Abstract
The intracellular environment imposes a variety of constraints on biochemical reaction systems that can substantially change reaction rates and equilibria relative to an ideal solution-based environment. One of the most notable features of the intracellular environment is its dense macromolecular crowding, which, among many other effects, tends to strongly enhance binding and assembly reactions. Despite extensive study of biochemistry in crowded media, it remains extremely difficult to predict how crowding will quantitatively affect any given reaction system due to the dependence of the crowding effect on numerous assumptions about the reactants and crowding agents involved. We previously developed a two dimensional stochastic off-lattice model of binding reactions based on the Green’s function reaction dynamics (GFRD) method in order to create a versatile simulation environment in which one can explore interactions among many parameters of a crowded assembly system. In the present work, we examine interactions among several critical parameters for a model dimerization system: the total concentration of reactants and inert particles, the binding probability upon a collision between two reactant monomers, the mean time of dissociation reactions, and the diffusion coefficient of the system. Applying regression models to equilibrium constants across parameter ranges shows that the effect of the total concentration is approximately captured by a low-order nonlinear polynomial model, while the other three parameter effects are each accurately captured by a linear model. Furthermore, validation on tests with multi-parameter variations reveals that the effects of these parameters are separable from one another over a broad range of variation in all four parameters. The simulation work suggests that predictive models of crowding effects can accommodate a wider variety of parameter variations than prior theoretical models have so far achieved.
I. INTRODUCTION
Physiological and biochemical conditions inside of a cell are very different from the well-mixed and dilute conditions in typical in vitro models [1]. One of the most significant features of the intracellular environment is its dense molecular crowding, with total concentrations in the cell generally in the range of 50–400 mg/ml [2]. This molecular crowding can enhance many important cellular processes, such as macromolecule association [3–6], protein folding [7,8], and enzyme activity [9,10]. Molecular crowding significantly increases equilibria and reaction rates for many cellular processes, with crowded and dilute conditions sometimes differing by more than a hundred-fold [11]. Theoretical and computational approaches have been used to predict how crowding influences biochemical reaction processes under a variety of assumptions [11–15]. Various theoretical models have treated crowding effects as a form of “correction” to classical mass-action models of idealized chemistry. For example, Kopelman [16] and Schnell and Turner [17] developed models for reaction systems in crowded media based on the concept of fractal reaction orders. Minton and collaborators developed a theoretical framework based on statistical thermodynamics models and scaled particle theory for modeling reaction thermodynamics and kinetics in terms of additive corrections to activity coefficients relative to idealized reaction rates [18–23]. Such models have made it possible to explore how many individual parameter variations in a system influence the magnitude and direction of crowding effects relative to idealized kinetics [18, 24, 25]. Such analytical approaches are crucial for gaining insight into how various parameters affect crowding, but they can be limited by the difficulty of analytically expressing and analyzing the diversity of possibly synergistic features of a system.
Simulation methods provide an alternative that can make predictions about arbitrarily complicated parameters or combinations of parameters in crowded media. Simulation methods are limited primarily by the accuracy of their underlying models and the computational cost required to apply them [15, 26]. For example, Monte Carlo lattice models provide high efficiency but with the price of oversimplified geometry that can lead them to overestimate the influence of crowding on binding chemistry [27–29]. Off-lattice models based on Brownian or Langevin dynamics provide a more realistic model of particle trajectories, but at greatly increased computational cost [30, 31]. These computational costs in turn limit one’s ability to thoroughly explore a parameter space. To attempt to better balance these competing factors, we previously developed a coarse-grained two dimensional stochastic off-lattice model (2DSOLM) [32] based on GFRD [33] for simulating binding kinetics in various molecular crowding conditions. The prior study confirmed that this coarse-grained model provides a reasonable qualitative representation of the effects of crowding on binding chemistry, exhibiting an expected enhancement of polymerization with increasing crowding by either reactant monomers or inert crowding agents. Although this simulator still yields a highly simplified representation of the cellular environment we wish to study, it provides a way to isolate specific features of crowded systems and examine how variations in those specific features, singly or in combination, would influence binding chemistry in an idealized model. Such an idealized model will provide a basis for moving towards more realistic computational models of biochemistry in the cell by helping us identify precisely which features of the cell must be explicitly incorporated into biochemical models, which can be adequately captured by simplified mathematical corrections, and which are superfluous to producing quantitatively accurate descriptions of reaction chemistry.
In the present work, we use the simulation method to explore how the parameters of 2DSOLM affect binding thermodynamics with the goal of identifying parameters that act independently of one another and can be fit to simple analytical models. We explore how dimerization propensity is affected by four adjustable parameters: the total concentration (C), defined as the ratio of all particle area to the investigated area; the probability of binding upon a collision between two reactant monomers (B); the mean time for dissociation event (M), defined as the inverse of the rate constant; and the diffusion coefficient for reactants and inert particles (D). We show that a simple regression model fit analytically to simulation results on variations of C, B, M, and D accurately predicts their variations across a broad range of the simulation parameter space. Because our model is a hypothetical homodimerization system in two dimensional space, the simulating conditions of our model are far from the actual conditions of the intracellular environment. Our model and simulation results, however, demonstrate an opportunity for which a predictive regression model can provide a means of controlling for crowding effects in simulations too large in particle numbers or too long in timescale to allow for an explicit crowding model.
II. METHOD
A. Simulation conditions and parameter values
We conducted simulations using a model of homodimerization reactions developed in our prior work [32], which models reactant monomers and dimers as well as inert crowding agents as rigid, circular particles. Simulation proceeds through GFRD [33], a discrete-event method for fast off-lattice particle simulation. We applied a hard reflective boundary condition with a square boundary (100 nm × 100 nm) and used a reactant monomer radius of 2.5 nm, as in our prior work [32]. These values are meant to approximate the size scale for a globular actin monomer [34] in a typical eukaryotic cell. Note, however, that while the motivation of this work is to improve models of biochemistry in the cell, the model itself is a highly simplified and is more properly regarded as an idealized model of a crowded system rather than a model of the cell per se.
In order to discover the individual parameter effects on the homodimerization reaction, we first assigned default values for all parameters and then tested variations in each single parameter individually with all others held at their default values. We established a baseline simulation parameter set with default parameter values of B=0.7, M=1ns, and D=13.9×10−11m2s−1. These default values were chosen based on our prior simulation study [32] to produce a reasonably strong crowding effect as well as to approximate the temperature and viscosity conditions of the cytoplasm [35, 36]. The model also has three additional adjustable parameters that were not explored here: the ratio of dimer area to monomer area ( ); the ratio of inert particle area to reactant monomer area ( ); and the threshold distance, a simulation parameter describing the maximum distance at which two particles can interact with each other (dth). For the present study, we set α=2 (dimer area is exactly twice that of an unbound monomer), β=1(crowding agents have equal size to reactant monomers), and dth=0.5nm (threshold distance is one fifth of the radius of a reactant monomer), values also chosen based on our prior simulation study [32] to provide a physically reasonable model of dimerization. Second, we separately varied the individual parameter values for B, M, and D with the default values for the other parameters to test these individual parameter effects on the binding chemistry. We simulated five B values (0.1, 0.3, 0.5, 0.7, 0.9), five M values (0.6, 0.8, 1.0, 1.2, 1.4ns) and five D values (3.9, 13.9, 23.9, 33.9, 43.9×10−11m2s−1). For each of these conditions, we simulated eight total concentrations (C) (0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45) with dimensionless units of fraction of total simulation area occupied by particles. To produce varying concentrations, we began with a fixed concentration of reactants of 0.1 and then added inert crowding particles to yield each higher value of total concentration (e.g., C=0.1 corresponds to purely reactant monomers while C=0.25 corresponds to concentration 0.1 of reactant monomers and 0.15 of inert crowding agents). Initially, all reactants are monomers. Initialization of particle positions is performed by establishing a grid of potential particle positions at the maximum possible packing density for whichever of reactant monomers and crowding agents occupies the larger total area and then randomly inserting particles into the corresponding grid positions. This protocol was developed because it makes it possible to initialize in highly crowded conditions where independent uniform placement of particles would usually result in overlapping particles. Each simulation was run for 25μs with 30 repetitions per simulation, with progress recorded every 0.15625μs. For each condition, we measured reaction progress by the mean number of dimers as a function of time across all simulations.
