Abstract
In this report, we present results of a genome-wide linkage scan using as a phenotype the number of externalizing symptoms associated with alcohol use disorders. Subjects were collected by the Collaborative Study on the Genetics of Alcoholism project from families in which at least three first degree relatives were affected by alcohol dependence. We use a novel non-parametric regression method based on kernel smoothing for our analysis. We report a statistically significant linkage close to the ADH gene cluster on Chromosome 4. We also obtain evidence for epistatic interaction between a region on Chromosome 1 and one on Chromosome 15. Although alcoholism as a covariate does not have any effect on the linkage scan, it has an effect on the epistatic interaction.
Keywords: non-parametric regression, linkage, externalizing symptoms
Alcohol dependence is a complex disorder that is influenced by genetic and environmental factors. A well-known link exists between externalizing behavior disorders such as conduct disorder and alcohol use disorders. For example, adults with alcohol dependence report an increased rate of externalizing behaviors [Cloninger et al., 1981; Hesselbrock et al., 1986; Babor et al., 1992]. In addition, children and adolescents who manifested conduct disorder are at heightened risk for the subsequent development of alcohol problems [Robins, 1966; Myers et al., 1995]. Twin studies have demonstrated that shared genetic factors are largely responsible for much of the co-occurrence childhood conduct disorders and adult alcohol dependence [Kendler et al., 1987; Slutske et al., 1998]. This relationship between externalizing symptoms and alcoholism is also seen in the Collaborative Study on the Genetics of Alcoholism (COGA) dataset. A higher prevalence of alcohol dependence and more severe alcohol problems are correlated with the most disruptive behavior [Bucholz et al., 2000].
The Collaborative Study on the Genetics of Alcoholism is a multicenter research program established to detect and map susceptibility genes for alcohol dependence and related phenotypes. Studies have revealed that many regions of the genome exhibit significant linkage with alcohol dependence [Reich et al., 1998; Foroud et al., 2000] as well as alocohol-related phenotypes such as a severity phenotype [Foroud et al., 1998], maximum number of drinks in a 24 hr period [Saccone et al., 2000], a combined phenotype of alcohol dependence or depression [Nurnberger et al., 2001] and habitual smoking in families of alcoholics [Bierut et al., 2004]. In addition, electro-physiological phenotypes related to alcoholism like event-related potentials (ERP) [Begleiter et al., 1998; Almasy et al., 2001; Porjesz et al., 2002b], electroencephalogram (EEG) waves [Porjesz et al., 2002a; Ghosh et al., 2003], event-related oscillations (ERO) [Jones et al., 2004] and a bivariate phenotype combining alcohol dependence and ERP [Williams et al., 1999] have also shown evidence of linkage on multiple chromosomes.
The COGA project sytematically ascertained through probands from alcoholism treatment facilities [Begleiter et al., 1995; Reich, 1998]. COGA sites providing data for this study include SUNY Downstate Medical Center, New York; University of Connecticut Health Science Center; Indiana University School of Medicine, University of Iowa School of Medicine, University of California School of Medicine, San Diego and Washington University School of Medicine, St. Louis. The data collection procedures were standardized at all sites [Begleiter et al., 1998]. The extensive SSAGA interview instrument was developed to obtain detailed information on symptoms related to alcohol use, abuse and dependence, other substance abuse and dependence, and different psychiatric conditions [Bucholz et al., 1994, 1995; Hesselbrock et al., 1999]. The genetic dataset comprises 262 extended families in which at least three members met the COGA (DSM-III-R and Feighner [Feighner et al., 1972]) criterion of alcohol dependence. Data on externalizing symptoms were available for 1,553 subjects ranging from 7 to 70 years of age.
Genotyping was performed at laboratories in Indiana University and in Washington University [Foroud et al., 2000]. For this study, a map comprising 405 markers, mostly microsatellites, was used. The average intermarker distance and the average heterozygosity of the markers were 10.9 cM and 0.74, respectively, although the marker coverage in some regions was denser than 5 cM. Most markers were tri- and tetranucleotide repeat polymorphisms. In order to be consistent and be able to compare with previous linkage studies performed in the COGA project, the program CRI-MAP [Lander and Green, 1987] was used to calculate marker order and distances. The maximum likelihood estimates of the allele frequencies were obtained using USERM13 [Boehnke, 1991].
