Abstract
The aetiology of leukaemias among children is believed to be distinct from that of adults, mainly due to the clearer role for early life exposures, including those in utero. However, few risk factors have been established, because of the challenge of studying a disease with relatively low incidence. Identified risk factors, including ionizing radiation, chemotherapeutic agents and specific genetic abnormalities, explain <10% of incidence(1,2). Although the causes for the remaining 90% are unknown, it is possible that genetic susceptibility factors, either alone or in conjunction with environmental factors, may be involved. In this paper, the authors (a) review the evidence surrounding genetic susceptibility factors, with emphasis on the genes' main effects; (b) review some recent developments in the Northern California Childhood Leukaemia Study (NCCLS) as a case study of design and practical considerations in genetic epidemiology research and (c) highlight both challenges and future directions in this exciting research area.
BACKGROUND
Generally, cancer is a disease of ageing – one in which the DNA of a normal cell accumulates enough uncorrected damage over time to become malignant. This accumulation of damage usually takes many years, and most cancers therefore occur during adulthood. Accordingly, it stands to reason that cancers occurring during childhood arise from cells that have some early cancer advantage, conferred at least in part by increased inherited susceptibility to damage by exogenous exposures. However, while a dramatic excess risk of childhood acute lymphocytic leukaemia (ALL) has been observed among monozygotic twins of ALL patients compared with dizygotic twins of ALL patients, recent studies indicate that this excess risk may be due more to intraplacental metastasis rather than a highly penetrant inherited risk allele(3). Thus, for ALL, as for other multi-factorial diseases(4), inherited risk alleles are likely to be low penetrance susceptibility alleles that must interact with environmental factors to modulate disease risk. Indeed, leukaemia-associated chromosomal rearrangements alone are insufficient for disease onset: common chromosomal rearrangements usually associated with ALL are found in ∼1% of children who do not have ALL(5), and in vitro and animal studies have shown that while expression of the TEL–AML1 fusion protein is sufficient to establish a covert pre-leukaemic clone, it is not sufficient to transform haematopoietic cell lines into leukaemic cells(6). Taken together, these points indicate a probable role for inherited susceptibility in at least one of the ‘hits’ in the minimal two-hit model for onset of ALL proposed by Greaves(7).
The term ‘genetic susceptibility’ refers to inherited factors that modulate disease risk, either via the factors' main effects, or more likely via their interaction with other inherited factors (gene–gene interactions) or exogenous exposures such as chemicals, dietary factors and infectious agents (gene–environment interactions). This is in contrast to the genetics of Mendelian diseases, which are more deterministic – that is, the presence of the genetic abnormality virtually guarantees the disease. The ‘penetrance’ of a genetic factor is another term that should be clarified. In epidemiological terms, penetrance is an indicator of the relative involvement of other factors in the genetic factor's causal pathway(s). As such, one can expect higher risk estimates associated with higher penetrance genetic factors, and vice versa. For example, in breast cancer risk, a mutation in the BRCA1 gene, which encodes an important DNA repair enzyme, is associated with a high relative risk (OR > 5), and is considered a high-penetrance susceptibility locus. Conceptually, the BRCA1 mutation can be thought of as a major part of a short causal pathway to disease. In contrast, susceptibility loci with low risk estimates (ORs ∼ 1.2–1.5) are considered low penetrance, and can be thought of conceptually as minor parts of longer causal pathways to disease. It is likely that most inherited cancer risk is due to common, low-penetrance loci.
REVIEW OF PUBLISHED STUDIES
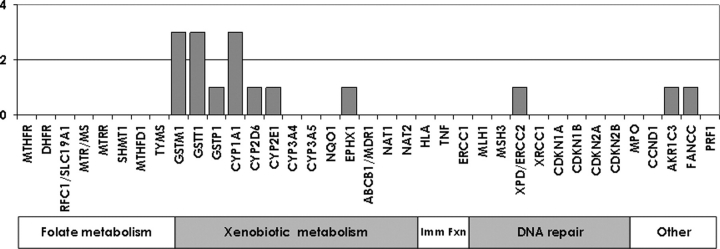
To date, childhood leukaemia genetic susceptibility literature has focused on genetically influenced mechanisms involved in protection against various external threats. Some examples of these candidate pathways are immune function, response to infection, folate-mediated one-carbon metabolism, membrane transport, detoxification and biotransformation of reactive intermediates derived from environmental carcinogens, trapping or decomposition of reactive oxygen species (ROS) and DNA repair enzymes. Functionally significant inherited polymorphisms in genes involved at critical junctions along these pathways may alter the way in which a child responds to environmental threats, thus influencing the risk of developing childhood leukaemia. A growing number of primary studies and meta-analyses(8,9) have been published linking variants in these genes with the risk of childhood ALL. Many of these genes were examined in three or fewer reports; for these genes, additional evidence needs to be gathered. Those genes are summarised briefly here with four or more reports. It should be noted that while many genes have been examined for childhood acute myelocytic leukaemia (AML) in one or two reports, none have been examined in four or more reports (Figure 1), and thus the following summary focuses only on childhood ALL.
Figure 1.
Frequency of reports of genes' main effects for childhood AML as of April 2008, by candidate pathway.
Published studies for childhood ALL
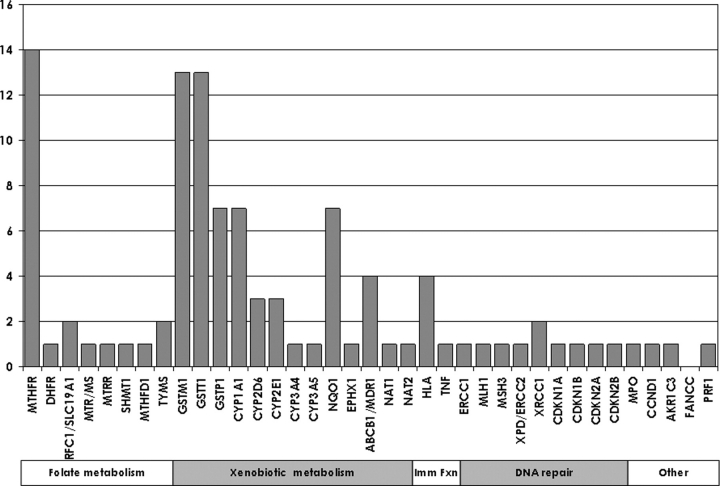
As of April 2008, 59 reports of gene main effects have been published for ALL. As shown in Figure 2, these papers have covered 36 genes in the candidate pathways noted earlier.
Figure 2.
Frequency of reports of genes' main effects for childhood ALL as of April 2008, by candidate pathway.
