Abstract
Type 2 diabetes results from severe insulin resistance coupled with a failure of β-cells to compensate by secreting sufficient insulin. Multiple genetic loci are involved in the development of diabetes, although the effect of each gene on diabetes susceptibility is thought to be small. MicroRNAs (miRNA) are non-coding 19–22 nucleotide RNA molecules that potentially regulate the expression of thousands of genes. To understand the relationship between miRNA regulation and obesity-induced diabetes, we quantitatively profiled ~220 miRNAs in pancreatic islets, adipose tissue, and liver from diabetes-resistant (B6) and diabetes-susceptible (BTBR) mice. More than half of the miRNAs profiled were expressed in all 3 tissues, with many miRNAs in each tissue showing significant changes in response to genetic obesity. Further, several miRNAs in each tissue were differentially responsive to obesity in B6 versus BTBR mice, suggesting that they may be involved in the pathogenesis of diabetes. In liver, there were ~40 miRNAs that were down-regulated in response to obesity in B6, but not BTBR mice, indicating that genetic differences between the mouse strains play a critical role in miRNA regulation. In order to elucidate the genetic architecture of hepatic miRNA expression, we measured the expression of miRNAs in genetically obese F2 mice. Approximately 10% of the miRNAs measured showed significant linkage (miR-eQTLs), identifying loci that control miRNA abundance. Understanding the influence that obesity and genetics exert on the regulation of miRNA expression will reveal the role miRNAs play in the context of obesity-induced type 2 diabetes.
Introduction
MicroRNAs (miRNAs) are endogenously expressed single-stranded non-coding RNAs of 19–22 nucleotides in length. Approximately 500 miRNAs are listed in the current mouse miRNA registry (microRNA.sanger.ac.uk). MiRNAs regulate gene expression by destabilizing target mRNAs through multiple rounds of RNA cleavage (Hutvagner and Zamore 2002), or by repressing the transcription of its target gene by binding to its complementary sequence, usually at the 3’UTR (Filipowicz et al. 2008; He and Hannon 2004).
Over-expression of miRNAs in human cells has been shown to down-regulate the expression level of hundreds of putative mRNA targets (Lim et al. 2005). MiRNAs are transcribed as long RNA precursor molecules (pri-miRNAs) that contain a stem-loop structure of ~70 nucleotides (Lee et al. 2002). Pri-miRNAs are processed in the nucleus by the RNase III enzyme Drosha and its partner, DGCR8/Pasha, to generate a 60–70 nucleotide hairpin-structured pre-miRNA (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). Pre-miRNAs are transported from the nucleus into the cytoplasm by the nuclear membrane protein, Exportin-5 (Yi et al. 2003). In the cytoplasm, the RNase III enzyme, Dicer, cleaves the pre-miRNA hairpin to yield short miRNA duplexes (Hutvagner et al. 2001). The miRNA duplexes are subsequently unwound to liberate single-stranded mature miRNAs by the RNA-induced Silencing Complex (Khvorova et al. 2003; Schwarz et al. 2003).
MiRNAs have been shown to be involved in multiple biological processes, including glucose homeostasis and lipid metabolism (Krutzfeldt and Stoffel 2006; Tang et al. 2008; Zhang and Farwell 2008). For example, over-expression of miR-375 was shown to inhibit insulin secretion from the mouse insulinoma cell line, Min6, by directly targeting myotrophin (Poy et al. 2004), which is an actin-binding protein (Bhattacharya et al. 2006). Actin is known to play an important role in insulin secretion (Wang and Thurmond 2009). Further, in the rat insulinoma cell line, Ins-1E, miR-375 over-expression resulted in decreased insulin gene expression, by targeting the PI3K-pathway gene, phosphoinositide-dependent protein kinase-1 (El Ouaamari et al. 2008). Recently, miR-34a over-expression was shown to decrease glucose-stimulated insulin secretion and mediate FFA-induced apoptosis in Min6 cells by targeting Vamp2 and Bcl2, respectively (Lovis et al. 2008). Over-expression of miR-9 in Ins-1E cells results in decreased expression of the transcription factor, Onecut-2, leading to increased expression of Granuphilin/Slp4 and increased insulin secretion (Plaisance et al. 2006).
In addition to playing important roles in pancreatic islets, miRNA-dependent regulation has been reported in liver and adipose tissue in various model systems. MiR-122a is abundantly expressed in mouse liver (Chang et al. 2004). Injection of anti-sense oligonucleotides against miR-122a leads to a significant reduction in hepatic steatosis and plasma cholesterol (Esau et al. 2006). Similar findings have been reported in primate liver (Elmen et al. 2008), suggesting that miR-122a plays an important role in lipid metabolism across multiple species. MiRNAs 103 and 143 have been reported to increase adipogenesis in 3T3L1 adipocytes by affecting several key lipid metabolism genes, including Pparg, Fabp4 and adiponectin (Xie et al. 2009). Taken together, these studies clearly demonstrate that miRNAs are critically involved in important metabolic processes in multiple tissues. To more fully understand miRNA-dependent regulation in our model of obesity-induced type 2 diabetes, we set out to quantitatively profile miRNA expression in pancreatic islets, liver, and adipose tissue.
