Abstract
Myxococcus xanthus, a Gram-negative soil bacterium, undergoes multicellular development when nutrients become limiting. Aggregation, which is part of the developmental process, requires the surface motility of this organism. One component of M. xanthus motility, the social (S) gliding motility, enables the movement of cells in close physical proximity. Previous studies demonstrated that the cell surface-associated exopolysaccharide (EPS) is essential for S motility and that the Dif proteins form a chemotaxis-like pathway that regulates EPS production in M. xanthus. DifA, a homologue of methyl-accepting chemotaxis proteins (MCPs) in the Dif system, is required for EPS production, S motility and development. In this study, a spontaneous extragenic suppressor of a difA deletion was isolated in order to identify additional regulators of EPS production. The suppressor mutation was found to be a single base pair insertion in cheW7 at the che7 chemotaxis gene cluster. Further examination indicated that mutations in cheW7 may lead to the interaction of Mcp7 with DifC (CheW-like) and DifE (CheA-like) to reconstruct a functional pathway to regulate EPS production in the absence of DifA. In addition, the cheW7 mutation was found to partially suppress a pilA mutation in EPS production in a difA+ background. Further deletion of difA from the pilA cheW7 double mutant resulted in a triple mutant that produced wild-type levels of EPS, implying that DifA (MCP-like) and Mcp7 compete for interactions with DifC and DifE in the modulation of EPS production.
INTRODUCTION
Myxococcus xanthus is a rod-shaped Gram-negative bacterium with a developmental cycle and surface motility known as gliding (Kaiser, 2003; Shimkets, 1999; Whitworth, 2008; Zusman et al., 2007). When nutrients are abundant, M. xanthus cells grow and divide as vegetative cells. Gliding enables cells to move and expand to new territories during the vegetative cell cycle. When nutrients become limiting, hundreds of thousands of cells can aggregate and organize into a multicellular structure known as a fruiting body. Within fruiting bodies, cells undergo differentiation to form dormant spherical myxospores that can survive harsh conditions including desiccation and heat. Gliding motility is essential for the aggregation and organization of M. xanthus fruiting bodies during the developmental process (Kaiser, 2003).
M. xanthus possesses two genetically distinct forms of gliding motility: adventurous (A) and social (S) (Hodgkin & Kaiser, 1979a, b). A motility enables the movement of well-isolated cells, whereas S motility is functional in cells in close proximity or in cell groups. The mechanism of A motility remains unresolved (Mignot, 2007; Mignot et al., 2007; Wolgemuth et al., 2002; Yu & Kaiser, 2007). S motility, analogous to bacterial twitching, requires the extension and retraction of type IV pili (Tfp) (Merz et al., 2000; Semmler et al., 1999; Skerker & Berg, 2001; Sun et al., 2000; Wu & Kaiser, 1995). In addition, the cell surface component known as exopolysaccharide (EPS) is required for M. xanthus S motility (Shimkets, 1986b, 1989; Yang et al., 2000). It has been proposed that EPS functions as anchor and trigger for Tfp retraction in M. xanthus (Li et al., 2003), and the attachment of an extended Tfp to EPS on a neighbouring cell triggers retraction to move one cell relative to another.
Bacterial motility is mostly directed such that cells may move toward favourable environments and away from unfavourable ones. This is achieved at the molecular level by the function of the chemotaxis regulatory pathway (Armitage et al., 2005; Bourret & Stock, 2002; Bren & Eisenbach, 2000). Signals detected by bacterial chemotaxis systems are typically soluble chemicals that interact with transmembrane chemoreceptors known as methyl-accepting chemotaxis proteins (MCPs). Conformational changes in MCPs upon ligand recognition are communicated to the histidine protein kinase CheA via the coupling protein CheW. In classical bacterial chemotaxis, MCP, CheW and CheA form a transmembrane ternary signalling complex such that environmental signals can modulate the kinase activity of the cytoplasmic CheA. CheA autophosphorylates on a conserved histidine and the phosphate can be subsequently transferred to a conserved aspartate on the CheY response regulator. The phosphorylated form of CheY (CheY-P) induces changes in bacterial motility behaviour to achieve chemotaxis or biased cell movement.
There are eight chemosensory pathways encoded by the M. xanthus genome (Kirby et al., 2008; Zusman et al., 2007). While the Frz chemosensory pathway clearly plays a central role in chemotaxis regulation, the functions of the remaining pathways are not as well understood and some may function to regulate processes other than chemotaxis (Kirby et al., 2008; Zusman et al., 2007). The work reported here was initiated with the Dif proteins, which form a chemotaxis-like pathway that regulates the production of EPS (Bellenger et al., 2002; Black & Yang, 2004; Yang et al., 1998, 2000). DifA is homologous to MCPs, DifC to CheW and DifE to CheA (Bellenger et al., 2002; Yang et al., 1998). DifA, DifC and DifE are positive regulators of EPS production that likely form a ternary signalling complex required for EPS production (Bellenger et al., 2002; Black & Yang, 2004; Black et al., 2006; Yang et al., 1998, 2000; Yang & Li, 2005). DifD and DifG, homologues of CheY and CheC, respectively, function as negative regulators of EPS production, possibly by modulation of signal flow through the core Dif pathway (Black & Yang, 2004; Black et al., 2006). It has been demonstrated recently that the input signals of the Dif chemotaxis pathway are likely mediated by Tfp, which function upstream to positively regulate EPS production (Black et al., 2006). Many questions remain regarding EPS production and regulation, and the mechanism by which the Dif proteins regulate EPS production in M. xanthus is unclear.
To identify additional EPS regulators, a difA suppressor mutant was isolated and investigated in this study. The suppressor mutation restored EPS production, as well as agglutination, S motility and development, to a difA mutant. Genetic analysis suggested that the suppressor mutation was located at the M. xanthus che7 locus. The suppressor was further identified as a single base pair insertion about two-thirds into the cheW7 ORF. This mutation, designated cheW7-1, shifts the reading frame of the mRNA transcript and generates an extended CheW7 protein. Our results demonstrated that the suppression of difA by cheW7 requires both the inactivation of cheW7 and the concurrent expression of mcp7. Since this suppression of difA by cheW7-1 requires Mcp7 and both DifC (CheW-like) and DifE (CheA-like), it is likely that the elimination of CheW7 and DifA led to an interaction of Mcp7 with DifC and DifE to recreate a functional pathway to restore EPS production. We propose a model in which the Mcp from the Che7 pathway substitutes for its homologue DifA in the Dif pathway.
METHODS
Bacterial strains and growth conditions.
M. xanthus strains and plasmids used in this study are listed in Table 1 and Supplementary Table S1. M. xanthus was grown and maintained at 32 °C on Casitone–yeast extract (CYE) agar plates or in CYE liquid medium (Campos & Zusman, 1975). Clone-fruiting (CF) agar plates were used to examine M. xanthus development (Hagen et al., 1978). The Escherichia coli strains used in this study, XL1-Blue (Stratagene) and DH5α λpir (Rubin et al., 1999), were grown and maintained at 37 °C on Luria–Bertani (LB) agar plates or in LB liquid medium (Miller, 1972). Unless otherwise noted, standard or regular agar plates contained 1.5 % agar. When necessary, kanamycin (100 μg ml−1) and oxytetracycline (15 μg ml−1) were used for selection purposes.
Table 1.