Folate-mediated one-carbon metabolism pathway
Folate, an essential micronutrient, plays a central role in preserving the balance between fidelity of DNA synthesis and availability of methyl groups for DNA methylation(10). Folate deficiency induces chromosomal damage and formation of fragile chromosomal sites, which are often associated with carcinogenesis(11), and maternal folate supplementation during pregnancy may reduce the risk of ALL in children(12). Furthermore, hyperdiploid cases, whose lymphocytes often have multiple copies of chromosome 21, are particularly responsive to methotrexate, the folate pathway inhibitor whose receptor is encoded on chromosome 21(13). Variants in more than a dozen genes, including those encoding the methylene tetrahydrofolate reductase (MTHFR) may alter folate metabolism and therefore the risk of childhood leukaemia. MTHFR has two low-function polymorphisms: 677C > T (rs1801133) and 1298A > C (rs1801131). A recent meta-analysis(8) of 10 studies(10,14–22) concluded that there was no significant decrease in ALL risk associated with 677C > T (TT versus CT + CC: OR = 0.88, 95% CI: 0.73–1.06), and four subsequent studies indicate either no association or a reduced risk associated with the variant(23–26); on balance, results indicate no effect or a modestly decreased risk associated with this marker. The 1298A > C marker of MTHFR was found in the same meta-analysis, to be unassociated with childhood ALL risk(8), a mixed effect mirrored by subsequent studies(24–26).
Xenobiotic metabolism and transport
In order to exert their effects, potentially harmful chemicals must gain entry into target cells and undergo cellular metabolic processes that alter activity. Enzymes involved in phase I (bioactivation) and phase II (detoxification) metabolism maintain a critical balance of activation and inactivation of a wide range of chemical exposures of relevance to childhood leukaemia, including drugs, chemical carcinogens, insecticides, petroleum products, nitrosamines, polycyclic aromatic hydrocarbons and other environmental pollutants(27). Major metabolic enzyme families include the phase I cytochrome P450 (CYP) and the phase II glutathione-S-transferase (GST) and N-acetyl transferase enzymes. In addition, membrane transporters such as those encoded by the multiple drug resistance (ABCB1/MDR1) gene act as efflux pumps to expel compounds from the cell and are strategically expressed in regions of the body that act as epithelial barriers or perform excretory functions(28).
In childhood leukaemia, the most commonly studied xenobiotic metabolism genes are those in the GST family (Figure 2), specifically, GST-mu-1 (GSTM1), GST-theta-1 (GSTT1) and GST-pi-1 (GSTP1). GSTM1 detoxifies polycyclic aromatic hydrocarbons, and GSTT1 metabolises epoxides and halomethanes; common deletions in both of these genes result in the loss of enzyme function(29). A meta-analysis(9) of seven studies published through July 2004(15,30–35) revealed no overall association of the GSTM1 and GSTT1 deletions with the risk of childhood ALL, and six subsequent studies(36–41) showed no, or only increased, risks for the GSTM1 marker and no significant risk change for the GSTT1 marker, suggesting no association overall. The most commonly studied variant in GSTP1 is I105V. A meta-analysis(9) including four studies published through July 2004(15,34,35,42) showed no overall association of GSTP1 with ALL, a finding supported by three subsequent studies(38,41,43).
There are a large number of genes in the CYP family, and the most commonly studied of them is CYP1A1. Results from the seven individual studies that have examined the 6235T > C variant in CYP1A1 with regard to childhood ALL risk are inconsistent, with four showing some evidence of association with the same allele(30,35,36,39), whereas the other three showed no effect(15,40,41). Fewer childhood ALL studies reported on two other CYP1A1 variants: just three reported on the 4889A > G(30,36,40) and two reported on the 4887T > A variant(30,40).
The phase II enzyme NAD(P)H dehydrogenase, quinone 1 (NQO1) detoxifies quinones and acts as an anti-oxidant, reducing the exposure of DNA to ROS. Seven reports of NQO1 and childhood ALL that were published through October 2007(41,44–49) were summarised in a recent meta-analysis(50), which found no childhood ALL association of rs1800566, a loss-of-function variant. Another variant in the NQO1 gene, rs4986998, has been reported in only two studies(44,48), both of which reported no association.
For ABCB1/MDR1, there are four inconsistent reports on the 3435C > T variant in childhood ALL risk(51–54). Although the evidence for this marker is equivocal, one of the published studies found evidence of a significant synergistic effect of MDR1 variants with exposure to insecticides on childhood ALL risk(53).
Immune function
Greaves proposed that ALL, particularly the pre-B cell ALL that usually occurs among children aged 2–5 y, is the result of a rare, abnormal response to non-specific common infections(7,55). As a malignancy of lymphocytes, ALL is likely to be influenced by elements of the immune system's normal function, which is to protect against infectious agents and tumour growth. Upon antigen-specific activation of T lymphocytes, a cascade of events leads to T cell proliferation, cytokine secretion and recruitment of other immune cells(56). In addition, through clonal selection of specific B cells, the immune system mounts adaptive responses to infection. A potential source of a second ‘hit’(7) may come from an abnormal T-cell response to infection involving inflammation and production of cytokines that result in a transient suppression of haematopoiesis; under this condition, as postulated by Greaves(57), the pre-leukaemic clone may expand due to its survival and proliferative advantages, and the rapid expansion of the pre-leukaemic clone may provide the opportunity for acquiring additional mutations leading to the development of ALL. As immune response plays a central role in this proposed mechanism, it is important to study genetic variation in genes involved in the immunological pathways, including genes that participate in cytokine production.
The human leukocyte antigen (HLA) is an obvious target, due to its critical role in the presentation of specific antigenic peptides derived from infectious agents. However, the major histocompatibility complex is complicated and traditional methods of assessing HLA types are time-consuming and costly, and therefore prohibitive for large epidemiological studies. Newer, more cost-effective genotyping methods have been developed to assess the HLA; however, these have not yet been widely deployed in epidemiological studies. Nevertheless, three HLA loci have been examined using the traditional methods. HLA-DRB4 and HLA-DQB1 were found to be significantly associated with risk in individual studies(58,59). Another locus, the HLA-DPB1 locus, was found to be associated with childhood ALL in two UK studies(60,61).
Summary
In summary, of the ∼30 000 genes in the human genome, fewer than 0.1% have been examined for childhood ALL risk. Even fewer of these genes have been studied for childhood AML risk. Only a handful of these genes have been examined in more than four reports. Even within the candidate pathways noted here, there remain a number of unstudied or under-studied high-priority candidates. Other candidate pathways with strong biological plausibility, including DNA repair mechanisms and immune function, have had very little coverage in the literature.
Furthermore, the coverage of the genes themselves has been incomplete. Often, just one or two single nucleotide polymorphisms (SNPs, single base changes) have been assessed within each gene. There is a great amount of heterogeneity within genes, and this is rarely, if ever, captured with the assessment of a single polymorphism. However, there are millions of known common SNPs (>5% frequency) within the human genome, and it is infeasible to assess these millions of polymorphisms within an epidemiological study to capture all the genetic variation within a gene or gene region. Genetic recombination events that occur during meiosis over many generations create regions of the genome that are in linkage disequilibrium (LD) with one another – that is, they are associated with one another. Thus, for variants at loci that are in LD with one another, one can examine a small number of these loci and use this information to estimate with a high degree of certainty the value at the others. This provides an elegant way of reducing the number of variants one would need to characterise a specific genetic region. The amount of LD between neighbouring markers can be estimated for different ethnic groups using data from large sequencing efforts such as the International HapMap Project (http://hapmap.org) and the SNP500CANCER effort (http://snp500cancer.nci.nih.gov). Two commonly used approaches are tagSNPs(62) and haplotype-tagging SNPs (htSNPs)(63). Both these approaches are implemented in several software programs, including HaploView (www.broad.mit.edu/mpg/haploview/).
