Abstract
We describe the validation of a serum-based test developed by Rules-Based Medicine which can be used to help confirm the diagnosis of schizophrenia. In preliminary studies using multiplex immunoassay profiling technology, we identified a disease signature comprised of 51 analytes which could distinguish schizophrenia (n = 250) from control (n = 230) subjects. In the next stage, these analytes were developed as a refined 51-plex immunoassay panel for validation using a large independent cohort of schizophrenia (n = 577) and control (n = 229) subjects. The resulting test yielded an overall sensitivity of 83% and specificity of 83% with a receiver operating characteristic area under the curve (ROC-AUC) of 89%. These 51 immunoassays and the associated decision rule delivered a sensitive and specific prediction for the presence of schizophrenia in patients compared to matched healthy controls.
Keywords: schizophrenia, serum-based test, diagnosis
Introduction
Current diagnostic approaches for schizophrenia are based on patient interviews, which entail a subjective assessment of clinical symptoms.1 There is increasing interest in the identification of molecular abnormalities that can be used to identify, stratify and monitor schizophrenia patients. Since longer periods of untreated psychosis are associated with poorer prognoses,2 an accurate test would enable early intervention and improve patient outcomes, providing significant reductions of patient morbidity and health care costs. In addition, such a test would open up the possibility to stratify more accurately the disease and could represent a novel translational medicine tool, crucial for the discovery and development of more efficacious therapies.
One approach in development of a test for schizophrenia is to find a biomarker signature that is capable of distinguishing schizophrenia patients from healthy controls. Many studies have identified molecules related to schizophrenia, but these are not likely to be useful as a disease test when used as single markers due to lack of specificity. Recent studies on other medical conditions using gene expression approaches have shown that multiplexed biomarkers can give reproducible results, which have proven useful in clinical applications.3
The recent development and application of multiplex immunoassay platforms allows the simultaneous measurement of many analytes from individual samples. The Rules-Based Medicine (Austin, TX, USA) DiscoveryMAPTM technology has already been applied successfully in numerous clinical studies targeting diseases such as epithelial ovarian cancer,4 scleroderma,5 coronary artery disease,6 myocardial infarction,7 autoimmune disorders8 and sickle cell anemia.9 This platform is also suitable for the development of sensitive and specific tests for use in medical practice. With this in mind, we have used the DiscoveryMAP platform to profile serum samples from schizophrenia and control subjects.
The first objective of this multicenter study was to identify a set of analytes which were altered reproducibly in schizophrenia patients compared to matched healthy controls. The second objective was to use this analyte signature to construct a diagnostic decision rule, and then validate the signature for its utility as a laboratory developed test using another large cohort of schizophrenia patient and control samples. Both phases of this study were conducted in the Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory at Rules-Based Medicine.
Methods
The present study consisted of two phases (Fig. 1). The first phase was aimed at selection of accurate and reproducible schizophrenia biomarkers from 181 assays comprising the Rules-Based Medicine DiscoveryMAP assay collection. Phase I resulted in the selection of 51 specific immunoassays to be used in assay validation. Phase II featured a refinement of the individual components of the multiplexed immunoassay, development of a decision rule for separating schizophrenia patients from normal controls, and validation of the decision rule using a cohort of 806 clinical samples. For biological validation of the decision rule, 480 of these samples were only analyzed during phase II of this study. The protocols for the study participants, clinical samples and test methods were carried out in compliance with the Standards for Reporting of Diagnostic Accuracy (STARD) initiative.10
Figure 1.
Overview of the test development process.
Abbreviation: Dx, Diagnosis.
