: 10.1073/pnas.0408128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shao F, Augustyn KE, Barton JK. J. Am. Chem. Soc. 2005;127:17445. doi: 10.1021/ja0563399. [DOI] [PubMed] [Google Scholar]
- 77.Liu T, Barton JK. J. Am. Chem. Soc. 2005;127:10160. doi: 10.1021/ja053025c. [DOI] [PubMed] [Google Scholar]
- 78.Osakada Y, Kawai K, Fujitsuka M, Majima T. Chem. Comm. 2008;23:2656. doi: 10.1039/b801876f. [DOI] [PubMed] [Google Scholar]
- 79.Nakatani K, Dohno C, Saito I. J. Am. Chem. Soc. 2000;122:5893. [Google Scholar]
- 80.Kodera H, Osakada Y, Kawai K, Majima T. Nat. Chem. 2009;1:156. doi: 10.1038/nchem.171. [DOI] [PubMed] [Google Scholar]
- 81.Okamoto A, Nakatani K, Saito I. J. Am. Chem. Soc. 2003;125:5066. doi: 10.1021/ja0294008. [DOI] [PubMed] [Google Scholar]
- 82.Hunter WN, Brown T, Kennard O. Nuc. Acids. Res. 1987;15:6589. doi: 10.1093/nar/15.16.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hunter WN, Brown T, Kneale G, Anand NN, Rabinovich D, Kennard O. J. Biol. Chem. 1987;262:9962. doi: 10.2210/pdb113d/pdb. [DOI] [PubMed] [Google Scholar]
- 84.Kumamoto S, Watanabe M, Kawakami N, Nakamura M, Yamanato K. Bioconj. Chem. 2008;19:65. doi: 10.1021/bc070097f. [DOI] [PubMed] [Google Scholar]
- 85.Takada T, Fujitsuka M, Majima T. Proc. Natl. Acad. Sci. USA. 2007;104:11179. doi: 10.1073/pnas.0700795104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elias B, Shao F, Barton JK. J. Am. Chem. Soc. 2008;130:1152. doi: 10.1021/ja710358p. [DOI] [PubMed] [Google Scholar]
- 87.Giese B, Wessely S. Chem. Comm. 2001;20:2108. doi: 10.1039/b106059g. [DOI] [PubMed] [Google Scholar]
- 88.Boal AK, Yavin E, Lukianova OA, O’Shea VL, David SS, Barton JK. Biochem. 2005;44:8397. doi: 10.1021/bi047494n. [DOI] [PubMed] [Google Scholar]
- 89.Gasper SM, Schuster GB. J. Am. Chem. Soc. 1997;119:12762. [Google Scholar]
- 90.Hickerson RP, Prat F, Muller JG, Foote CS, Burrows CJ. J. Am. Chem. Soc. 1999;121:9423. [Google Scholar]
- 91.Drummond TG, Hill MG, Barton JK. Nat. Biotech. 2003;21:1192. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 92.Inouye M, Ikeda R, Takase M, Tsuri T, Chiba J. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11606. doi: 10.1073/pnas.0502078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boal AK, Yavin E, Barton JK. J. Inorg. Biochem. 2007;101:1913. doi: 10.1016/j.jinorgbio.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gorodetsky AA, Dietrich LEP, Lee PE, Demple B, Newman DK, Barton JK. Proc. Natl. Acad. Sci. USA. 2008;105:3684. doi: 10.1073/pnas.0800093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Watanabe S, Kita A, Kobayashi K, Miki K. Proc. Natl. Acad. Sci. USA. 2008;105:4121. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gorodetsky AA, Ebrahim A, Barton JK. J. Am. Chem. Soc. 2008;130:2924. doi: 10.1021/ja7106756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rajski SR, Barton JK. Biochem. 2001;40:5556. doi: 10.1021/bi002684t. [DOI] [PubMed] [Google Scholar]
- 98.Nakatani K, Dohno C, Ogawa A, Saito I. Chem. Biol. 2002;9:361. doi: 10.1016/s1074-5521(02)00119-9. [DOI] [PubMed] [Google Scholar]
- 99.Kurbanyan K, Nguyen KL, To P, Rivas EV, Lueras AMK, Kosinski C, Steryo M, González A, Mah DA, Stemp EDA. Biochemistry. 2003;42:10269. doi: 10.1021/bi020713p. [DOI] [PubMed] [Google Scholar]