It should be noted that, in addition to SNPs, which are single nucleotide alterations in the genome, there are other types of genetic variations, including microsatellites (repeated sequences of nucleotides of varying length), insertions (nucleotide sequences that have been inserted into the genome), deletions (nucleotide sequences that have been deleted from the genome) and copy number variants (varying numbers of copies of certain genes or regions). With the exception of the GSTs, for which the commonly studied variants are deletions, the bulk of current efforts in childhood leukaemia have focused on SNPs.
NCCLS EXPERIENCE
Study design
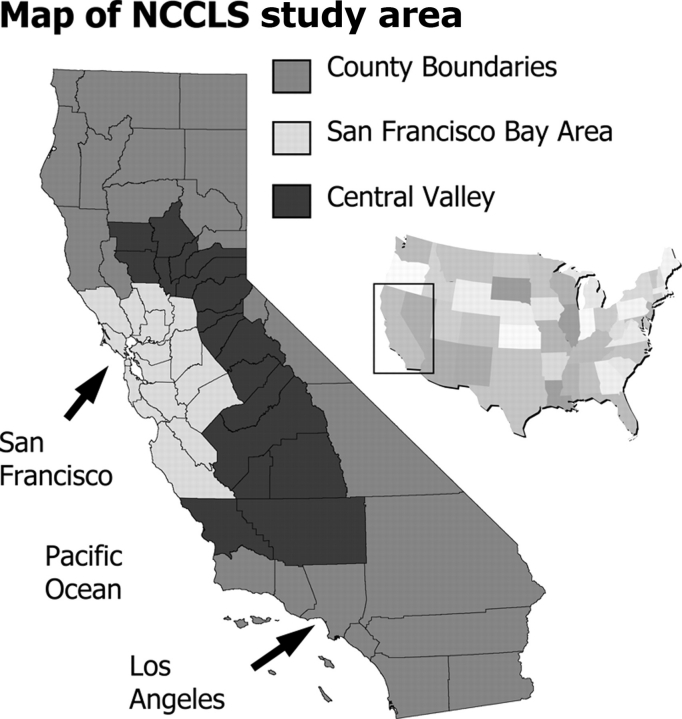
The NCCLS is a population-based case–control study ongoing since 1995. Incident cases under 15 y of age are rapidly ascertained (within 72 h) through eight collaborating paediatric treatment facilities serving the 35-county study area (Figure 3). Comparison of ascertained cases with population-based California Cancer Registry indicates that case ascertainment is 93.0–95.6% complete. Population-based control subjects are selected from the California statewide birth registry, individually matched to the index case on child's date of birth, sex, Hispanic status and maternal race (using a control:case ratio of 1:1 for early cases, later increased to 2:1). By the conclusion of subject accrual in March 2009, the NCCLS would have interviewed 1000 eligible, consenting cases (including 830 ALL 160 AML), and 1300 consented and enrolled controls. Of interest, 42% of this California study population is of Hispanic ethnicity.
Figure 3.
Study area for the NCCLS.
Biospecimen collection
The NCCLS collects a variety of biospecimens from cases and controls. The genetics efforts focus mainly on DNA extracted from buccal cell and archived newborn blood (ANB) specimens. From 1995 to March 2007, buccal cells were collected from child cases and controls using cytobrushes (three cytobrushes each) at the time of personal interview by trained interviewers. The brush head was immediately placed into a lysis/preservative solution and shipped at room temperature by overnight mail to the study laboratory. Buccal cell specimens have been successfully obtained from 95% of interviewed cases and controls. In April 2007, the cytobrush collection was replaced with Oragene saliva collection kits (DNA Genotek, Ottawa, Canada), due to the enhanced quality and quantity of DNA available from these kits as well as their ease of use and ability to stabilise DNA at room temperature. The Oragene sponge accessory kits were used to collect saliva from individuals who could not expectorate. ANB specimens, collected at birth on paper cards from children born in California (about 93% of cases and all controls), were requested from the California Department of Public Health, Genetic Disease Branch (CDPH-GDB, a state agency). Since the 1960s, four to six bloodspots, each containing ∼60 µl of blood, have been collected from each child born in the state of California on a single card and archived at −20 °C by the CDPH-GDB. The NCCLS has received a single ANB spot for 90% of the California-born cases and controls. Diagnostic pre-treatment bone marrow and/or peripheral blood specimens have been also collected from 86% of enrolled cases. However, due to the elevated number of transformed blasts likely to be present in peripheral blood specimens of cases, these do not serve as reliable source of germline DNA.
Large-scale genotyping
In 2005, with support from Children with Leukaemia Foundation, a UK charity, the authors embarked on an effort to genotype 464 childhood leukaemia cases (including 385 ALL cases) and 464 controls for 1536 SNPs in 183 candidate genes across five major pathways: immune function, folate metabolism, xenobiotic metabolism and transport, DNA repair and dietary/anti-oxidant pathways. They have selected htSNPs in the chosen genes, with special emphasis on haplotype structures in Hispanics, and included 95 ancestry informative markers to estimate genetic ancestry in study participants using structured association methods(64,65). Using GoldenGate technology from Illumina (San Diego, CA), these markers were genotyped in late 2007, and data from this effort became available in February 2008. Statistical analyses are ongoing.
CHALLENGES
Statistical power and sample size issues
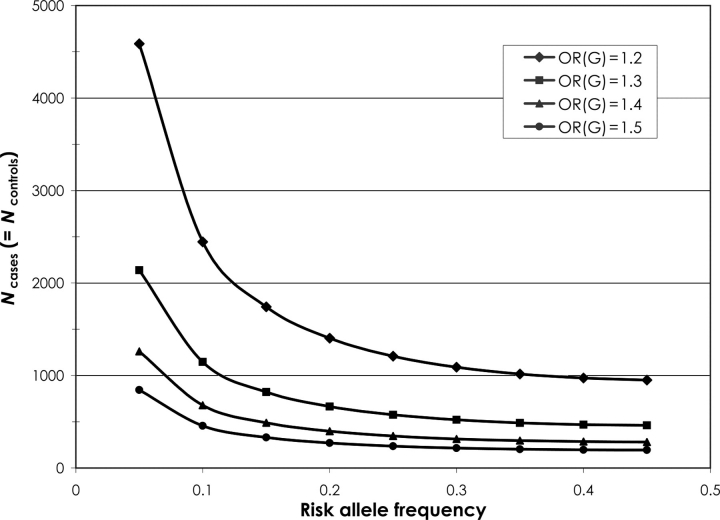
One major challenge facing genetic susceptibility research is the fact that the expected effect sizes for common, low-penetrance genetic markers are small, usually in the range of OR = 1.2–1.5. Detection of such effects for markers whose allele frequencies are 5–10% requires large sample sizes. As shown in Figure 4, to observe an association with a marker whose minor allele frequency is 40% and effect size is 1.5, 197 cases and 197 controls would be required. This number is reasonable for many epidemiological studies of childhood leukaemia. However, at the other end of the spectrum, for a less common marker (10% frequency) and a weaker effect size (OR = 1.2), detection would require 2445 cases and 2445 controls; achieving such a sample size would undoubtedly be a challenge for most epidemiological studies of childhood leukaemia. Indeed, most studies published to date present data for 100–300 cases. It should be noted that estimated sample sizes in Figure 4 do not account for multiple hypothesis testing or potential gene–environment interactions. Doing so would increase sample size requirements further.