Study participants
Subjects were recruited from the Departments of Psychiatry at the Universities of Cologne (cohort 1), Muenster (cohort 2), Magdeburg (cohorts 3 and 4), Rotterdam (cohort 5) and the US military (n = 110 Bipolar Disorder patients and n = 110 controls). Cohorts used for the marker selection phase were comprised of 250 first- and recent-onset schizophrenia patients and 230 control subjects (Table 1A). Schizophrenia patients of cohort 1 (n = 71), 2 (n = 46), 4 (n = 47) and 5 (n = 40) were antipsychotic-naïve and 32 out of 46 subjects from cohort 3 had not been treated with antipsychotic medication for more than 6 weeks prior to sample collection. Drug naïve patients are difficult to recruit since even large clinical facilities can only expect to diagnose about 20–30 such patients each year. To facilitate the future development of a test with differential diagnosis capability, we also carried out DiscoveryMAP analysis using samples from subjects within 30 days before their first contact with US military psychiatric services and who later received a confirmed diagnosis of bipolar disorder (BD) (n = 110, Table 1B). The cohort used to validate and implement the decision rule was comprised of samples from a mixture of first onset and chronic antipsychotic-treated schizophrenia patients along with healthy matched controls. The cohort originally consisted of a total of 838 subjects, 593 subjects diagnosed with schizophrenia and 245 matched healthy controls. During the laboratory testing of the samples, 32 samples were found to be of insufficient serum quantity, leaving a final validation population of 577 subjects diagnosed with schizophrenia and 229 healthy, matched subjects recruited at the Universities of Cologne, Muenster and Magdeburg (Table 2).
Table 1A.
Demographic details of subjects included in phase I (biomarker selection).
| Class | Cohort | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|---|
| Control | n | 59 | 46 | 45 | 40 | 40 |
| M/F | 31/28 | 35/11 | 27/18 | 33/07 | 26/14 | |
| Age* | 30 ± 8 | 27 ± 9 | 34 ± 12 | 27 ± 4 | 36 ± 11 | |
| BMI* | 23 ± 4 | na | 24 ± 4 | na | 24 ± 3 | |
| Schizophrenia first onset | n | 71 | 46 | 46 | 47 | 40 |
| M/F | 42/29 | 35/11 | 30/16 | 36/11 | 27/13 | |
| Age* | 31 ± 10 | 27 ± 9 | 35 ± 12 | 26 ± 8 | 35 ± 10 | |
| BMI* | 24 ± 5 | 22 ± 2 | 26 ± 5 | na | 25 ± 5 |
values are shown as mean ± sd.
Table 1B.
Demographic details of pre-symptomatic bipolar disorder and control subjects.
values are shown as mean ± sd.
Table 2.
Demographic details of 707 subjects used during phase II (51-plex validation). 99 patient follow up samples were available from cohort 2, yielding a total sample number of 806.
| Class | Cohort | 1 | 2 | 3 |
|---|---|---|---|---|
| Control | n | 72 | 84 | 73 |
| M/F | 31/40* | 41/43 | 51/22 | |
| Age* | 31 ± 9 | 37 ± 14 | 34 ± 11 | |
| BMI* | 24 ± 3 | na | 25 ± 4 | |
| Schizophrenia first onset | ||||
| Drug naïve | n | 132 | 18 | 56 |
| M/F | 78/54 | 14/4 | 36/20 | |
| Age* | 30 ± 9 | 28 ± 9 | 37 ± 11 | |
| BMI* | 23 ± 4 | 22 ± 3 | 25 ± 5 | |
| Treated | n | 130 | 71 | |
| M/F | 73/56* | 49/22 | ||
| Age* | 34 ± 12 | 26 ± 8 | ||
| BMI* | 25 ± 5 | 24 ± 4 | ||
| Schizophrenia chronic | ||||
| Drug free | n | 11 | ||
| M/F | 8/3 | |||
| Age* | 32 ± 9 | |||
| BMI* | 26 ± 6 | |||
| Treated | n | 60 | ||
| M/F | 32/28 | |||
| Age* | 33 ± 9 | |||
| BMI* | 26 ± 5 |
Demographic information for one patient not available.
Schizophrenia was diagnosed based on the Structured Clinical Interview for Diagnostic and Statistical Manual (DSM)-IV. Patients used for phase I of this study fulfilled the criteria of the paranoid subtype (DSM-IV 295.30). All diagnoses and clinical tests were performed by psychiatrists following Good Clinical Practice guidelines. Patients whose clinical diagnosis required revision at a later stage were excluded from the study. Control subjects used in phase I of this study were matched to the schizophrenia patients for age, gender and social demographics and were recruited from the same economic and geographical area of the university districts. Controls with a family history of mental disease or with other medical conditions such as type II diabetes, hypertension, cardiovascular or autoimmune diseases were excluded from the study. Pre-symptomatic BD patients and respective controls (n = 110) were selected from a US military serum bank comprising approximately 43 million sera, which facilitated matching for age, gender, ethnicity and lifestyle.