B. Fitting Keq to simulation results
We used the results of the simulations described in the preceding section to fit an equilibrium constant (Keq) to each condition. For the homodimerization reaction, the governing chemical reaction is given by Eq. (1):
| (1) |
where M is the reactant monomer, D is the reactant dimer, I is the inert crowding particle, K+ is the forward reaction rate, and K− is the reverse reaction rate. The equilibrium constant can be computed from Eq. (1) as follows:
| (2) |
where [Deq] is the concentration of dimers at the quasi-equilibrium state, [Meq] is the concentration of monomers at the quasi-equilibrium state, M0 is the number of initial monomers, Meq is the number of monomers at the quasi-equilibrium state, Deq is the number of dimers at the quasi-equilibrium state, and A is the square investigated area ((100nm)2). The inert particle in Eq. (1) cannot influence the homodimeriation reaction in an idealized mass-action model, because [I] drops out in Eq. (2). Therefore, Eq. (2) is only used to calculate the equilibrium constant from the simulation data on the assumption that 2DSOLM appropriately represents the crowding effect of all particles in the various test cases. In addition, we derived the quadratic equation to calculate the number of dimers at the quasi-equilibrium state (Deq) using the estimated Keq value from Eq. (2), shown in Eq. (3). The analytical solution of Eq. (3) is Eq. (4).
| (3) |
| (4) |
where estimates were based on averaging dimer counts at 5μs intervals from 5–25 μs. 5μs was judged a reasonable time to mix the system based on a series of additional simulations conducted to estimate the mixing time of the system under different parameter conditions. For these experiments, we placed a single reactant monomer at the center of the investigated area and measured its displacement from the center as a function of time for varying C and D values with all the other particles inert. These plots were used to establish a mixing time usable across simulation conditions for measuring steady-state dimer counts. Errorbars in all plots of simulation results were estimated from the standard deviations across the five 5μs time points and thirty repetitions per condition, giving 150 total measurements per condition. This data collection approach produces a reasonably large amount of data while allowing us to simulate the reaction for a relatively long time, providing a post hoc check that simulation time was sufficient to reach a steady-state.
To analyze the molecular crowding effect on the homodimerization reaction in 2DSOLM, we first calculated Keq values for the simulation results using Eq. (2) from C=0.1 to 0.45 at increments of 0.05. Second, we fit polynomials of degrees 0 through 12 for varying C and default values of all other parameters and measured the root mean value of the sum of square difference between the regression equation and simulations for each value of C with 10-fold cross-validation. We used the resulting best-fit to build a regression model of Keq in terms of C, assuming defaults for all other parameters. We also estimated dimer counts at quasi-equilibrium using Keq values from the regression model and Eq. (4).
To derive regression models of B, M, and D, we first calculated Keq values for the simulation results of different B, M, and D values separately using Eq. (2) from C=0.1 to 0.45 at increments of 0.05. Second, we fit three scaling factors, expressed as multiplicative functions of B, M, and D individually, so as to provide a least-squares best fit to the simulation estimates of Keq as a function of each parameter. We then multiplied the previously-built regression model of C by these three scaling factors, corresponding to an assumption that the crowding effect acts independently of these three parameters. Finally, we created a unified regression model by assuming that the effects of each of the four parameters on Keq are independent and multiplicative.
C. Evaluation of the regression model
To evaluate the unified regression model, we randomly selected 40 test cases for fixed reactant concentration 0.1 and other parameters selected uniformly at random the ranges C = 0.1 – 0.45, B = 0.1 – 0.9, M = 0.6 – 1.4ns, and D = 3.9 – 43.9 ×10−11m2s−1. For each test case, we compared the mean dimer counts from the simulation to the estimated dimer counts predicted by the unified regression model and Eq. (4). As an additional test of the performance of the model outside the parameter ranges considered in model-fitting, we conducted another 40 random trials in which we also varied the concentration of reactants (CR) uniformly between 0.1 and 0.45 for the second 40 test cases, choosing the concentration of inert particles (CI) uniformly from 0 to 0.45-CR and B, M, and D from the same ranges as above. We similarly compared these simulation results with estimates from the regression model. Simulation values are averages over 30 repetitions per data point.
In order to test whether boundary effects induced by the relatively small bounding box might influence the reactions, we conducted additional random parameter experiments using a 200nm × 200nm boundary area. For these experiments, we selected parameter values at random from the same ranges as in the preceding paragraph. As before, we conducted 40 test cases with fixed CR=0.1 and variable inert concentration from 0 to 0.35 and 40 test cases with CR uniformly between 0.1 and 0.45 and CI uniformly between 0 and 0.45-CR. We compared the results to the predictions from the regression model. Due to the larger number of particles per trial, we performed five repetitions per data point.
D. Generating movie files
To help illustrate the effects of various parameters on binding kinetics, we animated the dynamics of these particles in movie files showing the progress of individual homodimerization simulations. Each movie contains two different moving images, which are distinguished by different values for a single parameter while all other parameters use the common default values. We generated movies for the following comparisons: C=0.1 (0.1 CR + 0.0 CI) vs C=0.45 (0.1 CR + 0.35 CI), B=0.1 vs B=0.9, M=0.6ns vs M=1.4ns, and D=3.9×10−11m2s−1 vs D=43.9×10−11m2s−1. The first half of each movie shows the system in the initial (pre-equilibration) state, and the second half of the movie shows a quasi-equilibrium state [37]. High-resolution versions of the movies can be downloaded from: http://www.cs.cmu.edu/~russells/projects/crowding/crowding.html.