M. xanthus strains and plasmids used in this study
| Strain or plasmid | Genotype/description | Reference or source |
|---|---|---|
| Strains | ||
| DK1622 | Wild-type | Kaiser (1979) |
| DK10407 | ΔpilA : : TetRdifA+ | Wall et al. (1998) |
| LS308 | Tn5Ω1407 difA+ | Shimkets (1986a) |
| SW504 | ΔdifA-1 | Yang et al. (1998) |
| YZ101 | ΔdifA-1 cheW7-1 | This study |
| YZ601 | ΔdifA-2 | Xu et al. (2005) |
| YZ615 | ΔdifA-1 ΔdifE cheW7-1 | This study |
| YZ617 | ΔdifA-1 ΔdifC cheW7-1 | This study |
| YZ647 | ΔdifA-1cheW7-1 ΔpilA : : TetR | This study |
| YZ648 | ΔdifA-1 ΔpilA : : TetR | This study |
| YZ651 | ΔdifA-1 cheW7-1 MXAN4984 : : pWB513 | This study |
| YZ666 | ΔdifA-2 cheW7-1 | This study |
| YZ684 | ΔdifA-1 cheW7-1 che7 : : pWB521 | This study |
| YZ687 | ΔdifA-2 che7 : : pWB521 | This study |
| YZ688 | ΔdifA-2 che7 : : pWB515 | This study |
| YZ697 | ΔdifA-2 che7 : : pWB530 | This study |
| YZ1600 | ΔdifA-2 che7 : : pWB531 | This study |
| YZ1604 | ΔdifA-2 ΔcheW7 Δmcp7 | This study |
| YZ1605 | ΔdifA-2 ΔcheW7 Δmcp7 attB : : pWB426 | This study |
| YZ1612 | ΔdifA-2 ΔcheW7 Δmcp7 attB : : pWB433 | This study |
| YZ1613 | ΔdifA-1 cheW7-1 Δmcp7 | This study |
| YZ1614 | ΔdifA-2 attB : : pWB433 | This study |
| YZ1615 | ΔdifA-2 ΔcheW7 attB : : pWB435 | This study |
| YZ1617 | ΔdifA-1 cheW7-1 Δmcp7 attB : : pWB433 | This study |
| YZ1618 | difA+cheW7-1 ΔpilA : : TetR | This study |
| YZ1619 | ΔdifA-2 ΔcheW7 | This study |
| YZ1625 | ΔdifA-2 ΔcheW7 attB : : pWB433 | This study |
| BY132 | ΔdifA-1 cheW7-1 cpc7 : : magellan4 | This study |
| Plasmids | ||
| pZErO-2 | Cloning vector, KanR | Invitrogen |
| pMycoMar | magellan4 mutagenesis vector | Rubin et al. (1999) |
| pBJ113 | Cloning vector, KanR, galK | Julien et al. (2000) |
| pBY132 | magellan4 isolated from BY132, KanR | This study |
| pWB116 | ΔdifA-2 in-frame deletion, pBJ113 | Xu et al. (2005) |
| pWB425 | Expression vector, KanR, intP (Mx8 attP) | Unpublished |
| pWB426 | SacI–NotI che7 fragment in pWB425 | This study |
| pWB433 | mcp7 PCR fragment in pWB425 | This study |
| pWB435 | cheW7-1 PCR fragment in pWB425 | This study |
| pWB513 | Fragment ∼2.3 Mb from che7 in pZErO-2 | This study |
| pWB515 | SacI–NotI che7 fragment in pZErO-2 | This study |
| pWB521 | Fragment downstream of che7 in pZErO-2 | This study |
| pWB530 | SacI–PstI che7 fragment in pZErO-2 | This study |
| pWB531 | ClaI–NotI che7 fragment in pZErO-2 | This study |
| pWB534 | ΔcheW7 mcp7 in-frame deletion in pBJ113 | This study |
| pWB552 | ΔcheW7 in-frame deletion in pBJ113 | This study |
| pWB553 | Δmcp7 in-frame deletion in pBJ113 | This study |
Transposon mutagenesis.
M. xanthus cells were mutagenized as previously described (Youderian et al., 2003) by the mariner-based transposon magellan4 (Rubin et al., 1999), which contains the R6Kγ replication origin for E. coli and a gene conferring kanamycin resistance (KanR). Approximately 500 ng pMycoMar, a plasmid harbouring magellan4 (Rubin et al., 1999), was electroporated into M. xanthus (Kashefi & Hartzell, 1995). Mutagenized cells were allowed to recover at 32 °C and subsequently plated on CYE agar with kanamycin and Congo red (30 μg ml−1) to identify transposon insertion mutants and score transformants for EPS production.
The site of a transposon insertion in a mutant was determined as follows. Genomic DNA (1 μg) was digested with SacII, an enzyme that does not cut within magellan4. The resulting DNA was used for self-ligation at a final concentration of 20 ng μl−1. The ligation mix was used to transform DH5α λpir, an E. coli strain capable of propagating oriR6Kγ-based plasmids. Recovered plasmids from the transformants were sequenced using primers MarR1 and/or MarL1 (Youderian et al., 2003), and the transposon insertion sites were identified by comparison with the M. xanthus genome sequence (Goldman et al., 2006).
Plasmid construction.
To construct the plasmid for kanamycin resistance (KanR) linked to the che7 locus, a PCR fragment (chromosomal coordinates 8 512 566–8 513 278) (Arshinoff et al., 2007; Goldman et al., 2006) ∼2 kb downstream of the che7 locus was amplified and cloned into the EcoRV site of pZErO-2 (Invitrogen) to generate pWB521. To construct the plasmid for a KanR marker away from the che7 locus, another PCR fragment (chromosomal coordinates 6 232 354–6 233 024) (Arshinoff et al., 2007; Goldman et al., 2006) ∼2.3 Mb away from che7 was amplified and cloned into the EcoRV site of pZErO-2 to generate pWB513. The fragment in pWB513 included the 3′ end of MXAN4984 and downstream sequence.
To construct plasmids for in-frame deletions, fragments containing both upstream and downstream sequences of cheW7 and/or mcp7 were obtained by PCR. The relevant fragments were joined by a two-step overlap PCR and cloned into pBJ113 (Julien et al., 2000). The resulting plasmids pWB552 (ΔcheW7), pWB553 (Δmcp7) and pWB534 (ΔcheW7 Δmcp7) deleted coding sequences for amino acids 5–135 of CheW7, 96–845 of Mcp7, and amino acids 5 of CheW7 to 845 of Mcp7, respectively.
Plasmids containing different portions of cheW7 and/or mcp7 from the suppressor were constructed as follows. The strain BY132 (Table 1), which harbours an insertion of magellan4 in cpc7 (MXAN6961) at the che7 locus (Fig. 1), was derived from the suppressor strain YZ101 (ΔdifA cheW7-1) by transposon mutagenesis. The plasmid pBY132, which contains the magellan4 transposon and the full-length cheW7 and mcp7, was constructed by digestion of genomic DNA from BY132 with SphI and self-ligation. The 3.4 kb SacI–NotI fragment from pBY132 was cloned into the same sites in pZErO-2 to construct pWB515. The 2 kb SacI–PstI fragment from pWB515 was cloned into the same sites in pZErO-2 to generate pWB530. pWB531 was created by digesting pWB515 with ClaI and HindIII (site on the vector), treating with T4 DNA polymerase and religating. These three pZErO-based plasmids, pWB515, pWB530 and pWB531, can integrate into the M. xanthus chromosome by recombination with their homologous sequences at the che7 locus.