Data on three covariates were used in the analyses: age, sex and diagnosis of alcoholism by the COGA criteria noted above. Our analysis was based on 171 independent sib-pairs belonging to distinct families and having at least one parent genotyped [i.e., sibs with both parents missing were excluded from the analysis for obtaining more precise estimates of identity-by-descent (i.b.d.) scores]. To ensure independence of observations, only one sib-pair was chosen from each family (the sib-pair with the maximum number of marker genotypes available for both sibs). The sample comprises 49 male–male pairs, 89 male–female pairs and 33 female–female pairs. Most of the families (148, 86.5%) were Caucasian while 15 families (9%) were African-American.
A set of 24 externalizing symptoms (see Appendix A) related to anti-social behavioral traits was chosen. The number of these symptoms endorsed by each individual was summed to obtain a quantitative scale ranging from 0 to 24. In this study, we present results on a genome scan of the number of externalizing symptoms. Since there are gender and age differences with externalizing behaviors, we regress out the effects of sex and age from the phenotypic values using least squares linear regression. We analyze the data both with and without regressing out the effect of alcoholism.
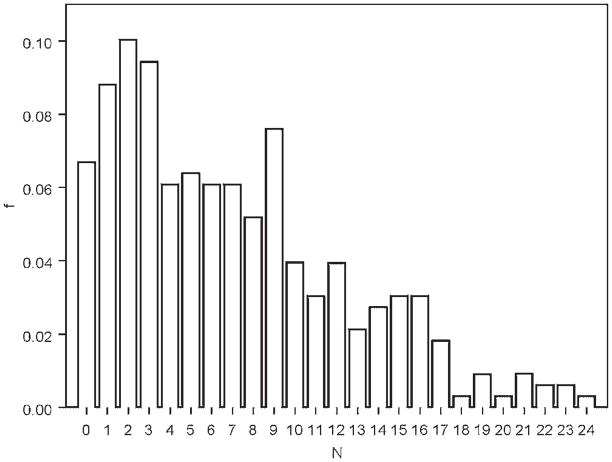
The probability distribution of the number of externalizing symptoms is very skewed (see Fig. 1) and hence deviant from normal. A possible better model fit is a truncated Poisson distribution. Thus, using likelihood based linkage methods which assume normality of traits may lead to misleading inferences. The classical Haseman-Elston regression [1972] and its extensions do not assume any specific probability distribution for the trait but assume a linear relationship between a function of the sib-pair trait values and the identity-by-descent scores at a marker locus. Since this relationship deteriorates with increase in dominance at the trait locus, a more prudent strategy is to estimate empirically the nature of the relationship between the regression variables [Ghosh and Majumder, 2000]. We use a modified version of the non-parametric regression proposed by Ghosh et al. [2003]. The major advantage of the non-parametric regression is that it does not involve any likelihood based framework which requires explicit specification of the probability distribution of the quantitative trait values, and hence is robust to violations in distributional assumptions like normality. The details of the non-parametric regression are provided in Appendix B.
Fig. 1.
Relative frequencies of the number of externalizing symptoms; N = number of symptoms; f = relative frequency.
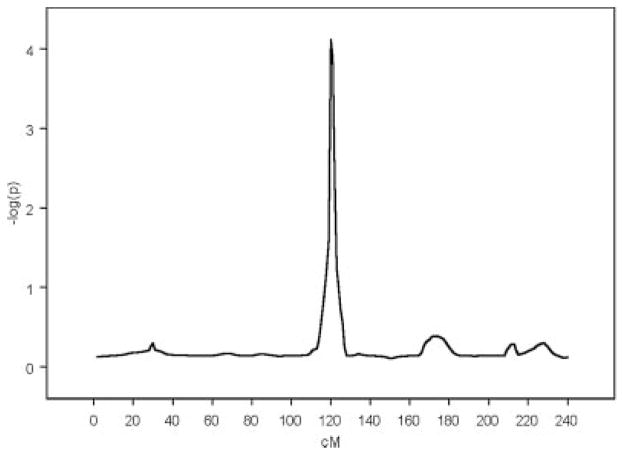
We performed the proposed non-parametric regression at 1 cM intervals throughout the genome. The only statistically significant (nominal P-value < 0.0001, a conservative correction equivalent to 100 independent tests would give genome-wide significance <0.01) peak (when alcoholism is not regressed out) is located at 120 cM on Chromosome 4 (see Fig. 2), near a marker in the ADH1C gene (124.9 cM). When the effect of alcoholism is regressed out as a covariate, we find that the linkage finding is hardly altered. The position of the peak is almost identical (within 3 cM) to that obtained when the effect of alcoholism is not regressed out. Moreover, the P-value corresponding to the peak is only changed marginally.
