Abstract
In a voltage-dependent sodium channel, the activation of voltage-sensors upon depolarization leads to the opening of pore gates. To elucidate the principles underlying this conformational coupling, we investigated a putative gating interface in domain III of the sodium channel using voltage-clamp fluorimetry and tryptophan-scanning mutagenesis. Most mutations have similar energetic effects on voltage-sensor activation and pore opening. However, several mutations stabilized the activated voltage-sensor while concurrently destabilizing the open pore. Mapping of these mutations onto a homology model of the sodium channel showed that most localize to hinge regions of the gating interface. Our analysis shows that these residues are involved in energetic coupling of the voltage-sensor and the pore when both are in the resting or activated conformations, supporting the notion that electromechanical coupling in a voltage-dependent ion channel involves the movement of rigid segments connected by elastic hinges.
Keywords: tryptophan mutagenesis, fluorescence, electrophysiology, voltage-sensor, sodium channels, gating, cooperativity
INTRODUCTION
Voltage-gated sodium channels respond to membrane depolarization by undergoing a series of conformational changes that culminate in channel opening and subsequent inactivation. Ionic fluxes through these channels underlie electrical signaling in many physiological processes particularly in the nervous system and the cardiovascular system. The voltage-dependent sodium channel shares a common architecture with other members of the voltage-gated ion channel family and comprises a central ion-conducting pore surrounded by four voltage-sensing domains1–3. The “outward” movement of charged residues in the voltage-sensors in response to a depolarization triggers a conformational change that is transmitted to the pore gates4–10. The structural mechanisms underlying conformational coupling of the voltage-sensor and pore gates is one of the central questions in ion channel biology11.
The principal subunit of the voltage-gated sodium channel is a large 260 kDa polypeptide and has four homologous domains (DI to DIV) which together form a functional channel12 (Figure 1a). Each domain of the sodium channel is equivalent to a single subunit of the voltage-dependent potassium channel. The S4 transmembrane segments in the sodium channel are highly conserved across the voltage-gated ion channel family and along with the S1–S3 segments, constitute the voltage-sensing domain of the channel 13 (Figure 1b). The S5 and S6 helices of the four domains together form the central pore of the channel. The high-resolution structures of the eukaryotic potassium channels, presumably representing the open conformation of the channel, reveal that the voltage-sensing domains make limited physical contacts with the pore 14,15. Studies in the Shaker potassium channel indicate that at least two putative pathways mediate the transmission of “information” from the voltage-sensor to the central pore 16–20. Scanning mutagenesis and domain swap experiments combined with detailed electrophysiological studies have highlighted the importance of the S4, the S4–S5 linker and the intracellular ends of the S5 and S6 segments 17,20,21. This pathway likely couples the voltage-sensor and the pore within the same subunit or domain. Tryptophan scanning along with statistical coupling analysis has uncovered another interface constituted in part by the S1 segment, which may be involved in coupling the voltage-sensing domain of one subunit to the pore of the neighboring subunit 18,19. These studies are beginning to unravel the molecular mechanisms underlying the propagation of structural change from the voltage-sensor to the pore of a voltage-dependent ion channel.
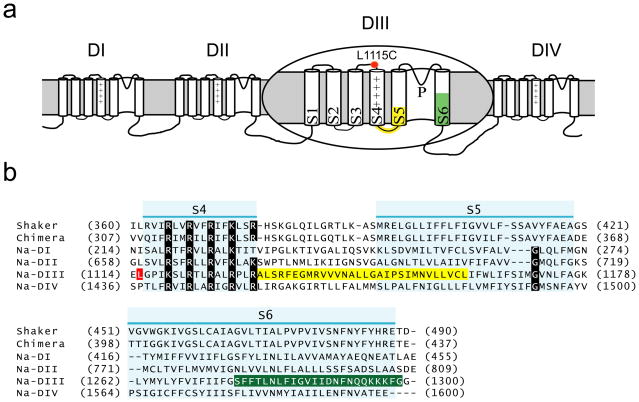
Figure 1. Membrane topology of a sodium channel and a sequence comparison of the skeletal muscle sodium channel with the Shaker and Kv 1.2/2.1 chimeric potassium channel.
(a) Domain III of the sodium channel is enlarged for clarity. The approximate location of the fluorescent probe at the L1115C position is marked by a red symbol. The mutated regions in the S4–S5 linker and N-terminus of the S5 are highlighted in yellow and those in the S6 segment are highlighted in green.
(b) Sequence alignment of the individual sodium channel domains with the two potassium channels. Only the region from the start of the S4 to the end of S6 without the extracellular pore loops are shown. The S4, S5, and S6 transmembrane segments are highlighted in light blue. The mutated regions are marked as described in (a).
In comparison to the voltage-gated potassium channels, little is known about the molecular determinants of electromechanical coupling in the sodium channels. The complexity of the sodium channel structure and an absence of functional symmetry make interpretation of structure-function data extremely challenging. Recently, Swartz and colleagues 22 have described an elegant methodology to dissect the functional behavior of parts of the sodium channel by transplanting them into a symmetric potassium channel template. Here, we have used a novel approach that combines site-specific fluorescence recordings with conductance measurements to examine the molecular basis of coupling between the voltage-sensor and the pore of the sodium channel. We have systematically mutated residues in the S4–S5 linker, parts of the S5 and S6 of domain III (Figure 1) to study the effect of these mutations on coupling interactions between voltage-sensor and pore of the sodium channel. Site-specific fluorescence recordings determined the effect of these mutations on the thermodynamic parameters of the domain III voltage-sensor whereas the effect on the pore conformation was ascertained by conductance measurements. Upon mapping a small set of critical residues on to a structural model of the sodium channel, we find that a majority of them are either in close proximity to or in the hinge regions of key helices such as the S4, S4–S5 linker, S5 and S6 segments. Two of these, arginine (R1135) at the beginning of S4–S5 linker and a phenylalanine (F1298) at the tail end of the S6 segment, are likely to form an interacting pair. Our analysis shows that these residues are involved in coupling interactions both in the closed and open conformation of the channel.
RESULTS
In this study, the role of the residues in a putative interaction surface likely to be involved in voltage sensor-pore coupling was assessed by tryptophan scanning mutagenesis approach. A mutation to tryptophan, which has the largest side chain volume amongst naturally occurring amino acids, has the potential to disrupt protein-protein interactions radically by introducing defects in the interfaces stabilizing the protein core 18,23–25. We introduced tryptophans in the rat skeletal muscle sodium channel (Nav 1.4), which is one of the better-characterized members of the sodium channel family. Fluorophores attached to cysteine residues proximal to the S4 segments of this channel were previously shown to track the movement of gating charges 26. Here, the conformational state of the voltage-sensor of domain III was monitored by a fluorophore attached to substituted cysteine at the L1115 position (Figure 1a) using a voltage-clamp fluorimetry setup. Thus, all the mutants were made in L1115C background, which is hereafter referred to as the wild type (WT). The conformation of the pore gates was monitored by conductance measurements in a two-electrode voltage-clamp setup. Mutations of residues in the S6-domain III are known to significantly alter the stability of the sodium channel pore 27. Furthermore, the mutations of charged residues in the voltage-sensor of domain III reduce the slope of conductance-voltage relationships suggesting that the movement of domain III gating charges is coupled to opening of the pore gates 28. Taken together, these studies suggest that any direct or indirect alteration in the conformational state of the pore helices of domain III is likely to be manifested in the conductance properties of the sodium channel.