Our laboratory has modeled the genetics of obesity-induced type 2 diabetes in two mouse strains, diabetes-resistant C57BL/6 (B6) mice and diabetes-susceptible BTBR T+ tf/J (BTBR) mice. When made morbidly obese by the leptin mutation (Lepob/ob), B6-ob/ob mice experience moderate and only transient hyperglycemia, due to a large expansion of β-cell mass, resulting in a 20–50 fold increase in plasma insulin levels (Clee et al. 2005; Keller et al. 2008). In contrast, BTBR-ob/ob mice experience severe hyperglycemia due to a failure to increase their circulating insulin levels. An in vivo measure of cellular replication showed that B6-ob/ob mice experience a ~3-fold increase in islet cell proliferation, whereas BTBR-ob/ob mice do not increase islet cellular replication in response to obesity (Keller et al. 2008).
Insulin-dependent glucose uptake is a measure of insulin signaling. Previous work has shown that insulin signaling in adipocytes (Nadler et al. 2000) and muscle (Flowers et al. 2007) is dramatically reduced in BTBR mice, relative to that measured in B6 mice. Type 2 diabetes is a progressive disease, involving multiple tissues, which ultimately leads to the loss of β-cells in the pancreatic islets.
Obesity-induced insulin resistance in peripheral tissues (e.g., muscle, adipose and liver) requires increasing amounts of insulin production and secretion from pancreatic islets, in order to maintain euglycemia. When insulin levels are insufficient, hyperglycemia results and β-cells die. To understand the potential role that miRNA-dependent regulation may have in this metabolic cycle of obesity, insulin resistance and β-cell dysfunction in the context of diabetes, we profiled the expression of ~220 miRNAs in islets, adipose tissue, and liver in our model system. Our quantitative profiling approach has allowed us to survey absolute miRNA expression across islet, adipose tissue, and liver, determining which miRNAs are commonly expressed and which show tissue-selectivity. We report that several miRNAs undergo significant changes in their expression in response to obesity in each tissue profiled. Finally, by profiling miRNAs in the liver of an F2-intercross derived from the B6 and BTBR parental mice, we identify a small group of miRNAs whose expression shows heritability, enabling us to identify the genomic loci that control miRNA expression levels in liver.
Materials and methods
Animals
Mice were either purchased from the Jackson Labs (Bar Harbor, Maine) or generated from our in-house colony. B6:BTBR F2-ob/ob were generated from B6:BTBR F1-ob/ob breeder pairs. All the mice were kept at the University of Wisconsin Biochemistry Department, and housed in an environmentally-controlled facility on a 12-h light/dark cycle (6 am–6 pm, respectively). Mice were provided free access to water at all times and to a standard rodent chow (Purina no. 5008) ab libitum. At 10 weeks of age, animals were sacrificed by decapitation and tissues collected in the following order: left lateral liver lobe, right gonadal fat pad (white adipose tissue) and pancreas. Islets were isolated from whole pancreas as described (Keller et al. 2008). All other tissues were flash-frozen in liquid nitrogen.
RNA collection
Total RNA from liver and adipose tissue was isolated by Trizol reagent according to manufacturer instructions. Islet RNA was purified using the mirVana miRNA Isolation Kit (Ambion) according to manufacturer directions. An Agilent Bioanalyzer 2100 was used to assess RNA quality for all samples, which typically showed a 28/18S ratio of ~1.5 or greater.
Experimental Design
We employed an experimental design that relies on pooling total RNA prior to hybridization followed by validation of interesting genes (validation described below in real time PCR). Kendziorski et al. have shown the power of pooling when resources (e.g. total RNA, number of chips) are limited (Kendziorski et al. 2005). The number of mice used to generate group-specific islet pools was 46, 22, 39 and 41 for B6-lean, B6-ob/ob, BTBR-lean and BTBR-ob/ob, respectively. All individuals within a pool contributed the same amount of total RNA, thereby insuring that no one mouse with an exceptionally high RNA yield would dominate the measurement of the pool. However, the pooling was not used for the miRNA profiling in adipose and liver, as sufficient RNA mass was available from individual mice. All procedures were approved by the University of Wisconsin Animal Care and Use Committee.
Real time PCR measurements of miRNA abundance
Approximately 220 miRNA were measured by LNA based-RT PCR as described (Raymond et al. 2005). Because the amount of islet RNA available from a single mouse was insufficient for LNA-based profiling, total RNA was pooled from up to 46 mice for each experimental animal group. Since, mass requirements were not limiting for adipose tissue and liver, 5 individual mice were separately profiled per group for these two tissues. Briefly, miRNA-specific real time primers were used to generate first-strand cDNA molecules. cDNA was added with SYBR Green master mix, followed by 40 cycles of real time PCR amplification. Standard curves were created for all miRNAs profiled allowing us to present miRNA abundance as copies/10 pg of total RNA; a typical mammalian cell contains 10–30 pg of total RNA. To confirm the LNA-based measured from the pooled islet samples, 10 randomly selected mice from each experimental group were chosen for the Taqman-based PCR study. cDNA was reverse transcribed from 10 ng of total RNA using specific miRNA primers according to the manufacturer instructions (ABI). MiRNA expression is shown as ΔCt, normalized by parallel measure of snoRNA-234, a house-keeping non-coding RNA molecule.