- 100.Perrier S, Hau J, Gasparutto D, Cadet J, Favier A, Ravanat JL. J. Am. Chem. Soc. 2006;128:5703. doi: 10.1021/ja057656i. [DOI] [PubMed] [Google Scholar]
- 101.Cadet J, Douki T, Ravanat J-L. Acc. Chem. Res. 2008;41:1075. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 102.Bjorklund CC, Davis WB. Biochemistry. 2007;46:10745. doi: 10.1021/bi700475b. [DOI] [PubMed] [Google Scholar]
- 103.Nuñez ME, Noyes KT, Barton JK. Chem. Biol. 2002;9:403. doi: 10.1016/s1074-5521(02)00121-7. [DOI] [PubMed] [Google Scholar]
- 104.Nuñez ME, Holmquist GP, Barton JK. Biochemistry. 2001;40:12465. doi: 10.1021/bi011560t. [DOI] [PubMed] [Google Scholar]
- 105.Merino EJ, Barton JK. Biochemistry. 2008;47:1511. doi: 10.1021/bi701775s. [DOI] [PubMed] [Google Scholar]
- 106.Nguyen KL, Steryo M, Kurbanyan K, Nowitzki KM, Butterfield SM, Ward SR, Stemp EDA. J. Am. Chem. Soc. 2000;122:3585. [Google Scholar]
- 107.Johansen ME, Muller JG, Xu XY, Burrows CJ. Biochemistry. 2005;44:5660. doi: 10.1021/bi047580n. [DOI] [PubMed] [Google Scholar]
- 108.Peng X, Pigli YZ, Rice PA, Greenberg MM. J. Am. Chem. Soc. 2008;130:12890. doi: 10.1021/ja805440v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu C-S, Schuster GB. J. Am. Chem. Soc. 2003;125:6098. doi: 10.1021/ja029333h. [DOI] [PubMed] [Google Scholar]
- 110.Lewis FD, Daublain P, Cohen B, Vura-Weiss J, Shafirovich V, Wasielewski MR. J. Am. Chem. Soc. 2007;129:15130. doi: 10.1021/ja076876o. [DOI] [PubMed] [Google Scholar]
- 111.O’Neill MA, Barton JK. Proc. Natl. Acad. Sci USA. 2002;99:16543. doi: 10.1073/pnas.012669599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shao F, Barton JK. J. Am. Chem. Soc. 2007;129:14733. doi: 10.1021/ja0752437. [DOI] [PubMed] [Google Scholar]
- 113.Grozema FC, Tonzani S, Berlin YA, Schatz GC, Siebbeles LDA, Ratner MA. J. Am. Chem. Soc. 2008;130:5157. doi: 10.1021/ja078162j. [DOI] [PubMed] [Google Scholar]
- 114.Davis WB, Ratner MA, Wasielewski MR. J. Am. Chem. Soc. 2001;123:7877. doi: 10.1021/ja010330z. [DOI] [PubMed] [Google Scholar]
- 115.Smalley JF, Sachs SB, Chidsey CED, Dudek SP, Sikes HD, Creager SE, Yu CJ, Feldberg SW, Newton MD. J. Am. Chem. Soc. 2004;126:14620. doi: 10.1021/ja047458b. [DOI] [PubMed] [Google Scholar]
- 116.Newton MD, Smalley JF. Phys. Chem. Chem. Phys. 2007;9:555. doi: 10.1039/b611448b. [DOI] [PubMed] [Google Scholar]
- 117.O’Neill MA, Barton JK. J. Am. Chem. Soc. 2004;126:13234. doi: 10.1021/ja0455897. [DOI] [PubMed] [Google Scholar]
- 118.Siriwong K, Voityuk AA, Newton MD, Rösch N. J. Phys. Chem. B. 2003;107:2595. [Google Scholar]
- 119.Kocherzhenko AA, Patwardhan S, Grozema FC, Anderson HL, Siebbeles LDA. J. Am. Chem. Soc. 2009;131:5522. doi: 10.1021/ja809174y. [DOI] [PubMed] [Google Scholar]
- 120.Wan CZ, Fiebig T, Kelley SO, Treadway CR, Barton JK, Zewail AH. Proc. Natl. Acad. Sci. USA. 1999;96:6014. doi: 10.1073/pnas.96.11.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kelley SO, Holmlin ER, Stemp EDA, Barton JK. J. Am. Chem. Soc. 1997;199:9861. [Google Scholar]
- 122.Lewis FD, Wu T, Xhang Y, Letsinger RL, Greenfield MR, Wasielewski MR. Science. 1997;277:673. doi: 10.1126/science.277.5326.673. [DOI] [PubMed] [Google Scholar]