Fig. 2.
Linkage results on Chromosome 4 for the phenotype defined as the number of externalizing symptoms.
There has been some evidence in previous COGA studies that a region (near 100 cM) on Chromosome 1 and a region on Chromosome 4 (around 57 cM) interact epistatically with a region (near 50 cM) on Chromosome 15 [Ghosh et al., 2003] for different alcohol-related phenotypes. Although the only statistically significant linkage was on Chromosome 4, recent theoretical and simulation studies [Culverhouse et al., 2002] have shown that epistatic effects may be present even in the absence of main effects. Hence we investigated the presence of epistasis between two pairs of regions: (i) 95–105 cM on Chromosome 1 and 45–55 cM on Chromosome 15; and (ii) 52–62 cM on Chromosome 4 and 45–55 cM on Chromosome 15. The details of the procedure for testing the presence of epistatic interactions are provided in Appendix B.
When alcoholism is not regressed out, we observe that the epistatic interaction: between each of the integer points on Chromosome 1 (95–105 cM) and each of the integer points on Chromosome 15 (45–55 cM) is significant at P < 0.01 (adjusted for multiple tests). In all these cases, epistasis (in terms of increase in Δ for Model II over Model I) explains about 8% of the variance in the squared difference in sib-pair externalizing symptoms values. However, none of these interactions are significant at P < 0.01 when the effect of alcoholism is regressed out. There was no evidence of epistatic interaction between the above mentioned regions on Chromosomes 4 and 15 either with or without regressing out the effect of alcoholism. We note here that since our linkage finding is around 120 cM on Chromosome 4, we tested for possible epistatic interactions between that region and the above mentioned regions on Chromosomes 1 and 15. However, none of these were statistically significant.
The linkage finding on Chromosome 4 is in the 4q22.3 region. This region contains the alcohol dehydrogenase gene cluster (ADH1A, ADH1B, ADH1C, ADH4–ADH7). Variants in the class I ADH genes affect enzyme kinetic properties and have been reported to reduce the risk of alcoholism in Chinese and Japanese populations [Edenberg and Bosron, 1997]. This region has produced a linkage finding with alcohol dependence in a native American Indian population [Long et al., 1998]. Previous COGA studies have also found evidence of linkage in this region with various alcohol-related phenotypes. Reich et al. [1998] obtained suggestive linkage evidence of a protective factor in the risk of alcoholism among unaffected sibs of alcoholic probands. A phenotype defined as the maximum number of drinks consumed in a 24 hr period [Saccone et al., 2000] which provides a quantitative measure to grade non-alcoholic individuals and is closely related to alcoholism diagnosis also produced a linkage peak in this region. In individuals at risk for alcoholism, the amplitude of the visual P300 component of event-related potentials (ERP) is significantly decreased [Porjesz et al., 1998]. Williams et al. [1999] analyzed a bivariate phenotype combining DSM-IV diagnosis and the amplitude of the P300 component of the CZ event-related potential. Their linkage scan gave a significant finding near the class I alcohol dehydrogenase subunit ADH1C. Oscillations within the electro-encephalogram (EEG) gamma and beta bands have been linked to sensory perception and memory and have been shown to be modified by anesthetic agents. Since these oscillations appear to be trait as opposed to state-related, Ghosh et al. [2003] used Beta 2 EEG waves as an endophenotype for studying the risk of developing alcohol dependence. They obtained a linkage signal in the same region on Chromosome 4.