miRNA interval mapping
Mice were genotyped using the Affymetrix mouse 5K SNP panel (www.affymetrix.com). For our intercross of B6 and BTBR, 1,973 SNPs reliably segregated for the founders and were used for gene mapping. Simple interval mapping (Lander and Botstein 1989) was performed using R/qtl (Broman et al. 2003), adjusting for sex as an interacting covariate and batch effects as an additive covariate. All miRNA traits were transformed to normal scores. Permutation thresholds were computed based on the 290 mice with miRNA data by simulating 10,000 independent normal samples. Only those miRNA that pass the 5% permutation threshold, and only chromosomes with at least one significant peak, are displayed in false-color image
Results
MiRNA expression survey in pancreatic islets, adipose tissue, and liver
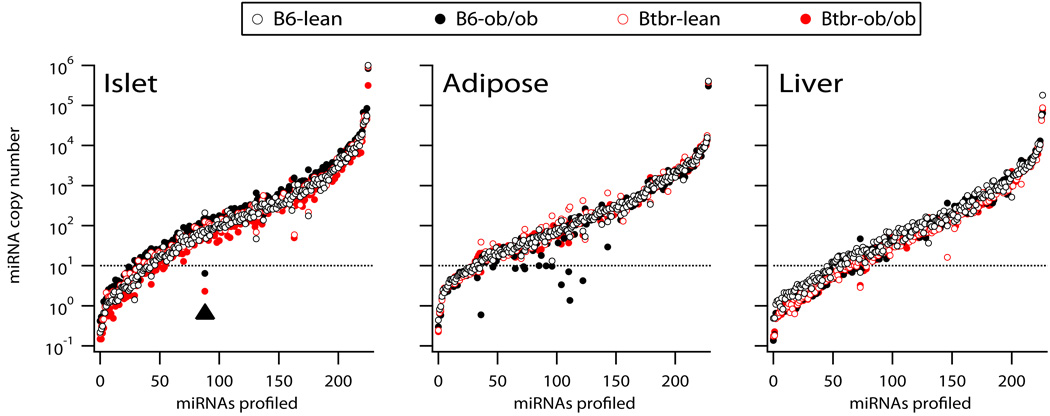
We quantitatively profiled miRNA abundance in three tissues collected from 4 groups of mice: B6-lean, B6-ob/ob, BTBR-lean and BTBR-ob/ob (all male and 10 weeks of age). A Locked Nucleic Acid (LNA) real-time PCR measurement was used to quantify miRNA expression levels. Due to RNA mass requirements for the miRNA profiling platform in islets, total RNA from islets was pooled from multiple individual mice for each of the 4 groups. RNA yields were not limiting in adipose tissue and liver and, therefore, 5 individual mice from each group were used for the profiling measurement in these tissues. Figure 1 illustrates the copy number for each miRNA profiled in islets, adipose tissue, and liver. Greater than 75% of the miRNAs profiled had ≥10 copies/cell across the 3 tissues. More than 50% of the miRNAs profiled were expressed in all tissues. A few miRNAs were highly expressed in all tissues; >1,000 copies/cell. Supplementary Table 1 provides a complete listing of all miRNAs profiled and is available for download at, http://attie.wisc.edu/zhaoetal2009/Zhao_et_al_2009_Supplementary_Table1.xls. In contrast to those miRNAs that were commonly expressed across islets, adipose tissue, and liver, some miRNAs showed dramatic tissue selectivity. For example, miR-127, 141, 153, 200a and 200c are highly expressed in islet (>2,000 copies/cell),>20-fold higher than in either adipose tissue or liver. MiR-375 showed the greatest tissue-selectivity for an islet-specific miRNA, which averaged ~60,000 copies/cell in islets, but was not expressed in liver and was expressed at low levels in adipose tissue (~100 copies/cell). Recent studies have shown that miR-375 plays a role in insulin expression (El Ouaamari et al. 2008), insulin secretion (Poy et al. 2004) and obesity-dependent β-cell proliferation (Poy et al. 2009).
Figure 1. Quantitative measure of ~220 miRNAs in pancreatic islets, adipose tissue, and liver in diabetes resistant and susceptible mice.
RNA was harvested from pancreatic islets, adipose tissue and liver collected from 10 week old B6-lean, B6-ob/ob, BTBR-lean or BTBR-ob/ob male mice. The expression of ~220 miRNAs was quantitatively determined by real time PCR. MiRNA abundance is shown as copy number/10 pg RNA, determined from standard curves established for each miRNA (see methods). For each tissue, the miRNAs are ranked from lowest to highest using the average expression level across the four experimental mouse groups. Horizontal dash line indicates a copy number of 10, below which we consider the miRNA is not expressed. Filled arrow head highlights miR-184 expression in islets that was dramatically reduced as a function of obesity in both B6 and BTBR mice. A complete listing of the miRNA expression values for all tissues is provided in Supplementary Table 1 (http://attie.wisc.edu/zhaoetal2009/Zhao_et_al_2009_Supplementary_Table1.xls).
MiRNAs 122a and 192 are abundantly (~50,000 and ~4,000 copies/cell, respectively) and selectively expressed in the liver. MiR-122a is not expressed in islet and expressed in adipose tissue ~200-fold less than in liver; miR-192 is expressed in islet and adipose tissue ~30-fold less than that expressed in liver. These results demonstrate that islet and liver have a small collection of miRNAs that are selectively expressed. Interestingly, there were no miRNAs that showed a strong selective expression in adipose tissue.