Figure 4.
Required sample size to detect genes' main effects (power = 0.80, α = 0.05, log-additive inheritance).
Several factors impose limitations on the sample size of genetic susceptibility studies of childhood leukaemia. First among these is the low incidence of disease: in the USA, the incidence rate is just 4.5 per 100 000 person-years(66), a figure comparable to those in other countries. Second, there is substantial disease heterogeneity: not only are there two major immunophenotypes (ALL and AML) but there are also a number of subtypes defined by the affected precursor cell type (e.g. B-cell ALL), cytogenetic factors (e.g. TEL–AML1, hyperdiploidy), age of onset (e.g. infant leukaemia, 0–12 months) and combinations thereof (common B-cell ALL (c-ALL), diagnosed between ages 2–5 and expressing surface antigens CD10 and CD19). It is possible that the aetiologies of each of these subtypes are distinct, and thus it may be necessary to look for risk factors specifically for these smaller subgroups. A third consideration is the availability of high-quality DNA samples. The earliest epidemiological studies may not have set out to collect DNA, and even if they did, the evolution of technology means that the implemented specimen collection and/or DNA isolation methods may not yield suitable DNA for modern genotyping methods.
Interactions
The modest effect sizes expected for common, low-penetrance markers are consistent with the hypothesised multi-factorial nature of childhood leukaemia(57), which strongly implicates the effects of gene–environment interactions on childhood leukaemia risk. The term ‘gene–environment interaction’ refers here to interactions of genes with any non-genetic exposures, including environmental factors (e.g. chemicals, infections, etc.) as well as behavioural/lifestyle factors (tobacco smoking, diet, etc.). It is impossible to clarify the role of genetic susceptibility in childhood leukaemia risk without also clarifying these interactions. Therefore, it is very important that epidemiological studies continue to collect high-quality environmental, lifestyle and other exposure data, in addition to genetic data. It should be noted that widespread testing for combined effects of genes and environmental factors, as well as gene–gene interactions, dramatically increases the number of tests performed, thus compounding the multiple testing problem. A plan for testing a limited number of interactions should be developed a priori, and replication of interactions detected in this way should be planned. Also, as noted, sample size requirements for detection of gene–environment interactions are even greater than those illustrated in Figure 4.
Maternal and paternal genetics
To date, most published studies have focused on the genetic susceptibility of the child. Many of these studies utilise a case–control design. In genetic epidemiology, family-based methods that take advantage of known transmission of genetic factors from parents to children can be used as an alternatives to the case–control approach. Most prominent among these is the transmission disequilibrium test (TDT)(67), which utilises genotyping data from the case and the cases' biological parents. This approach has the advantage that no genetic information is needed for controls, which can be useful if control DNAs are not available. A further benefit is that, relative to traditional case–control analyses, the TDT is less susceptible to confounding by genetic ancestry (population stratification) because each case is effectively compared with a matched hypothetical individual harbouring the two alleles not transmitted by the parents (the case's anti-self)(68). Accordingly, the hypothetical ‘control’ is ethnically matched to the case. Thus, it can be useful to have genotyping data for the cases' parents.
It should be noted that there are methods to address population stratification within a traditional case–control study. For example, one can assess a panel of SNPs known as ancestry informative markers and estimate the relative contributions of ancestral populations to an individual's genetic makeup(69,70). Such approaches do not require collection and genotyping of parental DNA.
Other studies have examined the joint effects of the child's genetic susceptibility with the mothers' environmental exposures. These approaches pay heed to the importance of the child's own susceptibility as well as the importance of this susceptibility in the context of the in utero environment. However, the in utero environment is also influenced by the mother's genetics. For example, the foetus' exposure to folate is influenced by not only the mother's folate intake but also by the mother's own folate metabolism, which may be modulated by genetic factors. Thus, when considering the impact of genetic susceptibility on childhood leukaemia risk, it is important to consider susceptibility conferred through maternal genotypes, which may modulate in utero exposures to the developing child. Methods have recently been developed that overcome the correlation between maternal and child's genotypes and permit examination of the effects of maternal genes on the child's disease risk(71,72). Future studies must strive to account for joint effects of maternal environmental exposures, child's genes and mother's genes.
Other challenges
As for any risk factor in epidemiology, one of the chief causal criteria is consistency of results. To examine the cumulative evidence of a particular association between a specific gene or gene region and childhood leukaemia, one must consider whether the finding replicates across multiple studies and study populations. Individual studies are often too under-powered to detect small effect sizes of variants that are not very common (∼5–10% frequency). Furthermore, with the number of genetic hypotheses being tested in any given study, it is unlikely that a single study will also have sufficient power to overcome the multiple comparisons issue. Thus, replication of findings is absolutely critical to understanding the role of genes in childhood leukaemia risk. In this regard, it is important that all available evidence be included in the cumulative assessment of a given association. Therefore, concerted efforts must be made to address publication bias, which occurs when positive findings are published but null findings are not(73).
FUTURE DIRECTIONS
Childhood Leukaemia International Consortium
In support of the need for larger sample sizes and the importance of replication studies for genetic, behavioural and environmental exposure findings, a consortium of epidemiological studies of childhood leukaemia has been developing since 2006. To date, the Childhood Leukaemia International Consortium (CLIC, www.clic.berkeley.edu) has been successful in recruiting over 20 investigators representing 14 epidemiological investigations and over 9000 cases of childhood leukaemia in 10 countries. CLIC is now an active consortium with funding provided by the U.S. National Cancer Institute and the Children with Leukaemia Foundation, a UK charity. CLIC members met in formal meetings in 2007 and 2008. Several working groups have been formed, i.e. pathology review, genotyping, environmental exposures, infant leukaemia, AML and birth characteristics (i.e. birthweight and foetal growth)(74,75). The first pooled analyses to be conducted will evaluate maternal alcohol use, maternal folate intake and MTHFR genes and risk of ALL.
The synergies afforded by CLIC will help overcome a number of the challenges noted earlier. By pooling of data across multiple studies, sample size concerns can be addressed, particularly for rarer subtypes of childhood leukaemia or for gene–environment interactions. There is also the opportunity for replication of findings, through joint or coordinated analysis and publication. This may be an effective way to address publication bias: if one study shows positive results but another is negative, joint publication of results can ensure that all the evidence is made available. Even if the overall conclusion is null, the larger sample size afforded by data from multiple studies can lend more credence to the findings, and would therefore be more appealing to a journal. Finally, CLIC may offer the opportunity for new collaborative research among complementary studies.