Serum samples
The medical faculty ethical committees of the respective research facilities approved the protocols of the study. Informed consent was given in writing by all participants recruited at universities and clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki. Blood samples were collected from all subjects between 8:00 and 12:00 hours into S-Monovette 7.5 mL serum tubes (Sarstedt; Numbrecht, Germany). The samples were left at room temperature for 2 hours to allow for blood coagulation and then centrifuged at 4000 × g for 5 minutes. The resulting supernatants were stored at −80 °C in Low Binding Eppendorf tubes (Hamburg, Germany).
DiscoveryMAP multiplex immunoassay profiling
Analytes were measured in 250 μL serum samples using the DiscoveryMAP multiplexed antigen immunoassays in the CLIA-certified laboratory at Rules-Based Medicine. Assays were calibrated using duplicate 8-point standard curves and raw intensity measurements were converted to absolute protein concentrations using proprietary software. Machine performance was verified using quality control samples at low, medium and high levels for each analyte. All standard and quality control samples were analyzed in a complex matrix to match the sample background. Serum samples were analyzed at optimized dilutions and analytes exceeding the highest concentrations on calibration curves were assigned the concentration of the highest standard, and those assayed below minimum concentrations were assigned the value 0.0. Assay reproducibility was assessed by reanalysis of the same samples approximately three months later by using Pearson’s correlation coefficients and monitoring the shift in average measurement levels.
Biomarker selection—phase I
The biomarker selection phase of the present study was aimed at identification of analytes that were altered reproducibly in schizophrenia compared to control subjects across independent cohorts (Fig. 1). Analytes were ranked based on the number of centers in which significant differences were observed using unpaired, two-tailed t-tests (P < 0.05). Analyte selection was guided by the following criteria: i) reproducibility (including the same directional change) in three or more centers, ii) high correlation (>0.8) and low average measurement shifts (<40%) in repeat measurement (see above) and iii) mean experimental values distant from the least detectable dose (>20 fold; LDD is defined as the average of the signal plus 3 standard deviations of 20 blank samples analyzed at the same time).
51-plex development and clinical validation—phase II
Efficient analysis of the 51 analytes required construction of new multiplexes. This procedure was guided by optimum dilution of serum and mixing of antibodies to give the most sensitive assays. The required dilutions of serum were 1:5, 1:50, 1:200, 1:10,000, and 1:200,000. The 1:5 dilution group consisted of 31 analytes which were divided into 4 multiplexes. For each higher dilution, only one multiplex was used, yielding a total of 8 new multiplexes for the 51 analytes. Once the multiplexes were created, large batches of reagents were manufactured to allow consistent testing of approximately 7000 samples. The reagents were validated using the following parameters: sensitivity, linearity, spike recovery, common serum matrix interferences, cross-reactivity, precision, correlation, freeze-thaw stability and short-term room temperature antigen stability.
Classification decision rule—design and optimization
To discriminate schizophrenia patients from controls using the markers selected in phase I of this study, we implemented a linear support vector machine (SVM) algorithm. This method minimized errors by counting each misclassified observation with a penalty parameter C. Specific penalty parameters were chosen for patients (CSZ) and controls (CNC), and the ratio F = CSZ/CNC was varied to modify the balance between sensitivity and specificity (visualised in a receiver operating characteristic (ROC) curve). Given a pair of parameters C and F, all elements of the data set were used to train the algorithm, and performance was measured using 10-fold cross validation (CV). The measured sensitivity and specificity calculated in each CV round were averaged and designated as the sensitivity and the specificity of the decision rule for the parameters C and F.
The optimization process was carried out using an in house developed code (Matlab 2009a). The search for optimal performance was performed among 3,100 pairs of parameters (C,F ) covering the following ranges: log2C = −10.0 to 0.0 with step 0.1 (100 values in total). log2F = −1.5 to +1.5 with step 0.1 (31 values in total).