Scanning mutagenesis in the S4–S5 linker and inner pore of DIII in the sodium channel
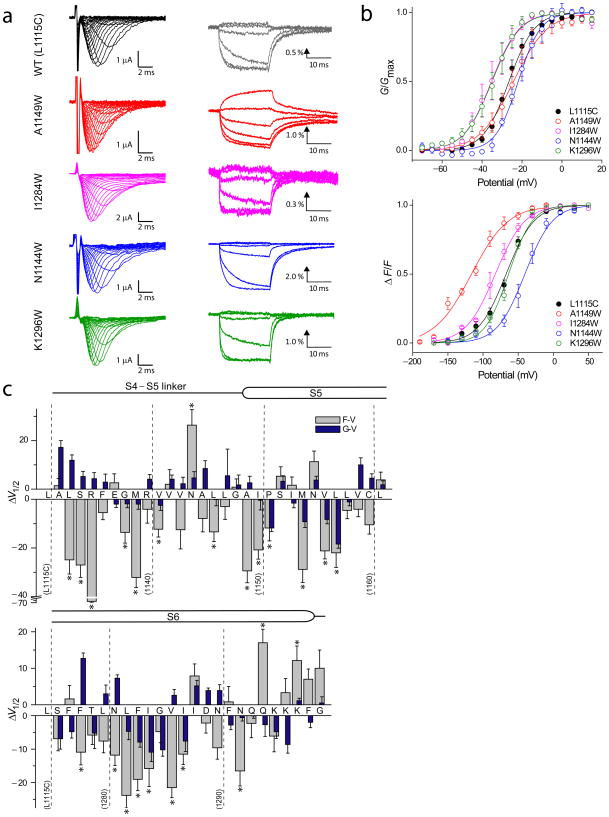
The tryptophan substitutions were introduced in the S4–S5 linker and parts of S5 and S6 towards the intracellular end (Figure 1b). These substitutions encompassed the gating interface identified by Lu and colleagues 17 in the Shaker potassium channel. Amongst the 54 tryptophan mutants, functional channel activity was observed in all but 10 mutants (Table I). Tryptophans at these 10 positions presumably disrupt protein packing which causes either a complete loss of channel function or low plasma membrane expression. In all such instances, substitution to alanine instead of tryptophan generated a functional channel and allowed us to probe the role of side chains at those sites. Figure 2a shows typical voltage-dependent ionic current and fluorescence traces of the WT and few of the mutants, representing a broad spectrum of functionally altered channels. The conductance-voltage (G-V) relationships were obtained from peak conductance values whereas the fluorescence-voltage (F-V) relationships represent the steady state fluorescence intensities (Figure 2b).
Figure 2. Voltage-dependent conductance and fluorescence responses in the mutant sodium channels.
(a) Families of ionic current and fluorescence traces corresponding to the WT (L1115C) and four representative tryptophan mutants. Ionic currents were elicited by pulsing to various test potentials (−100 to 65 mV at 5mV intervals) for 20 ms following a brief (50 ms) prepulse to −120 mV. Fluorescence traces were recorded by pulsing to various test potentials (−170 to 50 mV at 20 mV intervals) for 20 ms following a 50 ms prepulse to −120 mV. Each of fluorescence trace was obtained by averaging 10 trials with a 1 s inter pulse interval. Fluorescence traces are shown at 40 mV interval for clarity.
(b) Normalized G–V and F–V curves corresponding to the wild type (L1115C) and the tryptophan mutants shown in (a). The conductance was normalized to the peak conductance (Gmax) whereas the fluorescence response was normalized to maximum steady state fluorescence (F) values for each oocyte. The data points represent the mean ± standard error (S.E.) of at least three independent measurements and the smooth curves represent the best fits of the averaged data to a single Boltzmann function.
(c) The difference in the half-maximal responses (ΔV1/2) between the WT and mutants. The ΔV1/2 of both fluorescence (grey) and conductance (blue) for each of the mutants are shown. Error bars represent propagation of errors. Statistical analysis of ΔV1/2 of the F–V relationships was performed with a one-way ANOVA with Dunnett’s post-tests, *p<0.05.
Structure-function studies on the brain sodium channel show that the steady state activation curves of the inactivation deficient mutants are not significantly different compared to the wild type channels29. This is mainly because the activation rates of the wild type channels are about 4–5 fold faster than the inactivation rates26. The ratios of activation to inactivation time constants of the mutants were similar to those of the WT sodium channels (Supplementary Figure 4). This suggests that inactivation is unlikely to have a significant effect on our estimates of activation parameters derived from peak conductance values.
To quantitatively evaluate the effects of the perturbation, we fitted each of the G-V and the F-V curves to a single Boltzmann function and obtained the values of two parameters, the slope (z) and the voltage eliciting half-maximal response (V1/2) (Table I). The difference in V1/2 of the mutant relative to the WT was expressed as ΔV1/2 (ΔV1/2 = V1/2 Mutant − V1/2 WT) and these differences calculated from the F-V and G-V curves were summarized in Figure 2c. The plot reveals that more than half of the tryptophan mutants cause a statistically significant shift in the voltage range describing the fluorescence response compared to the WT. These data highlight the importance of the residues in this linker region in voltage-sensor activation and/or pore opening.