MiRNAs undergo significant change in abundance as a function of obesity and strain
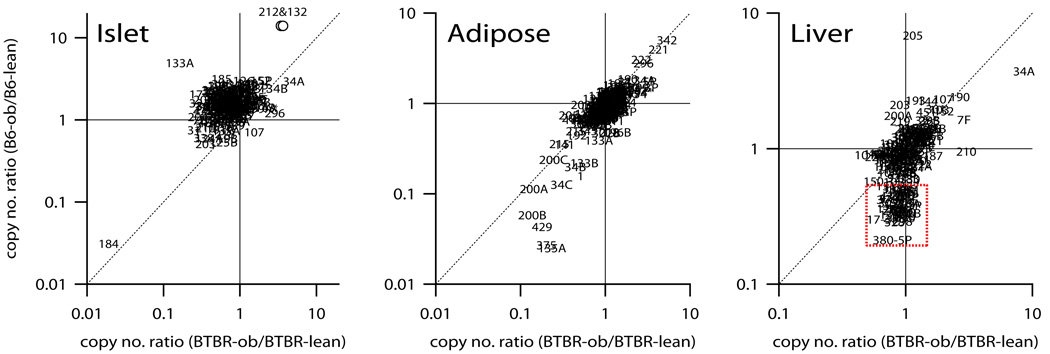
Obesity affected the expression of many miRNAs in all tissues. For example, in the islet, obesity strongly suppresses the expression of miR-184 (>20-fold) in B6 and BTBR mice (see filled arrow head Figure 1, islet panel). In adipose tissue, there were several miRNAs that were dramatically reduced in response to obesity in B6, but less so in BTBR mice (see filled black symbols <10 copies/cell, Figure 1, adipose panel). To highlight which miRNAs showed significant expression changes as a function of obesity, Figure 2 illustrates the ratio of the copy number of B6-ob/ob to B6-lean mice plotted against the same ratio for BTBR mice for islet, adipose tissue, and liver.
Figure 2. Obesity regulates miRNA expression in islet, adipose tissue, and liver.
The ratio of miRNA copy number measured in obese versus lean mice was determined for B6 (y-axis) and plotted against the same for BTBR (x-axis) in islets, adipose tissue, and liver. Values significantly different than 1.0 indicate a miRNA whose expression is altered in response to obesity. MiRNAs equally responsive to obesity in B6 and BTBR are those positioned along the diagonal for each tissue. MiRNAs are shown as their symbol number. The expression ratio values for miRNAs 132 and 212 in the islet were virtually the same, resulting in overlapping symbols (see open circles). Only miRNAs with expression levels ≥10 copies/cell are shown. Red dotted square highlights a group of ~40 miRNAs in the liver whose expression was reduced in response to obesity in B6 but not BTBR mice.
Of the miRNAs that were determined to be expressed at >10 copies/cell in islets, 12 were observed to undergo significant changes (>3-fold) as a function of obesity (Figure 2, left panel). MiRNAs 132, 212, 133a, 185, 152, 126-5p, 34a and 34b all show a >3-fold obesity-dependent increase in abundance in at least one mouse strain. As discussed below, several of these miRNAs have been shown to regulate β-cell glucose responsiveness and susceptibility to apoptosis.
The magnitude of the obesity-dependent increase observed for some miRNAs was much greater in islets derived from B6 mice than BTBR mice. For example, miRNAs 132 and 212 showed ~14-fold and 3-fold increase in B6 and BTBR mice, respectively. Whereas many miRNAs showed an obesity-dependent increase in expression, miR-184 abundance decreased 95% as a function of obesity in both strains. In addition to miR-184, miRNAs 31, 204 and 7b were decreased >67% in at least one mouse strain in response to obesity. Interestingly, miRNAs 204 and 7b were each changed as a function of obesity in BTBR, but not in B6.
Because pools of RNA samples collected from >20 mice were used for the islet profiling, we selected a handful of miRNAs to be measured on samples from 10 individual mice per experimental group (Supplementary Table 2). Nine miRNAs were chosen for this confirmation study. Of these, 7 had expression changes across the 4 mouse groups and were highly correlated (r > 0.96) with the microarray results in the pools.
In adipose tissue, ~190 miRNAs were expressed on average >10 copies/cell. Fifteen miRNAs increased > 3-fold as a function of obesity, including miRNAs 342, 221, and 222. The latter two miRNAs are modulated during adipocyte differentiation. Obesity caused >4-fold increase in miRNAs 342 and 221 in both B6 and BTBR mice. In contrast to the effect of obesity observed in the islets, many more miRNAs decreased than increased their expression in response to obesity in adipose tissue. MiRNAs 135a, 375, 429, 200b, 200a, 34c, 1, 34b, 133b, 200c, 215 and 141 all decreased >3-fold in response to obesity in at least one strain.
In liver, the expression levels of ~150 miRNAs were >10 copies/cell; 11 of these showed a significant expression change (>3-fold) in response to obesity in at least one mouse strain. MiRNAs 34a and 205 were increased in response to obesity, whereas miRNAs 151, 133a, 329, 201, 330, 17-3p, 298, 328 and 380-5p were decreased in response to obesity. Interestingly, miR-205 showed the greatest change in response to obesity, but only in B6, as miR-205 expression was unchanged as a function of obesity in BTBR mice (Figure 2, liver panel). Similarly, miR-380-5p showed ~5-fold reduction in B6 but no change in BTBR in response to obesity. The opposite strain effect was observed for miR-34a where the effect of obesity was greater in BTBR (~9-fold increase) than in B6 (~4-fold increase). These results demonstrate that the expression of several miRNAs in liver is altered as a function of obesity differently in B6 versus BTBR mice. In fact unique to the liver was a collection of ~43 miRNAs that showed a small obesity-dependent decrease in their expression (>40%) in B6 but not BTBR mice (Figure 2, liver panel). Several of these miRNAs are very abundant (~1,000 copies/cell), including miRNA 211, 330, 298, 151 and 328. Taken together, these results suggest that the regulation of hepatic miRNA expression in response to obesity is distinct between the diabetes-resistant B6 and the diabetes-susceptible BTBR mice.