We also computed the conditional probability C that a subject with a given score S is a schizophrenia patient. The computation of the conditional probability was based on the methodology developed by Vapnik.11 The conditional probabilities were used to augment the accuracy estimation of binary classification decision rules with various levels of confidence.
Results and Discussion
Schizophrenia biomarker selection—phase I
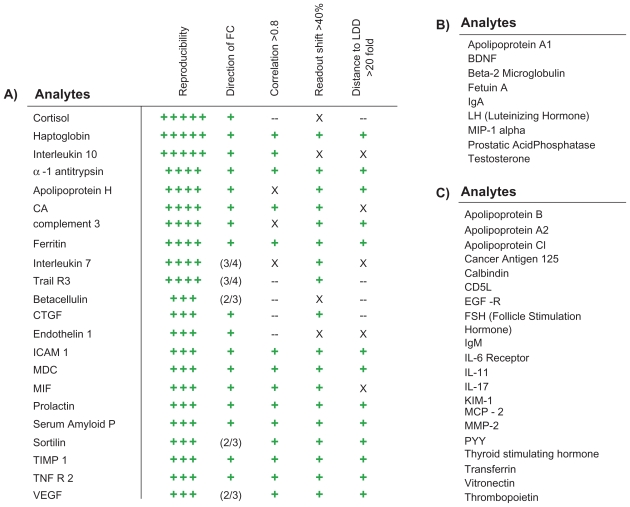
The first stage of the biomarker selection process resulted in the identification of 22 markers of the DiscoveryMAP assay platform which were altered in the schizophrenia population in three or more of the clinical centers (Fig. 2A). Technical reproducibility was assessed by repeating the measurements (n = 63 subjects) approximately 3 months later. This showed an average correlation of 0.83, an average measurement shift of 29% and an average log distance to the LDD of 1.29. In contrast, analytes that were not selected featured an average correlation of 0.65, measurement shift of 54% and log distance to the LDD of 0.70.
Figure 2.
Selection of analytes incorporated into the 51-plex. A) Selection of 22 analytes was guided by (1) reproducible changes across independent cohorts of schizophrenia patients and controls (green plus; P < 0.05 in 3–5 cohorts), (2) consistent directional fold-change (green plus), (3) good correlation (R > 0.8), (4) a low shift (<40%) between the first and second quality control measurements and (5) a high measurement to LDD ratio (>20:1) (X = lower, -- = not tested). B) 9 analytes were selected that are known to be involved in schizophrenia from the scientific literature or that we have identified as being differentially expressed using orthogonal platforms. C) 20 analytes were also selected which showed significant changes in bipolar disorder patients compared to controls.
Nine additional biomarkers were incorporated into the 51-plex due to their known association with schizophrenia or due to the fact that we identified significant changes in these analytes in studies of schizophrenia patients using orthogonal platforms (Fig. 2B). In addition, we compared 110 pre-symptomatic bipolar disorder patients with 110 matched controls and incorporated significant analytes (n = 20, parametric two-tailed t-test, P < 0.05) to facilitate the future development of a test with differential diagnosis capability (Fig. 2C).
Decision rule development and performance
New multiplexed immunoassays were developed which measured all 51 analytes. These were used to analyze a cohort of 806 subjects comprised of 577 schizophrenia and 229 control subjects. For technical validation, these also contained 227 samples which had been used previously during the marker selection phase (phase I, Fig. 1) of the study. The average correlation for markers with high measurement quality (indicated by “+” in the correlation column of Fig. 2) between phase I and phase II using 84 controls out of the 227 samples was 0.86 (range: 0.71–0.99), indicating good reproducibility. Two classification decision rules were constructed using SVM. The first decision rule (SVM-A) was optimized to discriminate schizophrenia patients from controls in the combined dataset of 806 subjects. This yielded a cross-validation classification accuracy of 83% (sensitivity 83%, specificity 83%, ROC-AUC 89%). Since the cohort contained multiple samples from 99 antipsychotic-treated patients, a second decision rule (SVM-B) was built using only the 707 unique samples. This decision rule also yielded a good separation between patients and controls with a cross-validation accuracy of 83% (sensitivity 83%, specificity 82%, ROC-AUC 88%). Fig. 3A displays the classification accuracy and corresponding regions of conditional probabilities which are further detailed in Figure 3B. The conditional probability of correct classification of schizophrenia and control subjects increased with higher or lower scores relative to the decision boundary (Figure 3D lower panel).