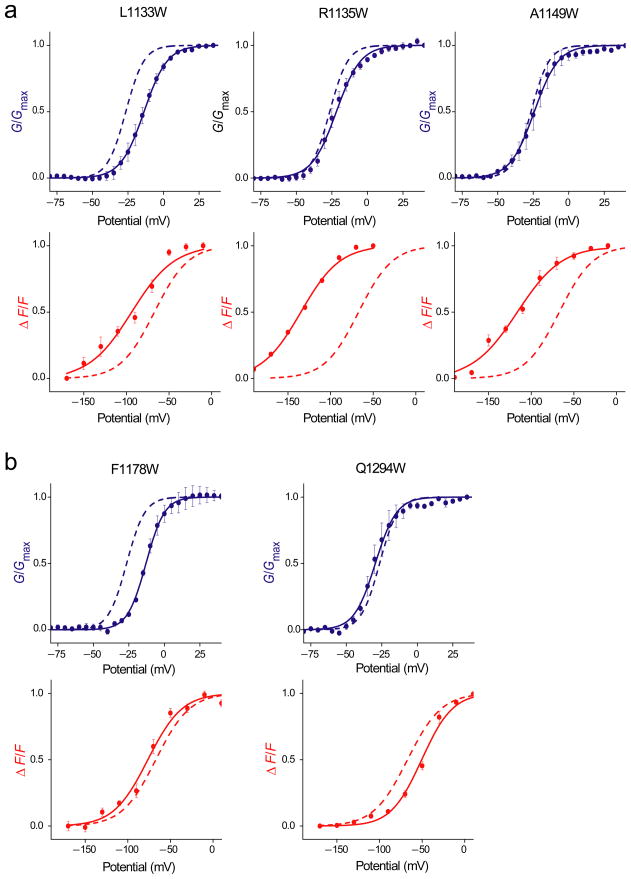
For the most part, the tryptophan substitutions cause much a larger effect on the F-V rather than the G-V parameters. This trend can be partly accounted by the fact that the fluorescence specifically monitors the movement of domain III voltage-sensor whereas conductance requires contribution from the pore segments of all four domains. Based on their functional effects, the mutants can be broadly categorized into two groups: a) those that shift the ΔV1/2 of both the G-V and F-V curves in the same direction (referred hereafter as class I mutations) and, b) those that shift ΔV1/2 of F-V and G-V curves in the opposite direction (class II mutations). Class I mutations have similar effects on the energetics of voltage-sensor activation and pore opening whereas the Class II mutations have opposite effects on those processes (Supplementary figure 1). For instance, tryptophan substitution at the L1133 position generates a class II mutant, causing a 12 mV rightward shift in the V1/2 of the G-V curve and a 25 mV leftward shift in the V1/2 of the F-V curve (Figure 4). This mutation stabilizes the voltage-sensor in the activated state and concurrently stabilizes the closed conformation of the pore. In the following sections, we will analyze these mutations and their implication in detail.
Figure 4.
Normalized G–V (blue) and F–V relationships (red) for prominent class II mutants. The data points represent the mean ± SE of at least three independent measurements and smooth curves represent the best fits of the averaged data to a single Boltzmann function. The fitted WT data is shown as dashed curves
(a) Class II mutants that cause a right shift in the G–V and a left shift in the F–V curve relative to the WT.
(b) Class II mutants that cause a left shift in the G–V and a right shift in the F–V curve relative to the WT.
An implicit assumption in this sort of analysis is that the mutations do not induce a novel conformational change but only affect the thermodynamic parameters that define the coupled system. The time course (at saturating potentials) and the direction of fluorescence change (after accounting for shifts in V1/2) in the mutants were similar to the WT (Supplementary Figure 5). Furthermore, the amplitudes of fluorescence signals of the mutants are broadly correlated with channel expression as estimated from the peak outward currents obtained during fluorescence recordings. To quantitate the effect of the mutations on fluorescence amplitudes, it is necessary to estimate the number of channels per cell and the fraction of labeled channels. Although, gating current measurements have been utilized to normalize for channel density, we cannot rule out the possibility that these mutations may also modify the total gating charge per channel by altering the structure. Recent studies on the Shaker potassium channel show that the total gating currents is highly sensitive to the position of the charge in a focused electric field30 and it is conceivable that a mutation could alter the total charge per channel by reshaping the electric field31 or by hindering charge movement. Given these considerations, we cannot completely rule out the possibility that either the fluorescence amplitudes or gating charge, in some instances, was modified by these mutations. Nevertheless, our overall fluorescence data is consistent with the notion that the probe is likely to track the same structural rearrangements in the WT and mutant channels.
Class I mutations
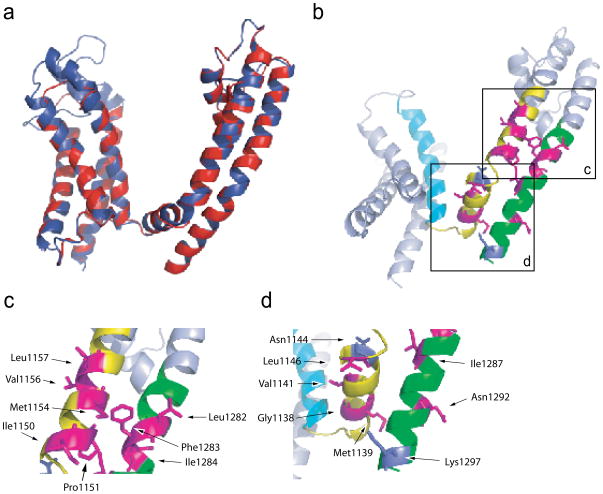
The voltage range of activation (V1/2) of both F-V and G-V curves were shifted to more negative voltages in fourteen mutants suggesting that the energy required to activate the voltage-sensor and pore opening is reduced (Figure 2c and Table I). In order to evaluate this data in a structural context, we generated a homology model of the sodium channel structure (Figure 3a, red; and Supplementary figure 2a and c) based on the crystal structure of Kv1.2/2.1 chimera (Figure 3a, blue; and Supplementary figure 2a) (PDB code: 2R9R; see Materials and Methods) 15. The model of Nav1.4 has two major differences: first, the sodium channel has a shorter S3–S4 linker compared to the chimeric potassium channel, and second, the break between the S4 segment and the S4–S5 linker occurs immediately following the fourth charge. In the eukaryotic potassium channel, the break is after one helical turn. Mapping those positions onto the structural model of the sodium channel (Figure 3b) shows that many of these residues (8 of the 14 residues) are clustered in the intracellular region of the S5 and S6 segments (Figure 3c). Residues in S5 (I1150, P1151, M1154, V1156 and L1157) face the clustered residues in S6 (L1282, F1283 and I1284) and away from the voltage-sensor of domain III. The side chains of these residues project towards the S4 and S4–S5 linker of the neighboring domain (Domain II) and, thus, the mutations at these positions may perturb inter-domain interactions leading to altered activation profiles. Residues in this region have been shown to be important in activation gating of the Shaker potassium channel 20. The remaining six residues are positioned throughout the S4–S5 linker and the inner portion of S6 (Figure 3d). In contrast to the fourteen residues that cause leftward shifts in the F-V and G-V curves, mutations at two positions cause rightward shifts in both curves. N1144 is at the C-terminal end of the S4–S5 linker while K1297 is at the end of the S6 helix (Figure 3d). These two mutations in all likelihood stabilize the closed conformation of the channel. While there are a few such exceptions, we assume that for most part large aromatic substitutions such as the introduction of tryptophans disrupt protein packing. Thus, our finding that a large fraction of the mutants destabilize the closed state of the channel may reflect increased steric clashes in the closed state of the channel.
Figure 3. Superposition of the homology model of sodium channel domain III with the structure of the Kv1.2/2.1 chimera and mapping the prominent class I positions on the homology model.
(a) A superposed view of sodium channel domain III and the crystal structure of the potassium channel chimera (Kv1.2/2.1) (PDB code: 2R9R). The sodium channel is shown in blue while the chimeric potassium channel is shown in red.