III. RESULTS
A. Reaction progress in 2DSOLM
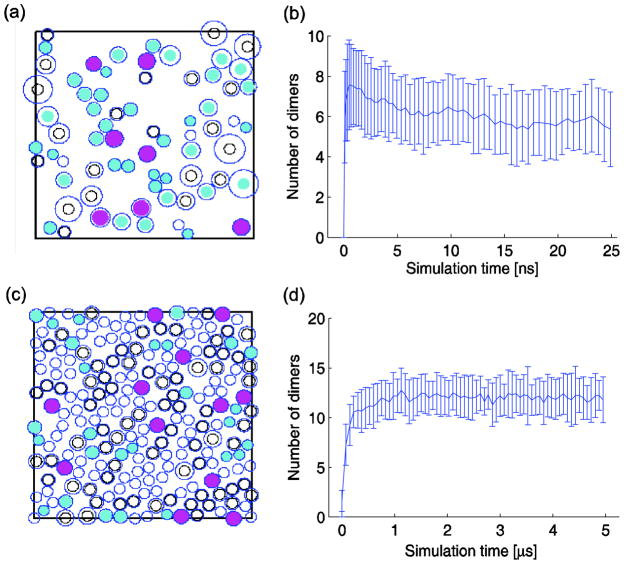
Fig. 1 illustrates the progress of typical simulations, illustrating a snapshot of the simulator at the quasi-equilibrium state (25μs) and the number of dimers versus time for default parameters (B=0.7, M=1ns, D=13.9×10−11m2s−1, α=2, β=1, and dth=0.5nm) under two crowding conditions: low crowding (C=0.15; 0.1 for reactants + 0.05 for inert particles, with simulation progress shown from 0 to 25ns in Fig. 1(b)) and high crowding (C=0.45; 0.1 for reactants + 0.35 for inert particles, with simulation progress shown from 0 to 5μs in Fig. 1(d)). Initially, all reactant particles are monomers, so the dimer count is zero at 0s. The system approaches the quasi-equilibrium state at roughly 5ns for the low crowding case and at 1μs for the high crowding case, and then shows fluctuation of dimer counts around an equilibrium value. There is approximately a 2-fold difference in the equilibrium level between these two scenarios, although more than two orders of magnitude difference in the kinetics of the reaction.
FIG. 1.
(Color online) Illustrations of the 2DSOLM reaction simulation for various crowded environments. (a) Snapshot of a low crowding case C=0.15 (0.1 for reactants + 0.05 for inert particles) at 25μs. Cyan (gray in print) circles represent reactant monomers, magenta (dark in print) circles represent reactant dimers, open circles represent inert particles, and the outer circle of a particle represents a diffusion limit circle in 2DSOLM. (b) Reaction progress from 0 to 25ns for the low-crowding condition of (a), (c) snapshot of a high crowding case C=0.45 (0.1 for reactants + 0.35 for inert particles) at 25μs. (d) Reaction progress from 0 to 5μs for the high-crowding condition of (c). The default parameter values (B=0.7, M=1ns, D=13.9×10−11m2s−1, α=2, β=1, and dth=0.5nm) are used with both low and high crowding cases.
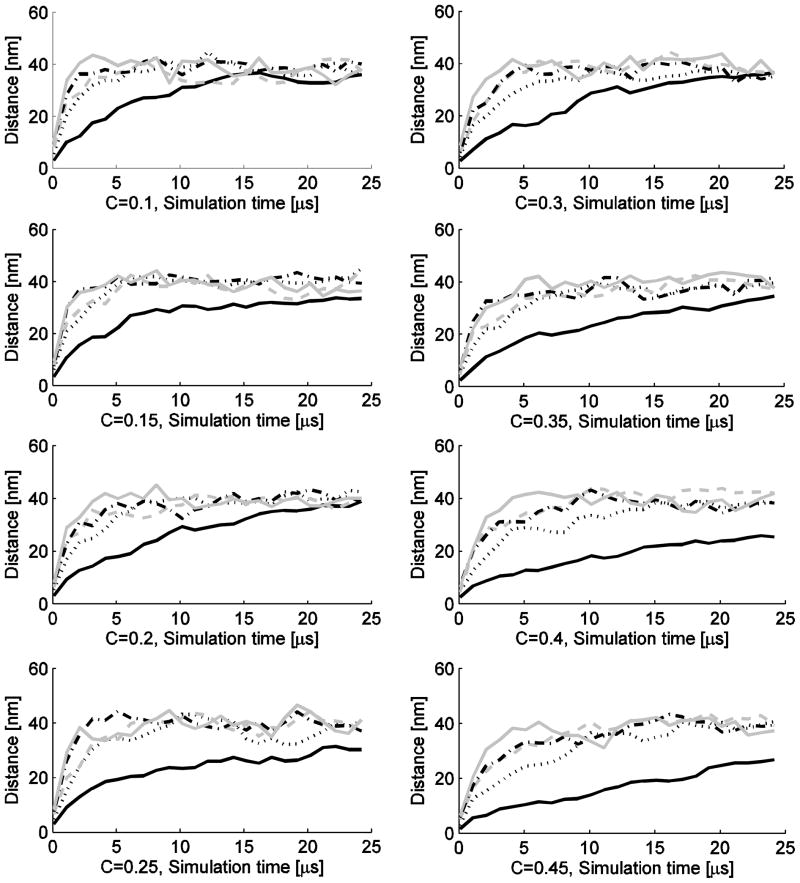
Given large differences in kinetics of the simulations, we ran a series of simulation experiments examining particle diffusion as a function of time to determine a reasonable upper bound on mixing time across parameter values, as described in Sec. II.B. The results are shown in Fig. 2. Based on the observed mixing times, we concluded that 5μs would be sufficient time to adequately mix at all but the lowest extreme of diffusion rate. We therefore estimate equilibrium dimer counts in subsequent experiments by averaging over 5μs intervals from 5–25μs per experiment.
FIG. 2.
Average distance from the origin versus time for a single reactant particle embedded with varying numbers of inert particles. The subparts of the figure each show the average displacement of the single reactant particle versus time for one value of C (0.1–0.45) and for five values of D: D=3.9 (black solid), 13.9 (black dotted), 23.9 (gray dashed), 33.9 (black dash-dot), 43.9 ×10−11m2s−1 (gray solid). The experiments measured the displacement of the reactant monomer from (0 – 25μs) with 30 repetitions per each simulation case. Errorbars are not depicted because they hinder readability of the figures.