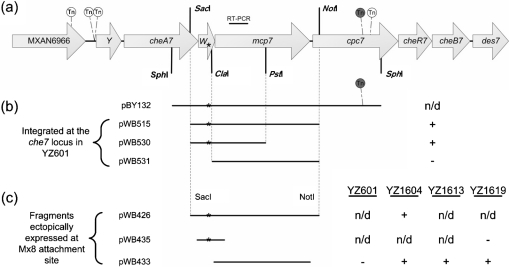
Fig. 1.
(a) Schematic of the M. xanthus che7 locus and transposon (Tn) insertions. The che7 locus shown here is ∼13 kb long. Gene lengths, Tn insertion sites and restriction sites are all drawn to scale. Shown are five independent Tn insertions that eliminated EPS production in the suppressor strain YZ101. These insertions starting from the left to right are designated MXAN6966 : : magellan4, cheY7-1 : : magellan4, cheY7-2 : : magellan4, cpc7-1 : : magellan4 and cpc7-2 : : magellan4 (Table 1 and Supplementary Table S1). Genomic DNA from strain BY132 (Table 1), which contains cpc7-1 : : magellan4 (shaded grey), was used to construct pBY132. The target for RT-PCR shown in Fig. 4 is shown above mcp7. (b) Indicated plasmids were transformed into YZ601 (ΔdifA) and integration occurred by homologous recombination at the che7 locus. ‘+’ and ‘−’ indicate that 5–10 % of the transformants were EPS+ or that 100 % of the transformants were EPS−, respectively, as determined by Congo red binding; ‘n/d’, not determined. (c) Listed expression plasmids were integrated at the Mx8 attachment site into the strains listed on the right: YZ601 (ΔdifA), YZ1604 (ΔdifA ΔcheW7 Δmcp7), YZ1613 (ΔdifA cheW7-1 Δmcp7) and YZ1619 (ΔdifA ΔcheW7). ‘+’ and ‘−’ indicate EPS+ or EPS−, respectively, as determined by Congo red binding. The asterisks mark the approximate location of the cheW7-1 mutation in all panels. cpc7 is predicted to encode a phycobilisome (PBS) lyase HEAT-like repeat protein (Arshinoff et al., 2007; Goldman et al., 2006). It is likely that there are promoters in the che7 locus, one upstream of MXAN6966 or cheY7 and another within cheW7 (see text for details).
The expression vector pWB425 is able to drive heterologous gene expression in M. xanthus from the nptII (kanamycin phosphotransferase) promoter (Beck et al., 1982). The construction and validation of pWB425 will be described elsewhere (W. P. Black and Z. Yang, unpublished results). pWB426 was created by inserting the same 3.4 kb SacI–NotI fragment from pWB515 into the same sites of pWB425. pWB433 and pWB435 were created by cloning PCR fragments containing either mcp7 or the mutant version of cheW7 (cheW7-1) into the EcoRV site of pWB425. Since pWB425 contains the Mx8 phage elements for chromosomal integration (Magrini et al., 1999; W. P. Black and Z. Yang, unpublished results), the three pWB425-based plasmids, pWB426, pWB433 and pWB435, may recombine with the Mx8 attachment site on the M. xanthus chromosome by site-specific recombination (Magrini et al., 1999). All plasmids constructed in this study were confirmed by restriction digestion and/or DNA sequencing.
M. xanthus strain construction.
pWB552 (ΔcheW7), pWB553 (Δmcp7), pWB534 (ΔcheW7 Δmcp7) and pWB116 (ΔdifA-2) were used to make the respective in-frame deletions by a procedure previously described (Black & Yang, 2004; Julien et al., 2000; Ueki et al., 1996). YZ101 (cheW7-1 ΔdifA-1) was the parental strain for YZ666 (cheW7-1 ΔdifA-2) and YZ1613 (cheW7-1 ΔdifA-1 Δmcp7). YZ601 (ΔdifA-2) was the parent of YZ1619 (ΔdifA-2 ΔcheW7) and YZ1604 (ΔdifA-2 ΔcheW7 Δmcp7). PCR analysis of genomic DNA was used to identify and verify all mutants. In brief, a PCR was performed using a three-primer system. Two of the primers flanked the deleted region and the third was internal to the gene of interest. The genomic DNAs of the wild-type and the intermediate (prior to the negative selection) were used as controls in these PCRs.
Markers linked to the che7 locus were established by electroporation of pWB521 into YZ101 and YZ601 to create strains YZ684 and YZ687. YZ651 was constructed by the insertion of a KanR marker ∼2.3 Mb away from che7 by electroporating pWB513 into YZ101. The transfer of an antibiotic marker or linkage from one strain to another was performed using genomic DNA transformation as described by Vlamakis et al. (2004). Basically, a recipient strain was transformed using genomic DNA from a donor by electroporation. The pilA cheW7-1 double mutant YZ1618 was constructed by transformation of DK10407 with genomic DNA from YZ684 and selection for kanamycin resistance. The pilA ΔdifA cheW7-1 triple mutant YZ647 was constructed by transformation of the suppressor strain YZ101 with genomic DNA from DK10407 and selection for oxytetracycline resistance.
Strains with various che7 fragments integrated at the che7 locus were constructed by electroporation of pWB515, pWB530 and pWB531 into YZ601 (ΔdifA-2) to create YZ688, YZ697 and YZ1600, respectively (Table 1, Fig. 1). To express various che7 fragments ectopically, pWB426, pWB433 and pWB435 were integrated at the Mx8 attachment site (attB) in various strains. YZ1604 transformed with pWB426 gave rise to YZ1605. pWB433 transformed into YZ1604, YZ601, YZ1613 and YZ1619 produced YZ1612, YZ1614, YZ1617 and YZ1625, respectively. The integration of pWB435 into YZ1619 produced YZ1615.
Analysis of mcp7 expression by RT-PCR.
Total RNA was isolated from cells in exponential growth using the RNeasy mini kit (Qiagen) and treated with RQ1 DNase (Promega) according to the manufacturer's instructions. Total RNA (200 ng) from each strain was subjected to RT-PCR using the Qiagen OneStep RT-PCR kit following the manufacturer's protocol plus Q-solution. A total of 15 % of each RT-PCR was examined by agarose gel electrophoresis.
Assessment of S motility and development.
Strains to be tested were grown in CYE liquid medium and harvested during the exponential growth phase. A 5 μl volume of cell suspension at 5×109 cells ml−1 was spotted onto CYE plates containing 0.4 % agar for analysis of S motility or onto standard CF plates for development. Results were documented after 5 days incubation at 32 °C.
Analysis of EPS production.
Plates containing Congo red or Calcofluor white were used to examine EPS production qualitatively (Dana & Shimkets, 1993; Ramaswamy et al., 1997). Briefly, 5 μl of a cell suspension at 5×109 cells ml−1 was spotted onto CYE plates supplemented with either Congo red (30 μg ml−1) or Calcofluor white (50 μg ml−1). These plates were incubated at 32 °C for 5 days before documentation. EPS+ strains produce reddish-orange colonies, whereas EPS− ones form yellow colonies on Congo red plates. Calcofluor white plates were examined under long-wave UV (365 nm) for fluorescence, which is indicative of dye binding and EPS production.