Heritability of hepatic miRNA expression
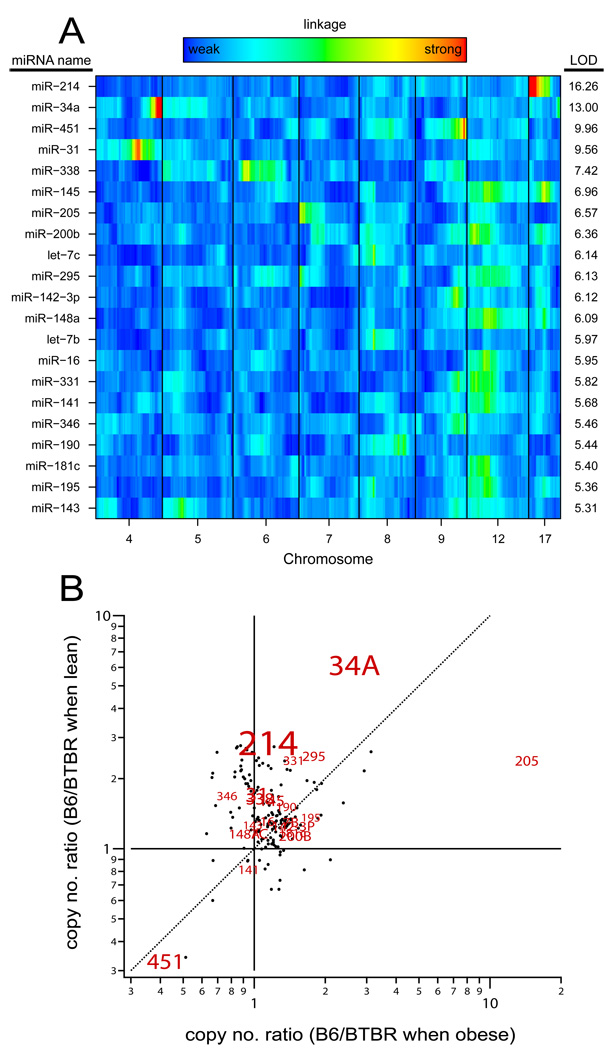
To map the gene loci controlling hepatic miRNA expression, we profiled the miRNAs in 290 liver samples from a population of F2 ob/ob mice. Figure 3A illustrates that 21 miRNAs achieved significant linkage (LOD > 5.3, genome-wide p-value < 0.05). Supplementary Table 3 provides a list of these 21 miRNAs with their genomic location as well as the position of peak linkage. Three of the 21 miRNAs (miR-34a, 31 and 295) mapped to the genomic region where they are physically located (cis-acting), whereas the others mapped to a locus that does not encode the miRNA (trans-acting). This suggests that hepatic expression of these 21 miRNAs may be under the control of regulatory factor(s). MiR-214, physically located in Chr 1, had the strongest linkage of all miRNAs, with LOD score of ~16 to a locus on Chr 17. In contrast to the trans linkage of miR-214, miR-34a, which is physically located on Chr 4, shows cis linkage with a LOD of ~13 to the same genomic region. This suggests that miR-34a expression may be affected by genetic variability at or near its genomic location. Interestingly, 9 miRNAs (145, 200b, 295 148a, 16 331, 141, 181 and 195) showed trans-linkage to a region on Chr 12, suggesting this may be a region generally involved in regulation miRNA expression. These results demonstrate that miRNA expression, like mRNA expression, is a heritable trait that can be used to reveal cis and trans-acting regulatory factors governing miRNA abundance.
Figure 3. Genetic architecture of hepatic miRNA expression.
Of ~220 miRNAs profiled in the liver of 290 F2-ob/ob mice, 21 showed significant linkage (LOD > 5.3, p-value <0.05). Panel A illustrates a heat map whereby the 21 miRNAs are ordered by their LOD score. Red indicates strong linkage, whereas blue indicates weaker linkage; green is intermediate. Panel B illustrates the copy number ratio of miRNA expression of B6 to BTBR mice when lean (y-axis) plotted against the copy number ratio of B6 to BTBR when obese (x-axis) from the profiling of parental liver samples. The miRNAs that showed significant linkage in the F2 mice are highlighted by their miRNA name, the size of which is proportional to their LOD score.
That hepatic miR-34a expression had strong cis-linkage (Figure 3A) and showed a larger change in expression in response to obesity in BTBR (~9-fold) than B6 (~4-fold) mice (Figure 2, right panel), suggests that those miRNAs that showed a strain difference in the parental profiling are likely to show linkage in the F2 mice. Figure 3B illustrates the effect of strain on hepatic miRNA expression in the parental liver samples, highlighting those that demonstrated significant linkage in the F2 livers. The results indicate that the miRNAs showing the strongest strain effect are more likely to also show linkage in the F2 liver samples.