Figure 3.
Testing of decision rules SVM-A and SVM-B. A) The top panel displays the percentage of accurately identified patients and controls in the respective conditional probability regions. Blue = schizophrenia. Green = healthy control. The lower sections show the percentage of total schizophrenia patients and controls in the respective conditional probability regions. B) Four regions with differing conditional probabilities are shown. Region III can be used as indeterminate zone where no classification is performed. C) ROC curve based on the SVM-A and SVM-B decision rule. D) Conditional probability curves based on SVM-A (top panel) and SVM-B (bottom panel) for schizophrenia patients (blue line) and controls (green line).
To obtain an unbiased estimate of classification performance and a biological validation of the schizophrenia analyte signature, we determined the performance of the decision rules in samples which had not been used for marker selection in phase I. Application of SVM-B (n = 480 subjects) yielded an overall classification accuracy of 84% (conditional probabilities are shown in Table 3). We also determined the classification accuracy of the SVM-B decision rule for four regions of conditional probabilities (Table 4). This resulted in an increase in accuracy of up to 96% for schizophrenia patients and up to 97% for controls in the highest probability regions (Table 4). When Region III was designated as indeterminate, 17% of the total number of subjects were excluded (SVM-A, 20% for SVM-B).
Table 3.
Classification performance of SVM-B in phase II (51-plex development). Accuracy estimates are shown for the entire set of samples from unique patients as well as for the subset of 480 samples which were not used during phase I of the study. The conditional probability estimate is the median of all conditional probabilities in the respective group.
| Group | Subgroup | SVM-B |
SVM-B (480 validation samples) |
||||
|---|---|---|---|---|---|---|---|
| n | Classification accuracy | Conditional probability | n | Classification accuracy | Conditional probability | ||
| Controls | 229 | 83% | 0.69 | 116 | 77% | 0.61 | |
| Schizophrenia | FE drug naive | 189 | 77% | 0.86 | 111 | 78% | 0.85 |
| FE treated | 201 | 85% | 0.91 | 173 | 86% | 0.91 | |
| Chronic | 71 | 96% | 0.94 | 71 | 96% | 0.94 | |
Abbreviation: FE, first episode.
Table 4.
Classification performance of SVM-B in phase II (51-plex development). Accuracy estimates are shown for the entire set of samples from unique patients as well as for the subset of 480 samples which were not used during phase I of the study. Individual estimates are given for four regions of conditional probabilities.
| Probability region | Conditional probability | SVM-B |
SVM-B (480 validation samples) |
||||
|---|---|---|---|---|---|---|---|
| n patients (%) | n controls (%) | Classification accuracy# | n patients (%) | n controls (%) | Classification accuracy# | ||
| Region I | 1.00–0.91 | 234 (49%) | 9 (4%) | 96% | 186 (51%) | 0 (0%) | 100% |
| Region II | 0.91–0.68 | 128 (27%) | 17 (7%) | 88% | 119 (32%) | 21 (18%) | 85% |
| Region IV* | 0.60–0.83 | 31 (6%) | 60 (26%) | 73% | 20 (5%) | 39 (34%) | 66% |
| Region V* | 0.83–0.84 | 2 (0.4%) | 84 (37%) | 97% | 1 (0.2%) | 21 (18%) | 95% |
The conditional probabilities in regions marked with an asterisks reflect those determined for controls.
Classification accuracies reflect the percentage of correct patient identifications in regions I and II and correct control identifications in regions IV and V.
Out of the 478 total schizophrenia patients included in phase II, 111 were suffering from a non-paranoid type of the disease (Table 5). SVM-A identified 95 (86%) of these patients correctly suggesting that the biomarker signature was present regardless of the schizophrenia subtype.