The dye Trypan blue and spectrometry were used to measure EPS production quantitatively as previously described (Black & Yang, 2004; Black et al., 2006). Strains to be tested were harvested at similar culture densities (∼3.5×108 cells ml−1). Cells were washed once in MOPS buffer [10 mM MOPS (pH 7.6), 2 mM MgSO4] containing 1 mM CaCl2 and resuspended at 2.5×108 cells ml−1 in the same buffer containing Trypan blue (5 μg ml−1). After incubation at 25 °C on a rotary shaker at 300 r.p.m. for 30 min, the cell and dye suspension was centrifuged and the absorbance of the cell-free supernatant was measured at 585 nm, the maximum wavelength for Trypan blue absorbance. EPS production was calculated from the amount of Trypan blue that was bound to the cells. All strains were compared with the wild-type, the EPS level of which was arbitrarily set as 1.
Agglutination, a very sensitive method to assess EPS production (Xu et al., 2005), was performed as follows. Strains to be tested were inoculated into CYE liquid medium in 24-well polystyrene tissue culture plates. After incubation at 32 °C on a rotary shaker at 300 r.p.m. for ∼18 h, the plate was removed and incubated overnight at room temperature in the dark without agitation. Agglutination was scored as positive if cells formed a clump that adhered to the bottom or the walls of the well.
RESULTS
Isolation of difA suppressors by restoration of S motility
The difA deletion mutant SW504 (Yang et al., 1998) was used as the parental strain for the isolation of difA suppressors. difA mutants, which are defective in S motility and EPS production, do not agglutinate or clump in liquid medium (Yang et al., 1998, 2000). To enrich for spontaneous difA suppressors, SW504 cells were cultured in test tubes in liquid medium and incubated at 32 °C for 5 days without agitation. Aliquots (5 μl) from the bottom of the test tubes were spotted on CYE plates containing 0.4 % agar and incubated at 32 °C. difA mutants form compact colonies on 0.4 % agar plates due to the lack of S motility (Shi & Zusman, 1993; Yang et al., 1998). Only cells that had regained S motility were expected to move away from the origin of inoculation. Cells from motile flares were purified as potential difA suppressors for further study.
A difA suppressor strain displays S motility, development and EPS production
A potential difA suppressor strain from a motile flare, designated YZ101, was examined for the suppression of the defects displayed by difA mutants. Colonies of the wild-type are rough and dry, whereas those of difA mutants are smooth and glossy on 1.5 % agar plates. Colonies of YZ101 on 1.5 % agar appeared more similar to those of difA mutants initially, but they acquired a more wild-type appearance after 5–7 days of incubation. Soft agar plates (0.4 % agar) as shown in Fig. 2 were used to examine S motility more specifically. The nearly identical colony sizes of YZ101 and the wild-type indicated that S motility was restored in the suppressor strain. Furthermore, YZ101 produced distinct fruiting bodies as did the wild-type (Fig. 2). This is in contrast to the difA mutant, which was unable to form fruiting bodies (Fig. 2) (Yang et al., 1998). EPS production was examined more directly by the binding of the fluorescent dye Calcofluor white (CW) on plates (Dana & Shimkets, 1993; Ramaswamy et al., 1997). YZ101 fluoresced on the CW plate like the wild-type (Fig. 2), indicating EPS production. In addition, the suppressor strain agglutinated similarly to the wild-type (Supplementary Table S2), providing further evidence for EPS production. Collectively, these results demonstrate that S motility and EPS production have been restored in YZ101 by a difA suppressor.
Fig. 2.
Proficiency of the difA suppressor strain (YZ101) in S motility, development and EPS production. Cell suspensions were spotted onto CYE plates with 0.4 % agar (upper panels), standard CF plates (middle panels) and standard CYE plates with Calcofluor white (50 μg ml−1) (lower panels) to examine S motility, development and EPS, respectively. Photographs were taken after incubation at 32 °C for 5 days. Strains: DK1622 (wild-type); YZ601 (ΔdifA); YZ101 (ΔdifA cheW7-1).
The difA suppressor is not linked to the dif locus
Since the suppressor strain (YZ101) was isolated without mutagenesis and its parental strain (SW504) harbours a difA in-frame deletion (Yang et al., 1998), the above results suggest that a difA bypass suppressor arose as a result of a spontaneous mutation. A bypass suppressor of difA could have resulted from a mutation at the dif locus in either difC or difE, both of which encode proteins that function downstream of DifA (Black et al., 2006; Yang et al., 1998). Genetic experiments were conducted to determine whether the suppressor resides at the dif locus. LS308, containing the Tn5 insertion Ω1407 linked to the wild-type dif locus (Lancero et al., 2002; Shimkets, 1986a; Yang et al., 2000), was used as the donor in Mx4-mediated transduction of the suppressor strain (YZ101). All resulting transductants, including those identified as difA− by immunoblotting with α-DifA antibodies (Xu et al., 2005), displayed S motility and development (Supplementary Table S2). These results suggest that the suppressor is not linked to the dif locus and must reside elsewhere on the chromosome. Since the parental strain contains a difA deletion, this must be a second-site or extragenic bypass suppressor of difA.
The suppression requires difC and difE
difC and difE were deleted from YZ101 to determine whether they were required for the suppression. The resulting mutants, YZ615 and YZ617, were found to lack development, S motility and EPS production (Supplementary Table S2). These results indicate that difC and difE are required for the suppression of difA in YZ101, and the suppressor likely restored functions upstream of DifC and DifE (Black & Yang, 2004; Black et al., 2006). One concern about the suppressor mutant was that the ΔdifA allele in the parental strain SW504 contained a deletion of the C terminus of DifA only (Yang et al., 1998) (Table 1). As a result, the predicted N-terminal transmembrane region is intact and could be involved in the suppression. The more complete difA deletion allele in YZ601 (Xu et al., 2005) (Table 1) was introduced into the suppressor strain (YZ101) to delete the entire difA coding region (see Methods). The resulting strain YZ666 was indistinguishable from YZ101 with regard to all three critical characteristics: S motility, development and EPS production (Supplementary Table S2). YZ601 (Xu et al., 2005) and YZ666, with the more complete difA deletion allele, were used in later experiments whenever possible once they were constructed. These findings indicate that YZ101 harbours a true difA bypass suppressor that requires DifC and DifE for the suppression. In other words, the difA suppressor appears to restore EPS production by signalling through the downstream part of the Dif pathway.
The difA suppressor mutation is linked to the M. xanthus che7 locus
After attempts with other approaches proved unsuccessful, YZ101 was mutagenized with the transposon magellan4 (Rubin et al., 1999; Youderian et al., 2003; Youderian & Hartzell, 2006) and screened for an EPS− phenotype to identify the suppressor mutation. Specifically, mutagenized cells were selected for kanamycin resistance (KanR) and screened for EPS− phenotype on plates with the dye Congo red (Black et al., 2006; Dana & Shimkets, 1993). Of the approximately 20 000 KanR colonies examined, about 70 were identified as EPS− due to the lack of or reduced Congo red binding. The transposon insertion sites in these EPS− mutants were identified by cloning and sequencing as described elsewhere (Youderian et al., 2003; Youderian & Hartzell, 2006). Seven of these insertions occurred in either difC or difE, confirming the earlier conclusion that difC and difE are required for the suppression. In addition, 21 of the insertions were found to reside at the M. xanthus eps locus (Lu et al., 2005). More importantly, five insertions occurred at the M. xanthus che7 locus (Fig. 1a), suggesting that the suppressor mutation in YZ101 may reside or require the genes at the che7 locus. These five che7 transposon insertions were introduced into both the wild-type strain DK1622 and the difA suppressor strain YZ101 by genomic DNA transformation (Vlamakis et al., 2004). All five insertions led to defects in Congo red binding in the YZ101 (suppressor) background, but not in DK1622 (wild-type) (Supplementary Tables S1 and S2). These findings indicate that the suppressor mutation in YZ101 is functionally and genetically associated with the che7 locus.
