Abstract
Functional protein analysis often calls for lengthy, laborious in vivo protein expression and purification, and can be complicated by the lack of stability of the purified protein. In this study, we demonstrate the feasibility of a simplified procedure for functional protein analysis on magnetic particles using cell-free protein synthesis of the catalytic subunit of human cAMP-dependent protein kinase as a HaloTag® fusion protein. The cell-free protein synthesis systems provide quick access to the protein of interest, while the HaloTag technology provides efficient, covalent protein immobilization of the fusion protein, eliminating the need for further protein purification and minimizing storage-related stability issues. The immobilized cPKA fusion protein is assayed directly on magnetic beads and can be used in inhibitor analyses. The combination of rapid protein synthesis and capture technologies can greatly facilitate the process of protein expression and activity screening, and therefore, can become a valuable tool for functional proteomics studies.
Keywords: cell-free expression, in vitro translation, HaloTag, protein immobilization, magnetic particles, PKA, kinase
Introduction
The field of proteomics requires protein expression for structural and functional studies. Cell-free protein expression systems, also known as in vitro translation systems, are alternatives to cell-based protein expression methods. The advantages of cell-free approaches include (1) no requirement for cell culture making them faster and easier to use, (2) the ability to express proteins that are difficult or toxic to express in cell culture, and (3) an open format that allows for the addition of auxiliary components such as labels that can facilitate downstream detection or applications such as structural analyses. They are amenable to the synthesis of many proteins in parallel allowing for the quick screening of constructs for protein expression. Also, any storage-related stability issues can be minimized by synthesizing the target protein immediately prior to analysis.
The most commonly used cell-free extracts are based on rabbit reticulocytes and E. coli extracts. Additionally, extracts made from wheat germ and insect cells are also commercially available. Some extracts function purely as translation systems and require the addition of RNA templates. Other extracts contain components for transcription, permitting the reactions to begin when a DNA template is present. These coupled transcription/translation systems (TNT® systems) eliminate the need to produce and purify RNA, further simplifying and speeding the cell-free protein expression process.
In vitro translation systems have been used to study protein function, especially to map domains involved in protein-protein interactions1. In this study we wanted to show the feasibility of using cell-free extracts to study enzymatically active proteins. We chose a human kinase, the catalytic subunit of human cAMP-dependent protein kinase (cPKA), as our model protein. PKA was chosen because sensitive PKA activity assays, known inhibitors and purified recombinant PKA, which could be used as a positive activity control, were obtainable.
Four commercially available cell-free extracts were used for cPKA protein expression and then compared with respect to expression levels and activity. These were the TNT® T7 Quick Coupled Transcription/ Translation System based on rabbit reticulocyte lysates (RR),1,2 the TNT® SP6 High-Yield Protein Expression System made from wheat germ embryo extracts (WG),3,4 the TNT® T7 Insect Cell Extract Protein Expression System prepared from lysates of Sf21 cells (ICE)5,6 and the bacterial S30 T7 High-Yield Protein Expression System (S30).7
To capture cPKA from the protein translation reaction, it was expressed as a fusion protein containing a purification tag. The HaloTag® Technology was chosen for this purpose because it enables rapid, specific, covalent and oriented binding of HaloTag fusion protein to solid surfaces.8,9 The HaloTag protein is a genetically engineered version of a hydrolase enzyme. The modified protein forms a covalent bond with its substrate, enabling protein labeling or immobilization when the substrate is attached to a label, such as a fluorophore, or solid surface, respectively. Therefore, when HaloTag protein (MW 34 kDa) is used as a fusion tag, the fusion protein can be labeled or captured. Vectors containing the HaloTag coding region and specifically designed for cell-free expression were used to express the cPKA fusion proteins.
In this study cPKA fusion protein was expressed in cell free reactions, monitored for detection levels, captured directly from the cell-free expression reaction and tested for activity (Fig. 1). The kinase was active and inhibited by known inhibitors. cPKA-HaloTag fusion protein expressed in wheat germ extracts has previously been shown to be functional.9 In this instance the kinase was assayed after covalent capture onto glass slides that had been activated with ligand specific for the HaloTag moiety.9 The present study expands this finding by comparing the expression and activity of cPKA in multiple cell-free extracts and examining its ability to be inhibited by known PKA inhibitors while attached to magnetic particles. The HaloTag technology has also been used for the functional immobilization of another kinase, a plant receptorlike kinase, for the purposes of ligand identification. 10 The HaloTag fusion of the plant receptor-like kinase, expressed in tobacco cells and captured on HaloLink resin, was shown to be active and useful for monitoring ligand-receptor interactions. 10 As cell-free systems are able to rapidly produce proteins in comparison to other protein expression methods, they are useful for rapid screening of multiple proteins. Iffland et al used in vitro translation in wheat germ extracts to produce multiple phosphodiesterase active site mutants.11 The His 6-tagged mutants were captured on nickel-chelating resin and characterized in enzymatic assays.11 Together these studies demonstrate the usefulness of combining cell-free synthesis methods, for rapid functional protein production, with immobilization technologies, to eliminate the need for lengthy purification, when conducting enzymatic activity assays.