Table 5.
Subtypes of the 478 schizophrenia patients investigated in phase II of this study.
| DSM-IV code | Subtype | n |
|---|---|---|
| 295.1 | Schizophrenia, disorganized type | 18 |
| 295.2 | Schizophrenia, catatonic type | 7 |
| 295.3 | Schizophrenia, paranoid type | 367 |
| 295.4 | Schizophreniform disorder | 26 |
| 295.6 | Schizophrenia, residual type | 3 |
| 295.7 | Schizoaffective disorder | 27 |
| 295.9 | Schizophrenia, undifferentiated type | 15 |
| 297.1 | Delusional disorder | 1 |
| Schizophrenia non specified | 14 |
We also investigated 80 subjects (baseline Positive and Negative Syndrome Scale [PANSS] of 66.0 ± 17.8) before and after 4–6 weeks of antipsychotic treatment which resulted in an overall reduction in symptoms of 13% (average reduction of 10.3 ± 17.3), as measured using the Positive and Negative Syndrome Scale (PANSS positive = 18% lower [average reduction of 3.7 ± 4.1], PANSS negative = 8% lower [average reduction of 1.6 ± 4.9], PANSS general = 12% lower [average reduction of 5.0 ± 8.5]).12 Interestingly, SVM-B was capable of identifying 85% of these patients at the first time-point and after the treatment period. There was an average correlation of 0.49 across all 51 analytes, supporting the stable identification capability of the decision rule. This suggested that schizophrenia patients in remission still feature schizophrenia-like serum profiles even after 4–6 weeks of treatment.
Conclusions
In this multicenter study, we discovered and validated a biomarker panel for schizophrenia based on biological and technical reproducibility of the molecular signature. All stages of the process, including conduction of the assays, analyte selection, biomarker panel refinement and development of the decision rule, were carried out in a CLIA-certified laboratory at Rules-Based Medicine. Biomarker selection was based on a large number of samples collected from antipsychotic naïve, acutely psychotic patients to facilitate relatively uniform conditions. Subjects were recruited from four independent clinical centers and samples were collected according to strict standard operating procedures to maximize reliability and accuracy of the results. Biological variability arising from the collection in different geographical regions may contribute to the generality of the present findings. Another factor which should be considered in this regard is that the samples were collected at the same centers for phases I and II of the study. However, as the data were generated from three independent centers and from independent subjects, it is likely that the findings will be generalizable to other clinical centers. As the assay progresses from beta site testing to analysis in different subpopulations, the performance against present clinical classification and observed prevalence and incidence must be monitored and differences will need examination.
The implementation of the 51 marker decision rule was based on a cohort comprised of both untreated and treated schizophrenia patients who were either experiencing a first episode of illness or who were chronically ill (54% of patients were on current antipsychotic treatment). This collection is likely to represent more closely the patient population encountered in clinical practice. High classification performance demonstrated that the decision rule could identify schizophrenia patients with high accuracy irrespective of the disease duration or treatment state. Interestingly, the biomarker signal was still apparent in subjects even after 4–6 weeks of successful treatment with antipsychotic medication. This suggests that the 51-plex is robust for identification of subjects with schizophrenia at different stages of the schizophrenia disease process. Further work is required for the development of a biomarker panel aiding in the monitoring of patient responses to treatment.
In summary, the present findings demonstrate the applicability of a rapid and non-invasive test to confirm the presence of schizophrenia. This first attempt to develop a molecular test with clinical utility for the diagnosis of schizophrenia was focused on the distinction of schizophrenia patients against healthy controls. For this application, we have developed a refined 51-plex assay panel and decision rule with a sensitivity and specificity of 83%. We anticipate that the 51-plex assay panel will result in the future development of a differential diagnostic test that can distinguish among various neuropsychiatric disorders such as schizophrenia, bipolar disorder and major depressive disorder. Therefore, the next stage towards clinical translation is to conduct a large scale clinical validation study using samples from diverse psychiatric patient populations and settings in a series of prospective studies with the Rules-Based Medicine assay platform.