- 123.Lewis FD, Wu T, Liu X, Letsinger RL, Greenfield SR, Miller SE, Wasielewski MR. J. Am. Chem. Soc. 2000;122:2889. [Google Scholar]
- 124.Lewis FD, Zhu H, Daublain P, Fiebig T, Raytchev M, Wang Q, Shafirovich V. J. Am. Chem. Soc. 2006;128:791. doi: 10.1021/ja0540831. [DOI] [PubMed] [Google Scholar]
- 125.Lewis FD, Zhu H, Daublain P, Cohen B, Wasielewski MR. Angew. Chem. Int. Ed. 2006;45:7982. doi: 10.1002/anie.200603455. [DOI] [PubMed] [Google Scholar]
- 126.Lewis FD, Zhu H, Daublain P, Sigmund K, Fiebig T, Raytchev M, Wang Q, Shafirovich V. Photochem. Photobiol. Sciences. 2008;7:534. doi: 10.1039/b719715b. [DOI] [PubMed] [Google Scholar]
- 127.Andreatta D, Pérez Lustres JL, Kovalenko SA, Ernsting NP, Murphy CJ, Coleman RS, Berg MA. J. Am. Chem. Soc. 2005;127:7270. doi: 10.1021/ja044177v. [DOI] [PubMed] [Google Scholar]
- 128.Dohno C, Stemp EDA, Barton JK. J. Am. Chem. Soc. 2003;125:9586. doi: 10.1021/ja036397z. [DOI] [PubMed] [Google Scholar]
- 129.Armitage B, Yu C, Devadoss C, Schuster GB. J. Am. Chem. Soc. 1994;116:9847. [Google Scholar]
- 130.Sanii L, Schuster GB. J. Am. Chem. Soc. 2000;122:11545. [Google Scholar]
- 131.Vicic DA, Odom DT, Núñez ME, Gianolio DA, McLaughlin LW, Barton JK. J. Am. Chem. Soc. 2000;122:8603. [Google Scholar]
- 132.Dotse AK, Boone EK, Schuster GB. J. Am. Chem. Soc. 2000;122:6825. [Google Scholar]
- 133.Kawai K, Osakada Y, Takada T, Fujitsuka M, Majima T. J. Am. Chem. Soc. 2004;126:12843. doi: 10.1021/ja0475813. [DOI] [PubMed] [Google Scholar]
- 134.Kawai K, Osakada Y, Fujitsuka M, Majima T. Chem. Eur. J. 2008;14:3721. doi: 10.1002/chem.200701835. [DOI] [PubMed] [Google Scholar]
- 135.Kawai K, Takada T, Nagai T, Cai X, Sugimoto A, Fujitsuka M, Majima T. J. Am. Chem. Soc. 2003;125:16198. doi: 10.1021/ja038309g. [DOI] [PubMed] [Google Scholar]
- 136.Kawai K, Osakada Y, Fujitsuka M, Majima T. J. Phys. Chem. B. 2008;112:2144. doi: 10.1021/jp075326+. [DOI] [PubMed] [Google Scholar]
- 137.Arkin MR, Stemp EDA, Holmlin RE, Barton JK, Hörmann A, Olson EJC, Barbara PF. Science. 1996;273:475. doi: 10.1126/science.273.5274.475. [DOI] [PubMed] [Google Scholar]
- 138.Wan CZ, Fiebig T, Schiemann O, Barton JK, Zewail AH. Proc. Natl. Acad. Sci. USA. 2000;97:14052. doi: 10.1073/pnas.250483297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lewis FD, Liu J, Zuo X, Hayes RT, Wasielewski MR. J. Am. Chem. Soc. 2003;125:4850. doi: 10.1021/ja029390a. [DOI] [PubMed] [Google Scholar]
- 140.Senthilkumar K, Grozema FC, Guerra CF, Bilkelhaupt FM, Lewis FD, Berlin YA, Ratner MA, Siebbeles LDA. J. Am. Chem. Soc. 2005;127:14894. doi: 10.1021/ja054257e. [DOI] [PubMed] [Google Scholar]
- 141.Santhosh U, Schuster GB. J. Am. Chem. Soc. 2002;124:10986. doi: 10.1021/ja026932f. [DOI] [PubMed] [Google Scholar]
- 142.O’Neill MA, Barton JK. In: Topics in Current Chemistry. Schuster GB, editor. Vol. 237. Berlin: Springer; 2004. p. 67. [Google Scholar]
- 143.Boussicault F, Robert M. Chem. Rev. 2008;108:2622. doi: 10.1021/cr0680787. [DOI] [PubMed] [Google Scholar]
- 144.Seidel CAM, Shulz A, Sauer MHM. J. Phys. Chem. 1996;100:5541. [Google Scholar]
- 145.Caruso T, Carotenuto M, Vasca E, Peluso A. J. Am. Chem. Soc. 2005;127:15040. doi: 10.1021/ja055130s. [DOI] [PubMed] [Google Scholar]
- 146.Sheu C, Foote CS. J. Am. Chem. Soc. 1995;117:6439. [Google Scholar]