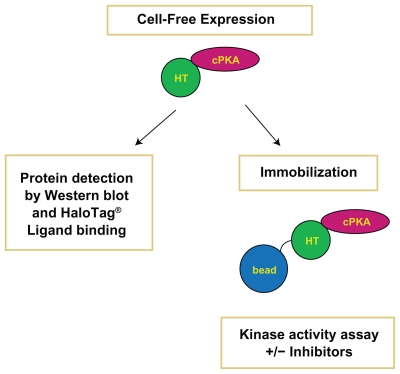
Figure 1.
Schematic of the Protein Analysis Process. Cell-free expression of the cPKA fusion protein was confirmed by western blot analysis and binding to a fluorescent HaloTag ligand. Kinase activity was measured after immobilization from cell-free reactions in the presence and absence of cPKA inhibitors. The “bead” in the figure represents the HaloLink magnetic resin and the “HT” refers to the HaloTag protein at the amino-terminus of the cPKA protein.
Materials and Methods
DNA template preparation
The catalytic subunit of human cAMP-dependent protein kinase (cPKA; NM_002730) was cloned into vectors designed for expression in cell-free systems (Fig. 2; Promega, Madison, WI, USA). The cloning vectors used were pFN19K and pFC20K (HaloTag) T7 SP6 Flexi® Vectors for expression in the RR and WG systems; pFN18 K (HaloTag) T7 Flexi Vector was used for expression in the S30 system. These vectors have the HaloTag coding sequences at the amino-terminus (pFN18 and pFN19) or at the carboxy-terminus (pFC20) of the target protein insertion site. For expression in the ICE system, derivatives of the pF25K ICE T7 Flexi Vector (Promega, Madison, WI, USA) were constructed with the HaloTag gene at the amino-terminus or carboxy-terminus. The pF25K vector has untranslated regions from the baculovirus polyhedrin gene at the 5’ and 3’ ends of the transcriptional unit to enhance translational efficiency specifically in the insect cell extract.5,12 The Flexi System was used for cloning (Promega, Madison, WI, USA).13,14 The plasmid templates were purified using the PureYield™ Plasmid MidiPrep System (Promega, Madison, WI, USA) followed by ethanol precipitation and resuspension in nuclease-free water.
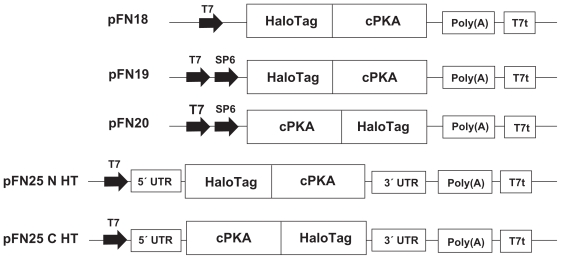
Figure 2.
Features of Cell-Free Expression Vectors. The cPKA gene was cloned into Flexi Vectors specific for cell-free expression. All of the vectors possessed the HaloTag gene to generate synthesis of a fusion protein with the tag at the N or C-terminus. Elements depicted include the promoters for T7 and SP6 polymerases (T7 and SP6), untranslated regions (UTRs) located at the 5’ and 3’ of the protein coding regions in pF25-derived vectors pF25 N HT and pF25 C HT, a synthetic poly(A) site and the T7 polymerase terminator sequence (T7t). pFN19 and pFC20 plasmids directed expression in RR and WG. pF25 N HT and pF25 C HT templates were used with the ICE system, while S30 reactions used the pFN18 plasmid. The sequences are not drawn to scale.
Cell-free protein expression
Four cell-free systems were used to express the cPKA fusion proteins. These were the TNT T7 Quick Coupled Transcription/Translation System based on rabbit reticulocyte lysates (RR),1,2 the TNT SP6 High-Yield Protein Expression System made from wheat germ embryo extracts (WG),3,4 the TNT T7 Insect Cell Extract Protein Expression System prepared from lysates of Sf21 cells (ICE)5,6 and the bacterial S30 T7 High-Yield Protein Expression System (S30).7 All systems were from Promega, Madison, WI, USA. The TNT systems are eukaryotic coupled transcription/translation systems that are initiated by the addition of DNA templates. The prokaryotic S30 product is also driven by DNA templates. The reactions were assembled and incubated as recommended in the manufacturer’s instructions for each system. Larger 500 μl reactions were prepared and after incubation, aliquoted and frozen at −70°C.