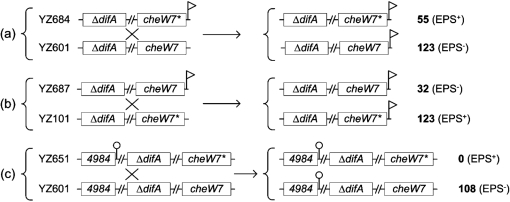
Genetic mapping was used to determine whether the suppressor mutation was located at the che7 locus. Using pWB521, a KanR marker (Fig. 1a) was inserted ∼1040 bp downstream of des7 in the intergenic region of two convergent ORFs by homologous recombination. This insertion in the suppressor (YZ101) and a ΔdifA mutant (YZ601) generated YZ684 and YZ687, respectively (Table 1; see Methods for details). As was expected, YZ684 was EPS+ and YZ687 was EPS− (Supplementary Table S2). Genetic crosses indicated in Fig. 3 were performed by genomic transformation, which has been developed recently (Vlamakis et al., 2004). When YZ684 (ΔdifA cheW7-1 che7 : : pWB521) was crossed with YZ601 (ΔdifA) (Fig. 3a), 55 of the 178 transformants became EPS+ as determined by Congo red binding. These results indicated that the suppressor mutation is linked to the che7 locus in the suppressor strain. When YZ687 (ΔdifA che7 : : pWB521) was crossed with the suppressor strain (YZ101), 32 of the 155 transformants lost the ability to produce EPS (Fig. 3b), indicating that the wild-type allele of the suppressor gene is linked to che7. As a control, a KanR marker was inserted ∼2.3 Mb away from the che7 locus in YZ101 to generate YZ651 (ΔdifA cheW7-1 MXAN4984 : : pWB513) (see Methods). When YZ651 was crossed with YZ601 (ΔdifA) by genomic transformation, all 108 transformants were EPS− (Fig. 3c), demonstrating that genomic DNA transformation is a valid method to establish genetic linkages in M. xanthus. These results confirmed that the suppressor mutation(s) was located within or near the che7 gene cluster and that the suppression was not caused by mutations at multiple loci.
Fig. 3.
Genetic crosses by transformation of a recipient strain using genomic DNA from a donor strain (Vlamakis et al., 2004). A genetic cross between two strains is indicated by a cross (×). For each cross (a, b and c), indicated on the left are the strains and the genotypes for both the donor (upper) and the recipient (lower). cheW7 represents the wild-type allele and cheW7* represents the suppressor allele (cheW7-1). 4984 represents MXAN4984. The flag denotes the che7-linked KanR marker used for selection in the cross. The lollipop denotes the MXAN4984-linked KanR marker used for selection in the cross. A break (//) between two genes indicates a chromosomal distance of ∼275 kb between difA and cheW7 or ∼2 Mb between MXAN4984 and difA. The two possible genotypes from each cross are shown on the right. The number of transformants with each genotype as determined by the phenotypes (EPS+ or EPS−) is shown on the far right (see Methods).
The suppressor allele is located in cheW7
Different DNA fragments from the suppressor strain were cloned and examined for suppressor activity by transformation into YZ601 (ΔdifA) (see Methods). These plasmids can integrate to create merodiploid strains by homologous recombination at the che7 locus. Suppression was scored as positive if a fraction of the transformants was able to bind Congo red on plates. It was observed that about 5 and 10 % of the transformants from pWB515 and pWB530, respectively (Fig. 1b), were able to bind Congo red. In addition, pWB426, which contains both cheW7 and mcp7 (Fig. 1b), conferred EPS production upon all transformants of YZ1604 (ΔdifA ΔcheW7 Δmcp7), confirming the presence of the suppressor mutation on the fragment in pWB426 (Fig. 1c). The SacI–PstI fragment with suppressor activity in pWB530 was sequenced, and one single cytosine–guanine base pair insertion was identified 75 bp upstream of the ClaI site in cheW7 in this 2019 bp fragment (Fig. 1). CheW7 is predicted to be 148 amino acids long (Arshinoff et al., 2007; Goldman et al., 2006) and the insertion mutation occurred in the 108th codon of cheW7. This frameshift mutation is predicted to result in a polypeptide of 228 aa that extends 256 nt into the coding region of mcp7 out of frame. The N-terminal 107 aa of the mutant protein would be identical to the wild-type CheW7 and the rest has no homology to known proteins. We designated the suppressor mutation as cheW7-1.
Inactivation of cheW7 and concurrent expression of mcp7 are sufficient to suppress difA
To examine whether the cheW7-1 suppressor allele is a gain-of-function mutation, the cheW7 coding region was deleted from a difA deletion mutant (see Methods). The resulting ΔdifA ΔcheW7 double mutant (YZ1619) lacked EPS production as determined by Congo red binding on plates (Supplementary Table S2); this would be consistent with cheW7-1 as a gain-of-function mutation because difA was not suppressed by the cheW7 deletion as a loss-of-function mutation. However, when the cheW7-1 allele was expressed in YZ1619 (ΔdifA ΔcheW7) using pWB435 (Table 1), the resulting strain failed to produce EPS (Fig. 1c), suggesting that cheW7-1 is unlikely to be a gain-of-function mutation.
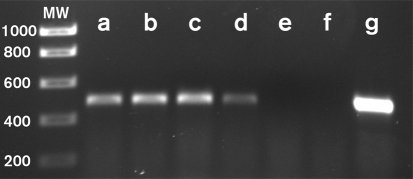
We considered the possibility that difA suppression may require the inactivation of cheW7 and the concurrent expression of other downstream genes such as mcp7 (Fig. 1). The ΔcheW7 allele in YZ1619 might have simply affected the expression of downstream genes. RT-PCR analysis indicated that the expression of mcp7 was indeed lower in YZ1619 (ΔdifA ΔcheW7) than in the suppressor strain or the wild-type (Fig. 4, compare lanes a, c and d). When mcp7 was deleted from the difA suppressor strain (YZ101), the resulting strain (YZ1613) was defective in EPS production (Supplementary Table S1), indicating that mcp7 is indeed required for difA suppression by cheW7-1. The results here also suggest that there is a promoter for mcp7 inside cheW7. The presence of this promoter also explains why a small percentage of the merodiploid transformants (Fig. 1b) from pWB515 and pWB530 showed the suppressor phenotype.
Fig. 4.
Analysis of mcp7 expression by RT-PCR. The same amounts of total RNA from M. xanthus strains were subjected to RT-PCR analysis as described in Methods. MW, DNA standards (molecular mass in base pairs indicated on the left). (a) DK1622 (wild-type); (b) YZ601 (ΔdifA); (c) YZ101 (ΔdifA cheW7-1); (d) YZ1619 (ΔdifA ΔcheW7); (e) YZ1613 (ΔdifA cheW7-1 Δmcp7); (f) YZ1604 (ΔdifA ΔcheW7 Δmcp7); (g) YZ1612 (ΔdifA ΔcheW7 Δmcp7 attB : : pWB433). No samples yielded any band in PCR-only reaction controls (data not shown). The sequences of the two primers used for the RT-PCR were 5′-TCCATGACGGAGATGAACGCCA-3′ and 5′-GCCAGGGAATTGATTTGCTCGGA-3′. See Fig. 1(a) for the location of the target for RT-PCR.