B. Parameter effect on binding chemistry: C
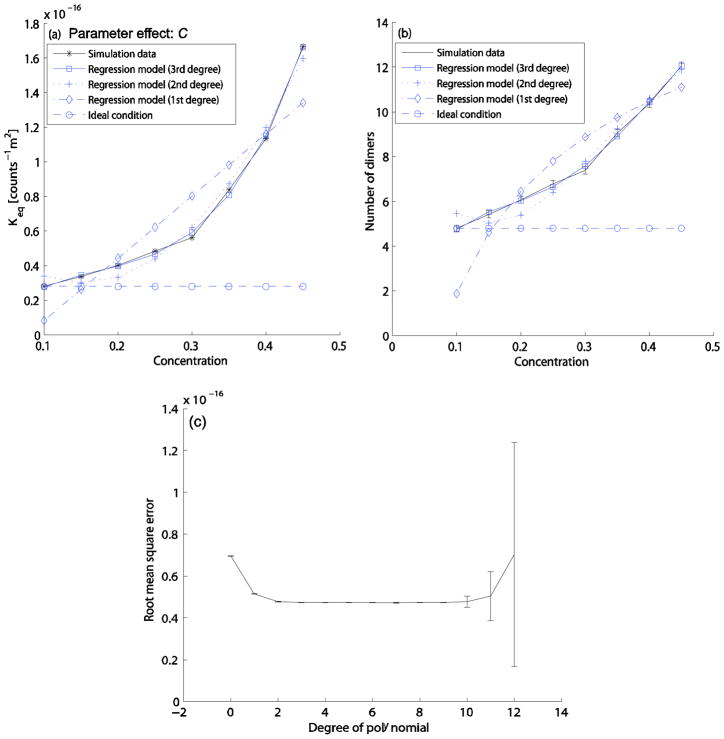
In analyzing effects of parameter variations, we first attempted to determine a model of the effect of C on Keq. We calculated the average Keq from Eq. (2) and the simulation data from C=0.1 to C=0.45, with all other parameters at default values. All particles are reactants at C=0.1, and inert particles are added as C increases. The calculated Keq values for different concentrations are shown in Fig. 3(a), represented by a solid line with star marks. We fit the Keq curve by polynomial regression of varying degrees, minimizing sum-of-square errors for each degree with 10-fold cross validation. Fig. 3(a) shows best-fit first-, second-, and third-order regression models, along with the idealized Keq assuming no crowding effect. We see negligible improvement in regression quality past degree three; beyond degree nine, cross-validated accuracy begins to drop, suggestive of overfitting (Fig. 3(c)). The resulting best-fit third-order model is given in Eq. (5):
| (5) |
FIG. 3.
Effect of parameter C on the homodimerization reaction. For the simulation data, all particles are reactants at C=0.1, and then inert particles are added as the total concentration increases. C values are tested from 0.1 to 0.45 as the increment of 0.05. The other parameter values for this test case are set to the default parameter values (B=0.7, M=1ns, D=13.9×10−11m2s−1, α=2, β=1, and dth=0.5nm). (a) Keq curves: simulation data (solid line with star marks), 3rd degree fitted curve (solid line with square marks), 2nd degree fitted curve (dotted line with plus marks), 1st degree fitted curve (dash-dot line with diamond marks), idealized values assuming no crowding effect (dashed line with circle marks). (b) Quasi-equilibrium dimer count curves: simulation data (solid line with errorbars), predicted data curve from the 3rd degree fitted curve in (a) (solid line with square marks), predicted data curve from the 2nd degree fitted curve in (a) (dotted line with plus marks), predicted data curve from the 1st degree fitted curve in (a) (dash-dot line with diamond marks), idealized values assuming no crowding effect (dashed line with circle marks). (c) Root mean square errors of fit for the C regression model as a function of model degree. Each fit is based on 10-fold cross validation using 100 iterations, with all data points randomly shuffled for every iteration.
Note that because C is dimensionless, as it is the area ratio of all particles to the simulation space, we must multiply the formula by (1counts−1m2) to match the physical dimension. Fig. 3(b) shows dimer counts at pseudo-equilibrium for the homodimerization reaction from the simulation data; the expected constant dimer count assuming no crowding effect (or infinitely dilute conditions); and the calculated data from the first, second, and third degree polynomials of Fig. 3(a) as determined by Eq. (4), providing an alternative way of visualizing the quality of the fit.
C. Parameter effects on binding chemistry: B, M, and D
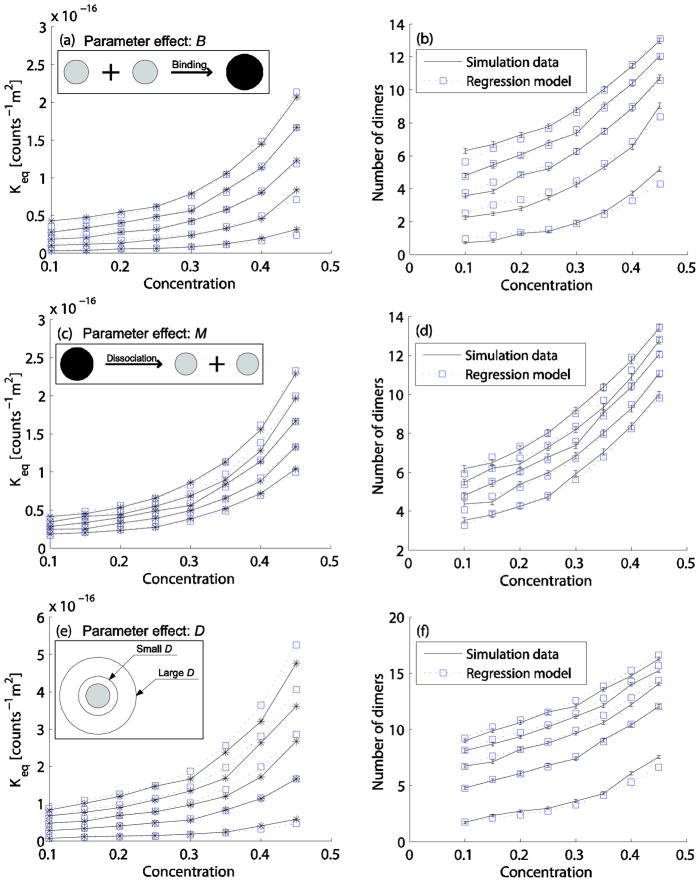
We next calculated the Keq values of the simulation data for different B, M, and D values (Fig. 4) with fixed reactant monomer concentration 0.1, varying C from 0.1 to 0.45 and all other parameters set to the defaults. Fig. 4(a) shows Keq for B values 0.1 (bottom), 0.3, 0.5, 0.7, and 0.9 (top). The figure also shows a linear regression fit to the resulting Keq curves, which was combined multiplicatively with the previous third-order C regression model in Eq. (5) to yield the following two-parameter regression curve:
| (6) |
FIG. 4.
Effect of parameters B, M, and D on the homodimerization reaction. The first column (a, c, e) shows the Keq curves of the average simulation data (solid line with star marks) and fitted data (dotted line with square marks) from Eq. (6–8, respectively). The second column (b, d, f) shows the dimer counts for simulation data (errorbar) and predicted data (a dotted line with square marks) from Eq. (6–8, respectively) and Eq. (4). (a,b) B values from 0.1 (bottom), 0.3, 0.5, 0.7, 0.9 (top), and C from 0.1 (all reactants) to C=0.45 (0.1 for reactants and 0.35 for inert particles) with other parameter values set to the default values (M=1ns, D=13.9×10−11m2s−1, α=2, β=1, and dth=0.5nm). (c,d) M values from 0.6 (bottom), 0.8, 1.0, 1.2, 1.4 ns (top) with other conditions the same as in (a,b). (e,f) D values from 3.9 (bottom), 13.9, 23.9, 33.9, 43.9×10−11m2s−1 (top) with other conditions the same as for (a,b).