Immunoblot analysis
Protein synthesis was assessed using western blot analysis. One microliter of translation reaction was added to 9 μl of SDS loading buffer and loaded onto 4%–20% Tris-Glycine SDS PAGE gels (Invitrogen, Carlsbad, CA, USA). The gels were transferred to PVDF paper using the iBlot™ Dry Blotting System (Invitrogen, Carlsbad, CA, USA). Protein size standards included ProSieve® Color Protein Markers (Lonza, Rockland, ME, USA) and SeeBlue® Plus2 Prestained Standards (Invitrogen, Carlsbad, CA, USA). The blots were blocked in TBST (TBS with 0.05% Tween 20) with 3% blot-qualified BSA (Promega, Madison, WI, USA). PKA expression was detected using anti-PKA polyclonal antibody (Millipore, Billerica, MA, USA) and an anti-HaloTag polyclonal antibody (Promega, Madison, WI, USA). The secondary antibody was alkaline phosphatase conjugated donkey anti-rabbit (Promega, Madison, WI, USA). All antibodies were diluted in TBST for incubation with the PVDF membranes. The membranes were rinsed with TBS before developing with the colorimetric reagent Western Blue® Stabilized Substrate for Alkaline Phosphatase (Promega, Madison, WI, USA). Gels that were to be probed with the antibody to PKA also included a lane with a highly purified recombinant catalytic subunit of bovine PKA (Promega, Madison, WI, USA).
HaloTag TMR ligand binding assay
The HaloTag Technology enables covalent protein labeling and immobilization.8,9 In this case, a ligand containing a TMR fluorophore (HaloTag TMR Ligand; Promega, Madison, WI) was used to covalently label the HaloTag fusion proteins synthesized in the translation reactions. Ten microliters of each translation reaction was incubated with 5 μl of PBS and 5 μl of 50 μM TMR Ligand for 30 minutes at room temperature. Two microliters of the labeling reaction, equivalent to one microliter of the starting translation reaction, was mixed with SDS loading buffer and loaded onto a 4%–20% Tris-Glycine SDS PAGE gel (Invitrogen, Carlsbad, CA). After electrophoresis, the gel was imaged using the Typhoon Variable Mode Imager 9410 (GE Healthcare, Piscataway, NJ) with a green laser (532 nm) and a 580BP30 emission filter. Data was processed using the program ImageQuant (GE Healthcare, Piscataway, NJ). For concentration estimates, purified recombinant GST-HaloTag protein of known concentration was diluted in PBS, incubated with TMR Ligand for 30 minutes at room temperature, and used as a standard.
Immobilization on HaloLink™ magnetic beads
The substrate for the HaloTag protein has also been linked to surfaces to covalently capture fusion proteins.8,9 HaloLink Magnetic Beads (Promega, Madison, WI) were used to covalently capture the PKA fusion proteins out of the translation reactions. For immobilization, 100 μl of magnetic particle slurry were transferred to 1.5 ml microfuge tubes in a magnetic separation stand and washed three times with 1 ml of wash buffer (TBS + 0.05% IgePal CA-630; Sigma Aldrich, St. Louis, MO) before final resuspension in 100 μl of the wash buffer. Fifty microliters of the protein synthesis reaction and an additional 50 μl of wash buffer were added to the magnetic particles, for a total volume for 200 μl. Immobilization was allowed to proceed for 30 minutes at room temperature with shaking (900 rpm in an orbital benchtop shaker). After the initial incubation, the unbound material was collected and the particles were washed three times with 1 ml of wash buffer. The particles were then washed three times with 1 ml of kinase buffer, prepared by making a 1X dilution of the 5X Kinase Buffer provided with the ProFluor® PKA Assay (see below). The particles were resuspended in 200 μl of kinase buffer and immediately used in the kinase activity assay.
PKA activity assay and inhibitor test
The ProFluor PKA Assay system (Promega, Madison, WI) was used to assay for PKA kinase activity. The assay was essentially conducted following the protocol in the manufacturer’s technical manual. Three buffers were prepared: (1) 1X kinase buffer (40 mM Tris-HCl pH 7.5, 20 mM MgCl2, 0.1 mg/ml BSA), (2) reaction buffer (1X kinase buffer with 1 μl rhodamine 110 peptide substrate and 10 μl 10 mM ATP per ml), and (3) termination buffer (40 mM Tris-HCl pH 7.5, 100 mM EDTA, 0.1 mg/ml BSA with 20 μl protease reagent per ml). Five to 25 μl of magnetic particles with immobilized PKA fusion protein covalently attached were distributed into wells of a black 96 well plate. Care was taken to make sure the particles in the 200 μl of kinase buffer were well dispersed before pipetting into wells. The volume in each well was adjusted to 25 μl as necessary with 1X kinase buffer. Twenty-five microliters of reaction buffer were then added, for a final ATP concentration of 50 μM in the activity reaction. The plate was covered and incubated for 20 minutes at room temperature with shaking (on a plate shaker at 600 rpm). Twenty five microliters of termination buffer were added. Again the plate was covered and incubated for 30 minutes at room temperature with shaking. The fluorescent signal was measured at excitation and emission wavelengths of 485 and 527 nm, respectively, using a plate fluorescence reader.
For the inhibitor test, 100 μM stocks were made in 10% DMSO or in 1X kinase buffer depending on individual solubility characteristics. The activity assay was conducted as above except that after pipetting particles into the well, the volume was brought to only 20 μl with kinase buffer. Then 5 μl of the 100 μM inhibitor stock was added to the particles bringing the final concentration of the inhibitor in the 50 μl kinase reaction to 10 μM. Controls lacking ATP, to prevent kinase activity and therefore determine maximal fluorescent output, were also performed in parallel. The highly purified recombinant catalytic subunits of bovine PKA (Promega) and human PKA (Sigma) were used as positive controls. The kinase inhibitors were from Promega (Madison, WI; PKI and U0126), Sigma Aldrich (St. Louis, MO; Staurosporine) and EMD (San Diego, CA; all other inhibitors).