- 147.Thorp HH. In: Topics in Current Chemistry. Schuster GB, editor. Vol. 237. Berlin: Springer; 2004. p. 159. [Google Scholar]
- 148.Johnston DH, Glasgow KC, Thorp HH. J. Am. Chem. Soc. 1995;117:8933. [Google Scholar]
- 149.Shinde SS, Maroz A, Hay MP, Anderson RF. J. Am. Chem. Soc. 2009;131:5203. doi: 10.1021/ja8087339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sistare MF, Codden SJ, Heimlich G, Thorp HH. J. Am. Chem. Soc. 2000;122:4742. [Google Scholar]
- 151.Lewis FD, Liu X, Hayes RT, Wasielewski MR. J. Am. Chem. Soc. 2000;122:12037. [Google Scholar]
- 152.Kurnikov IV, Tong GSM, Madrid M, Beratan DN. J. Phys. Chem. B. 2002;106:7. [Google Scholar]
- 153.Saito I, Takayama M, Sugiyama H, Nakatani K. J. Am. Chem. Soc. 1995;117:6406. [Google Scholar]
- 154.Sugiyama H, Saito I. J. Am. Chem. Soc. 1996;118:7063. [Google Scholar]
- 155.Adhikary A, Khanduri D, Sevilla MD. J. Am. Chem. Soc. 2009;131:8614. doi: 10.1021/ja9014869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hall DB, Holmlin RE, Barton JK. Nature. 1996;382:731. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- 157.Saito I, Nakamura T, Nakatani K, Yoshioka Y, Yamaguchi K, Sugiyama H. J. Am. Chem. Soc. 1998;120:12686. [Google Scholar]
- 158.Voityuk AA, Jortner J, Bixon M, Rösch N. Chem. Phys. Lett. 2000;324:430. [Google Scholar]
- 159.Dandliker PJ, Nuñez ME, Barton JK. Biochemistry. 1998;37:6491. doi: 10.1021/bi980041w. [DOI] [PubMed] [Google Scholar]
- 160.Yavin E, Stemp EDA, O’Shea VL, David SS, Barton JK. Proc. Natl. Acad. Sci. USA. 2006:3610. doi: 10.1073/pnas.0600239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Treadway CR, Hill MG, Barton JK. Chem. Phys. 2002;281:409. [Google Scholar]
- 162.Anne A, Demaille C. J. Am. Chem. Soc. 2008;130:9822. doi: 10.1021/ja801074m. [DOI] [PubMed] [Google Scholar]
- 163.Wong ELS, Gooding JJ. Anal. Chem. 2006;78:2138. doi: 10.1021/ac0509096. [DOI] [PubMed] [Google Scholar]
- 164.Gorodetsky AA, Buzzeo MC, Barton JK. Bioconj. Chem. 2008;19:2285. doi: 10.1021/bc8003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ikeda R, Akaishi A, Chiba J, Inouye M. ChemBioChem. 2007;8:2219. doi: 10.1002/cbic.200700522. [DOI] [PubMed] [Google Scholar]
- 166.Hartwich G, Caruana DJ, de Lumley-Woodyear T, Wu Y, Campbell CN, Heller A. J. Am. Chem. Soc. 1999;121:10803. [Google Scholar]
- 167.Lewis FD, Kalgutkar RS, Wu Y, Liu X, Liu J, Hayes RT, Miller SE, Wasielewski MR. J. Am. Chem. Soc. 2000;122:12346. [Google Scholar]
- 168.LeBard DN, Lilichenko M, Matyushov DV, Berlin YA, Ratner MA. J. Phys. Chem. B. 2003;107:14509. [Google Scholar]
- 169.Lewis FD, Liu J, Weigel W, Rettig W, Kurnikov IV, Beratan DN. Proc. Natl. Acad. Sci. USA. 2002;99:12536. doi: 10.1073/pnas.192432899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Huynh MHV, Meyer TJ. Chem. Rev. 2007;107:5004. doi: 10.1021/cr0500030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Stubbe J, Nocera DG, Yee CS, Chang MCY. Chem. Rev. 2003;103:2167. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 172.Reece SY, Hodgkiss JM, Stubbe J, Nocera DG. Phil. Trans. R. Soc. B. 2006;361:1351. doi: 10.1098/rstb.2006.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Seyedsayamdost MR, Yee CS, Reece SY, Nocera DN, Stubbe J. J. Am. Chem. Soc. 2006;128:1562. doi: 10.1021/ja055927j. [DOI] [PubMed] [Google Scholar]