Increasing B yields more dimers at the quasi-equilibrium state because it increases the probability of binding between two reactant monomers upon collisions. Fig. 4(b) shows the measured and predicted equilibrium dimer counts resulting from this curve. Fig. 4(c) shows Keq for M values 0.6 (bottom), 0.8, 1.0, 1.2, and 1.4ns (top), along with the regression model
| (7) |
derived by multiplicative combination of the C model with a linear best-fit regression model for M. Increasing M yields more dimers at the quasi-equilibrium state due to reduced dissociation rates. Fig. 4(d) shows the measured and predicted equilibrium dimer counts from the regression models for the same variations in M and C. Fig. 4(e) shows Keq for D values 3.9 (bottom), 13.9, 23.9, 33.9, and 43.9×10−11m2s−1 (top) using a best-fit linear model of D combined with the previous third-order C model:
| (8) |
Increasing D produces more dimers at the quasi-equilibrium state by increasing the collision frequency of reactant monomers and thus the forward reaction rate. Fig. 4(f) shows the measured and predicted equilibrium dimer counts from the regression models for the same variations in D and C. The relative scaling factor terms in Eq. (6–8) are dimensionless because each term is divided by the default values. Therefore, Eq. (6–8) are multiplied by (1counts−1m2) to match the physical dimension. The figures demonstrate that the contributions of B, M, and D to the crowding effect can each individually be treated as independent from the contribution of C.
D. Evaluating a unified regression model of C, B, M, and D
We next sought to test whether it is possible to build a unified model of all four parameters (C, B, M, and D) from their single-parameter fits based on the assumption that they independently contribute to the homodimerization reaction equilibrium. We therefore constructed a unified regression model of Keq for C, B, M, and D follows:
| (9) |
We then evaluated how well the model would predict simultaneous changes in all four parameters. To evaluate Eq. (9), we simulated two different test conditions. We first simulated 40 different test cases in which we randomly chose the concentration of inert particles (CI: from 0–0.35), B (from 0.1–0.9), M (from 0.6–1.4ns), and D (from 3.9–43.9 ×10−11m2s−1) values at a fixed concentration of reactants (CR=0.1). Table I provides the parameter values for the forty test cases. The other parameter values were set to the default values: (α=2,β=1, and dth=0.5nm). Fig. 5(a) plots the average simulation values versus predicted values from Eq. (4) and Eq. (9). The simulation values for each test case are derived from 30 different simulation runs with 5 different time progression points (5, 10, 15, 20, 25μs). The figure shows a close match between the regression model of Eq. (9) and the simulations across a broad range of crowding levels, reaction rates, and diffusion rates.
TABLE I.
Test cases for the evaluation of the regression model assuming fixed reactant concentration in a 100nm × 100nm area. We fixed the concentration of reactants as 0.1 as in the experiments used to derive the regression model and randomly selected the other parameter values from their parameter ranges: CI (0 – 0.35), B(0.1 – 0.9), M(0.6 – 1.4ns), D(3.9 – 43.9 ×10−11m2s−1).
| Test | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| CR | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| CI | 0.08 | 0.32 | 0.16 | 0.29 | 0.23 | 0.17 | 0.34 | 0.28 | 0.18 | 0.07 |
| B | 0.19 | 0.68 | 0.56 | 0.67 | 0.5 | 0.88 | 0.17 | 0.18 | 0.3 | 0.83 |
| M | 0.84 | 0.91 | 0.98 | 1.29 | 1.19 | 1.21 | 1.07 | 0.98 | 1.24 | 1.13 |
| D | 2.53E-10 | 2.50E-10 | 2.42E-10 | 3.99E-10 | 3.35E-10 | 2.96E-10 | 3.71E-10 | 4.31E-10 | 1.55E-10 | 3.10E-10 |
| Test | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| CR | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| CI | 0.04 | 0.07 | 0.27 | 0 | 0.01 | 0.14 | 0.06 | 0.35 | 0.2 | 0.24 |
| B | 0.26 | 0.36 | 0.24 | 0.42 | 0.61 | 0.87 | 0.33 | 0.2 | 0.62 | 0.73 |
| M | 1.07 | 1.33 | 0.91 | 0.66 | 0.73 | 0.78 | 0.66 | 0.64 | 0.89 | 0.64 |
| D | 8.12E-11 | 4.32E-10 | 8.56E-11 | 1.95E-10 | 2.05E-10 | 2.63E-10 | 2.09E-10 | 1.35E-10 | 8.95E-11 | 1.58E-10 |
| Test | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 |
| CR | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| CI | 0.24 | 0.17 | 0.18 | 0.16 | 0.06 | 0.11 | 0.05 | 0.01 | 0.34 | 0.08 |
| B | 0.22 | 0.67 | 0.13 | 0.67 | 0.55 | 0.65 | 0.54 | 0.27 | 0.24 | 0.44 |
| M | 1.32 | 1.02 | 0.96 | 0.88 | 1.23 | 0.84 | 1.38 | 1.32 | 0.98 | 0.99 |
| D | 3.35E-10 | 4.23E-10 | 3.86E-10 | 1.94E-10 | 4.34E-10 | 8.48E-11 | 1.65E-10 | 2.62E-10 | 2.60E-10 | 6.74E-11 |
| Test | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 |
| CR | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| CI | 0.08 | 0.03 | 0.09 | 0.13 | 0.31 | 0.16 | 0.01 | 0.04 | 0.24 | 0.3 |
| B | 0.86 | 0.83 | 0.19 | 0.59 | 0.81 | 0.7 | 0.71 | 0.4 | 0.48 | 0.48 |
| M | 0.8 | 0.85 | 0.71 | 0.68 | 1.23 | 1.01 | 0.61 | 0.8 | 0.86 | 0.71 |
| D | 1.69E-10 | 1.04E-10 | 4.38E-10 | 1.78E-10 | 1.56E-10 | 3.89E-10 | 2.11E-10 | 1.63E-10 | 1.28E-10 | 3.81E-10 |
FIG. 5.
Comparison of dimer counts between simulation data and predicted data for simultaneous variation in C, B, M, and D. Predicted data are calculated by Eq. (4) and Eq. (9). (a) test cases for fixed reactant concentration (CR) of 0.1 (Table I) in a 100nm × 100nm area. Concentrations of inert particles (CI), B, M, D are randomly selected, with CI from 0–0.35, B from 0.1–0.9, M from 0.6–1.4ns, and D from 3.9–43.9 ×10−11m2s−1. (b) test cases for varying reactant concentration (CR) from 0.1–0.45 (Table II) in a 100nm × 100nm area. CI, B, M, and D are also randomly selected, with CI from 0 – (0.45-CR), B from 0.1–0.9, M from 0.6–1.4ns, and D from 3.9–43.9 ×10−11m2s−1. (c) test cases for fixed reactant concentration (CR) of 0.1 (Table III) in a 200nm × 200nm area. CI, B, M, and D are randomly selected from the same parameter ranges as in (a). (d) test cases for varying reactant concentration (CR) from 0.1–0.45 (Table IV) in a 200nm × 200nm area. CI, B, M, and D are randomly selected from the same parameter ranges as in (b).The other parameter values are set to the default values: (α=2,β=1, and dth=0.5nm).
To examine how well the model would perform outside of the training parameter set, we simulated 40 additional test cases for which we randomly chose the concentration of reactants (CR:0.1–0.45) and then varied the total concentration and other parameters as in the preceding test cases: concentration of inert particles [CI: from 0 – (0.45-CR)], B (from 0.1–0.9), M (from 0.6–1.4ns), and D (from 3.9–43.9 ×10−11m2s−1). The parameter values for these forty test cases are shown in Table II. The other parameter values are set to the default values: (α=2,β=1, and dth=0.5nm). Fig. 5(b) plots the average simulation values versus predicted values from the regression model of Eq. (4) and Eq. (9). The simulation values of each test case are derived from 30 different simulation runs with 5 different time progression points (5, 10, 15, 20, 25μs). The figure reveals high accuracy for low to moderate reactant concentrations. The regression model does, however, slightly overestimate the equilibrium dimer counts under conditions in which the equilibrium favors exceptionally high dimer concentration (more than approximately 40).