Results
Protein expression and levels
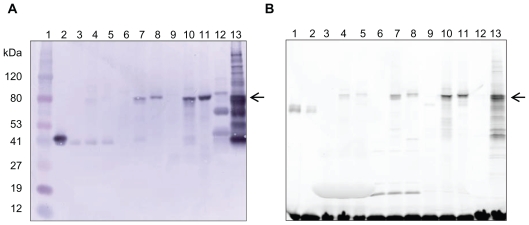
The catalytic subunit of human cAMP-dependent protein kinase was expressed from the appropriate vector in each of four cell-free systems, made from rabbit reticulocyte, wheat germ, insect cell or E. coli extracts. The kinase was expressed as a HaloTag fusion protein with the tag at the aminoterminus or carboxy-terminus. Synthesis of the 74 kDa fusion protein was confirmed by western blot analysis using antibodies specific for PKA and HaloTag protein. The blot probed with antibody to PKA is shown in Figure 3A. Similar results were obtained when the anti-HaloTag antibody was used (data not shown). Protein was detectable in all four systems, though expression levels varied between the systems, with the S30 producing the greatest amount of protein and the rabbit reticulocyte lysate system the least. Therefore, the protein from the RR system was more easily visualized when blotted separately from the S30 system to allow for longer development time (Fig. 4A). Both amino-terminal and carboxy-terminal fusion proteins were successfully expressed in three systems: the RR, ICE and WG systems. In the S30 system the HaloTag-cPKA was expressed as an amino-terminal fusion protein from the pFN18 vector, the vector recommended for use with this system. It was also expressed from the pFN19 vector, but not from the pFC20 vector (data not shown).
Figure 3.
Protein Detection. A) Western Blot with anti-PKA antibody. One μl of each protein synthesis reaction was loaded per lane. The arrow points to the expressed cPKA fusion protein, which has a predicted molecular weight 74 kDa. Lane 1, MW markers; Lane 2, recombinant bovine heart PKA catalytic subunit; Lane 3, RR no DNA; Lane 4, RR pFN19; Lane 5, RR pFC20; Lane 6, ICE no DNA; Lane 7, ICE N-terminal fusion; Lane 8, ICE C-terminal fusion; Lane 9, WG no DNA; Lane 10, WG pFN19; Lane 11, WG pFC20; Lane 12, S30 no DNA; Lane 13, S30 pFN18. Proteins of unknown identity were reproducibly detected by immunoblot in the S30 reaction (see Lane 12). B) Fluorimage of the fusion protein fluorescently labeled with HaloTag TMR ligand. Each lane contains the equivalent of 1 μl of protein expression reaction. The arrow points to the expressed cPKA fusion protein, which has a predicted molecular weight 74 kDa. Lanes 1 and 2, purified GST-HaloTag protein (MW 62 kDa); Lane 3, RR no DNA; Lane 4, RR pFN19; Lane 5, RR pFC20; Lane 6, ICE no DNA; Lane 7, ICE N-terminal fusion; Lane 8, ICE C-terminal fusion; Lane 9, WG no DNA; Lane 10, WG pFN19; Lane 11, WG pFC20; Lane 12, S30 no DNA; Lane 13, S30 pFN18. Free, unbound HaloTag TMR ligand is visible at the bottom of the gel.
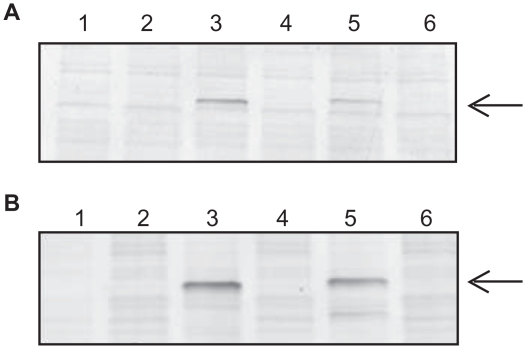
Figure 4.
Detection of cPKA Fusion Proteins Expressed in RR Before and After Immobilization. A) Western Blot Analysis. The amount of fusion protein before and after immobilization was determined by gel electrophoresis followed by immunoblotting with antibody specific for PKA at a 1:1000 fold dilution. A volume equivalent to 1 μl of the protein synthesis reaction was loaded per lane. Lanes 1 and 2: no DNA template negative control reaction; Lanes 3 and 4: pFN19-cPKA; Lanes 5 and 6: pFC20-cPKA. The starting materials from the TNT reactions are in lanes 1, 3 and 5, and the post-immobilization supernatants (“flowthroughs”) are in lanes 2, 4 and 6. B) HaloTag TMR Ligand Binding Assay. The starting material and post-immobilization supernatants were incubated with HaloTag TMR Ligand as indicated in the Experimental Section. A volume equivalent to 1 μl of the starting reaction was loaded per lane in the same lane order listed in Panel A. A comparison of protein levels in lanes 3 and 4, and lanes 5 and 6, is made to determine binding of the fusion protein to the magnetic beads. The arrow points to the expressed cPKA fusion protein.