Discussion
MicroRNAs have emerged as important regulators of gene expression in obesity and diabetes (Krutzfeldt and Stoffel 2006; Tang et al. 2008). However, to date there has not been a comprehensive survey of miRNA expression in multiple tissues from an animal model of obesity-induced type 2 diabetes. In this study we provide a quantitative measurement of the expression of ~220 miRNAs in islets, adipose tissue, and liver from diabetes resistant (B6) and diabetes susceptible (BTBR) mice.
To the best of our knowledge, this is the first study to survey miRNA expression in isolated pancreatic islets in the context of obesity and diabetes. In addition to islets, we report on the relative expression of miRNAs in adipose tissue and liver, identifying miRNAs that are commonly expressed across these 3 tissues, as well as those that show dramatic tissue-specific expression patterns. Further, we report on many miRNAs that are differentially regulated by obesity and/or genetic differences that exist between B6 and BTBR mice. Finally, by profiling hepatic miRNA expression in an F2-intercross, we demonstrate that miRNA abundance, like mRNA abundance, is a heritable trait, giving rise to miR-eQTLs, or genomic loci that govern their expression. The information provided by this study will aid in understanding the relationship between miRNA regulation and type 2 diabetes.
In islets, the expression of two miRNAs (132 and 212) each increased ~14 and ~4-fold in response to obesity in B6 and BTBR mice, respectively. This obesity-dependent increase in miRNA expression observed in B6 mice was the largest of any miRNA profiled in islets, adipose tissue, or liver. MiRNAs 132 and 212 are genomically located on Chr 11, ~200 base pairs apart, suggesting they may be generated from a common pri-miRNA molecule (Vo et al. 2005), explaining why obesity regulates their expression equally. Interestingly, these two miRNAs are either not expressed or expressed at a low level in liver and adipose tissue, suggesting that obesity-dependent regulation is unique to islets. Previous work has shown that they are highly expressed in neurons and may regulate neuronal differentiation (Vo et al. 2005; Wayman et al. 2008). MiR-132 has also been shown to modulate the circadian clock in the superchiasmatic nucleus via the MAPK/CREB-dependent pathway (Cheng et al. 2007). Interestingly, a circadian clock has been identified in islets that regulates the expression of several key islet genes, including Glut2, Ins-1, Gck, Sst, and Stx1a (Allaman-Pillet et al. 2004). Further investigation would be needed to determine whether the obesity-dependent regulation of miR-132 regulates the islet circadian clock. Finally, miRNAs 132 and 212 were shown to be highly up-regulated in ovarian granulosa cells in response to the hormone hCG (Fiedler et al. 2008). It would be interesting to determine if changes in circulating levels of hCG that is known to occur during pregnancy (Braunstein et al. 1976), induces the expression of these miRNAs in pancreatic islets.
There are two consensus cAMP-response element binding (CREB) sites located proximal to the genomic position of miRNAs 132 and 212 (Vo et al. 2005). Consistent with these CREB sites playing a role in their regulation, we have found that incubating islets with IBMX or forskolin, compounds that are known to elevate cellular cAMP levels (Barnett et al. 1994), results in a ~2-fold increase in the expression of miRNAs 132 and 212 (unpublished observations). Further, the incretin hormone Glp-1, also known to elevate cellular cAMP levels (Thorens and Waeber 1993), induces the expression of these miRNAs in mouse islets and Ins-1 cells (Shang et al., manuscript in preparation). Glp-1 stimulates glucose-stimulated insulin secretion, stabilizes insulin mRNA levels, promotes β-cell replication and suppresses apoptosis in β-cell lines and isolated islets (Drucker 2003), all of which has resulted in Glp1 offering a new target for the treatment of diabetes. Interestingly, the receptor for Glp1 (Glp1R) contains a coding SNP between B6 and BTBR, converting a highly-conserved histidine at position 47 to arginine. While we do not know if this amino acid substitution alters the activity of Glp1R, our data suggests that the difference in the magnitude of the obesity-dependent induction of miRNAs 132 and 212 reflects differential Glp-1 responsiveness between B6 and BTBR mice, which may contribute to the increased susceptibility to diabetes of the BTBR mice.
Fatty acids have been shown to affect miRNA expression in the mouse insulinoma cell line, Min6 (Lovis et al. 2008). The expression of miR-34a was increased >2-fold when Min6 cells were treated with 1 mM palmitate. Interestingly, we found that the expression of miR-34a was elevated ~3 and ~5-fold as a function of obesity in B6 and BTBR mice, respectively. The greater obesity-dependent induction of miR-34a expression in BTBR mice may in part reflect increased plasma lipids, which we have previously reported for BTBR-ob/ob mice (Clee et al. 2005). Lovis and colleagues (Lovis et al. 2008) also report that miR-34a over-expression in Min6 cells lead to decreased glucose-stimulated insulin secretion targeting Vamp2 and increased apoptosis by targeting Bcl2.