We have previously conducted a genome-wide linkage scan of conduct disorder diagnoses in the COGA dataset, using standard non-parametric, multipoint methods of linkage analyses (e.g., as implemented in Aspex) for affected sibling pairs [Dick et al., 2004]. The chromosomal regions yielding evidence of linkage in that study (on chromosomes 2 and 19) were not overlapping with the significant linkage peak from this study. We note here that the conduct disorder variables contained in the set of externalizing symptoms are identical to the SSAGA diagnosis protocol. Accordingly, our data illustrate that different findings can emerge when analyzing phenotypes capturing different information, and when utilizing different methods of analysis (that is, methods for analyzing quantitative traits versus binary traits).
The sample comprised Caucasian as well as African-American families. While population stratification cannot result in an inflated rate of false positives in sib-pair linkage analyses, it may lead to false negatives. To investigate this possibility, we also performed our linkage scan on the subset of 148 Caucasian sib-pairs, but found that the linkage findings were identical. Although we used CRI-MAP for the purpose of consistency with previous COGA studies, we note that the Decode map is estimated from a larger number of meioses and hence, likely to be more precise. We found that the COGA map was marginally expanded compared to the Decode map, but the map orders were identical. When we repeated our linkage analyses with the Decode map, the results remained unchanged with the only linkage finding being in the same region on Chromosome 4, albeit with a marginally lower P-value. We also realize that larger sibships carry more information on linkage than independent sib-pairs. However, a direct extension of the proposed non-parametric method to sibships involves modeling of the covariance structure between the squared phenotypic difference of different sib-pairs within a sibship, which compromises on the model-free framework of the method. We are currently exploring a non-parametric modification of the linear regression procedure proposed by Ghosh and Reich [2002] based on a so-called “contrast function” of the quantitative trait values of the sibs within each sibship.
As discussed earlier, a possible parametric probability model for the phenotype is Poisson. Hence, it may be more appropriate to perform a Poisson regression instead of a linear regression while eliminating the effects of covariates. We assumed a conditional Poisson distribution of the externalizing symptoms count given age, sex and alcoholism and estimated the residuals using maximum likelihood. However, the non-parametric regression method based on these residuals did not improve on the linkage results described above. While the only significant linkage finding remained in the same region in the ADH gene cluster on Chromosome 4, the P-value of the linkage peak was less extreme. A possible reason for the Poisson regression not performing as well as the linear regression may be due to the fact that the conditioning covariates (sex and alcoholism) are binary in nature and hence a Poisson distribution may not be the optimal fit for the conditional distribution of the phenotype. We also note that a Box–Cox transformation on the residuals to induce normality in the trait did not change or improve on the linkage findings. This can be explained by the fact that the non-parametric regression is not sensitive to the actual distribution of the quantitative trait.
Since the proposed Δ statistic does not consider the direction of relationship between squared sib-pair trait difference and estimated i.b.d. scores, there may be concern that the rate of false positives is inflated due to a random positive relationship between the variables under the null hypothesis of no linkage. One way to circumvent this problem is to ensure that the rank correlation between the variables is negative at the significant linkage regions. We found that the rank correlations were significantly negative (evaluated by an asymptotic normal test) near our linkage peak.
We note that our linkage analysis after regressing out the effect of alcoholism yielded very similar results to those obtained without regressing out the effect of alcoholism. However, the epistatic interactions between regions on Chromosomes 1 and 15 were found only when the effect of alcoholism was not regressed out. A plausible model explaining this phenomenon is that the above regions on Chromosomes 1 and 15 may have negligible marginal genetic effects on alcoholism defined by the COGA criterion, but may have a stronger interaction effect. Thus, the epistatic effects of the two regions (for externalizing symptoms and alcoholism) may cancel out when the effect of alcoholism is regressed out from the number of externalizing symptoms. With the current availability of SNPs in this region, we are carrying out further association and functional studies on these regions to have a better understanding of the role of the genes in this region in modulating externalizing symptoms and its association with alcoholism.