Protein levels were also assessed using a fluorescent assay specific for HaloTag protein. For this assay, the translation reactions were incubated with a TMR ligand specific for HaloTag protein. Covalent binding of the ligand to the HaloTag fusion protein results in the attachment of one fluorescent label per protein molecule and makes it possible to assess protein expression levels when compared to labeled protein standards. The reactions were electrophoresed on polyacrylamide gels which were imaged to detect the fluorescent signal (Figs. 3B and 4B). The relative levels of the proteins agreed with the ranking obtained with both western analyses. Also, using known amounts of purified GST-HaloTag protein as a standard, the concentration of protein was determined (Table 1). In addition to facilitating sensitive detection and measurement, the ligand-binding assay also served as a functional assay for the HaloTag portion of the fusion protein. The ability of the fusion protein to bind the fluorescent ligand provided evidence that the HaloTag part of the protein was expressed in an active form.
Table 1.
Estimated protein expression levels.
| Cell-free extract | Estimated protein concentration (ng/μl) |
|
|---|---|---|
| HaloTag® at N-terminus | HaloTag® at C-terminus | |
| Rabbit Reticulocyte | 5 | 3 |
| Insect Cell | 23 | 10 |
| Wheat Germ | 83 | 65 |
| E. coli S30 | 230 | ND |
The fusion protein concentration was estimated by comparing fluorescent signal emitted by HaloTag TMR ligand bound to a known amount of HaloTag-GST fusion protein to the signal from the cell-free expressed HaloTag-cPKA fusion proteins. One molecule of fluorescent HaloTag TMR ligand binds per protein, so the calculations were made based on the number of moles of protein and then converted to concentration in terms of mass.
Abbreviation: ND: not done.
Immobilization and activity assay
cPKA kinase activity was assayed using the Pro-Fluor PKA Assay System, a fluorescent assay based on the phosphorylation of a rhodamine 110 peptide substrate. In the absence of phosphorylation the peptide is proteolytically cleaved to release the highly fluorescent rhodamine 110; phosphorylation prevents this cleavage. Thus there is an inverse relationship between the fluorescent signal and the amount of kinase present.
Before assaying for kinase activity, it was necessary to isolate the fusion protein from the cell-free reaction. This was accomplished by specific immobilization onto HaloLink Magnetic Beads. These paramagnetic particles are chemically modified with chloroalkane, a substrate for HaloTag protein, to provide a solid support for covalent attachment of HaloTag fusion proteins. To assay for activity, 50 μl of protein synthesis reactions were incubated with the particles as described in the Experimental Section.
After the initial binding step, protein binding was verified by the lack of protein in the supernatant, as assessed by western blotting and TMR ligand binding (Fig. 4). In Figure 4 Panels A and B, the reduced amount of protein in lanes 4 and 6, as compared to lanes 3 and 5, respectively, indicate the successful and efficient attachment of the fusion protein to the magnetic beads in the RR system. Comparison of the TMR fluorescent signal in the total reaction and the post-binding supernatant indicated that 90% of the fusion protein bound to the magnetic beads for all systems except the S30, where 80% bound (data not shown).
The particles were washed with binding and kinase buffers and then resuspended in 200 μl of kinase buffer. The activity of the immobilized cPKA fusion protein was assayed in a 96-well plate by adding various volumes of particles per well and using the ProFlour PKA Assay. Activity was detected for the rabbit reticulocyte lysate, insect cell extract and wheat germ extract reactions (Table 2). The activity in the S30 reactions could not be assessed due to bead aggregation, most likely the result of extensive protein binding. Translation reactions that lacked DNA template served as the negative controls. Magnetic particles incubated with these negative control reactions showed the maximal fluorescent signal expected in the absence of kinase activity. Wells with kinase captured on particles had lower signals indicating the presence of kinase activity. The activity was highest for the insect cell and wheat germ expressed proteins. When 25 μl of particles from these reactions were added per well, the maximal amount of kinase activity detectable with the ProFluor PKA Assay was observed.
Table 2.
ProFluor PKA activity assay.
| Extract | Volume of particle solution (μl) | Fluorescent signal |
||
|---|---|---|---|---|
| No DNA | HaloTag® at N-terminus | HaloTag® at C-terminus | ||
| Rabbit Reticulocyte | 5 | 135447 | 110315 | 125433 |
| 25 | 105567 | 65218 | 91560 | |
| Insect Cell | 5 | 97799 | 26406 | 25442 |
| 25 | 92087 | 506 | 476 | |
| Wheat Germ | 5 | 105628 | 34406 | 28210 |
| 25 | 84489 | 656 | 579 | |
| E. coli S30 | 5 | 119561 | NT | ND |
| 25 | 112126 | NT | ND | |
cPKA fusion proteins were expressed in all four cell-free extracts. Reactions without added DNA templates were used as negative controls. After immobilization of fusion protein onto magnetic particles, the particles were resuspended in 200 μl 1X Kinase Buffer. The indicated amount of particle solution was added per well for the kinase assay. All reactions were done in duplicate and the signal averaged. The fluorescent signal is inversely correlated with kinase activity.