(b) Class I mutants that stabilize the open pore are in magenta whereas those that destabilize the open conformation are highlighted in purple. The other mutated regions in the S4–S5 linker and S5 are in green while those in the S6 are in yellow.
(c) Enlarged view of the lower part of S5 and the middle part of S6 near the gating hinge.
(d) Enlarged view of the S4–S5 linker and the bottom part of S6.
Class II mutations
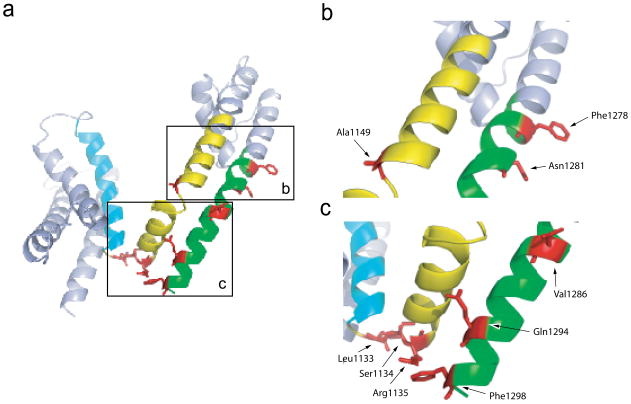
Of the 54 sites probed in this study, tryptophan mutations in seven sites corresponding to L1133, S1134, R1135, A1149, F1278, N1281 and V1286 showed left shift in the F-V and a right shift in the G-V relationships. Thus, the mutations at these positions cause voltage-sensors to activate more easily while concurrently making the opening of the pore more difficult. Figure 4a represents the F-Vs and G-Vs of four of the mutants along with those of the WT. Right shifts of G-V curves were in the range of 2.6 to 12.7 mV and left shifts of F-V relationships were in the range of 10.9 to 66.9 mV (Table I). A stretch of three residues (L1133W, S1134 and R1135) occurs at the hinge linking the S4 segment with the S4–S5 linker (Figure 5a and c). Remarkably, three other residues are also either in the hinge or in close proximity. A1149 is at the hinge between the S4–S5 linker and S5 segment (Figure 5b) and V1286 is in close proximity to this bend but in the S6 helix (Figure 5c). F1278 is near the putative gating hinge on the S6 helix (Figure 5b). Localization of these mutations to the hinges in the structural model implies that these regions may play a unique role in the gating process.
Figure 5. Mapping the prominent class II mutants on to the structural model of sodium channel domain III.
(a) The sites of class II mutations are highlighted in red. The voltage-sensing S4 segment is in cyan. The other mutated positions in the S4–S5 linker and S5 are shown in yellow whereas those in the S6 segment are shown in green.
(b) Enlarged view of the lower part of S5 and the middle part of S6 near the gating hinge.
(c) Enlarged view of the S4–S5 linker and the bottom part of the S6.
Mutations at three positions (Q1294, K1296 and F1298) in the lower half of the S6 helix caused a rightward shift in the F-V and a left shift in the G-V relationships bringing their voltage-range closer to each other (Figure 4b). Of these three, the F-V curve of Q1294W showed the largest depolarizing shift. A simplistic interpretation is that the tryptophan mutations at these positions increase the strength of coupling between the voltage-sensor and the pore domain. As a result, the functional properties of these two domains resemble each other. Conversely, we can infer that the class II mutations, which stabilize the activated voltage-sensor and destabilize the open pore, decouple the voltage-sensor and pore movements. We will revisit these interpretations in the discussion section.
Probing a putative interaction pair
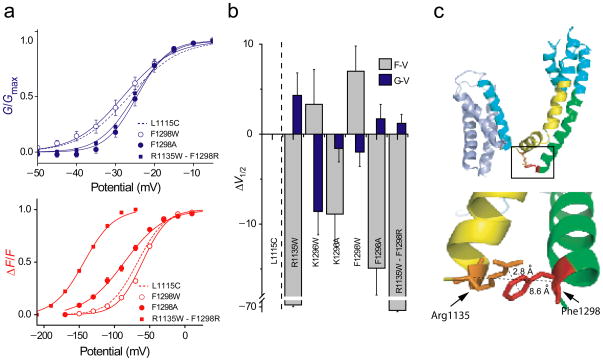
The crystal structure of both Kv1.2 and the Kv1.2/2.1 chimera shows that the S4–S5 linker is juxtaposed to the S6 helix of the same subunit 14,15. The S4–S5 linker is almost parallel to the membrane plane and positioned over the S6 inner helix forming an interaction surface with one face of the S6 helix. Studies in the HERG potassium channel 32 have shown that positively charged residues in the S4–S5 linker are likely to interact with the S6 segment of the potassium channel. Sequence alignment of different members of the voltage-gated channel family shows that the tail end of the S6 segment is rich in negatively charged residues or aromatic groups. In our structural model of the sodium channel, two phenylanines (F1298 and F1136) were observed to be in proximity to positive charges (R1135 and K1296) on S4–S5 linker in the tertiary structure (Figure 6c) raising the possibility that these residues form a cation-π type of interaction 33. Tryptophan substitutions at both these positions showed a trend consistent with increased coupling. Tryptophans have a higher electron cloud density compared to phenylalanines which would contribute to a stronger cation-π interaction 34. In the model, the side chains of R1135 and F1298 are pointing towards each other and their α-carbons are within 8.6 Å of each other, close enough to interact. To test this possibility, we first mutated these residues to alanine. Interestingly, the opposite shifts in F-V and G-V curves observed in F1298W were reversed in F1298A (Figure 6a and b), suggesting that F1298, but not K1296 (Figure 6b), may interact with R1135 in the S4–S5 linker. To further probe this putative interaction between R1135 and F1298, we switched the positions of aromatic and cationic side chains to create R1135W-F1298R double mutant. The F-V and G-V curves of this double mutant were similar to the R1135W single mutant and not to the WT suggesting that these two groups are unable to form an electrostatic interaction pair. Note that cation-π interaction is geometry dependent-cation interacts in en face orientation as opposed to edge-on orientation with the aromatic side chain 33. It is possible that switching the positions of the interacting side-chains does not create an optimal orientation for such an interaction. Moreover, R1135 is in close proximity to other class II (L1133 and S1134) mutations and is in the putative hinge connecting the S4 to the S4–S5 linker. Substitution of arginine by a bulky tryptophan in this crucial putative hinge may introduce steric perturbations that prevent movements necessary for voltage-sensor-pore coupling. Thus, to fully examine the interaction between R1135 and F1298, it will be necessary to use a less disruptive substitutions such as those involving the use of non-natural amino acids 35.
Figure 6. The G–V and F–V curves upon perturbation of putative interaction pairs and mapping the positions of an interaction pair on to the homology model of sodium channel domain III.
(a) Normalized G–V (top panel) and F–V (bottom panel) curves for the WT (dashed line), F1298A (filled circle), F1298W mutant (open circle), and R1135W-F1298R double mutant (filled square). The data points represent the mean ± SE of at least three independent experiments, and the smooth curves represent the best fits of the averaged data to a Boltzmann function.