Genome-wide association studies
One area for potential new collaborative research within CLIC is a genome-wide association study (GWAS). Studies to date have examined a few genes, or selected genes within a candidate pathway, which typically entails genotyping of 1–20 markers in selected regions of the genome. While these candidates are considered high-priority regions due to their positions at junctures in critical biological processes, it is very likely that there are genes in novel, unexplored regions that have particular relevance for childhood leukaemia risk. A GWAS could address this shortcoming. A GWAS entails genotyping of hundreds of thousands of markers across the genome, allowing exploration of regions beyond current candidates. Indeed, in prostate cancer, GWAS scans in multiple populations have consistently turned up a novel region of association on chromosome 8, at 8q24(76). It is entirely possible that such novel regions may turn up in a GWAS for childhood leukaemia.
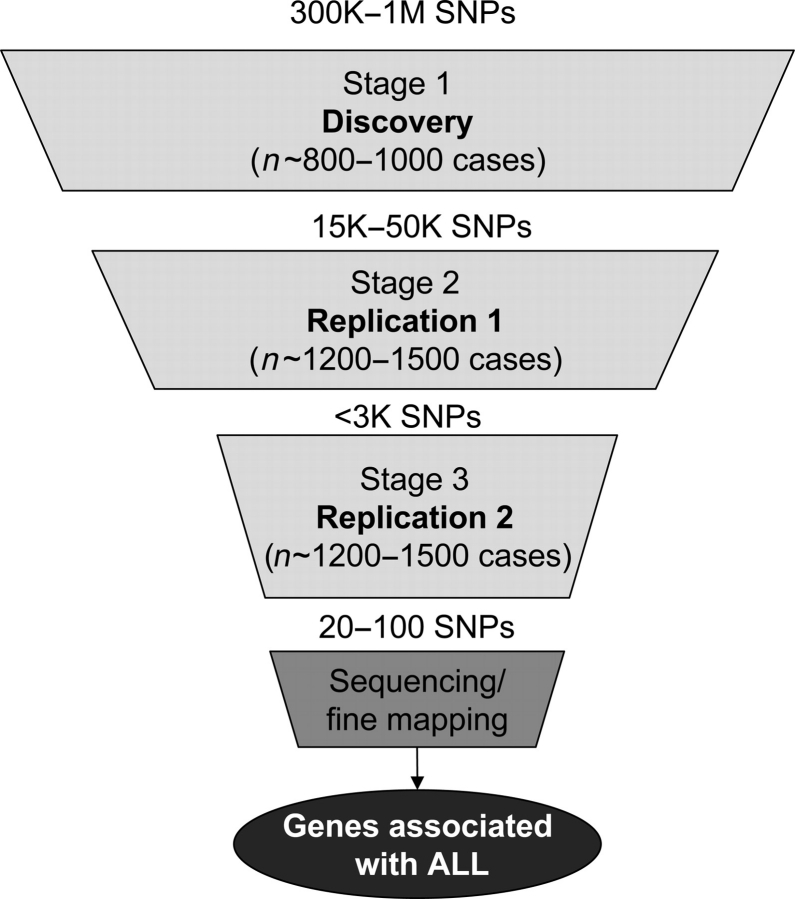
Conducting a GWAS is a major scientific endeavour. The DNA samples must be of high quality, and often amplified DNA cannot be used. The cost per specimen is high, and there is a tremendous multiple comparisons problem that must be considered. As such, tiered study designs such as that depicted in Figure 5 have been proposed to address these and other concerns and help ensure reliable results(77). The numbers presented in Figure 5 represent a realistic plan for childhood ALL, wherein three separate stages of genotyping would occur. A GWAS in the Stage 1 samples would be followed by genotyping within the Stage 2 samples of a smaller number of SNPs representing the positive results from Stage 1; positive results from Stage 2 would be genotyped in the Stage 3 samples. Results that survive replication across all three stages would be considered high-priority regions, and could be followed up with fine marker mapping, sequencing and laboratory studies to identify potential causal regions. CLIC is an ideal setting in which to conduct a GWAS, given that the large sample sizes required for this staged approach preclude any individual study from conducting a convincing GWAS on its own.
Figure 5.
A tiered replication approach for GWAS in childhood ALL.
CONCLUSION
The evidence base for genetic susceptibility to childhood leukaemia is small, but growing. To date, based on single-marker studies, a few genes appear interesting, including MTHFR and the GST family of genes. A number of studies with high-quality DNA specimens are now beginning to publish results from high-priority candidate genes, including those of immune function, xenobiotic metabolism, DNA repair, folate metabolism and oxidative stress. More complete methods of assessing genetic variation, including htSNPs, are available and are being utilised by these studies. In addition, the advent of GWAS scans holds much promise for identifying novel susceptibility regions for childhood leukaemia. Also the formation of CLIC offers a unique opportunity to advance the elucidation of risk factors—both genetic and non-genetic—for childhood leukaemia. The next 5 y will see enormous growth in the knowledge of the genetic epidemiology of childhood leukaemia.
FUNDING
This work was supported by grants from the United States National Institute of Environmental Health Sciences (grants R01ES009137 and P42ES04715) and the Children with Leukaemia Foundation.
ACKNOWLEDGEMENTS
The authors wish to acknowledge Drs Joseph Wiemels, Lisa Barcellos, Catherine Metayer, Jeffrey Chang, Ghislaine Scelo, Kevin Urayama, and other collaborators in the NCCLS. Thanks also to Drs Muin Khoury and Stephen Chanock, advisors to the NCCLS, whose guidance has been critical in developing the genetics research both within the study and beyond.