Abbreviations: NT: not tested. ND: not done.
Inhibitor testing
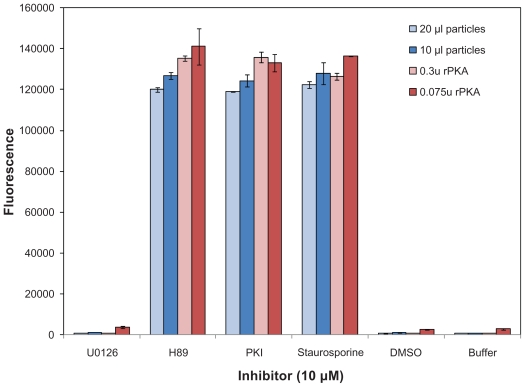
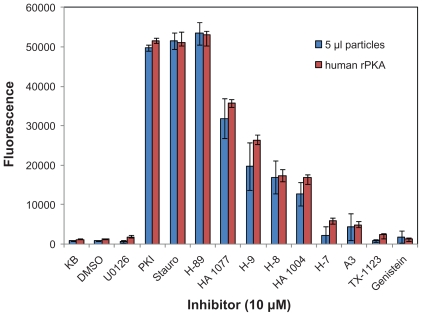
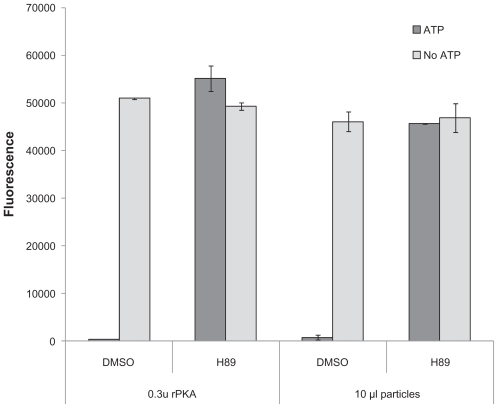
After determining that the cell-free expressed kinases were expressed and active, they were tested with known inhibitors of PKA. The HaloTagcPKA N-terminal fusion protein expressed in the WG system was tested for its ability to be inhibited by 3 potent PKA inhibitors (i.e. Ki’s reportedly in low nM range) and one inhibitor, U0126, of an unrelated kinase (MEK) as a negative control. Two volumes of particles were tested, 10 μl and 20 μl. Also, as a positive activity control, two concentrations of highly purified recombinant bovine cAMP-dependent protein kinase catalytic subunit (rPKA) were included in the experiment. Inhibition was observed for both the cell-free expressed and recombinant protein upon incubation with the 3 potent inhibitors, but not with the U0126 inhibitor (Fig. 5). The HaloTag-cPKA N-terminal fusion protein expressed in the insect cell extract system was also similarly inhibited by these three inhibitors and additional known PKA inhibitors of lesser potency (i.e. Ki’s reportedly in low μM range; Fig. 6). When the assay was carried out in the absence of added kinase or ATP, the fluorescent signal was maximal (Fig. 7 and data not shown). This demonstrated that reduced fluorescence was ATP and kinase dependent and not due to assay interference by the inhibitors (e.g. inhibition of the protease used in the ProFluor PKA Assay to cleave the unphosphorylated peptide).
Figure 5.
Inhibition of HaloTag-cPKA expressed in Wheat Germ Extract. cPKA fusion protein expressed from pFN19 in Wheat Germ extract was immobilized onto magnetic beads and assayed for activity using the fluorescent ProFluor PKA Assay, in the presence of the kinase inhibitors H-89, PKI and staurosporine. The final concentration of inhibitor in the reaction was 10 μM. Two amounts of particle solution were used, 10 μl and 20 μl. The assay was also done with rPKA at two concentrations. As negative controls the kinase was incubated with Kinase Buffer (Buffer), DMSO (1% final concentration) or an inhibitor U0126 for an unrelated kinase. Each inhibitor was tested in duplicate and the average fluorescence is depicted. The bars represent the signal from each of the duplicate wells (i.e. the range).
Figure 6.
Inhibition of HaloTag-cPKA expressed in Insect Cell Extract. HaloTag-cPKA protein was expressed from the pF25 N HT plasmid in the TNT T7 Insect Cell Extract. After immobilization, 5 μl of particle suspension were added per well of the 96 well plate. Reactions containing positive control human rPKA (0.001 units, 2.3 ng) were also performed. The final concentration of all compounds was 10 μM. Control reactions contain only kinase buffer (KB) or DMSO (1% final concentration). All reactions were done in triplicate. The average fluorescent signal is plotted and the error bars represent ± 1 S.D.
Figure 7.