(b) The difference in half-maximal responses (ΔV1/2) between the WT relative to the mutants R1135W, K1296W, K1296A, F1298W and R1135W-F1298R. The ΔV1/2 of both fluorescence (grey) and conductance (blue) for each of the mutants are shown. The error bars represent propagation of errors.
(c) The side chains of residues R1135 and F1298 are shown on the homology model. Inset, the region around the tail end of S6 and the beginning of S4–S5 linker is shown enlarged.
DISCUSSION
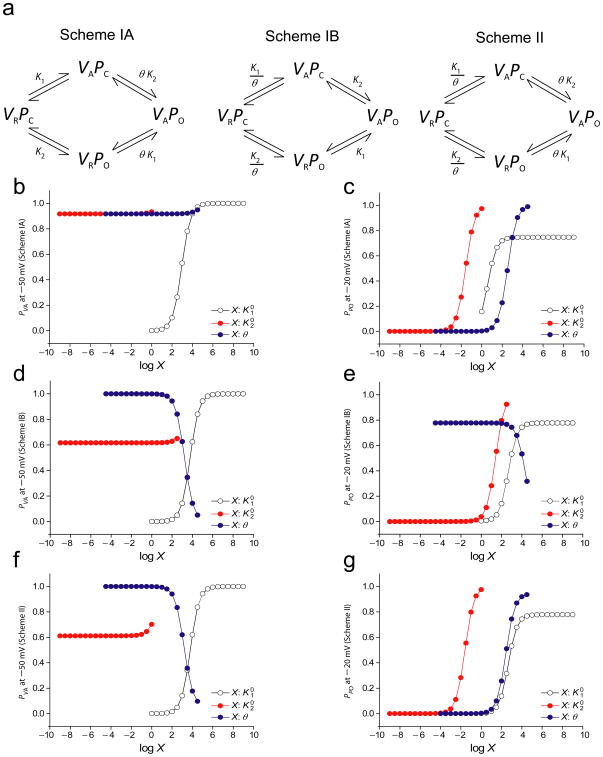
By combining tryptophan-scanning mutagenesis with voltage-clamp fluorimetry, we have identified a number of key residues that are likely to be involved in conformational coupling of the domain III voltage-sensor to the pore of the Nav 1.4 channel. Class I mutants modify the energetics of voltage-sensor activation and opening of the pore in the same direction. To understand how a single amino acid substitution can cause such effects on the voltage-sensor and pore, we first consider two simple canonical models of a cooperative system where the two domains interact either when both are in their resting state or in their activated state (Scheme IA and Scheme IB in Figure 7a). The effect of changing each of the three thermodynamic parameters on the activation probabilities of the voltage-sensor and pore was tested by using numerical simulations (details in Supplementary material XX). The plots in Figures 7b–e show that the probabilities of voltage-sensor activation and pore opening for Schemes IA and IB increase when either K1or K2 increases. When the interaction term (θ) is increased, the voltage-sensor and pore opening probabilities for Scheme IA also increase whereas those probabilities for Scheme IB were reduced. To evaluate the generality of these trends, we analyzed the derivatives of the probability expressions (Supplementary Table I and II). A positive value of the derivative with respect to a parameter indicates that the probability value will increase as the parameter value is increased. The derivatives of voltage-sensor activation and pore opening also show that both these probabilities move in the same direction when any one of the thermodynamic terms is modified.
Figure 7. Two general models describing the coupled activation process involving the voltage-sensor and pore of a voltage-dependent ion channel.
(a) Schemes IA and IB represent the canonical models of cooperativity, where a direct interaction between the open pore and the activated voltage-sensor mediates coupling of the two structural domains. Scheme II represents alternate model of voltage-sensor-pore coupling, where the closed pore interacts with the resting voltage-sensor and the open pore interacts with the activated voltage-sensor. K1 and K2 are the intrinsic activation constants of the voltage-sensor and the pore and θ is the coupling parameter. The intrinsic activation constants can be expressed as: and where and represent the contribution of chemical interactions to the activation process of the voltage-sensor and the pore, respectively.
(b) and (c) The probabilities of voltage-sensor activation and pore opening when the three thermodynamic parameters are modified in Scheme IA. The expressions for PVA and PPO from Scheme I (Eqs. 1 and 2) were fitted to the WT F–V and G–V curves respectively, to obtain the values for the different parameters: , z1 = 3.494, , z2= 4.219, θ = 999.916. and θ were varied over 10 orders of magnitude and the response to these changes on the PVA values at −50mV and PPO values at −20mV were plotted in (b) and (c) respectively.
(d) and (e) The probabilities of voltage-sensor activation and pore opening when the three thermodynamic parameters are modified in Scheme IB. The initial thermodynamic parameters , z1= 0.936, , z2= 3.914, θ = 1003.561 were obtained by fitting the expressions for PVA and PPO in Scheme IB (Eqs. 3 and 4) to the WT F–V and G–V curves respectively. and θ were varied over 10 orders of magnitude and the response to these changes on the PVA values at −50mV and PPO values at −20mV were plotted in (d) and (e) respectively.
(f) and (g) The probabilities of voltage-sensor activation and pore opening when the three thermodynamic parameters are modified in Scheme II. The expressions for PVA and PPO from Scheme II (Eqs. 3 and 4) were fitted to the WT F–V and G–V curves respectively, to obtain the values for the different parameters: , z1= 0.949, , z2= 3.905, θ= 999.913. and θ were varied over 10 orders of magnitude and the response to these changes on the PVA values at −50mV and PPO values at −20mV were plotted in (d) and (e) respectively.
The above analysis shows that if a mutation shifts both the probability curves in the same direction, we cannot attribute the effect of a mutation on any one of thermodynamic terms in Scheme IA or IB. Nonetheless, we can make some inferences regarding the role of these amino acids based on their position in the structural model of the sodium channel. A majority of the class I mutations relatively stabilize the open state of the channel and a large fraction of these are in the S5 and S6 segments which constitutes the pore domain. In KcsA structure36, which serves as a model of the closed pore, the four S6 helices are tightly packed at the bundle crossing. Studies in the Shaker and the HERG channel show that the mutations in these regions disrupt the tight packing and destabilize the closed pore5,27,37–40. These findings suggest that the introduction of tryptophan in the S5 and S6 region mostly destabilizes the packing in the closed state of the sodium channel.