- 174.Fiebig T, Wan C, Zewail AH. ChemPhysChem. 2002;3:781. doi: 10.1002/1439-7641(20020916)3:9<781::AID-CPHC781>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 175.Gervasio FL, Boero M, Parrinello M. Angew. Chem. Int. Ed. 2006;45:5606. doi: 10.1002/anie.200602106. [DOI] [PubMed] [Google Scholar]
- 176.Anderson RF, Shinde SS, Maroz A. J. Am. Chem. Soc. 2006;128:15966. doi: 10.1021/ja0658416. [DOI] [PubMed] [Google Scholar]
- 177.Kobayashi K, Yamagami R, Tagawa S. J. Phys. Chem. B. 2008;112:10752. doi: 10.1021/jp804005t. [DOI] [PubMed] [Google Scholar]
- 178.Yamagami R, Kobayashi K, Tagawa S. J. Am. Chem. Soc. 2008;130:14772. doi: 10.1021/ja805127e. [DOI] [PubMed] [Google Scholar]
- 179.Huber R, Fiebig T, Wagenknecht H-A. Chem. Comm. 2003;15:1878. doi: 10.1039/b305732a. [DOI] [PubMed] [Google Scholar]
- 180.Stemp EDA, Arkin M, Barton JK. J. Am. Chem. Soc. 1997;119:2921. [Google Scholar]
- 181.Schiemann O, Turro NJ, Barton JK. J. Phys. Chem. B. 2000;104:7214. [Google Scholar]
- 182.Kumar A, Sevilla MD. J. Phys. Chem. B. 2009;113:11359. doi: 10.1021/jp903403d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Shafirovich V, Dourandin A, Geacintov NE. J. Phys. Chem. B. 2001;105:8431. [Google Scholar]
- 184.Takada T, Kawai K, Fujitsuka M, Majima T. Chem. Eur. J. 2005;11:3835. doi: 10.1002/chem.200500052. [DOI] [PubMed] [Google Scholar]
- 185.Sobolewski AL, Domcke W. Phys. Chem. Chem. Phys. 2004;6:2763. [Google Scholar]
- 186.Sobolewski AL, Domcke W, Hattig C. Proc. Natl. Acad. Sci. USA. 2005;102:17903. doi: 10.1073/pnas.0504087102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Abu-Riziq A, Grace L, Nir E, Kabelac M, Hobza P, de Vries MS. Proc. Natl. Acad. Sci. USA. 2005;102:20. doi: 10.1073/pnas.0408574102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Schwalb NK, Temps F. J. Am. Chem. Soc. 2007;129:9272. doi: 10.1021/ja073448+. [DOI] [PubMed] [Google Scholar]
- 189.Crespo-Hernández CE, de La Harpe K, Kohler B. J. Am. Chem. Soc. 2008;130:10844. doi: 10.1021/ja802183s. [DOI] [PubMed] [Google Scholar]
- 190.Brillouin L. In: Horizons in Biochemistry. Kasha M, Pullman B, editors. New York City: Academic Press; 1962. p. 295. 1962. [Google Scholar]
- 191.Sekiguchi I-S, Sekiguchi T. Phys. Rev. Lett. 2007;99:228102. doi: 10.1103/PhysRevLett.99.228102. [DOI] [PubMed] [Google Scholar]
- 192.Baba Y, Sekiguchi T, Shimoyama I, Hirao N, Nath KG. Phys. Rev. B. 2006;74:205433. [Google Scholar]
- 193.Zakjevskii VV, King SJ, Dolgounitcheva O, Zakrzewski VG, Ortiz JV. J. Am. Chem. Soc. 2006;128:13350. doi: 10.1021/ja064621p. [DOI] [PubMed] [Google Scholar]
- 194.Close DM, Øhman KT. J. Phys. Chem. A. 2008;112:11207. doi: 10.1021/jp805308p. [DOI] [PubMed] [Google Scholar]
- 195.Gabelica V, Rosu F, Tabarin T, Kinet C, Rodolphe A, Broyer M, De Pauw E, Dugourd P. J. Am. Chem. Soc. 2007;129:4706. doi: 10.1021/ja068440z. [DOI] [PubMed] [Google Scholar]
- 196.Hübsch A, Enders RG, Cox DL, Singh RRP. Phys. Rev. Lett. 2005;94:178102. doi: 10.1103/PhysRevLett.94.178102. [DOI] [PubMed] [Google Scholar]
- 197.Haruna K-I, Iida H, Tanabe K, Nishimoto S-I. Org. Biomolec. Chem. 2008;6:1613–1617. doi: 10.1039/b800295a. [DOI] [PubMed] [Google Scholar]