Dependence of the Kinase Signal on ATP. Control reactions for the ProFluor PKA Assay were done in the absence of ATP to confirm the dependency of the signal on ATP and the lack of signal interference by components in the reactions. This figure shows the results for 1% DMSO and 10 μM H-89 in 1% DMSO with both recombinant rPKA and cell-free expressed cPKA fusion protein expressed in insect cell extract from the pF25 N HT vector. The average of triplicate wells is plotted with error bars representing ± 1 S.D.
Discussion
Protein expression and detection
Selection of a cell-free system to express a protein of interest is best determined empirically, therefore, the cPKA was tested in parallel in four different cell-free systems to determine the optimal system(s) for this particular study. Expression of human cPKA was demonstrated in all four systems at levels within the expected range for each of the individual cell-free products.2,3,5,7 The cPKA was expressed as a HaloTag fusion protein, with the tag at either the amino-or carboxy-terminus, and the HaloTag functionality was used for the covalent capture of the kinase from the complex cell-free extract mixtures prior to activity analysis. Total protein expression was analyzed by using antibody against the cPKA or HaloTag proteins, and active HaloTag protein was detected by labeling with a fluorescent ligand. The latter method was also used for protein quantitiation, using a fluorescently labeled HaloTag protein of known concentration as a reference standard. Relative protein expression levels were in agreement using both detection methods, suggesting the differences in HaloTag activity were due to expression levels not inactive protein. Fluorescent labeling was more sensitive in the RR reactions and more specific in S30 reactions, providing an advantage over immunoblotting.
Activity analysis
As components in the crude reaction mixtures interfere with the activity assay (data not shown), the kinase was detected after specific immobilization on magnetic particles. The greatest activity was detected on particles bound with cPKA from the ICE and WG systems. The fluorescent signal from the ProFluor PKA assay was maximally reduced when 25 μl of particles were assayed, indicating the presence of kinase activity at a level at or above the highest amount measurable by the assay. In contrast, 25 μl of particles from the RR reactions showed less activity. In this protocol, 100 μl of bead slurry was used to capture protein out of 50 μl of translation reaction. The manufacturer’s stated capacity of this volume of particles (200 μg fusion protein/100 μl slurry) has not been reached, so it may be possible to achieve higher activity by using the same amount of particles to capture fusion protein from a greater volume of rabbit reticulocyte translation reaction. On the other hand, after incubation with the S30 translation reactions containing DNA template particles aggregated so no reliable measurement was obtained. As the aggregation was not observed with negative control reactions that did not express the fusion protein, this aggregation was most likely due to the excessive amount of captured fusion protein. Our observation suggests that prior optimization of reaction volume to particle ratio might be advantageous in these situations.
Kinase captured from the insect cell and wheat germ extracts was active at levels that permitted easy and rapid assay. The kinase could be assayed directly on the magnetic particle and did not have to be released or further purified. The amount of activity was similar for both extracts and for both fusion orientations (Table 2). Since more fusion protein was synthesized in the wheat germ system (Table 1), the specific activity (activity per mass) seemed to be higher for cPKA produced in the insect cell extract system. The insect cell extract is made from Sf21 (Spodoptera frugiperda) cells, a cell line that often used for expression of large quantities of soluble, active proteins.15 It will be interesting to see if this is also an advantage of the cell-free ICE system in future studies.
Inhibitor analysis
The cPKA fusion proteins with N-terminal HaloTag from two systems, the WG and ICE systems, were tested with and found to be inhibited by known PKA inhibitors. The same inhibitors were also demonstrated to be active against the positive control protein, recombinant highly purified bovine PKA in solution. Therefore, the protein produced in and immobilized from cell-free systems was not just active, but exhibited the expected biologically relevant activity. This suggested that the cell-free expressed protein kinase could substitute for highly purified kinase in this application.
Initial inhibition experiments were done with known strong PKA inhibitors, which included a peptide inhibitor PKI (noncompetitive with ATP), a competitive inhibitor, H-89, with specificity for PKA, and a general kinase inhibitor, staurosporine (Figs. 5 and 6). When the inhibitor panel was then extended to include less potent inhibitors (Fig. 6), the assay was able to distinguish between inhibitors of higher and lower potencies. Therefore this approach might be used for small compound screening to identify inhibitors of unknown specificity and/or potency. To identify competitive inhibitors the assay could be done at increased and decreased ATP concentrations; also at decreased ATP concentrations weaker inhibitors would be more effective.
Combining cell-free protein synthesis and HaloTag technologies for functional protein analysis
In this paper, cell-free expression and HaloTag technologies were integrated and adapted to facilitate quick enzyme activity assays. The cell-free protein synthesis systems provided quick access to the protein of interest while the HaloTag technology provided efficient, covalent protein immobilization of the fusion protein, eliminating the need for further protein purification and minimizing storage-related stability issues. Efficient capture of the protein of interest was critical when dealing with the lower concentrations of protein that can be present in cell-free systems and this was accomplished by using the HaloTag technology. The covalent immobilization ensured that the fusion protein was not lost during subsequent wash steps. Both of these features increased the amount of protein captured on particles and therefore available for the downstream kinase assay. This led to robust signals and a reduction in the amount of material needed per assay.