Poy et al. were the first to show that miR-375 is abundantly expressed in pancreatic islets (Poy et al. 2004). Here, we show that miR-375 was the second most abundantly expressed miRNA in islets and further, was expressed ~600-fold higher in islets than in adipose tissue and was not expressed in liver. Recently, whole-body miR-375 knockout (375KO) mice have been generated (Poy et al. 2009). The mice have elevated plasma glucose levels due to a reduction in β-cell mass and an increase in the number of α-cells per islet. Further, when challenged with severe obesity (Lepob/ob), 375KO mice do not show enhanced β-cell proliferation, which is necessary to compensate for the peripheral insulin resistance resulting from the obesity. Our data reveals that miR-375 expression in islets is ~50% lower in BTBR-ob/ob mice than in B6-ob/ob mice. We recently reported that obesity does not stimulate β-cell proliferation in BTBR mice, whereas B6 mice demonstrate a ~3-fold increase in response to obesity (Keller et al. 2008). Perhaps the loss of miR-375 expression in the islets of BTBR-ob/ob mice is involved in their inability to increase β-cell replication in response to obesity.
Dietary and genetically-imposed obesity has previously been shown to regulate miRNA expression in mouse adipose tissue (Xie et al. 2009). Microarray chips were used to profile adipose tissue collected from either chow or high-fat diet-fed B6 mice, as well as adipose tissue from B6-lean and B6-ob/ob mice. The expression of several miRNAs changed in response to either obesity challenge, including miRNAs 107, 103, 30c, 30a-5p, 222, and 221. Furthermore, these same miRNAs were differentially expressed in differentiated versus non-differentiated 3T3-L1 pre-adipocytes. Our data supports an obesity-dependent change in the expression of these miRNAs (Figure 2, adipose tissue panel). Moreover, we did not see a strain difference between B6 and BTBR for the influence of obesity, suggesting that these miRNAs are not involved in the pathogenesis of diabetes, but rather play a critical role in adipogenesis, in agreement with Xie et al. (Xie et al. 2009). Sun et al. reported that the let-7 miRNA family was highly expressed in adipocytes and further, was up-regulated during 3T3-L1 adipogenesis (Sun et al. 2009). However, while our data supports the finding that the let-7 miRNA family is highly expressed in adipose tissue (let-7 a, b, c, d were all > 8,000 copies/cell), we did not see a change in their expression as a function of obesity in either B6 or BTBR mice (Supplementary Table 1).
Of the 3 tissues profiled in our study, the liver contained the greatest number of miRNAs whose expression patterns showed a strain difference in their response to obesity. The expression of ~43 miRNAs showed a small decrease (>60%) in response to obesity in B6 mice, but not in BTBR mice, suggesting that strain plays an important role in the regulation by obesity of hepatic miRNA expression. In order to reveal the specific gene loci mediating this regulation, we profiled miRNAs in the liver of a sub-set of F2-ob/ob mice generated as part of a larger ongoing study (Keller et al., manuscript in preparation).
We found that ~10% of the miRNAs profiled in the liver of the F2 mice showed heritability. Twenty-one miRNAs have a miR-eQTL across the genome (LOD > 5.3, p-value <0.05). The most significant linkage was observed for miR-214, which mapped to a locus on Chr 17 with a LOD score of ~16. Previous work has shown that the transcription factor Twist1 regulates the expression of miR-214 (Lee et al. 2009). MiRNA-214 is genomically located on Chr 1, therefore its linkage to Chr 17 is trans, indicating the presence of a regulatory element. Given that Twist1 is genomically located on Chr 12, our data suggests a new regulatory element is involved in miR-214 regulation. Interestingly, of the 21 miRNAs to show significant linkage, all but 3 mapped to loci in trans, suggesting that the expression of many miRNAs are under the control of specific regulatory elements.
The 3 miRNAs mapping in cis were miRNAs 34a, 31 and 295. MiR-34a had the second most significant miR-eQTL, mapping to Chr 4 with a LOD score of ~13. The tumor suppressor gene, p53 has been shown to directly regulate miR-34a expression (Chang et al. 2007). However, the p53 binding site in the promoter for miR-34a does not include any SNPs between B6 and BTBR, suggesting once again, that our data has revealed novel regulatory pathways that control hepatic miRNA expression. Finally, six out of the 21 miRNAs illustrated in Figure 3A, had trans-linkage to a region on Chr 12. Using a lower LOD threshold of 3.0, 209 miRNAs show linkage across the genome, 26% of which have a miR-eQTL on Chr 12. This suggests that Chr 12 may contain a general regulatory factor involved in the processing of up to 25% of the miRNAs expressed in liver.
To the best of our knowledge, our study is the first to report the genetic architecture of hepatic miRNA expression. Not only does this reveal specific genetic loci governing the control of miRNAs, we believe that it may allow us to identify which mRNAs are targets for specific miRNAs. For each miRNA that showed significant linkage, we were able to identify hundreds of mRNA transcripts that showed linkage to the same genomic region as the miRNA. This will enable us to utilize the linkage data from the two profiling studies to identify putative mRNA targets for a given miRNA. Future work will utilize the linkage profiles for miRNAs and co-mapping mRNAs to construct causal transcriptional networks to identify direct versus indirect mRNAs for a particular miRNA.
In conclusion, our study provides a quantitative measure ~220 miRNAs in tissues (islets, adipose and liver) that play a key role in glucose homeostasis in the context of obesity-induced type 2 diabetes. We identified many miRNAs that undergo a significant change in expression in response to obesity and/or genetic differences between diabetes resistant (B6) and diabetes susceptible (BTBR) mice. We also report for the first time that hepatic miRNA expression is a heritable trait, which can be used to identify putative mRNA targets. It is estimated that the expression of up to 30% of all genes may be effected by miRNAs (Brennecke et al. 2005; Lewis et al. 2005). Additionally, a miRNA can potentially influence the expression of hundreds to thousands of mRNA transcripts. Elucidating the regulatory mechanisms that mediate obesity and genetic-dependent regulation of miRNA expression will aid in understanding the role(s) miRNAs play in the context of obesity-induced type 2 diabetes.