REFERENCES
- 1.Greaves M. F., Alexander F. An infectious etiology for common acute lymphoblastic leukemia in childhood? Leukemia. 1993;7:349–360. [PubMed] [Google Scholar]
- 2.Buffler P. A., Kwan M. L., Reynolds P., Urayama K. Y. Environmental and genetic risk factors for childhood leukemia: appraising the evidence. Cancer Invest. 2005;23:60–75. [PubMed] [Google Scholar]
- 3.Greaves M. F., Maia A. T., Wiemels J. L., Ford A. M. Leukemia in twins: lessons in natural history. Blood. 2003;102:2321–2333. doi: 10.1182/blood-2002-12-3817. [DOI] [PubMed] [Google Scholar]
- 4.Hunter D. J. Gene-environment interactions in human diseases. Nat. Rev. Genet. 2005;6:287–298. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 5.Mori H., Colman S. M., Xiao Z., Ford A. M., Healy L. E., Donaldson C., Hows J. M., Navarrete C., Greaves M. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc. Natl. Acad. Sci. USA. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreasson P., Schwaller J., Anastasiadou E., Aster J., Gilliland D. G. The expression of ETV6/CBFA2 (TEL/AML1) is not sufficient for the transformation of hematopoietic cell lines in vitro or the induction of hematologic disease in vivo. Cancer Genet. Cytogenet. 2001;130:93–104. doi: 10.1016/s0165-4608(01)00518-0. [DOI] [PubMed] [Google Scholar]
- 7.Greaves M. Aetiology of acute leukaemia. Lancet. 1997;349:344–349. doi: 10.1016/s0140-6736(96)09412-3. [DOI] [PubMed] [Google Scholar]
- 8.Pereira T. V., Rudnicki M., Pereira A. C., Pombo-de-Oliveira M. S., Franco R. F. 5,10-Methylenetetrahydrofolate reductase polymorphisms and acute lymphoblastic leukemia risk: a meta-analysis. Cancer Epidemiol. Biomarkers Prev. 2006;15:1956–1963. doi: 10.1158/1055-9965.EPI-06-0334. [DOI] [PubMed] [Google Scholar]
- 9.Ye Z., Song H. Glutathione s-transferase polymorphisms (GSTM1, GSTP1 and GSTT1) and the risk of acute leukaemia: a systematic review and meta-analysis. Eur. J. Cancer. 2005;41:980–989. doi: 10.1016/j.ejca.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Xiao Z., Greaves M. F., Buffler P., Smith M. T., Segal M. R., Dicks B. M., Wiencke J. K., Wiemels J. L. Molecular characterization of genomic AML1-ETO fusions in childhood leukemia. Leukemia. 2001;15:1906–1913. doi: 10.1038/sj.leu.2402318. [DOI] [PubMed] [Google Scholar]
- 11.Blount B. C., Mack M. M., Wehr C. M., MacGregor J. T., Hiatt R. A., Wang G., Wickramasinghe S. N., Everson R. B., Ames B. N. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. USA. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson J. R., Gerald P. F., Willoughby M. L., Armstrong B. K. Maternal folate supplementation in pregnancy and protection against acute lymphoblastic leukaemia in childhood: a case-control study. Lancet. 2001;358:1935–1940. doi: 10.1016/S0140-6736(01)06959-8. [DOI] [PubMed] [Google Scholar]
- 13.Pui C. H., Relling M. V., Downing J. R. Acute lymphoblastic leukemia. N. Engl. J. Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 14.Franco R. F., Simoes B. P., Tone L. G., Gabellini S. M., Zago M. A., Falcao R. P. The methylenetetrahydrofolate reductase C677T gene polymorphism decreases the risk of childhood acute lymphocytic leukaemia. Br. J. Haematol. 2001;115:616–618. doi: 10.1046/j.1365-2141.2001.03140.x. [DOI] [PubMed] [Google Scholar]
- 15.Balta G., Yuksek N., Ozyurek E., Ertem U., Hicsonmez G., Altay C., Gurgey A. Characterization of MTHFR, GSTM1, GSTT1, GSTP1, and CYP1A1 genotypes in childhood acute leukemia. Am. J. Hematol. 2003;73:154–160. doi: 10.1002/ajh.10339. [DOI] [PubMed] [Google Scholar]
- 16.Chiusolo P., et al. Methylenetetrahydrofolate reductase genotypes do not play a role in acute lymphoblastic leukemia pathogenesis in the Italian population. Haematologica. 2004;89:139–144. [PubMed] [Google Scholar]
- 17.Krajinovic M., Lamothe S., Labuda D., Lemieux-Blanchard E., Theoret Y., Moghrabi A., Sinnett D. Role of MTHFR genetic polymorphisms in the susceptibility to childhood acute lymphoblastic leukemia. Blood. 2004;103:252–257. doi: 10.1182/blood-2003-06-1794. [DOI] [PubMed] [Google Scholar]
- 18.Chatzidakis K., Goulas A., Athanassiadou-Piperopoulou F., Fidani L., Koliouskas D., Mirtsou V. Methylenetetrahydrofolate reductase C677T polymorphism: association with risk for childhood acute lymphoblastic leukemia and response during the initial phase of chemotherapy in Greek patients. Pediatr. Blood Cancer. 2006;47:147–151. doi: 10.1002/pbc.20574. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira E., Alves S., Quental S., Ferreira F., Norton L., Costa V., Amorim A., Prata M. J. The MTHFR C677T and A1298C polymorphisms and susceptibility to childhood acute lymphoblastic leukemia in Portugal. J. Pediatr. Hematol. Oncol. 2005;27:425–429. doi: 10.1097/01.mph.0000177513.81465.94. [DOI] [PubMed] [Google Scholar]
- 20.Schnakenberg E., Mehles A., Cario G., Rehe K., Seidemann K., Schlegelberger B., Elsner H. A., Welte K. H., Schrappe M., Stanulla M. Polymorphisms of methylenetetrahydrofolate reductase (MTHFR) and susceptibility to pediatric acute lymphoblastic leukemia in a German study population. BMC Med. Genet. 2005;6:23. doi: 10.1186/1471-2350-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thirumaran R. K., Gast A., Flohr T., Burwinkel B., Bartram C., Hemminki K., Kumar R. MTHFR genetic polymorphisms and susceptibility to childhood acute lymphoblastic leukemia. Blood. 2005;106:2590–2591. doi: 10.1182/blood-2005-04-1719. author reply 2591–2592. [DOI] [PubMed] [Google Scholar]
- 22.Zanrosso C. W., Hatagima A., Emerenciano M., Ramos F., Figueiredo A., Felix T. M., Segal S. L., Guigliani R., Muniz M. T., Pombo-de-Oliveira M. S. The role of methylenetetrahydrofolate reductase in acute lymphoblastic leukemia in a Brazilian mixed population. Leuk. Res. 2006;30:477–481. doi: 10.1016/j.leukres.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Giovannetti E., Ugrasena D. G., Supriyadi E., Vroling L., Azzarello A., de Lange D., Peters G. J., Veerman A. J., Cloos J. Methylenetetrahydrofolate reductase (MTHFR) C677T and thymidylate synthase promoter (TSER) polymorphisms in Indonesian children with and without leukemia. Leuk. Res. 2007;32:19–24. doi: 10.1016/j.leukres.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 24.Reddy H., Jamil K. Polymorphisms in the MTHFR gene and their possible association with susceptibility to childhood acute lymphocytic leukemia in an Indian population. Leuk. Lymphoma. 2006;47:1333–1339. doi: 10.1080/10428190600562773. [DOI] [PubMed] [Google Scholar]