To further examine the role of mcp7 expression in difA suppression, we constructed pWB433, a plasmid that can express mcp7 in M. xanthus from the Mx8 attachment site (Fig. 1, Table 1). When this plasmid was transformed into YZ1619 (ΔdifA ΔcheW7), the resulting strain produced EPS at a similar level to the suppressor strain (Fig. 1c). Similar results were obtained when this mcp7 expression plasmid was transformed into YZ1604 (ΔdifA ΔcheW7 Δmcp7) and YZ1613 (ΔdifA cheW7-1 Δmcp7) (Fig. 1c). On the other hand, expression of mcp7 using the same plasmid (pWB433) in a difA deletion only strain (YZ601) did not lead to EPS production that could be detected by Congo red binding on plates (Fig. 1c). Since the expression of mcp7 in various strains was confirmed by RT-PCR (Fig. 4), these results established the importance of cheW7 inactivation and appropriate levels of mcp7 expression in the suppression of difA.
cheW7-1 partially bypasses the requirement for Tfp
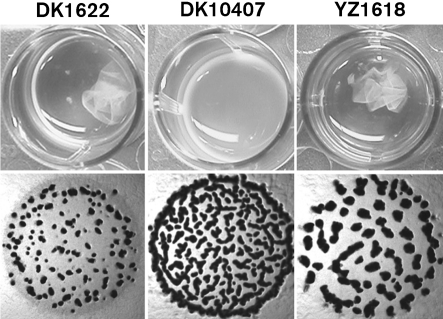
Previous studies indicated that Tfp function upstream of the Dif chemotaxis pathway as positive regulators of EPS production (Black et al., 2006). The suppression of difA by cheW7-1 led us to examine whether cheW7-1 could suppress the EPS defects of Tfp− mutants in an epistasis test. The cheW7-1 mutation was introduced into a pilA mutant to construct the cheW7-1 pilA double mutant YZ1618 (see Methods). As was expected, the pilA mutant DK10407 lacked agglutination and formed less organized fruiting bodies (Fig. 5) (Bonner et al., 2006; Dana & Shimkets, 1993; Wu et al., 1998). In contrast, YZ1618 was proficient in agglutination (Fig. 5), indicating EPS production (Dana & Shimkets, 1993; Shimkets, 1986b; Xu et al., 2005). The development of YZ1618 (pilA cheW7-1) was also markedly improved over DK10407 (pilA) (Fig. 5). However, YZ1618 showed no observable Congo red binding in plate assays, indicating that YZ1618 produces only low levels of EPS (Supplementary Table S2). These results indicate that the cheW7-1 mutation is epistatic to and partially suppresses pilA in EPS production.
Fig. 5.
Partial suppression of pilA by cheW7-1. Agglutination (upper panels) and development (lower panels) were examined as described in Methods. Strains: DK1622 (wild-type); DK10407 (pilA); YZ1618 (pilA cheW7-1). No EPS production by either DK10407 or YZ1618 could be detected by dye binding (Supplementary Table S2).
DifA and Mcp7 may compete with each other for interactions with DifC and DifE
Genetic experiments clearly demonstrated that Tfp are upstream of DifA in the regulation of EPS production (Black et al., 2006). Yet cheW7-1 fully suppresses difA (Figs 1, 2 and 6) but only partially suppresses pilA (Fig. 5, Supplementary Table S2). The lack of full suppression of pilA suggested that the presence of DifA interferes with the suppression of pilA by cheW7-1. difA was therefore deleted from the cheW7-1 pilA double mutant. As shown in Fig. 6, the resulting triple mutant (YZ647) produced similar levels of EPS to the wild-type (DK1622) and the difA suppressor strain (YZ101). This validated the notion that DifA is detrimental to the suppression of pilA by cheW7-1. Since DifA (MCP-like) forms a ternary complex with DifC and DifE (Fig. 7) (Yang & Li, 2005), and since Mcp7 along with DifC and DifE is essential for the suppression of difA by cheW7-1, these results are consistent with competition between DifA (MCP-like) and Mcp7 for interactions with DifC and DifE. The results here also further validated the epistasis of cheW7-1 to pilA.
Fig. 6.
Deletion of difA from the cheW7-1 pilA double mutant increases EPS production to wild-type levels. EPS production was analysed using the liquid Trypan blue binding assay (see Methods). Values for all strains were normalized to the wild-type. Strains: DK1622 (wild-type); YZ601 (ΔdifA); DK10407 (pilA); YZ648 (ΔdifA pilA); YZ101 (ΔdifA cheW7-1); YZ647 (ΔdifA cheW7-1 pilA).
Fig. 7.
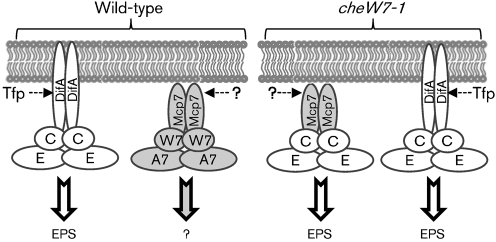
Schematic of the Dif and Che7 pathways and their interactions in the suppressor strain. In the wild-type (left), both DifA and Mcp7 interact with their cognate partners. Signal input into the Dif pathway is mediated by Tfp with an output of EPS production. Neither the input signal nor the output response of the Che7 pathway is currently known. In cheW7 mutants (right), Mcp7 and DifA may compete for interactions with DifC and DifE to regulate EPS production. DifA likely has a higher affinity for DifC and DifE than Mcp7. When both DifA and CheW7 are eliminated, Mcp7 may stably interact with DifC and DifE to regulate EPS production.
DISCUSSION
Here we report the identification of the cheW7-1 mutation as an extragenic suppressor of difA deletion in M. xanthus EPS production. The suppressor strain was isolated by enrichment through agglutination and restoration of S motility. The mutation was mapped and determined to be a single base pair insertion about two-thirds into cheW7. Instead of a gain-of-function mutation as we had initially thought, the suppressor allele turned out to be a loss-of-function mutation. The suppression of difA requires Mcp7 from the Che7 system as well as DifC (CheW-like) and DifE (CheA-like) from the Dif system. These observations suggest that while Mcp7 and DifA preferentially interact with their cognate CheWs and CheAs, they exhibit competitive interactions with DifC and DifE. In the suppressor strain, Mcp7 can substitute for DifA to form a stable and functional signalling complex with DifC and DifE to regulate EPS production in M. xanthus (Fig. 7).