- 198.Vladimirov E, Ivanova A, Rösch N. J. Phys. Chem. B. 2009;113:4425. doi: 10.1021/jp809774q. [DOI] [PubMed] [Google Scholar]
- 199.Yonemoto EH, Saupe GB, Schmehl RH, Hubig SM, Riley RL, Iverson BL, Mallouk TE. J. Am. Chem. Soc. 1994;116:4786. [Google Scholar]
- 200.Berlin YA, Grozema FC, Siebbeles LDA, Ratner MA. J. Phys. Chem. C. 2008;112:10988. [Google Scholar]
- 201.Page CC, Moser CC, Chen X, Dutton PL. Nature. 1999;402:47. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 202.Gray HB, Winkler JR. Q. Rev. Biophys. 2003;36:341. doi: 10.1017/s0033583503003913. [DOI] [PubMed] [Google Scholar]
- 203.Balabin IA, Beratan DN, Skourtis SS. Phys. Rev. Lett. 2008;101:158102. doi: 10.1103/PhysRevLett.101.158102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Rösch N, Voityuk AAIn. In: Topics in Current Chemistry. Schuster GB, editor. Vol. 237. Berlin: Springer; 2004. p. 37. [Google Scholar]
- 205.Sponer J, Riley KE, Hobza P. Phys. Chem. Chem. Phys. 2008;10:2595. doi: 10.1039/b719370j. [DOI] [PubMed] [Google Scholar]
- 206.Wang XF, Chakraborty T. Phys. Rev. Lett. 2006;97:106602. doi: 10.1103/PhysRevLett.97.106602. [DOI] [PubMed] [Google Scholar]
- 207.Giese B, Amaudrut; Köhler A-K, Sporman M, Wessely S. Nature. 2001;412:318. doi: 10.1038/35085542. [DOI] [PubMed] [Google Scholar]
- 208.Voityuk AA. J. Chem. Phys. 2008;128:115101. doi: 10.1063/1.2841421. [DOI] [PubMed] [Google Scholar]
- 209.Migliore A, Corni S, Varsano D, Klein ML, Di Felice R. J. Phys. Chem. B. 2009 doi: 10.1021/jp904295q. ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Troisi A, Orlandi G. Chem. Phys. Lett. 2001;344:509. [Google Scholar]
- 211.Troisi A, Orlandi G. J. Phys. Chem. B. 2002;106:2093. [Google Scholar]
- 212.Mallajosyula SS, Gupta A, Pati SK. J. Phys. Chem. A. 2009;113:3955. doi: 10.1021/jp8101942. [DOI] [PubMed] [Google Scholar]
- 213.Reha D, Barford W, Harris S. Phys. Chem. Chem. Phys. 2008;10:5437. doi: 10.1039/b719619a. [DOI] [PubMed] [Google Scholar]
- 214.Kubař T, Elstner M. J. Phys. Chem. B. 2008;112:8788. doi: 10.1021/jp803661f. [DOI] [PubMed] [Google Scholar]
- 215.Bongiorno A. J. Phys. Chem. B. 2008;112:13945. doi: 10.1021/jp801872e. [DOI] [PubMed] [Google Scholar]
- 216.Kubař T, Elstner M. J. Phys. Chem. B. 2009;113:5653. doi: 10.1021/jp901888r. [DOI] [PubMed] [Google Scholar]
- 217.Renger T, Marcus RA. J. Phys. Chem. A. 2003;107:8404. [Google Scholar]
- 218.Shih C, Museth AK, Abrahamsson M, Blanco-Rodriguez AM, Di Bilio AJ, Sudhamsu J, Crane BR, Ronayne KL, Towrie M, Vicek A, Richards JH, renger, marcusWinkler JR, Gray HB. Science. 2008;320:1760. doi: 10.1126/science.1158241. [DOI] [PubMed] [Google Scholar]
- 219.Jorner J, Bixon M, Langenbacher T, Michel-Beyerle ME. Proc. Natl. Acad. Sci USA. 1998;95:12759. doi: 10.1073/pnas.95.22.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Xu B, Tao N. Science. 2003;301:1221. doi: 10.1126/science.1087481. [DOI] [PubMed] [Google Scholar]
- 221.Hihath J, Chen F, Zhang P, Tao N. J. Phys. Condens. Matter. 2007;19:215202. [Google Scholar]
- 222.Mallajosyula SS, Lin JC, Cox DL, Pati SK, Singh RRP. Phys. Rev. Lett. 2008;101:176805. doi: 10.1103/PhysRevLett.101.176805. [DOI] [PubMed] [Google Scholar]