In contrast to class I mutations, the class II mutations cause opposite effects on the activation of the voltage-sensor and pore opening in the sodium channel. These “Janus-faced” mutants are reminiscent of the ILT mutant in the Shaker potassium channel 16,41. The voltage-sensor in ILT mutant is stabilized in the activated state whereas the open pore is highly destabilized. Our derivative analysis (Supplementary Table I and II) unequivocally shows that the opposing effects on the voltage-sensor and pore cannot be rationalized by changing a single parameter with Schemes IA & IB. We consider an alternate model of a cooperative system (Scheme II, Figure 7a), where the voltage-sensor and the pore interact with each other when both of them are in the doubly resting and doubly activated state. θ is the coupling term between the pore and voltage-sensor. We define this type of interaction as “conserved interactions” as opposed to “unconserved interactions” of the type depicted in Scheme IA & IB. Conserved interactions result in no net gain or loss of interaction energy going from the doubly resting to doubly activated state and achieve conformational coupling by destabilizing the intermediate states.
The probability expressions of activation for the voltage-sensor and the pore in scheme II is;
| Eqn. 1 |
| Eqn. 2 |
The WT F-V and G-V data were fitted to these probability expressions to obtain the initial estimates of the three thermodynamic parameters for numerical simulations. The plots in Figures 7f and g show the effect of changing each of these parameters on the probabilities of voltage-sensor activation and pore opening. Note that the probability of voltage-sensor activation decreases with increasing values of θ whereas that of pore opening increases. Analysis of derivatives of the above probability expressions also shows that a change in interaction term but not the intrinsic equilibrium constants has opposite effects on the probability terms under conditions when K2 <1 and K1>1 (Supplementary Table II). Thus in Scheme II, a loss of coupling would lead to a right shift in the G-V curve and a left-shift in the F-V curve. The assumption, K2 < 1 implies that the pore domain is intrinsically more stable in the closed or resting conformation. This is consistent with scanning mutagenesis studies on the pore segments 39,40. Our second assumption, K1> 1 implies that the voltage-sensor is intrinsically more stable in its activated conformation (in absence of membrane potential). The crystal structure of the isolated voltage-sensor domain of the prokaryotic voltage-dependent potassium channel, KvAP superposes extremely well (rms deviation 2.08 Å) with the voltage-sensor domain of KV1.2/2.1 15. The channel in the Kv 1.2/2.1 structure is in the open or activated conformation, thus indicating that the isolated voltage-sensor domain of KvAP is likely to be in the activated state.
Thus, the scheme II provides a plausible framework to rationalize the effects of class II mutations on the voltage-sensor movement and pore domain with a change in a single parameter. The three schemes that we consider here encompass all the possible ways two domains can be positively coupled to each other. If we assume that a single mutation alters only one thermodynamic term, then derivative analysis demonstrates that the class II mutations must modify interactions between the voltage-sensor and pore, both in the resting and activated states. This suggests that class II mutations play a unique role in coupling the voltage-sensor to the pore domain.
Interestingly, many of our class II mutants map close to the sites in the Shaker potassium channel that cause opposite shifts in the voltage-sensor activation and pore opening (Supplementary Figure 6). For instance, A1149 maps close to the hinge connecting the S4–S5 linker to S5 segment on the sodium channel. E395 and L398 occupy the equivalent position in the Shaker potassium channel. These residues are also facing the hinge connecting the S4 with S4–S5 linker in the neighboring subunit and may be involved in intersubunit coupling. Class II mutants F1278 and N1281 in S6 helix of domain III and map in vicinity of sites L472 in the Shaker potassium channels. S479 in the Shaker channel is in the same region as V1286 in the domain III of the sodium channel. These findings suggest that many of the structural mechanisms involved in electromechanical coupling are likely to be conserved between the members of voltage-gated ion channel superfamily.
Intriguingly, the majority of mutants with class II phenotype in the Shaker channel occur near the hinge regions (supplementary Figure XX). Although there is an uncertainty in the sodium channel due to a lack of experimentally derived structure, a surprisingly large number of the class II mutations were also localized to the hinge regions connecting the various helices such as the S4, S4–S5 linker, S5 segments in the sodium channel homology model. Calculations of angle bending potentials in hinge regions of lysozyme 42,43 and a number of other proteins 44–47 have shown that these potential functions are parabolic with respect to angular displacement. Thus, bending at the hinges generates a restoring force, which acts on the neighboring segments and thereby propagates conformational change. The conserved interactions, by definition, are elastic interactions and hinge-bending potentials will be emblematic of such interactions. Introduction of bulky tryptophan groups at or near the hinge regions may prevent hinge-bending motions and may account for the large number of class II hits at those regions. Based on these findings and previous studies on the Shaker channel, we speculate that the hinge regions play an important role in conformational coupling between the voltage-sensor and pore in voltage-dependent ion channels. The importance of hinge residues in electromechanical coupling suggests that the conformational propagation underlying voltage-dependent gating is likely to involve rigid body motions of helices rather than a wavelike spread such as those envisioned for the ligand gated ion channels 48–50.
METHODS
Molecular biology and oocyte expression
Tryptophan and alanine mutations in this study were all generated in a mutant (L1115C) rat skeletal muscle sodium channel (rNav 1.4) background. The region encompassing domain III was subcloned into the pBluescript (KS+) cloning vector by digesting the L1115C mutant sodium channel in the PBSTA vector with the enzymes Aat II and BspEI. A unique silent restriction site (Bam HI) was inserted into this region by overlap PCR method. Tryptophan or alanine mutations were generated by using QUIKCHANGE mutagenesis kit (Stratagene, CA) or by overlap PCR method. Mutations were confirmed by sequencing the entire cloning cassette. The mutated domain III region was reinserted back in to the original vector. The mutant clones were screened by using the unique restriction enzyme. For cRNA preparation, the mutant plasmids were linearized by Not I digestion. cRNA was generated by in vitro transcription using T7 RNA polymerase kit (Ambion, Inc., CA).
Electrophysiology and Fluorescence measurements
Ionic conductance measurements were performed with two electrode voltage clamp (OC-725C Oocyte Clamp, Warner, CT) at room temperature (22 ± 3°C). The oocytes were clamped at a holding potential of −80 mV for at least 5 min prior to recording. All current recordings were obtained within 12–36 hours of cRNA injection. The currents were recorded in an external solution containing 105 mM Na-Mes. In many instances, tetrodotoxin (Sigma Chemical Co.) was added to a final concentration of 10–50 nM in the external solution to obtain better voltage control of inward sodium currents.
Fluorescence signals were typically recorded 3–5 days after injection. To minimize series resistance errors, the external and internal solution contained 105 mM NMG-Mes 51. The channel was labeled by incubating the oocytes with 10 μM TMRM (10mM stock in DMSO) in a depolarizing solution for 30 minutes on ice (XX). After washing the excess unbound label, the oocytes were stored in dark in 105 mM NMG containing external solution. The fluorescence recordings were obtained in two setups both combining a cut-open oocyte voltage-clamp (Dagan Corp. MN) with an upright epifluorescence microscope (Olympus; BX50WI) using a LumPlanFL 40X water-immersion objective (0.8 N.A.). In one setup, the light source was a 150 W tungsten lamp (Newport Corp., CT) supplied by low noise power supply (Kepco, NY) and in the other, a 150 W super-quiet xenon arc lamp (Hamamatsu Inc., Japan) was used. The photocurrents generated by PIN photodiodes in response to emitted light were amplified using either an Axopatch 1B patch clamp amplifier (Axon Instruments, CA) or a Photomax-200 amplifier (Dagan Corp., MN).