Acknowledgements
This study was instigated and supported by Rules-Based Medicine, Psynova Neurotech Ltd and the Stanley Medical Research Institute (SMRI). We want to thank Anke Dudeck, Jeanette Schadow, Dr. Wolfgang Jordan, Dr. Bernd Hahndorf, Dr. Florian Kästner, Dr. Anya Pedersen, Dr. Ansgar Siegmund, Dr. Katja Kölkebeck, Torsten Schoenborn, Dr. Christoph W. Gerth, Dr. Christian Mauss, Dr. Brit M. Nolden, Dr. M. A. Neatby, Sandra Pietsch and Christin Oheim for their participation in sample characterization and collection. We would like to thank Dr. Fuller Torrey for his support and suggestions. We would also like to thank Michael G. Walker, Ph.D. for suggestions concerning data analysis. Most of all, thanks to all patients and healthy volunteers for their selfless donation of samples used in this study.
Footnotes
Disclaimers
The views expressed are those of the authors and should not be construed to represent the positions of the Department of the Army or Department of Defense. None of the authors have any associations, financial or otherwise, that may present a conflict of interest. This effort was funded by the Stanley Medical Research Institute, Bethesda, MD, and the Department of the Army. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.McGorry PD, Mihalopoulos C, Henry L, Dakis J, Jackson HJ, Flaum M, et al. Spurious precision: procedural validity of diagnostic assessment in psychotic disorders. Am J Psychiatry. 1995 Feb;152(2):220–3. doi: 10.1176/ajp.152.2.220. [DOI] [PubMed] [Google Scholar]
- 2.Singh SP. Outcome measures in early psychosis; relevance of duration of untreated psychosis. Br J Psychiatry Suppl. 2007 Aug;50:S58–S63. doi: 10.1192/bjp.191.50.s58. [DOI] [PubMed] [Google Scholar]
- 3.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009 Feb 19;360(8):790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 4.Bertenshaw GP, Yip P, Seshaiah P, Zhao J, Chen TH, Wiggins WS, et al. Multianalyte profiling of serum antigens and autoimmune and infectious disease molecules to identify biomarkers dysregulated in epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2008 Oct;17(10):2872–81. doi: 10.1158/1055-9965.EPI-08-0464. [DOI] [PubMed] [Google Scholar]
- 5.Duan H, Fleming J, Pritchard DK, Amon LM, Xue J, Arnett HA, et al. Combined analysis of monocyte and lymphocyte messenger RNA expression with serum protein profiles in patients with scleroderma. Arthritis Rheum. 2008 May;58(5):1465–74. doi: 10.1002/art.23451. [DOI] [PubMed] [Google Scholar]
- 6.Gurbel PA, Kreutz RP, Bliden KP, DiChiara J, Tantry US. Biomarker analysis by fluorokine multianalyte profiling distinguishes patients requiring intervention from patients with long-term quiescent coronary artery disease: a potential approach to identify atherosclerotic disease progression. Am Heart J. 2008 Jan;155(1):56–61. doi: 10.1016/j.ahj.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 7.Escobar GP, Lindsey ML. Multi-Analyte Profiling of Post-Myocardial Infarction Plasma Samples. FASEB J. 2007;21(746.11) [Google Scholar]
- 8.Delaleu N, Immervoll H, Cornelius J, Jonsson R. Biomarker profiles in serum and saliva of experimental Sjogren’s syndrome: associations with specific autoimmune manifestations. Arthritis Res Ther. 2008;10(1):R22. doi: 10.1186/ar2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SP, Ataga KI, Zayed M, Manganello JM, Orringer EP, Phillips DR, et al. Phase I study of eptifibatide in patients with sickle cell anaemia. Br J Haematol. 2007 Nov;139(4):612–20. doi: 10.1111/j.1365-2141.2007.06787.x. [DOI] [PubMed] [Google Scholar]
- 10.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Standards for Reporting of Diagnostic Accuracy. Clin Chem. 2003 Jan;49(1):1–6. doi: 10.1373/49.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Vapnik V. Statistical Learning Theory. John Wiley & Sons; 1998. [Google Scholar]
- 12.Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13(2):261–76. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]