Acknowledgments
The Collaborative Study on the Genetics of Alcoholism (COGA) Co-Principal Investigators: L. Bierut, H. Edenberg, V. Hesselbrock, B. Porjesz) includes nine different centers where data collection, analysis, and storage take place. The nine sites and Principal Investigators and Co-Investigators are: University of Connecticut (V. Hesselbrock); Indiana University (H. Edenberg, J. Nurnberger Jr., P.M. Conneally, T. Foroud); University of Iowa (R. Crowe, S. Kuperman); SUNY HSCB (B. Porjesz); Washington University in St. Louis (L. Bierut, J. Rice, A. Goate); University of California at San Diego (M. Schuckit); Howard University (R. Taylor); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy). Zhaoxia Ren serves as the NIAAA Staff Collaborator. This national collaborative study is supported by the NIH Grant U10AA08403 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA). This work has also been supported by the NIH grant R01-TW-6604 from the Fogarty International Center. The authors are grateful to Professor Partha Majumder for useful statistical inputs and to Joseph Mullaney for management of the data on externalizing symptoms. In memory of Henri Begleiter and Theodore Reich, Principal and Co-Principal Investigators of COGA since its inception and one of the founders of modern psychiatric genetics, we acknowledge his immeasurable and fundamental scientific contributions to COGA and the field.
Grant sponsor: National Institute on Alcohol Abuse and Alcoholism (NIAAA); Grant number: U10AA08403; Grant sponsor: Fogarty International Center; Grant number: R01-TW-6604.
APPENDIX A
List of the Externalizing Symptoms
1 = ‘hit or throw things while drinking’
2 = ‘physical fights while drinking’
3 = ‘play hooky from school (truancy)’
4 = ‘suspended or expelled from school’
5 = ‘run away from home overnight’
6 = ‘tell many lies or used alias’
7 = ‘steal (2+) from family/home’
8 = ‘shoplift (2+) from store/other people’
9 = ‘stolen/forged (3+) since age 15’
10 = ‘vandalism’
11 = ‘start fights’
12 = ‘get into fights (3+) since age 15’
13 = ‘4 or more traffic tickets’
14 = ‘non-traffic arrests’
15 = ‘jail for something other than drugs/alc’
16 = ‘challenge parents when young’
17 = ‘enjoy outsmarting authority’
18 = ‘breaking/entering into car/house’
19 = ‘fencing/selling/run numbers since age 15’
20 = ‘hit/physic attack/throw things snc age15’
21 = ‘quit, no new job lined up (3+) since age15’
22 = ‘ignored feelings of others since age 15’
23 = ‘frequently lost temper or often felt irritable/angry/resentful’
24 = ‘often blame others for your troubles’
APPENDIX B
Non-Parametric Regression
The non-parametric regression model of yj and πjp at any arbitrary point p on the genome is given by
where yj is the squared difference in the number of externalizing symptoms value for the jth sib-pair, πjp is the multipoint estimated i.b.d. score at a point p on the genome for the jth sib-pair computed using the linear regression method proposed by Fulker et al. [1995] and Ψ is a real valued function of πjp and ejs are random errors.
The functional form of Ψ is estimated using a kernel-smoothing technique [Silverman, 1986]. The kernel function used is
| (1) |
The predictor of yj is given by
| (2) |
where h is the “optimal” window length in the kernel smoothing procedure.
To assess the significance of our regression, we use a diagnostic measure [Ghosh et al., 2003] as follows: Δ = 1 − Σj{yj − Ψ(πj)}2/Σj{yj − yM}2, where yM is the mean of the yjs. One has to use resampling techniques such as bootstrap to obtain empirical thresholds under the null hypothesis of no linkage.
Testing for Epistatic Interactions
We consider two models:
where πjp1 and πjp2 are estimated marker i.b.d. scores at two positions on the genome, p1 and p2, respectively, and Ψ1, Ψ2, and Ψ3 are real-valued functions, which are estimated sequentially using kernel smoothing [see Ghosh and Majumder [2000] for details] based on the kernel function given in Equation (1). A significant increase in Δ for Model II over Model I would indicate the presence of epistatic interaction. Empirical null thresholds are determined using resampling techniques.