Data acquisition and analyses
Both ionic conductance and fluorescence signals were sampled at 250 kHz with a Digidata 1322A and 1440 interface, respectively (MDS Analytical Technologies, CA). For ionic current measurements, linear and membrane capacity currents were subtracted online using a P/4 procedure with a subtraction holding potential at −120 mV. The ionic current signals were low-pass filtered at 1 kHz. The interval between the each potential trace was 70 ms. Fluorescence traces were recorded without subtraction and low pass filtered at 10 KHz. Each fluorescence trace represents an average of 10 recordings with 1 s interpulse interval. Electrophysiology and fluorescence data were acquired using PClamp 10 acquisition software (MDS Analytical Technologies, CA), and analysis was performed using Clampfit (MDS Analytical Technologies, CA) and Excel (Microsoft, WA). The figures were prepared in Origin (MicroCal, MA), Illustrator (Microsoft, WA) or PYMOL (DeLano Scientific LLC). Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by the post-hoc Dunnett’s test using GraphPad Prism (San Diego, CA).
Conductance-voltage and fluorescence-voltage relationships were fitted to a single Boltzmann function:
where z is the valence and V1/2 is the half maximal voltage. K is the Boltzmann constant, T is the temperature and e is the electronic charge.
Structural modeling
The homology model of the sodium channel domains was developed using the crystal structure of the KV1.2/2.1 chimera (PDB ID code 2R9RB). The sequence of the putative sodium channel domains were first aligned to that of the KV1.2/2.1 chimera using ClustalW 52, operated on default parameters (Supplementary figure 3). A separate structure-sequence alignment was also performed using MODELLER 9v4 53. These alignments were combined to generate the final alignment. The homology models of the four sodium channel domains were developed using MODELLER 9v4. For each domain, 100 candidate models were generated. The final model for each domain was selected on the basis of the DOPE (discrete optimized potential energy) scores and the stereochemical quality of the candidate models, as assessed from the Ramachandran plots. The models of the four domains were organized symmetrically around the axis of K+ conduction by using appropriate transformation of coordinates to generate the final tetrameric structure. Energy minimization of this tetrameric structure was performed using the ZMM program 54. No steric clashes were observed within the domains or at the interface between the domains. In the above modeling exercise, the loop regions between the S5 helices and the pore helices were not modeled. This loop which is longest in DI (and relatively shorter in DIII) forms a gap aligned block in the multiple sequence alignment. Moreover, since these loops were not structurally important for the purpose of our study, their modeling was not considered necessary.
Supplementary Material
Acknowledgments
This project was supported by funds from the National Institutes of Health (GM084140-01), AHA Scientist Development Award (0535214N), and the Shaw Scientist award to B. C. We thank Dr. Marcel Goldschen for help with preparing the supplementary figure 6 and Kristina Schuldt for excellent technical assistance. We also thank the members of Chanda laboratory for their comments and discussions.
Footnotes
AUTHOR CONTRIBUTIONS
Y.M. and M.A-.M. contributed equally to this work. Y.M. and M.A-.M. performed all the experiments and analyzed the data. S.C. created the structural model. S.C. and B.C generated the coupling models. Y.M, M.A-.M and B.C wrote the paper.
Reference List
- 1.Noda M, et al. Primary Structure of Electrophorus-Electricus Sodium-Channel Deduced from Cdna Sequence. Nature. 1984;312:121–127. doi: 10.1038/312121a0. [DOI] [PubMed] [Google Scholar]
- 2.Papazian DM, Schwarz TL, Tempel BL, Jan YN, Jan LY. Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene from Drosophila. Science. 1987;237:749–753. doi: 10.1126/science.2441470. [DOI] [PubMed] [Google Scholar]
- 3.Tempel BL, Papazian DM, Schwarz TL, Jan YN, Jan LY. Sequence of a probable potassium channel component encoded at Shaker locus of Drosophila. Science. 1987;237:770–775. doi: 10.1126/science.2441471. [DOI] [PubMed] [Google Scholar]
- 4.Yang N, George AL, Jr, Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- 5.Perozo E, Cortes DM, Cuello LG. Structural rearrangements underlying K+-channel activation gating. Science. 1999;285:73–78. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong CM, Bezanilla F. Currents related to movement of the gating particles of the sodium channels. Nature. 1973;242:459–461. doi: 10.1038/242459a0. [DOI] [PubMed] [Google Scholar]
- 7.Glauner KS, Mannuzzu LM, Gandhi CS, Isacoff EY. Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature. 1999;402:813–817. doi: 10.1038/45561. [DOI] [PubMed] [Google Scholar]
- 8.Cha A, Snyder GE, Selvin PR, Bezanilla F. Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature. 1999;402:809–813. doi: 10.1038/45552. [DOI] [PubMed] [Google Scholar]
- 9.del Camino D, Holmgren M, Liu Y, Yellen G. Blocker protection in the pore of a voltage-gated K+ channel and its structural implications. Nature. 2000;403:321–325. doi: 10.1038/35002099. [DOI] [PubMed] [Google Scholar]
- 10.Holmgren M, Smith PL, Yellen G. Trapping of organic blockers by closing of voltage-dependent K+ channels: evidence for a trap door mechanism of activation gating. J Gen Physiol. 1997;109:527–535. doi: 10.1085/jgp.109.5.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horn R. Conversation between voltage sensors and gates of ion channels. Biochemistry. 2000;39:15653–15658. doi: 10.1021/bi0020473. [DOI] [PubMed] [Google Scholar]
- 12.Stuhmer W, et al. Structural parts involved in activation and inactivation of the sodium channel. Nature. 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- 13.Strong M, Chandy KG, Gutman GA. Molecular evolution of voltage-sensitive ion channel genes: on the origins of electrical excitability. Mol Biol Evol. 1993;10:221–242. doi: 10.1093/oxfordjournals.molbev.a039986. [DOI] [PubMed] [Google Scholar]