- 223.Hihath J, Xu B, Zhang P, Tao N. Proc. Natl. Acad. Sci. USA. 2005;102:16979. doi: 10.1073/pnas.0505175102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Dulic D, Tuukkanen S, Chung C-L, Isambert A, Lavie P, Filoramo A. Nanotech. 2009;20:115502. doi: 10.1088/0957-4484/20/11/115502. [DOI] [PubMed] [Google Scholar]
- 225.Barton JK, Nuckols C. Unpublished results [Google Scholar]
- 226.Takada T, Kawai K, Cai X, Sugimoto A, Fujitsuka M, Majima T. J. Am. Chem. Soc. 2004;126:1125. doi: 10.1021/ja035730w. [DOI] [PubMed] [Google Scholar]
- 227.Takada T, Kawai K, Fujitsuka M, Majima T. J. Am. Chem. Soc. 2006;128:11012. doi: 10.1021/ja0641554. [DOI] [PubMed] [Google Scholar]
- 228.Jortner J, Bixon M, Voityuk AA, Rösch N. J. Phys. Chem. A. 2002;106:7599. [Google Scholar]
- 229.Sartor V, Boone E, Schuster GB. J. Phys. Chem. B. 2001;105:11057. [Google Scholar]
- 230.Yoo J, Delaney S, Stemp EDA, Barton JK. J. Am. Chem. Soc. 2003;125:6640. doi: 10.1021/ja034326u. [DOI] [PubMed] [Google Scholar]
- 231.Kendrick T, Giese B. Chem. Comm. 2002;18:2016. doi: 10.1039/b205928b. [DOI] [PubMed] [Google Scholar]
- 232.Giese B, Wessely S. Angew. Chem. Int. Ed. 2000;39:3490. doi: 10.1002/1521-3773(20001002)39:19<3490::aid-anie3490>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 233.Bixon M, Jortner J. J. Am. Chem. Soc. 2001;123:12556. doi: 10.1021/ja010018p. [DOI] [PubMed] [Google Scholar]
- 234.Bixon M, Jortner J. Chem. Phys. 2002;271:393. [Google Scholar]
- 235.Bixon M, Jortner J. Chem. Phys. 2006;326:252. [Google Scholar]
- 236.Liu C-S, Hernandez R, Schuster GB. J. Am. Chem. Soc. 2004;126:2877. doi: 10.1021/ja0378254. [DOI] [PubMed] [Google Scholar]
- 237.Goldsmith RH, DeLeon O, Wilson TM, Finkelstein-Shapiro D, Ratner MA, Wasielewski MR. J. Phys. Chem. A. 2008;112:4410. doi: 10.1021/jp801084v. [DOI] [PubMed] [Google Scholar]
- 238.Yu ZG, Song X. Phys. Rev. Lett. 2001;86:6018. doi: 10.1103/PhysRevLett.86.6018. [DOI] [PubMed] [Google Scholar]
- 239.Berlin YA, Burin AL, Ratner MA. J. Am. Chem. Soc. 2001;123:260. doi: 10.1021/ja001496n. [DOI] [PubMed] [Google Scholar]
- 240.Kelley SO, Jackson NM, Hill MG, Barton JK. Angew. Chem., Int. Ed. 1999;38:941. doi: 10.1002/(SICI)1521-3773(19990401)38:7<941::AID-ANIE941>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 241.Hartwich G, Caruana DJ, de Lumley-Woodyear T, Wu Y, Campbell CN, Heller A. J. Am. Chem. Soc. 1999;121:10803. [Google Scholar]
- 242.Traub MC, Brunschwig BS, Lewis NS. J. Phys. Chem. B. 2007;111:6676. doi: 10.1021/jp065520g. [DOI] [PubMed] [Google Scholar]
- 243.Adhikary A, Kumar A, Khanduri D, Sevilla MD. J. Am. Chem. Soc. 2008;130:10282. doi: 10.1021/ja802122s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Jakobsson M, Stafström S. J. Chem. Phys. 2008;129:125102. doi: 10.1063/1.2981803. [DOI] [PubMed] [Google Scholar]
- 245.Schuster GB. Acc. Chem. Res. 2000;33:253. doi: 10.1021/ar980059z. [DOI] [PubMed] [Google Scholar]
- 246.Conwell EM, Rakhmanova SV. Proc. Natl. Acad. Sci. USA. 2000;97:4556. doi: 10.1073/pnas.050074497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kuznetsov AM. Charge Transfer in Physics, Chemistry and Biology: Physical Mechanisms of Elementary Processes and an Introduction to the Theory. Amsterdam: Gordon and Breach Science Publishers; 1995. p. 2. [Google Scholar]