TABLE II.
Test cases for the evaluation of the regression model allowing varying reactant concentrations in a 100nm × 100nm area. We randomly selected all parameter values from the following parameter ranges: CR (0.1 – 0.45), CI [0 – (0.45-CR)], B(0.1 – 0.9), M(0.6 – 1.4ns), D(3.9 – 43.9 ×10−11m2s−1).
| Test | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| CR | 0.38 | 0.32 | 0.1 | 0.43 | 0.19 | 0.24 | 0.25 | 0.3 | 0.43 | 0.43 |
| CI | 0.05 | 0.08 | 0.03 | 0.01 | 0.02 | 0.12 | 0.08 | 0.04 | 0.02 | 0.01 |
| B | 0.53 | 0.54 | 0.69 | 0.16 | 0.54 | 0.24 | 0.17 | 0.31 | 0.18 | 0.33 |
| M | 1.11 | 1.07 | 1.32 | 1.34 | 1.24 | 1.21 | 0.61 | 0.72 | 0.6 | 0.63 |
| D | 3.04E-10 | 2.70E-10 | 4.30E-10 | 2.16E-10 | 3.22E-10 | 4.47E-11 | 2.68E-10 | 1.49E-10 | 2.10E-10 | 3.25E-10 |
| Test | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| CR | 0.42 | 0.23 | 0.4 | 0.38 | 0.19 | 0.14 | 0.37 | 0.13 | 0.12 | 0.1 |
| CI | 0.02 | 0.02 | 0.02 | 0 | 0.26 | 0.24 | 0.05 | 0.04 | 0.33 | 0.19 |
| B | 0.13 | 0.46 | 0.17 | 0.52 | 0.63 | 0.9 | 0.18 | 0.59 | 0.55 | 0.64 |
| M | 0.85 | 0.68 | 1.29 | 1.31 | 1.17 | 1.19 | 0.76 | 1.32 | 0.82 | 1.24 |
| D | 2.08E-10 | 9.03E-11 | 2.11E-10 | 3.65E-10 | 3.64E-10 | 2.63E-10 | 9.49E-11 | 3.35E-10 | 2.02E-10 | 3.37E-10 |
| Test | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 |
| CR | 0.12 | 0.37 | 0.35 | 0.2 | 0.43 | 0.26 | 0.15 | 0.3 | 0.39 | 0.36 |
| CI | 0.24 | 0.07 | 0.05 | 0.04 | 0 | 0.04 | 0.06 | 0.12 | 0.04 | 0.06 |
| B | 0.1 | 0.47 | 0.88 | 0.13 | 0.36 | 0.36 | 0.18 | 0.46 | 0.76 | 0.2 |
| M | 0.77 | 0.69 | 0.65 | 0.86 | 1.4 | 1.14 | 0.86 | 0.85 | 0.64 | 1.36 |
| D | 6.87E-11 | 9.56E-11 | 1.53E-10 | 2.24E-10 | 6.94E-11 | 5.59E-11 | 3.07E-10 | 2.75E-10 | 5.95E-11 | 5.77E-11 |
| Test | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 |
| CR | 0.42 | 0.25 | 0.24 | 0.17 | 0.22 | 0.11 | 0.44 | 0.22 | 0.11 | 0.31 |
| CI | 0.02 | 0 | 0.03 | 0.21 | 0.21 | 0.19 | 0.01 | 0.08 | 0.2 | 0.1 |
| B | 0.24 | 0.68 | 0.61 | 0.62 | 0.78 | 0.75 | 0.83 | 0.85 | 0.48 | 0.11 |
| M | 1.39 | 0.99 | 1.12 | 0.96 | 1.21 | 1.18 | 1.16 | 1.1 | 1.13 | 0.87 |
| D | 1.83E-10 | 5.44E-11 | 3.42E-10 | 3.36E-10 | 4.26E-10 | 2.87E-10 | 2.40E-10 | 4.04E-10 | 4.16E-10 | 4.08E-11 |
We simulated 80 additional test cases sampled from the same parameter ranges using a 200nm × 200nm bounding area in order to explore whether boundary effects significantly impact the fit of the regression model. The parameter values for these eighty test cases are shown in Table III and Table IV, respectively. Fig. 5(c) plots the average simulation values versus predicted values from the regression model of Eq. (4) and Eq. (9) for the fixed reactant concentration case and Fig. 5(d) for the variable reactant concentration case. The results appear comparable to those of Fig. 5(a) and (b). The regression model and simulations provide close matches for low to moderate reactant concentrations, but the regression model slightly overestimates the equilibrium dimer counts under conditions in which the equilibrium favors exceptionally high dimer concentration (more than approximately 160). There is no noticable difference in quality of fit between the two sizes of simulation.
TABLE III.
Test cases for the evaluation of the regression model assuming fixed reactant concentration in a 200nm × 200nm area. We fixed the concentration of reactants as 0.1 as in the experiments used to derive the regression model and randomly selected the other parameter values from their parameter ranges: CI (0 – 0.35), B(0.1 – 0.9), M(0.6 – 1.4ns), D(3.9 – 43.9 ×10−11m2s−1).
| Test | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| CR | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| CI | 0.06 | 0.32 | 0.28 | 0.01 | 0.13 | 0.32 | 0.22 | 0.05 | 0.01 | 0.01 |
| B | 0.69 | 0.35 | 0.78 | 0.25 | 0.41 | 0.69 | 0.16 | 0.75 | 0.19 | 0.78 |
| M | 1.28 | 1.37 | 0.8 | 0.9 | 0.62 | 1.15 | 1.25 | 1.23 | 1.08 | 0.67 |
| D | 3.37E-10 | 9.54E-11 | 1.73E-10 | 2.16E-10 | 3.60E-10 | 9.69E-11 | 1.40E-10 | 5.25E-11 | 2.18E-10 | 3.32E-10 |
| Test | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| CR | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| CI | 0.23 | 0.18 | 0.14 | 0.09 | 0.05 | 0 | 0.18 | 0.09 | 0.35 | 0.31 |
| B | 0.82 | 0.85 | 0.88 | 0.11 | 0.48 | 0.79 | 0.18 | 0.45 | 0.72 | 0.48 |
| M | 1.17 | 1.02 | 0.85 | 1.1 | 0.7 | 0.8 | 1.18 | 1.28 | 0.98 | 1.36 |
| D | 1.68E-10 | 4.76E-11 | 1.24E-10 | 4.02E-10 | 3.19E-10 | 2.48E-10 | 3.60E-10 | 2.04E-10 | 2.47E-10 | 7.54E-11 |
| Test | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 |
| CR | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| CI | 0.17 | 0.09 | 0.08 | 0.3 | 0.23 | 0.29 | 0.21 | 0.09 | 0.22 | 0.11 |
| B | 0.31 | 0.32 | 0.38 | 0.59 | 0.87 | 0.55 | 0.58 | 0.62 | 0.27 | 0.62 |
| M | 1.14 | 1.28 | 1.22 | 1.02 | 1.29 | 1.18 | 0.95 | 0.94 | 0.84 | 1.08 |
| D | 2.64E-10 | 1.34E-10 | 4.77E-11 | 1.89E-10 | 2.81E-10 | 3.49E-10 | 1.71E-10 | 1.49E-10 | 1.24E-10 | 2.95E-10 |
| Test | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 |
| CR | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| CI | 0.17 | 0.33 | 0.01 | 0 | 0.35 | 0.03 | 0.07 | 0.2 | 0.12 | 0.15 |
| B | 0.22 | 0.9 | 0.47 | 0.5 | 0.55 | 0.82 | 0.39 | 0.21 | 0.33 | 0.74 |
| M | 1.22 | 1.18 | 1.08 | 1.11 | 1.17 | 1.01 | 1.02 | 0.92 | 0.86 | 1.08 |
| D | 4.17E-10 | 2.18E-10 | 1.07E-10 | 3.06E-10 | 2.40E-10 | 2.32E-10 | 3.94E-10 | 5.72E-11 | 1.84E-10 | 2.34E-10 |
TABLE IV.