- 25.Kim N. K., Chong S. Y., Jang M. J., Hong S. H., Kim H. S., Cho E. K., Lee J. A., Ahn M. J., Kim C. S., Oh D. Association of the methylenetetrahydrofolate reductase polymorphism in Korean patients with childhood acute lymphoblastic leukemia. Anticancer Res. 2006;26:2879–2881. [PubMed] [Google Scholar]
- 26.Petra B. G., Janez J., Vita D. Gene-gene interactions in the folate metabolic pathway influence the risk for acute lymphoblastic leukemia in children. Leuk. Lymphoma. 2007;48:786–792. doi: 10.1080/10428190601187711. [DOI] [PubMed] [Google Scholar]
- 27.Strange R. C., Fryer A. A. The glutathione S-transferases: influence of polymorphism on cancer susceptibility. IARC Sci. Publ. 1999;148:231–249. [PubMed] [Google Scholar]
- 28.Brockmoller J., Cascorbi I., Henning S., Meisel C., Roots I. Molecular genetics of cancer susceptibility. Pharmacology. 2000;61:212–227. doi: 10.1159/000028403. [DOI] [PubMed] [Google Scholar]
- 29.Coles B. F., Kadlubar F. F. Detoxification of electrophilic compounds by glutathione S-transferase catalysis: determinants of individual response to chemical carcinogens and chemotherapeutic drugs? Biofactors. 2003;17:115–130. doi: 10.1002/biof.5520170112. [DOI] [PubMed] [Google Scholar]
- 30.Krajinovic M., Labuda D., Richer C., Karimi S., Sinnett D. Susceptibility to childhood acute lymphoblastic leukemia: influence of CYP1A1, CYP2D6, GSTM1, and GSTT1 genetic polymorphisms. Blood. 1999;93:1496–1501. [PubMed] [Google Scholar]
- 31.Saadat I., Saadat M. The glutathione S-transferase mu polymorphism and susceptibility to acute lymphocytic leukemia. Cancer Lett. 2000;158:43–45. doi: 10.1016/s0304-3835(00)00504-8. [DOI] [PubMed] [Google Scholar]
- 32.Alves S., Amorim A., Ferreira F., Norton L., Prata M. J. The GSTM1 and GSTT1 genetic polymorphisms and susceptibility to acute lymphoblastic leukemia in children from north Portugal. Leukemia. 2002;16:1565–1567. doi: 10.1038/sj.leu.2402543. [DOI] [PubMed] [Google Scholar]
- 33.Davies S. M., Bhatia S., Ross J. A., Kiffmeyer W. R., Gaynon P. S., Radloff G. A., Robison L. L., Perentesis J. P. Glutathione S-transferase genotypes, genetic susceptibility, and outcome of therapy in childhood acute lymphoblastic leukemia. Blood. 2002;100:67–71. doi: 10.1182/blood.v100.1.67. [DOI] [PubMed] [Google Scholar]
- 34.Barnette P., et al. High-throughput detection of glutathione S-transferase polymorphic alleles in a pediatric cancer population. Cancer Epidemiol. Biomarkers Prev. 2004;13:304–313. doi: 10.1158/1055-9965.epi-03-0178. [DOI] [PubMed] [Google Scholar]
- 35.Canalle R., Burim R. V., Tone L. G., Takahashi C. S. Genetic polymorphisms and susceptibility to childhood acute lymphoblastic leukemia. Environ. Mol. Mutagen. 2004;43:100–109. doi: 10.1002/em.20003. [DOI] [PubMed] [Google Scholar]
- 36.Joseph T., Kusumakumary P., Chacko P., Abraham A., Radhakrishna Pillai M. Genetic polymorphism of CYP1A1, CYP2D6, GSTM1 and GSTT1 and susceptibility to acute lymphoblastic leukaemia in Indian children. Pediatr. Blood Cancer. 2004;43:560–567. doi: 10.1002/pbc.20074. [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Zhang L., Feng J., Wang H., Zhu S., Hu Y., Li Y. Genetic polymorphisms analysis of glutathione S-transferase M1 and T1 in children with acute lymphoblastic leukemia. J. Huazhong Univ. Sci. Technolog. Med. Sci. 2004;24:243–244. doi: 10.1007/BF02832001. [DOI] [PubMed] [Google Scholar]
- 38.Zielinska E., Zubowska M., Bodalski J. Polymorphism within the glutathione S-transferase P1 gene is associated with increased susceptibility to childhood malignant diseases. Pediatr. Blood Cancer. 2004;43:552–529. doi: 10.1002/pbc.20059. [DOI] [PubMed] [Google Scholar]
- 39.Aydin-Sayitoglu M., Hatirnaz O., Erensoy N., Ozbek U. Role of CYP2D6, CYP1A1, CYP2E1, GSTT1, and GSTM1 genes in the susceptibility to acute leukemias. Am. J. Hematol. 2006;81:162–170. doi: 10.1002/ajh.20434. [DOI] [PubMed] [Google Scholar]
- 40.Pakakasama S., Mukda E., Sasanakul W., Kadegasem P., Udomsubpayakul U., Thithapandha A., Hongeng S. Polymorphisms of drug-metabolizing enzymes and risk of childhood acute lymphoblastic leukemia. Am. J. Hematol. 2005;79:202–205. doi: 10.1002/ajh.20404. [DOI] [PubMed] [Google Scholar]
- 41.Clavel J., et al. Childhood leukaemia, polymorphisms of metabolism enzyme genes, and interactions with maternal tobacco, coffee and alcohol consumption during pregnancy. Eur. J. Cancer Prev. 2005;14:531–540. doi: 10.1097/00008469-200512000-00007. [DOI] [PubMed] [Google Scholar]
- 42.Krajinovic M., Labuda D., Sinnett D. Glutathione S-transferase P1 genetic polymorphisms and susceptibility to childhood acute lymphoblastic leukaemia. Pharmacogenetics. 2002;12:655–658. doi: 10.1097/00008571-200211000-00010. [DOI] [PubMed] [Google Scholar]
- 43.Gatedee J., Pakakassama S., Muangman S., Pongstaporn W. Glutathione S-transferase p1 genotypes, genetic susceptibility and outcome of therapy in Thai childhood acute lymphoblastic leukemia. Asian Pac. J. Cancer Prev. 2007;8:294–296. [PubMed] [Google Scholar]
- 44.Wiemels J. L., Pagnamenta A., Taylor G. M., Eden O. B., Alexander F. E., Greaves M. F. A lack of a functional NAD(P)H:quinone oxidoreductase allele is selectively associated with pediatric leukemias that have MLL fusions. United Kingdom Childhood Cancer Study Investigators. Cancer Res. 1999;59:4095–4099. [PubMed] [Google Scholar]
- 45.Krajinovic M., Sinnett H., Richer C., Labuda D., Sinnett D. Role of NQO1, MPO and CYP2E1 genetic polymorphisms in the susceptibility to childhood acute lymphoblastic leukemia. Int. J. Cancer. 2002;97:230–236. doi: 10.1002/ijc.1589. [DOI] [PubMed] [Google Scholar]
- 46.Kracht T., et al. NQO1 C609T polymorphism in distinct entities of pediatric hematologic neoplasms. Haematologica. 2004;89:1492–1497. [PubMed] [Google Scholar]
- 47.Sirma S., et al. NAD(P)H:quinone oxidoreductase 1 null genotype is not associated with pediatric de novo acute leukemia. Pediatr. Blood Cancer. 2004;43:568–570. doi: 10.1002/pbc.20098. [DOI] [PubMed] [Google Scholar]
- 48.Eguchi-Ishimae M., Eguchi M., Ishii E., Knight D., Sadakane Y., Isoyama K., Yabe H., Mizutani S., Greaves M. The association of a distinctive allele of NAD(P)H:quinone oxidoreductase with pediatric acute lymphoblastic leukemias with MLL fusion genes in Japan. Haematologica. 2005;90:1511–1515. [PubMed] [Google Scholar]
- 49.Lanciotti M., et al. Genetic polymorphism of NAD(P)H:quinone oxidoreductase is associated with an increased risk of infant acute lymphoblastic leukemia without MLL gene rearrangements. Leukemia. 2005;19:214–216. doi: 10.1038/sj.leu.2403613. [DOI] [PubMed] [Google Scholar]