As each of the technologies is not lengthy, data could be obtained quickly. The cell-free reactions were incubated one to four hours depending on the particular extract systems used. The immobilization step and subsequent washes proceeded for one hour. Assaying the fusion protein while still bound to the particles eliminated the need to cleave the kinase from the HaloTag protein. The particular activity assay used in these experiments, the ProFluor PKA Assay required 1.5 hours. Therefore, the method allows all steps be completed in the same day, from protein synthesis to activity measurement.
These results demonstrate the feasibility of using cell-free expression and immobilization technologies for rapid activity screening when purified enzyme is not available or necessary. It is being used to confirm the synthesis of other active kinases in the ICE system (data not shown) and the process should be adaptable to proteins other than kinases as well. The approach will be also useful to rapidly produce, capture and screen several different proteins, or their variants, in parallel without having to purify large numbers of proteins.
Conclusions
This article describes an approach for expressing functional proteins in cell-free systems for downstream activity analysis. The addition of a fusion tag, the HaloTag protein, allowed for efficient immobilization directly from the protein synthesis reaction and the subsequent activity assay of the immobilized protein. In this way, the catalytic subunit of human cAMP-dependent protein kinase was shown to be expressed in an active form and to be inhibited by known PKA inhibitory compounds. The combination of these rapid protein synthesis and capture technologies facilitated and simplified the process of protein expression and activity screening. The “on-demand” synthesis and purification may provide an alternative to extensive protein purification and the stability issues that can accompany long-term storage of the purified protein. This could offer an advantage especially when multiple proteins or variants need to be produced and assayed.
Acknowledgements
We thank Jim Hartnett, Jami English and Sarah Wheeler for vector construction and cloning and Dr. Said Goueli for helpful discussions about kinase assays and inhibitors.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Arduengo M, Schenborn E, Hurst R. The Role of Cell-Free Rabbit Reticulocyte Expression Systems in Functional Proteomics. In: Kudlicki W, Katzen W, Bennett R, editors. Cell-Free Expression. Vol. 2007. Austin, TX: Landes Bioscience; 2007. pp. 1–18. [Google Scholar]
- 2.Litterer L. Expressing Mammalian Proteins Using Insect and Rabbit Cell-Free Lysates. Promega Notes. 2009;101:19–21. [Google Scholar]
- 3.Hurst R, Creswell D, Slater MR, Schenborn E. TNT® SP6 High-Yield Protein Expression System: More protein from a coupled transcription/translation system. Promega Notes. 2006;93:15–8. [Google Scholar]
- 4.Zhao KQ, Hurst R, Slater MR, Bulleit RF. Functional protein expression from a DNA based wheat germ cell-free system. J Struc Funct Genomics. 2007;8:199–208. doi: 10.1007/s10969-007-9035-2. [DOI] [PubMed] [Google Scholar]
- 5.Leippe D, Creswell D, Hartnett J, Schenborn E. Cell-Free Protein Expression with the TNT® T7 Insect Cell Extract Protein Expression System. Promega Notes. 2008;100:11–2. [Google Scholar]
- 6.Ezure T, Suzuki T, Higashide S, et al. Cell-free protein synthesis system prepared from insect cells by freeze-thawing. Biotechnol Prog. 2006;22:1570–7. doi: 10.1021/bp060110v. [DOI] [PubMed] [Google Scholar]
- 7.Zhao KQ, Creswell D, Slater M. The S30 T7 High-Yield Protein Expression System. Promega Notes. 2008;100:9–10. [Google Scholar]
- 8.Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [DOI] [PubMed] [Google Scholar]
- 9.Nath N, Hurst R, Hook B, et al. Improving protein array performance: focus on washing and storage conditions. J Proteome Res. 2008;7:4475–82. doi: 10.1021/pr800323j. [DOI] [PubMed] [Google Scholar]
- 10.Shinohara H, Matsubayashi Y. Functional immobilization of plant receptor-like kinase onto microbeads towards receptor array construction and receptor-based ligand fishing. The Plant Journal. 2007;52:175–84. doi: 10.1111/j.1365-313X.2007.03204.x. [DOI] [PubMed] [Google Scholar]
- 11.Iffland A, Kohls D, Low S, et al. Structural Determinants for Inhibitor Specificity and Selectivity in PDE2 A Using the Wheat Germ in Vitro Translation System. Biochemistry. 2005;44:8312–25. doi: 10.1021/bi047313h. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T, Ito M, Ezure T, et al. Performance of expression vector, pTD1, in insect cell-free translation system. J Biosci Bioeng. 2006;102:69–71. doi: 10.1263/jbb.102.69. [DOI] [PubMed] [Google Scholar]
- 13.Slater M. The Flexi Vector Systems: The Easy Way to Clone. Promega Notes. 2006;93:8–10. [Google Scholar]
- 14.Blommel PG, Martin PA, Wrobel RL, Steffen E, Fox BG. High efficiency single step production of expression plasmids from cDNA clones using the Flexi Vector cloning system. Protein Expr Purif. 2006;47:562–70. doi: 10.1016/j.pep.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Kost TA, Condreay JP, Jarvis DL. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat Biotechnol. 2005;23:567–75. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]