Test cases for the evaluation of the regression model allowing varying reactant concentrations in a 200nm × 200nm area. We randomly selected all parameter values from the following parameter ranges: CR (0.1 – 0.45), CI [0 – (0.45-CR)], B(0.1 – 0.9), M(0.6 – 1.4ns), D(3.9 – 43.9 ×10−11m2s−1).
| Test | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| CR | 0.24 | 0.22 | 0.13 | 0.38 | 0.14 | 0.34 | 0.3 | 0.21 | 0.38 | 0.29 |
| CI | 0.09 | 0.15 | 0.24 | 0.06 | 0.01 | 0.01 | 0.04 | 0.02 | 0.06 | 0.04 |
| B | 0.51 | 0.3 | 0.8 | 0.29 | 0.39 | 0.74 | 0.58 | 0.77 | 0.28 | 0.35 |
| M | 0.68 | 1.39 | 0.95 | 1.09 | 1.01 | 0.91 | 0.92 | 0.86 | 1.28 | 1.29 |
| D | 1.49E-10 | 2.22E-10 | 1.58E-10 | 1.78E-10 | 2.53E-10 | 1.11E-10 | 3.28E-10 | 6.52E-11 | 2.65E-10 | 9.39E-11 |
| Test | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| CR | 0.17 | 0.43 | 0.34 | 0.12 | 0.19 | 0.33 | 0.33 | 0.24 | 0.16 | 0.23 |
| CI | 0.22 | 0.01 | 0.06 | 0.24 | 0.13 | 0.09 | 0 | 0.13 | 0.17 | 0.19 |
| B | 0.63 | 0.32 | 0.82 | 0.26 | 0.76 | 0.22 | 0.6 | 0.7 | 0.39 | 0.48 |
| M | 0.64 | 1 | 0.95 | 1.36 | 0.68 | 1.27 | 1.32 | 1.28 | 1.28 | 0.88 |
| D | 1.49E-10 | 8.66E-11 | 4.23E-10 | 2.39E-10 | 4.11E-10 | 2.98E-10 | 1.91E-10 | 1.30E-10 | 3.63E-10 | 6.06E-11 |
| Test | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 |
| CR | 0.42 | 0.32 | 0.42 | 0.11 | 0.16 | 0.25 | 0.39 | 0.24 | 0.33 | 0.14 |
| CI | 0.03 | 0.03 | 0.02 | 0.05 | 0.07 | 0 | 0.05 | 0.18 | 0 | 0.19 |
| B | 0.68 | 0.78 | 0.63 | 0.31 | 0.33 | 0.78 | 0.12 | 0.8 | 0.87 | 0.42 |
| M | 1.25 | 1.06 | 1.26 | 0.67 | 1.02 | 0.63 | 0.65 | 1.33 | 0.71 | 1.31 |
| D | 1.22E-10 | 3.97E-10 | 3.56E-10 | 4.28E-10 | 2.89E-10 | 2.38E-10 | 2.67E-10 | 4.04E-10 | 1.25E-10 | 2.83E-10 |
| Test | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 |
| CR | 0.44 | 0.43 | 0.16 | 0.4 | 0.24 | 0.23 | 0.32 | 0.33 | 0.42 | 0.45 |
| CI | 0 | 0.01 | 0.02 | 0.05 | 0.07 | 0.19 | 0.11 | 0.09 | 0.02 | 0 |
| B | 0.49 | 0.68 | 0.13 | 0.28 | 0.67 | 0.41 | 0.38 | 0.24 | 0.36 | 0.11 |
| M | 1.26 | 1.17 | 1.13 | 0.99 | 0.91 | 1.08 | 0.7 | 0.91 | 1.12 | 1.26 |
| D | 1.65E-10 | 4.37E-10 | 2.87E-10 | 1.80E-10 | 1.37E-10 | 2.63E-10 | 5.93E-11 | 3.47E-10 | 4.05E-10 | 2.34E-10 |
IV. DISCUSSION
We have conducted a series of simulations to test the effects of various binding parameters on effective equilibria of a simple model of binding chemistry under various crowded conditions. We further showed through the use of regression models that the parameter effects we examine act as independent contributions to the dimerization equilibrium over a broad four-dimensional parameter space. Our model is consistent with previous theoretical studies [18, 22, 23, 25] in finding that total concentration acts non-linearly to affect the binding equilibrium constant, but further shows that a third-degree polynomial (Eq. (5)) accurately describes this non-linear effect over a concentration range spanning physiologically relevant intracellular crowding levels. Furthermore, our model showed the other parameters examined (B, M, and D) influence the binding chemistry linearly and independently from one another and C across the range of crowded conditions examined. This result provides support for a strategy of using simulation-assisted studies to develop empirical and analytical models of crowding effects under broad parameter domains, models that can in turn be used where crowding simulations are too computationally intensive to be practical, such as in models of complicated assembly processes [38].
Our model does, however, begin to slightly overestimate the crowding effect at very high reactant concentrations, where unusually high dimer concentrations occur. We believe this effect suggests a need for further corrections in the model for the equilibrium dimer counts. In addition, there are several additional parameters important to crowding that we would not expect to act linearly nor independently, such as our simulation parameters α, β, and dth. Dealing with such parameters is likely to require a more involved study of covariance among parameter subgroups and multiparameter regression to build a more comprehensive and accurate model.