The M. xanthus genome encodes eight independent chemosensory pathways based on homology and chromosomal organization (Kirby et al., 2008; Zusman et al., 2007) (also see the brief description of chemotaxis pathways in the Introduction). The inherent similarities may be expected to result in interactions among these pathways. Earlier studies have indeed uncovered such interactions in M. xanthus. The Frz chemosensory pathway is well known for regulating the reversal frequency of both A- and S-motile cells (Ward & Zusman, 1999). It was discovered recently that the reversal of S-motile cells is inversely correlated with velocity, as fast-moving S-motile cells reverse less frequently than their slow-moving counterparts (Vlamakis et al., 2004). The Che4 chemotaxis system was found to regulate this dependence of reversal upon velocity in S motility (Vlamakis et al., 2004). In other words, the S-motility engine, presumably Tfp (Wu & Kaiser, 1995; Zusman et al., 2007), is a target of both Frz and Che4 pathways. In addition, the Dif system is known to regulate reversals of well-isolated or A-motile cells in studies of tactic responses to phosphatidylethanolamine (PE) (Bonner et al., 2005; Kearns & Shimkets, 2001). This indicates that the A-motility engine is the target of regulation by both Dif and Frz chemosensory pathways. Earlier studies have also revealed that the Dif pathway is required for excitation while the Frz pathway mediates adaptation in PE taxis (Bonner et al., 2005; Kearns & Shimkets, 2001). Although it is unclear how these pathways converge on their common regulatory target, some clues have been provided by a recent study of interactions between Frz and Dif. Xu et al. (2008) demonstrated that signalling through the Dif pathway clearly influences the methylation of FrzCD, the MCP in the Frz pathway. This reveals at least one possible mechanism for Dif–Frz interaction at the molecular level. Irrespective of the underlying mechanisms, these studies provide clear evidence for biologically meaningful interactions among chemosensory pathways in M. xanthus in vivo.
The results in this paper provide the first report, to our knowledge, of genetic suppression of one chemotaxis system by another in M. xanthus. The genetic evidence presented here supports the model that Mcp7 forms a signalling complex with DifC and DifE to regulate EPS in the suppressor strain (Fig. 7). Previous observations had hinted at roles of Che7 in EPS regulation and S motility. Kirby and co-workers noted that certain che7 mutations in a pilQ (leaky) mutant background led to further deterioration of S motility and development (Kirby et al., 2008; Wall et al., 1999), possibly attributable to changes in the level of EPS production (Xu et al., 2005). Even for pilQ+ or wild-type strains, only a fraction (∼30 %) of wild-type cells bears assembled pilus at any given time (Palsdottir et al., 2009; Wu et al., 1997). The partial suppression of pilA by cheW7-1 and the defects of che7 mutants in the leaky pilQ background suggest that the Che7 chemosensory system plays a role in EPS regulation under certain conditions.
We suggest that Mcp7 substitutes for its homologue DifA in the Dif pathway to regulate EPS in response to signals different from those for DifA (Fig. 7). It is possible that the suppressor strain genetically mimics the wild-type under particular physiological conditions yet to be defined. It may be assumed that under such conditions, there is no significant input signal into DifA, as mirrored by a difA or a pilA mutant (Black et al., 2006). In addition, these conditions may lead to transcription of mcp7 and downstream genes but not cheW7 and those upstream (Fig. 1). In this context, Mcp7 (de)methylation may play roles in EPS regulation, because the two insertions in cpc7 (Fig. 1) suggest the requirement for CheR7 and CheB7 for the suppression. Further experimentation is undoubtedly necessary to determine the roles that Mcp7 and Che7 play in EPS regulation.
Finally, the cheW7-1 mutation also validated our previous model that Tfp function upstream of the Dif chemotaxis-like pathway to regulate EPS production (Black et al., 2006). This model predicts that a difA bypass suppressor should suppress the EPS− phenotype of Tfp− mutants. Although a cheW7-1 pilA double mutant produced low levels of EPS detectable only by agglutination (Fig. 5), a difA cheW7-1 pilA triple mutant had a similar amount of EPS to the wild-type and the difA cheW7-1 double mutant (Fig. 6). This provides further evidence that Tfp function upstream of the Dif system to provide input signals in EPS regulation (Black et al., 2006).
Acknowledgments
We thank Drs Larry Shimkets, Pamela Bonner and John Kirby for helpful discussions. We are grateful to Dr Heidi Kaplan for kindly providing material assistance. This work was supported by National Institutes of Health grants GM54666 to W. S. and GM071601 to Z. Y. National Science Foundation grant MCB-0135434 to Z. Y. also partially supported this work.
Abbreviations
EPS, exopolysaccharide
MCP, methyl-accepting chemotaxis protein
S motility, social motility
Tfp, type IV pili
Footnotes
Two supplementary tables, listing additional Myxococcus xanthus strains used in this study and phenotypes of selected M. xanthus strains and progenies from selected crosses, are available with the online version of this paper.
References
- Armitage, J. P., Holland, I. B., Jenal, U. & Kenny, B. (2005). “Neural networks” in bacteria: making connections. J Bacteriol 187, 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshinoff, B. I., Suen, G., Just, E. M., Merchant, S. M., Kibbe, W. A., Chisholm, R. L. & Welch, R. D. (2007). Xanthusbase: adapting wikipedia principles to a model organism database. Nucleic Acids Res 35, D422–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, E., Ludwig, G., Auerswald, E. A., Reiss, B. & Schaller, H. (1982). Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene 19, 327–336. [DOI] [PubMed] [Google Scholar]
- Bellenger, K., Ma, X., Shi, W. & Yang, Z. (2002). A CheW homologue is required for Myxococcus xanthus fruiting body development, social gliding motility, and fibril biogenesis. J Bacteriol 184, 5654–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, W. P. & Yang, Z. (2004). Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J Bacteriol 186, 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, W. P., Xu, Q. & Yang, Z. (2006). Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol Microbiol 61, 447–456. [DOI] [PubMed] [Google Scholar]
- Bonner, P. J., Xu, Q., Black, W. P., Li, Z., Yang, Z. & Shimkets, L. J. (2005). The Dif chemosensory pathway is directly involved in phosphatidylethanolamine sensory transduction in Myxococcus xanthus. Mol Microbiol 57, 1499–1508. [DOI] [PubMed] [Google Scholar]
- Bonner, P. J., Black, W. P., Yang, Z. & Shimkets, L. J. (2006). FibA and PilA act cooperatively during fruiting body formation of Myxococcus xanthus. Mol Microbiol 61, 1283–1293. [DOI] [PubMed] [Google Scholar]
- Bourret, R. B. & Stock, A. M. (2002). Molecular information processing: lessons from bacterial chemotaxis. J Biol Chem 277, 9625–9628. [DOI] [PubMed] [Google Scholar]
- Bren, A. & Eisenbach, M. (2000). How signals are heard during bacterial chemotaxis: protein–protein interactions in sensory signal propagation. J Bacteriol 182, 6865–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos, J. M. & Zusman, D. R. (1975). Regulation of development in Myxococcus xanthus: effect of 3′:5′-cyclic AMP, ADP, and nutrition. Proc Natl Acad Sci U S A 72, 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana, J. R. & Shimkets, L. J. (1993). Regulation of cohesion-dependent cell interactions in Myxococcus xanthus. J Bacteriol 175, 3636–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, B. S., Nierman, W. C., Kaiser, D., Slater, S. C., Durkin, A. S., Eisen, J. A., Ronning, C. M., Barbazuk, W. B., Blanchard, M. & other authors (2006). Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci U S A 103, 15200–15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, D. C., Bretscher, A. P. & Kaiser, D. (1978). Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol 64, 284–296. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J. & Kaiser, D. (1979a). Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol Gen Genet 171, 167–176. [Google Scholar]
- Hodgkin, J. & Kaiser, D. (1979b). Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol Gen Genet 171, 177–191. [Google Scholar]
- Julien, B., Kaiser, A. D. & Garza, A. (2000). Spatial control of cell differentiation in Myxococcus xanthus. Proc Natl Acad Sci U S A 97, 9098–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, D. (1979). Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci U S A 76, 5952–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, D. (2003). Coupling cell movement to multicellular development in myxobacteria. Nat Rev Microbiol 1, 45–54. [DOI] [PubMed] [Google Scholar]
- Kashefi, K. & Hartzell, P. L. (1995). Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol Microbiol 15, 483–494. [DOI] [PubMed] [Google Scholar]
- Kearns, D. B. & Shimkets, L. J. (2001). Lipid chemotaxis and signal transduction in Myxococcus xanthus. Trends Microbiol 9, 126–129. [DOI] [PubMed] [Google Scholar]
- Kirby, J. R., Berleman, J. E., Muller, S., Li, D., Scott, J. C. & Wilson, J. M. (2008). Chemosensory signal transduction systems in Myxococcus xanthus. In Myxobacteria: Multicellularity and Differentiation, pp. 135–147. Edited by D. E. Whitworth. Washington, DC: American Society for Microbiology.