References
- Almasy L, Porjesz B, Blangero J, Goate A, Edenberg HJ, Chorlian DB, Kuperman S, et al. Genetics of event-related brain potentials in response to a semantic priming paradigm in families with a history of alcoholism. Am J Hum Genet. 2001;68:128–135. doi: 10.1086/316936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babor TF, Hofmann M, DelBoca FK, Hesselbrock V, Meyer RE, Dolinsky ZS, Rounsaville B. Types of alcoholics. I. Evidence for an empirically derived typology based on indicators of vulnerability and severity. Arch Gen Psychiatry. 1992;49:599–608. doi: 10.1001/archpsyc.1992.01820080007002. [DOI] [PubMed] [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li TK, Schuckit MA, Edenberg HJ, et al. The Collaborative Study on the Genetics of Alcoholism. Alcohol Health Res World. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B, Reich T, Edenberg HJ, Goate A, Blangero J, Almasy L, et al. Quantitative trait loci analysis of human event-related brain potentials: P3 voltage. Electroencephalogr Clin Neurophysiol. 1998;108:244–250. doi: 10.1016/s0168-5597(98)00002-1. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Rice JP, Goate A, Hinrichs AL, Saccone NL, Foroud T, Edenberg HJ, et al. A genomic scan for habitual smoking in families of alcoholics: Common and specific genetic factors in substance dependence. Am J Med Genet Part A. 2004;124A:19–27. doi: 10.1002/ajmg.a.20329. [DOI] [PubMed] [Google Scholar]
- Boehnke M. Allele frequency estimation from data on relatives. Am J Hum Genet. 1991;48:22–25. [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Reich T, et al. A new semi-structured psychiatric interview for use in genetic linkage studies: A report of the reliability of SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Hesselbrock VM, Shayka JJ, Nurnberger JI, Jr, Schuckit MA, Schmidt I, Reich T. Reliability of individual diagnostic criterion items for psychoactive substance dependence and the impact on diagnosis. J Stud Alcohol. 1995;56:500–505. doi: 10.15288/jsa.1995.56.500. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Hesselbrock VM, Heath AC, Kramer JR, Schuckit MA. A latent class analysis of antisocial personality disorder symptom data from a multi-center family study of alcoholism. Addiction. 2000;95(4):553–567. doi: 10.1046/j.1360-0443.2000.9545537.x. [DOI] [PubMed] [Google Scholar]
- Cloninger CR, Bohman M, Sigvardsson S Inheritance of alcohol abuse. Cross-fostering analysis of adopted men. Arch Gen Psychiatry. 1981;38:861–868. doi: 10.1001/archpsyc.1981.01780330019001. [DOI] [PubMed] [Google Scholar]
- Culverhouse R, Suarez BK, Lin J, Reich T. A perspective on epistasis: Limits of models displaying no main effect. Am J Hum Genet. 2002;70:461–471. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Li TK, Edenberg HJ, Hesselbrock V, Kramer J, Kuperman S, Porjesz B, et al. A Genome-Wide Screen for Genes Influencing Conduct Disorder. Mol Psychiatry. 2004;9:81–86. doi: 10.1038/sj.mp.4001368. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Bosron WF. Alcohol dehydrogenases. In: Guengerich FP, editor. Comprehensive toxicology, Vol. 3: Biotransformation. Chapter 3.08. New York: Pergamon; 1997. [Google Scholar]
- Feighner JP, Robins E, Guze SB, Woodruff RA, Jr, Winokur G, Munoz R. Diagnostic criteria for use in psychiatric research. Arch Gen Psychiatry. 1972;26:57–63. doi: 10.1001/archpsyc.1972.01750190059011. [DOI] [PubMed] [Google Scholar]
- Foroud T, Bucholz KK, Edenberg HJ, Goate A, Neuman RJ, Porjesz B, Koller DL, et al. Linkage of an alcoholism-related severity phenotype to chromosome 16. Alcohol Clin Exp Res. 1998;22:2035–2042. [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice JP, Flury L, Koller DL, Bierut LJ, et al. Alcoholism susceptibility loci: Confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24:933–945. [PubMed] [Google Scholar]