In evaluating how this work can inform models for cellular biochemistry, it is important to bear in mind that it is a highly simplified representation of binding in crowded media. The most obvious simplification is the use of a two-dimensional model for what is in reality a three-dimensional system. While the prior work [32] confirmed that the model qualitatively captures the expected effects of crowding on binding thermodynamics, it would not be expected to provide a close quantitative match to a three-dimensional model. We cannot exclude the possibility that there may even be significant qualitative differences between crowding in two and three dimensions. Similarly, the model omits many factors found in a true cellular system that are likely to influence binding processes, such as a fine compartmentalization structure, the presence of membranes that might act as scaffolds for reactions, chaperones or other agents that assist or prevent various reaction types, and various mechanisms for active transport within the cell. Likewise, there are other parameters even of our coarse-grained model one could vary and these factors may prove more difficult to capture by simple regression models. By omitting many such complications, the present approach allows us to isolate a few specific factors and parameters likely to be relevant to reaction kinetics in the cell and determine precisely how they affect reactions and how they might be efficiently modeled. Here, the approach showed specifically that a simple regression model accurately captures the dependence of binding equilibrium on several independent parameters of a crowding model. Such observations can help us on the path towards improved simulations of cellular biochemistry in the face of finite computational resources and many unknowns. Their results, however, cannot in themselves be regarded as realistic models of the cell. Considerable additional work will be needed to fully identify precisely which factors are needed for realistic models of cellular biochemistry, to develop methods to simulate these factors efficiently, and to verify that these methods provide accurate quantitative descriptions of biochemistry in vivo.
Acknowledgments
We thank Dr. Robert Murphy and the Center for Bioimage Informatics at Carnegie Mellon University for providing the use of their computer cluster for this research. This work is supported by U.S. National Science Foundation award #0346981, U.S. National Institutes of Health award #1R01AI076318, and the Korea Science and Engineering Foundation (M06-2004-000-10505).
Footnotes
PACS number(s): 83.10.Rs, 87.17.Aa, 82.39.Rt, 87.15.R-
References
- 1.Minton AP. J Cell Sci. 2006;119:2863. doi: 10.1242/jcs.03063. [DOI] [PubMed] [Google Scholar]
- 2.Minton AP. J Biol Chem. 2001;276:10577. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 3.Hatters DM, Minton AP, Howlett GJ. J Biol Chem. 2002;277:7824. doi: 10.1074/jbc.M110429200. [DOI] [PubMed] [Google Scholar]
- 4.del Alamo M, Rivas G, Mateu MG. J Virol. 2005;79:14271. doi: 10.1128/JVI.79.22.14271-14281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drenckhahn D, Pollard TD. J Biol Chem. 1986;261:12754. [PubMed] [Google Scholar]
- 6.Lindner RA, Ralston GB. Biophys Chem. 1997;66:57. doi: 10.1016/s0301-4622(97)00011-2. [DOI] [PubMed] [Google Scholar]
- 7.Tokuriki N, Kinjo M, Negi S, Hoshino M, Goto Y, Urabe I, Yomo T. Protein Sci. 2004;13:125. doi: 10.1110/ps.03288104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McPhie P, Ni Y, Minton AP. J Mol Biol. 2006;361:7. doi: 10.1016/j.jmb.2006.05.075. [DOI] [PubMed] [Google Scholar]
- 9.Jiang M, Guo Z. J Am Chem Soc. 2007;129:730. doi: 10.1021/ja065064+. [DOI] [PubMed] [Google Scholar]
- 10.Wenner JR, Bloomfield VA. Biophys J. 1999;77:3234. doi: 10.1016/S0006-3495(99)77154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimmerman SB, Minton AP. Annu Rev Biophys Biomol Struct. 1993;22:27. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 12.Minton AP. Curr Opin Biotechnol. 1997;8:65. doi: 10.1016/s0958-1669(97)80159-0. [DOI] [PubMed] [Google Scholar]
- 13.Minton AP. Mol Cell Biochem. 1983;55:119. doi: 10.1007/BF00673707. [DOI] [PubMed] [Google Scholar]
- 14.Hall D, Minton AP. Biochim Biophys Acta. 2003;1649:127. doi: 10.1016/s1570-9639(03)00167-5. [DOI] [PubMed] [Google Scholar]
- 15.LeDuc PR, Schwartz R. Cell Biochem Biophys. 2007;48:16. doi: 10.1007/s12013-007-0012-y. [DOI] [PubMed] [Google Scholar]
- 16.Kopelman R. Science. 1988;241:1620. doi: 10.1126/science.241.4873.1620. [DOI] [PubMed] [Google Scholar]
- 17.Schnell S, Turner TE. Progress in Biophysics & Molecular Biology. 2004;85:235. doi: 10.1016/j.pbiomolbio.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Minton AP. Biopolymers. 1981;20:2093. [Google Scholar]
- 19.Lebowitz JL, Helfand E, Praestgaard E. J Chem Phys. 1965;43:774. [Google Scholar]
- 20.Gibbons RM. Mol Phys. 1969;17:81. [Google Scholar]
- 21.Gibbons RM. Mol Phys. 1970;18:809. [Google Scholar]
- 22.Hall D, Minton AP. Biophys Chem. 2002;98:93. doi: 10.1016/s0301-4622(02)00087-x. [DOI] [PubMed] [Google Scholar]
- 23.Hall D, Minton AP. Biophys Chem. 2004;107:299. doi: 10.1016/j.bpc.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 24.Chatelier RC, Minton AP. Biopolymers. 1987;26:507. doi: 10.1002/bip.360260405. [DOI] [PubMed] [Google Scholar]
- 25.Hu Z, Jiang J, Rajagopalan R. Biophys J. 2007;93:1464. doi: 10.1529/biophysj.107.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ridgway D, Broderick G, Ellison MJ. Curr Opin Biotechnol. 2006;17:1. doi: 10.1016/j.copbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Apeltsin L, Puskar K, LeDuc PR, Schwartz R. (Unpublished) [Google Scholar]
- 28.Puskar K, Ta’asan S, Schwartz R, LeDuc PR. Cell Biochem Biophys. 2006;45:195. doi: 10.1385/cbb:45:2:195. [DOI] [PubMed] [Google Scholar]
- 29.Puskar K, Apeltsin L, Ta’asan S, Schwartz R, LeDuc PR. Mech Chem Biosystems. 2004;1:123. [PubMed] [Google Scholar]
- 30.Elcock AH. Proc Natl Acad Sci USA. 2003;100:2340. doi: 10.1073/pnas.0535055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izvekov S, Voth GA. J Phys Chem B. 2005;109:2469. doi: 10.1021/jp044629q. [DOI] [PubMed] [Google Scholar]
- 32.Lee B, LeDuc PR, Schwartz R. Phys Rev E. 2008;78:031911. doi: 10.1103/PhysRevE.78.031911. [DOI] [PubMed] [Google Scholar]
- 33.von Zon JS, ten Wolde PR. J Chem Phys. 2005;123:234910. doi: 10.1063/1.2137716. [DOI] [PubMed] [Google Scholar]
- 34.Sheterline P, Clayton J, Sparrow JC. Protein Profile: Actin. 4. Oxford university press; New York: 1998. p. 14. [Google Scholar]
- 35.Bicknese S, Periasamy N, Shohet SB, Verkman AS. Biophys J. 1993;65:1272. doi: 10.1016/S0006-3495(93)81179-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sengers JV, Watson JTR. J Phys Chem Ref Data. 1986;15:1291. [Google Scholar]
- 37.See EPAPS Document No. [number will be inserted by publisher] for the movie files in AVI format.
- 38.Sweeney B, Zhang T, Schwartz R. Biophys J. 2008;94:772. doi: 10.1529/biophysj.107.107284. [DOI] [PMC free article] [PubMed] [Google Scholar]