- 50.Guha N., Chang J. S., Chokkalingam A. P., Wiemels J. L., Smith M. T., Buffler P. A. NQO1 polymorphisms and de novo childhood leukemia: a HuGE review and meta-analysis. Am. J. Epidemiol. 2008 doi: 10.1093/aje/kwn246. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hattori H., Suminoe A., Wada M., Koga Y., Kohno K., Okamura J., Hara T., Matsuzaki A. Regulatory polymorphisms of multidrug resistance 1 (MDR1) gene are associated with the development of childhood acute lymphoblastic leukemia. Leuk. Res. 2007;31:1633–1640. doi: 10.1016/j.leukres.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 52.Jamroziak K., Mlynarski W., Balcerczak E., Mistygacz M., Trelinska J., Mirowski M., Bodalski J., Robak T. Functional C3435T polymorphism of MDR1 gene: an impact on genetic susceptibility and clinical outcome of childhood acute lymphoblastic leukemia. Eur. J. Haematol. 2004;72:314–321. doi: 10.1111/j.1600-0609.2004.00228.x. [DOI] [PubMed] [Google Scholar]
- 53.Urayama K. Y., Wiencke J. K., Buffler P. A., Chokkalingam A. P., Metayer C., Wiemels J. L. MDR1 gene variants, indoor insecticide exposure, and the risk of childhood acute lymphoblastic leukemia. Cancer Epidemiol. Biomarkers Prev. 2007;16:1172–1177. doi: 10.1158/1055-9965.EPI-07-0007. [DOI] [PubMed] [Google Scholar]
- 54.Semsei A. F., et al. Association of some rare haplotypes and genotype combinations in the MDR1 gene with childhood acute lymphoblastic leukaemia. Leuk. Res. 2008;32:1214–1220. doi: 10.1016/j.leukres.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 55.Greaves M. Molecular genetics, natural history and the demise of childhood leukaemia. Eur. J. Cancer. 1999;35:173–185. doi: 10.1016/s0959-8049(98)00433-x. [DOI] [PubMed] [Google Scholar]
- 56.Martin A. M., Athanasiadis G., Greshock J. D., Fisher J., Lux M. P., Calzone K., Rebbeck T. R., Weber B. L. Population frequencies of single nucleotide polymorphisms (SNPs) in immuno-modulatory genes. Hum. Hered. 2003;55:171–178. doi: 10.1159/000073201. [DOI] [PubMed] [Google Scholar]
- 57.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat. Rev. Cancer. 2006;6:193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 58.Dorak M. T., Owen G., Galbraith I., Henderson N., Webb D., Mills K. I., Darke C., Burnett A. K. Nature of HLA-associated predisposition to childhood acute lymphoblastic leukemia. Leukemia. 1995;9:875–878. [PubMed] [Google Scholar]
- 59.Dearden S. P., et al. Molecular analysis of HLA-DQB1 alleles in childhood common acute lymphoblastic leukaemia. Br. J. Cancer. 1996;73:603–609. doi: 10.1038/bjc.1996.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor G. M., Dearden S., Payne N., Ayres M., Gokhale D. A., Birch J. M., Blair V., Stevens R. F., Will A. M., Eden O. B. Evidence that an HLA-DQA1-DQB1 haplotype influences susceptibility to childhood common acute lymphoblastic leukaemia in boys provides further support for an infection-related aetiology. Br. J. Cancer. 1998;78:561–565. doi: 10.1038/bjc.1998.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor G. M., Dearden S., Ravetto P., Ayres M., Watson P., Hussain A., Greaves M., Alexander F., Eden O. B. Genetic susceptibility to childhood common acute lymphoblastic leukaemia is associated with polymorphic peptide-binding pocket profiles in HLA-DPB1*0201. Hum. Mol. Genet. 2002;11:1585–1597. doi: 10.1093/hmg/11.14.1585. [DOI] [PubMed] [Google Scholar]
- 62.Carlson C. S., Eberle M. A., Rieder M. J., Yi Q., Kruglyak L., Nickerson D. A. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gabriel S. B., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 64.Hoggart C. J., Shriver M. D., Kittles R. A., Clayton D. G., McKeigue P. M. Design and analysis of admixture mapping studies. Am. J. Hum. Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Falush D., Stephens M., Pritchard J. K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.U.S. Cancer Statistics Working Group: United States Cancer Statistics 2002 Incidence and Mortality. 2005 U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute. [Google Scholar]
- 67.Terwilliger J. D., Ott J. A novel polylocus method for linkage analysis using the lod-score or affected sib-pair method. Genet. Epidemiol. 1993;10:477–482. doi: 10.1002/gepi.1370100625. [DOI] [PubMed] [Google Scholar]
- 68.Thomas D. C. New York, NY: Oxford University Press; 2004. Statistical Methods in Genetic Epidemiology. [Google Scholar]
- 69.Collins-Schramm H. E., Chima B., Morii T., Wah K., Figueroa Y., Criswell L. A., Hanson R. L., Knowler W. C., Silva G., Belmont J. W., Seldin M. F. Mexican American ancestry-informative markers: examination of population structure and marker characteristics in European Americans, Mexican Americans, Amerindians and Asians. Hum. Genet. 2004;114:263–271. doi: 10.1007/s00439-003-1058-6. [DOI] [PubMed] [Google Scholar]
- 70.Yang N., Li H., Criswell L. A., Gregersen P. K., Alarcon-Riquelme M. E., Kittles R., Shigeta R., Silva G., Patel P. I., Belmont J. W., Seldin M. F. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum. Genet. 2005;118:382–392. doi: 10.1007/s00439-005-0012-1. [DOI] [PubMed] [Google Scholar]
- 71.Groves F., Auvinen A., Hakulinen T. Haemophilus influenzae type b vaccination and risk of childhood leukemia in a vaccine trial in Finland. Ann. Epidemiol. 2000;10:474. doi: 10.1016/s1047-2797(00)00110-1. [DOI] [PubMed] [Google Scholar]
- 72.Mellemkjaer L., Alexander F., Olsen J. H. Cancer among children of parents with autoimmune diseases. Br. J. Cancer. 2000;82:1353–1357. doi: 10.1054/bjoc.1999.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dickersin K. The existence of publication bias and risk factors for its occurrence. JAMA. 1990;263:1385–1389. [PubMed] [Google Scholar]
- 74.Milne E., Laurvick C. L., Blair E., Bower C., de Klerk N. Fetal growth and acute childhood leukemia: looking beyond birth weight. Am. J. Epidemiol. 2007;166:151–159. doi: 10.1093/aje/kwm065. [DOI] [PubMed] [Google Scholar]
- 75.Tower R. L., Spector L. G. The epidemiology of childhood leukemia with a focus on birth weight and diet. Crit. Rev. Clin. Lab. Sci. 2007;44:203–242. doi: 10.1080/10408360601147536. [DOI] [PubMed] [Google Scholar]
- 76.Witte J. S. Multiple prostate cancer risk variants on 8q24. Nat. Genet. 2007;39:579–580. doi: 10.1038/ng0507-579. [DOI] [PubMed] [Google Scholar]
- 77.Kraft P., Cox D. G. Study designs for genome-wide association studies. Adv. Genet. 2008;60:465–504. doi: 10.1016/S0065-2660(07)00417-8. [DOI] [PubMed] [Google Scholar]