- 248.Maniadis P, Kalosakas G, Rasmussen KØ, Bishop AR. Phys. Rev. B. 2003;68:174304. doi: 10.1103/PhysRevE.72.021912. [DOI] [PubMed] [Google Scholar]
- 249.Komineas S, Kalosakas G, Bishop AR. Phys. Rev. E. 2002;65:061905. doi: 10.1103/PhysRevE.65.061905. [DOI] [PubMed] [Google Scholar]
- 250.Chang C-M, Castro Neto AH, Bishop AR. Chem. Phys. 2004;303:189. [Google Scholar]
- 251.Conwell EM. Proc. Natl. Acad. Sci. USA. 2005;102:8795. doi: 10.1073/pnas.0501406102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Conwell EM, Bloch SM. J. Phys Chem. B. 2006;110:5801. doi: 10.1021/jp0553986. [DOI] [PubMed] [Google Scholar]
- 253.Conwell EM, Park J-H, Choi H-Y. J. Phys. Chem. B. 2005;109:9760. doi: 10.1021/jp044485f. [DOI] [PubMed] [Google Scholar]
- 254.Conwell EM, Bloch SM, McLaughlin PM, Basko DM. J. Am. Chem. Soc. 2007;129:9175. doi: 10.1021/ja0691472. [DOI] [PubMed] [Google Scholar]
- 255.Conwell EM, Basko DM. J. Phys. Chem. B. 2006;110:23603. doi: 10.1021/jp064373j. [DOI] [PubMed] [Google Scholar]
- 256.Middleton CT, de La Harpe K, Su C, Law YK, Crespo-Hernández CE, Kohler B. Ann. Rev. Phys. Chem. 2009;60:217. doi: 10.1146/annurev.physchem.59.032607.093719. [DOI] [PubMed] [Google Scholar]
- 257.Schwalb NK, Temps F. Science. 2008;322:243. doi: 10.1126/science.1161651. [DOI] [PubMed] [Google Scholar]
- 258.Crespo-Hernández CE, Cohen B, Kohler B. Nature. 2005;436:1141. doi: 10.1038/nature03933. [DOI] [PubMed] [Google Scholar]
- 259.Buchvarov I, Wang Q, Raytchev M, Trifonov I, Fiebig T. Proc. Natl. Acad. Sci. USA. 2007;104:4794. doi: 10.1073/pnas.0606757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kadhane U, Holm AIS, Hoffmann SV, Nielsen S. Phys. Rev. E. 2008;77:021901. doi: 10.1103/PhysRevE.77.021901. [DOI] [PubMed] [Google Scholar]
- 261.Takaya T, ; Su C, de La Harpe K, Crespo-Hernández CE, Kohler B. Proc. Natl. Acad. Sci. USA. 2008;105:10285. doi: 10.1073/pnas.0802079105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lange AW, Herbert JM. J. Am. Chem. Soc. 2009;131:3913. doi: 10.1021/ja808998q. [DOI] [PubMed] [Google Scholar]
- 263.Hatcher E, Balaeff A, Keinan S, Venkatramani R, Beratan DN. J. Am. Chem. Soc. 2008;130:11752. doi: 10.1021/ja802541e. [DOI] [PubMed] [Google Scholar]
- 264.Uskov DB, Burin AL. Phys. Rev. B. 2008;78:073106. [Google Scholar]
- 265.Voityuk AA. J. Chem. Phys. 2005;122:204904. doi: 10.1063/1.1924551. [DOI] [PubMed] [Google Scholar]
- 266.Sen S, Andreatta D, Ponomarev SY, Beveridge DL, Berg MA. J. Am. Chem. Soc. 2009;131:1724. doi: 10.1021/ja805405a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Voityuk AA. J. Chem. Phys. 2008;128:045104. doi: 10.1063/1.2823015. [DOI] [PubMed] [Google Scholar]
- 268.Friedman KA, Heller A. J. Phys. Chem. B. 2001;105:11859. [Google Scholar]
- 269.Heller A. Farad. Disc. 2000;116:1. [Google Scholar]
- 270.Merino EJ, Barton JK. Biochemistry. 2007;46:2805. doi: 10.1021/bi062024+. [DOI] [PubMed] [Google Scholar]
- 271.Merino EJ, Barton JK. Biochemistry. 2009;48:660. doi: 10.1021/bi801570j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Steenken S, Jovanovic SV. J. Am. Chem. Soc. 1997;119:617. doi: 10.1021/ja003823x. [DOI] [PubMed] [Google Scholar]
- 273.Crespo-Hernández CE, Close DM, Gorb L, Leszczynski J. J. Phys. Chem. B. 2007;111:5386. doi: 10.1021/jp0684224. [DOI] [PubMed] [Google Scholar]