- Fulker DW, Cherny SS, Cardon LR. Multipoint interval mapping of quantitative trait loci using sib-pairs. Am J Hum Genet. 1995;56:1224–1233. [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Majumder PP. A two-stage variable stringency semi-parametric method for mapping quantitative trait loci with the use of genome-wide scan data on sib pairs. Am J Hum Genet. 2000;66:1046–1061. doi: 10.1086/302815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Reich T. Integrating sibship data for mapping quantitative trait loci. Ann Hum Genet. 2002;66:169–182. doi: 10.1017/S0003480002001070. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Begleiter H, Porjesz B, Chorlian D, Edenberg HJ, Foroud T, Goate A, et al. Linkage mapping of Beta 2 EEG waves via non-parametric regression. Am J Med Genet. 2003;118:66–71. doi: 10.1002/ajmg.b.10057. [DOI] [PubMed] [Google Scholar]
- Hesselbrock VM, Hesselbrock MN, Workman-Daniels KL. The effect of major depression and antisocial personality on alcoholism: Core use and motivational patterns. J Stud Alcohol. 1986;47(3):207–212. doi: 10.15288/jsa.1986.47.207. [DOI] [PubMed] [Google Scholar]
- Hesselbrock MN, Easton C, Bucholtz KK, Schuckit M, Hesselbrock VM. A validity study of the SSAGA—A comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Jones K, Porjesz B, Reich T, Almasy L, Bierut L, Goate A, Hinrichs A, et al. Linkage and linkage disequilibrium of evoked EEG oscillations with CHRM2 receptor gene polymorphisms: Implications for human brain dynamics and cognition. Int J Pschophysiol. 2004;53(2):75–90. doi: 10.1016/j.ijpsycho.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry. 1987;60:929–937. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, et al. Evidence for genetic linkage to alcohol dependence on Chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Myers MG, Brown SA, Mott MA. Preadolescent conduct disorder behaviors predict relapse and progression of addiction for adolescent alcohol and drug abusers. Alcohol Clin Exp Res. 1995;19:1528–1536. doi: 10.1111/j.1530-0277.1995.tb01019.x. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Foroud T, Flury L, Su J, Meyer ET, Hu K, Crowe R, et al. Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry. 2001;158:718–724. doi: 10.1176/appi.ajp.158.5.718. [DOI] [PubMed] [Google Scholar]
- Porjesz B, Begleiter H, Reich T, Van Eerdewegh P, Edenberg HJ, Foroud T, Goate A, et al. Amplitude of visual P3 event-related potential as a phenotypic marker for a predisposition to alcoholism: Preliminary results from the COGA Project. Collaborative Study on the Genetics of Alcoholism. Alcohol Clin Exp Res. 1998;22(6):1317–1323. [PubMed] [Google Scholar]
- Porjesz B, Almasy L, Edenberg HJ, Wang K, Chorlian D, Foroud T, Goate A, et al. Linkage disequilibrium between the beta frequency of the human EEG and a GABA A receptor gene locus. Proc Natl Acad Sci USA. 2002a;99:3729–3733. doi: 10.1073/pnas.052716399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porjesz B, Begleiter H, Wang K, Almasy L, Chorlian D, Stimus AT, Kuperman S, et al. Linkage and linkage disequilibrium mapping of ERP and EEG phenotypes. Biol Psychol. 2002b;61(1–2):229–248. doi: 10.1016/s0301-0511(02)00060-1. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Robins L. Deviant children grown up: A social and psychiatric study of sociopathic personality. Baltimore: Williams\& Wilkins; 1966. [Google Scholar]
- Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, et al. A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet. 2000;96:632–637. doi: 10.1002/1096-8628(20001009)96:5<632::aid-ajmg8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Silverman BW. Density estimation for statistics and data analysis. London: Chapman and Hall; 1986. [Google Scholar]
- Slutske WS, Heath AC, Dinwiddie SH, Madden PAF, Bucholz KK, Dunne MP, Statham DJ, et al. Common genetic risk factors for conduct disorder and alcohol dependence. J Abnorm Psychol. 1998;107:362–374. doi: 10.1037//0021-843x.107.3.363. [DOI] [PubMed] [Google Scholar]
- Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, Goate A, et al. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and event-related potentials. Am J Hum Genet. 1999;65:1148–1160. doi: 10.1086/302571. [DOI] [PMC free article] [PubMed] [Google Scholar]