- 14.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 15.Long SB, Tao X, Campbell EB, Mackinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 16.Smith-Maxwell CJ, Ledwell JL, Aldrich RW. Uncharged S4 residues and cooperativity in voltage-dependent potassium channel activation. J Gen Physiol. 1998;111:421–439. doi: 10.1085/jgp.111.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Z, Klem AM, Ramu Y. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J Gen Physiol. 2002;120:663–676. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong KH, Miller C. The lipid-protein interface of a Shaker K(+) channel. J Gen Physiol. 2000;115:51–58. doi: 10.1085/jgp.115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SY, Banerjee A, Mackinnon R. Two separate interfaces between the voltage sensor and pore are required for the function of voltage-dependent K(+) channels. PLoS Biol. 2009;7:e47. doi: 10.1371/journal.pbio.1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soler-Llavina GJ, Chang TH, Swartz KJ. Functional interactions at the interface between voltage-sensing and pore domains in the Shaker K(v) channel. Neuron. 2006;52:623–634. doi: 10.1016/j.neuron.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Labro AJ, et al. Kv channel gating requires a compatible S4-S5 linker and bottom part of S6, constrained by noninteracting residues. J Gen Physiol. 2008;132:667–680. doi: 10.1085/jgp.200810048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosmans F, Martin-Eauclaire MF, Swartz KJ. Deconstructing voltage sensor function and pharmacology in sodium channels. Nature. 2008;456:202–208. doi: 10.1038/nature07473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li-Smerin Y, Hackos DH, Swartz KJ. alpha-helical structural elements within the voltage-sensing domains of a K(+) channel. J Gen Physiol. 2000;115:33–50. doi: 10.1085/jgp.115.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perozo E. Structure and packing orientation of transmembrane segments in voltage-dependent channels. Lessons from perturbation analysis. J Gen Physiol. 2000;115:29–32. doi: 10.1085/jgp.115.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monks SA, Needleman DJ, Miller C. Helical structure and packing orientation of the S2 segment in the Shaker K+ channel. J Gen Physiol. 1999;113:415–423. doi: 10.1085/jgp.113.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chanda B, Bezanilla F. Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J Gen Physiol. 2002;120:629–645. doi: 10.1085/jgp.20028679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarov-Yarovoy V, et al. Molecular determinants of voltage-dependent gating and binding of pore-blocking drugs in transmembrane segment IIIS6 of the Na(+) channel alpha subunit. J Biol Chem. 2001;276:20–27. doi: 10.1074/jbc.M006992200. [DOI] [PubMed] [Google Scholar]
- 28.Capes D, Arcisio-Miranda M, Bezanilla F, Chanda B. Measuring the contribution of S4 charges on gating currents of a sodium channel. Biophys J. 2009;93:252a. [Google Scholar]
- 29.West JW, et al. A cluster of hydrophobic amino acid residues required for fast Na(+)-channel inactivation. Proc Natl Acad Sci U S A. 1992;89:10910–10914. doi: 10.1073/pnas.89.22.10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahern CA, Horn R. Focused electric field across the voltage sensor of potassium channels. Neuron. 2005;48:25–29. doi: 10.1016/j.neuron.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Chanda B, Bezanilla F. A common pathway for charge transport through voltage-sensing domains. Neuron. 2008;57:345–351. doi: 10.1016/j.neuron.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Tristani-Firouzi M, Chen J, Sanguinetti MC. Interactions between S4-S5 linker and S6 transmembrane domain modulate gating of HERG K+ channels. J Biol Chem. 2002;277:18994–19000. doi: 10.1074/jbc.M200410200. [DOI] [PubMed] [Google Scholar]
- 33.Ma JC, Dougherty DA. The Cation-π Interaction. Chem Rev. 1997;97:1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- 34.Ryzhov V, Dunbar RC, Cerda B, Wesdemiotis C. Cation-pi effects in the complexation of Na+ and K+ with Phe, Tyr, and Trp in the gas phase. J Am Soc Mass Spectrom. 2000;11:1037–1046. doi: 10.1016/s1044-0305(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 35.Mecozzi S, West AP, Jr, Dougherty DA. Cation-pi interactions in aromatics of biological and medicinal interest: electrostatic potential surfaces as a useful qualitative guide. Proc Natl Acad Sci U S A. 1996;93:10566–10571. doi: 10.1073/pnas.93.20.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 37.Hackos DH, Chang TH, Swartz KJ. Scanning the intracellular S6 activation gate in the shaker K+ channel. J Gen Physiol. 2002;119:521–532. doi: 10.1085/jgp.20028569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding S, Horn R. Effect of S6 tail mutations on charge movement in Shaker potassium channels. Biophys J. 2003;84:295–305. doi: 10.1016/S0006-3495(03)74850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yifrach O, Mackinnon R. Energetics of pore opening in a voltage-gated K(+) channel. Cell. 2002;111:231–239. doi: 10.1016/s0092-8674(02)01013-9. [DOI] [PubMed] [Google Scholar]
- 40.Wynia-Smith SL, Gillian-Daniel AL, Satyshur KA, Robertson GA. hERG gating microdomains defined by S6 mutagenesis and molecular modeling. J Gen Physiol. 2008;132:507–520. doi: 10.1085/jgp.200810083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ledwell JL, Aldrich RW. Mutations in the S4 region isolate the final voltage-dependent cooperative step in potassium channel activation. J Gen Physiol. 1999;113:389–414. doi: 10.1085/jgp.113.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brooks B, Karplus M. Harmonic dynamics of proteins: normal modes and fluctuations in bovine pancreatic trypsin inhibitor. Proc Natl Acad Sci U S A. 1983;80:6571–6575. doi: 10.1073/pnas.80.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCammon JA, Gelin BR, Karplus M, Wolynes PG. The hinge-bending mode in lysozyme. Nature. 1976;262:325–326. doi: 10.1038/262325a0. [DOI] [PubMed] [Google Scholar]
- 44.McCammon JA, Gelin BR, Karplus M. Dynamics of folded proteins. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 45.Oi VT, et al. Correlation between segmental flexibility and effector function of antibodies. Nature. 1984;307:136–140. doi: 10.1038/307136a0. [DOI] [PubMed] [Google Scholar]
- 46.Mao B, Pear MR, McCammon JA, Quiocho FA. Hinge-bending in L-arabinose-binding protein. The “Venus’sflytrap” model. J Biol Chem. 1982;257:1131–1133. [PubMed] [Google Scholar]
- 47.Colonna-Cesari F, et al. Interdomain motion in liver alcohol dehydrogenase Structural and energetic analysis of the hinge bending mode. J Biol Chem. 1986;261:15273–15280. [PubMed] [Google Scholar]
- 48.Auerbach A. Gating of acetylcholine receptor channels: brownian motion across a broad transition state. Proc Natl Acad Sci U S A. 2005;102:1408–1412. doi: 10.1073/pnas.0406787102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitra A, Tascione R, Auerbach A, Licht S. Plasticity of acetylcholine receptor gating motions via rate-energy relationships. Biophys J. 2005;89:3071–3078. doi: 10.1529/biophysj.105.068783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purohit P, Mitra A, Auerbach A. A stepwise mechanism for acetylcholine receptor channel gating. Nature. 2007;446:930–933. doi: 10.1038/nature05721. [DOI] [PubMed] [Google Scholar]
- 51.Muroi Y, Chanda B. Local anesthetics disrupt energetic coupling between the voltage-sensing segments of a sodium channel. J Gen Physiol. 2009;133:1–15. doi: 10.1085/jgp.200810103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 54.Zhorov BS. Vector method for calculating derivatives of energy of atom-atom interactions of complex molecules according to generalized coordinates. J Struct Chem. 1981;22:4–8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.