- Lancero, H., Brofft, J. E., Downard, J., Birren, B. W., Nusbaum, C., Naylor, J., Shi, W. & Shimkets, L. J. (2002). Mapping of Myxococcus xanthus social motility dsp mutations to the dif genes. J Bacteriol 184, 1462–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Sun, H., Ma, X., Lu, A., Lux, R., Zusman, D. & Shi, W. (2003). Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc Natl Acad Sci U S A 100, 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, A., Cho, K., Black, W. P., Duan, X. Y., Lux, R., Yang, Z., Kaplan, H. B., Zusman, D. R. & Shi, W. (2005). Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol Microbiol 55, 206–220. [DOI] [PubMed] [Google Scholar]
- Magrini, V., Creighton, C. & Youderian, P. (1999). Site-specific recombination of temperate Myxococcus xanthus phage Mx8: genetic elements required for integration. J Bacteriol 181, 4050–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz, A. J., So, M. & Sheetz, M. P. (2000). Pilus retraction powers bacterial twitching motility. Nature 407, 98–102. [DOI] [PubMed] [Google Scholar]
- Mignot, T. (2007). The elusive engine in Myxococcus xanthus gliding motility. Cell Mol Life Sci 64, 2733–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot, T., Shaevitz, J. W., Hartzell, P. L. & Zusman, D. R. (2007). Evidence that focal adhesion complexes power bacterial gliding motility. Science 315, 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Palsdottir, H., Remis, J. P., Schaudinn, C., O'Toole, E., Lux, R., Shi, W., McDonald, K. L., Costerton, J. W. & Auer, M. (2009). Three-dimensional macromolecular organization of cryofixed Myxococcus xanthus biofilms as revealed by electron microscope tomography. J Bacteriol 191, 2077–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy, S., Dworkin, M. & Downard, J. (1997). Identification and characterization of Myxococcus xanthus mutants deficient in calcofluor white binding. J Bacteriol 179, 2863–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, E. J., Akerley, B. J., Novik, V. N., Lampe, D. J., Husson, R. N. & Mekalanos, J. J. (1999). In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc Natl Acad Sci U S A 96, 1645–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmler, A. B., Whitchurch, C. B. & Mattick, J. S. (1999). A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145, 2863–2873. [DOI] [PubMed] [Google Scholar]
- Shi, W. & Zusman, D. R. (1993). The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc Natl Acad Sci U S A 90, 3378–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets, L. J. (1986a). Role of cell cohesion in Myxococcus xanthus fruiting body formation. J Bacteriol 166, 842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets, L. J. (1986b). Correlation of energy-dependent cell cohesion with social motility in Myxococcus xanthus. J Bacteriol 166, 837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets, L. J. (1989). The role of the cell surface in social and adventurous behaviour of myxobacteria. Mol Microbiol 3, 1295–1299. [DOI] [PubMed] [Google Scholar]
- Shimkets, L. J. (1999). Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu Rev Microbiol 53, 525–549. [DOI] [PubMed] [Google Scholar]
- Skerker, J. M. & Berg, H. C. (2001). Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci U S A 98, 6901–6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., Zusman, D. R. & Shi, W. (2000). Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr Biol 10, 1143–1146. [DOI] [PubMed] [Google Scholar]
- Ueki, T., Inouye, S. & Inouye, M. (1996). Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183, 153–157. [DOI] [PubMed] [Google Scholar]
- Vlamakis, H. C., Kirby, J. R. & Zusman, D. R. (2004). The Che4 pathway of Myxococcus xanthus regulates type IV pilus-mediated motility. Mol Microbiol 52, 1799–1811. [DOI] [PubMed] [Google Scholar]
- Wall, D., Wu, S. S. & Kaiser, D. (1998). Contact stimulation of Tgl and type IV pili in Myxococcus xanthus. J Bacteriol 180, 759–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, D., Kolenbrander, P. E. & Kaiser, D. (1999). The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J Bacteriol 181, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, M. J. & Zusman, D. R. (1999). Motility in Myxococcus xanthus and its role in developmental aggregation. Curr Opin Microbiol 2, 624–629. [DOI] [PubMed] [Google Scholar]
- Whitworth, D. E. (2008). Myxobacteria: Multicellularity and Differentiation. Washington, DC: American Society for Microbiology.
- Wolgemuth, C., Hoiczyk, E., Kaiser, D. & Oster, G. (2002). How myxobacteria glide. Curr Biol 12, 369–377. [DOI] [PubMed] [Google Scholar]
- Wu, S. S. & Kaiser, D. (1995). Genetic and functional evidence that Type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol 18, 547–558. [DOI] [PubMed] [Google Scholar]
- Wu, S. S., Wu, J. & Kaiser, D. (1997). The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol Microbiol 23, 109–121. [DOI] [PubMed] [Google Scholar]
- Wu, S. S., Wu, J., Cheng, Y. L. & Kaiser, D. (1998). The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol Microbiol 29, 1249–1261. [DOI] [PubMed] [Google Scholar]
- Xu, Q., Black, W. P., Ward, S. M. & Yang, Z. (2005). Nitrate-dependent activation of the Dif signaling pathway of Myxococcus xanthus mediated by a NarX–DifA interspecies chimera. J Bacteriol 187, 6410–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q., Black, W. P., Cadieux, C. L. & Yang, Z. (2008). Independence and interdependence of Dif and Frz chemosensory pathways in Myxococcus xanthus chemotaxis. Mol Microbiol 69, 714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. & Li, Z. (2005). Demonstration of interactions among Myxococcus xanthus Dif chemotaxis-like proteins by the yeast two-hybrid system. Arch Microbiol 183, 243–252. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Geng, Y., Xu, D., Kaplan, H. B. & Shi, W. (1998). A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol Microbiol 30, 1123–1130. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Ma, X., Tong, L., Kaplan, H. B., Shimkets, L. J. & Shi, W. (2000). Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J Bacteriol 182, 5793–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian, P. & Hartzell, P. L. (2006). Transposon insertions of magellan-4 that impair social gliding motility in Myxococcus xanthus. Genetics 172, 1397–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian, P., Burke, N., White, D. J. & Hartzell, P. L. (2003). Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol Microbiol 49, 555–570. [DOI] [PubMed] [Google Scholar]
- Yu, R. & Kaiser, D. (2007). Gliding motility and polarized slime secretion. Mol Microbiol 63, 454–467. [DOI] [PubMed] [Google Scholar]
- Zusman, D. R., Scott, A. E., Yang, Z. & Kirby, J. R. (2007). Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol 5, 862–872. [DOI] [PubMed] [Google Scholar]