Supplementary Material
Acknowledgements
This work was supported by grants from CNPq Brazil, NIDDK grants 66369 and 58037, NIGMS grant 69430 and 76274, an ADA Mentor-based Fellowship #7-07-MN-02 and in-kind support from Rosetta Inpharmatics and Merck. We wish to thank the Rosetta Gene Expression Laboratory for their expertise in expression profiling of hundreds of tissues samples, Adam Steinberg for advice with our illustrations and Yuerong Zhu for development of analytical tools.
References
- Allaman-Pillet N, Roduit R, Oberson A, Abdelli S, Ruiz J, Beckmann JS, Schorderet DF, Bonny C. Circadian regulation of islet genes involved in insulin production and secretion. Molecular and cellular endocrinology. 2004;226:59–66. doi: 10.1016/j.mce.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Barnett DW, Pressel DM, Chern HT, Scharp DW, Misler S. cAMP-enhancing agents "permit" stimulus-secretion coupling in canine pancreatic islet beta-cells. The Journal of membrane biology. 1994;138:113–120. doi: 10.1007/BF00232639. [DOI] [PubMed] [Google Scholar]
- Bhattacharya N, Ghosh S, Sept D, Cooper JA. Binding of myotrophin/V-1 to actin-capping protein: implications for how capping protein binds to the filament barbed end. The Journal of biological chemistry. 2006;281:31021–31030. doi: 10.1074/jbc.M606278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein GD, Rasor J, Danzer H, Adler D, Wade ME. Serum human chorionic gonadotropin levels throughout normal pregnancy. American journal of obstetrics and gynecology. 1976;126:678–681. doi: 10.1016/0002-9378(76)90518-4. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS biology. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics (Oxford, England) 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA biology. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Molecular cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clee SM, Nadler ST, Attie AD. Genetic and genomic studies of the BTBR ob/ob mouse model of type 2 diabetes. American journal of therapeutics. 2005;12:491–498. doi: 10.1097/01.mjt.0000178781.89789.25. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Drucker DJ. Glucagon-like peptide-1 and the islet beta-cell: augmentation of cell proliferation and inhibition of apoptosis. Endocrinology. 2003;144:5145–5148. doi: 10.1210/en.2003-1147. [DOI] [PubMed] [Google Scholar]
- El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E. miR-375 targets 3'-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes. 2008;57:2708–2717. doi: 10.2337/db07-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell metabolism. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Fiedler SD, Carletti MZ, Hong X, Christenson LK. Hormonal regulation of MicroRNA expression in periovulatory mouse mural granulosa cells. Biology of reproduction. 2008;79:1030–1037. doi: 10.1095/biolreprod.108.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Flowers JB, Oler AT, Nadler ST, Choi Y, Schueler KL, Yandell BS, Kendziorski CM, Attie AD. Abdominal obesity in BTBR male mice is associated with peripheral but not hepatic insulin resistance. American journal of physiology. 2007;292:E936–E945. doi: 10.1152/ajpendo.00370.2006. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes & development. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science (New York, N.Y. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science (New York, N.Y. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Keller MP, Choi Y, Wang P, Davis DB, Rabaglia ME, Oler AT, Stapleton DS, Argmann C, Schueler KL, Edwards S, Steinberg HA, Chaibub Neto E, Kleinhanz R, Turner S, Hellerstein MK, Schadt EE, Yandell BS, Kendziorski C, Attie AD. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome research. 2008;18:706–716. doi: 10.1101/gr.074914.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell metabolism. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. The EMBO journal. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic acids research. 2009;37:123–128. doi: 10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lovis P, Roggli E, Laybutt DR, Gattesco S, Yang JY, Widmann C, Abderrahmani A, Regazzi R. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes. 2008;57:2728–2736. doi: 10.2337/db07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler ST, Stoehr JP, Schueler KL, Tanimoto G, Yandell BS, Attie AD. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11371–11376. doi: 10.1073/pnas.97.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, Regazzi R. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. The Journal of biological chemistry. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, Stoffel M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA (New York, N.Y. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 Regulates 3T3-L1 Adipogenesis. Molecular endocrinology (Baltimore, Md. 2009;23:925–931. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Tang G, Ozcan S. Role of microRNAs in diabetes. Biochimica et biophysica acta. 2008;1779:697–701. doi: 10.1016/j.bbagrm.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorens B, Waeber G. Glucagon-like peptide-I and the control of insulin secretion in the normal state and in NIDDM. Diabetes. 1993;42:1219–1225. doi: 10.2337/diab.42.9.1219. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Thurmond DC. Mechanisms of biphasic insulin-granule exocytosis - roles of the cytoskeleton, small GTPases and SNARE proteins. Journal of cell science. 2009;122:893–903. doi: 10.1242/jcs.034355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & development. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Farwell MA. microRNAs: a new emerging class of players for disease diagnostics and gene therapy. J Cell Mol Med. 2008;12:3–21. doi: 10.1111/j.1582-4934.2007.00196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.