Abstract
The M2 protein of influenza A viruses forms a tetrameric pH-activated proton-selective channel that is targeted by the amantadine class of anti-viral drugs. Its ion channel function has been extensively studied by electrophysiology and mutagenesis; however, the molecular mechanism of proton transport is still elusive, and the mechanism of inhibition by amantadine is controversial. We review the functional data on proton channel activity, molecular dynamics simulations of the proton conduction mechanism, and high-resolution structural and dynamical information of this membrane protein in lipid bilayers and lipid-mimetic detergents. These studies indicate that elucidation of the structural basis of M2 channel activity and inhibition requires thorough examination of the complex dynamics of the protein and the resulting conformational plasticity in different lipid bilayers and lipid-mimetic environments.
A. Function of the M2 proton channel of influenza A viruses
The M2 protein of influenza A and B viruses forms tetrameric proton channels that are important for the viral life cycle. After the virus enters the infected cell by endocytosis, the M2 proton channel opens in response to the low pH of the endosome, allowing proton flux into the virus, which triggers the dissociation of the viral RNA from the matrix proteins and the fusion of the viral and endosomal membranes. These events release the viral RNA to the cytoplasm for replication by the host cell (1). In a later stage of virus replication, the M2 protein maintains the high pH of the trans-Golgi network and prevents premature conformational changes of hemagglutinin in viruses with a high pH optimum of hemagglutinin-induced fusion (2).
The influenza A M2 (AM2) protein contains a short N-terminal periplasmic domain, a transmembrane (TM) domain, and a C-terminal cytoplasmic tail (Fig. 1). It is one of the smallest ion channel proteins and thus an excellent system for elucidating the structure-function relation of ion channels. Extensive mutagenesis, electrophysiology (3, 4) and sedimentation equilibrium experiments (5) have been conducted to characterize the function and stability of AM2 (for recent reviews, see (6, 7)). The AM2 proton channel is also the target of the amantadine class of drugs, one of only two classes of anti-influenza drugs currently available. However, the efficacy of amantadine dropped by two orders of magnitude between 2002 and 2007, although the 2008 seasonal flu strains were largely sensitive to amantadine. The resistance mainly resulted from the S31N mutation in the M2 TM domain (8). Thus, elucidating the mechanism of amantadine inhibition of AM2 has great public health relevance.
Fig. 1.
Amino acid sequence of influenza A/Udorn/72 M2 protein. The TM domain containing the crucial His37 and Trp41 residues (red) are underlined.
Recently, several high-resolution structural studies have been carried out to determine the structural basis of AM2 proton conductance and inhibition. In this article, we summarize the main functional data of AM2 and high-resolution structural information available on the TM domain, to promote future investigations of this intriguing and far from understood membrane protein.
Proton transport, selectivity, and activation
The proton conductance of AM2 has been well studied both in vivo and in vitro since its discovery as an ion channel (3). The gold standard of M2 functional assays is the replication of live viruses in infected cells, followed by the “silver standard” of whole-cell conductance data in Xenopus laevis oocytes. The wild-type protein is activated below pH 6.2 and its current is sensitive to amantadine. The single-channel activity under a −130 mV voltage is ~0.16 µA/ng protein at pH 6.2 for the A/Udorn/72 strain of M2 (4), which corresponds to 12 aA per channel, a conductance of ~100 aS, or ~100 protons per second. Extraction of single-channel currents requires quantification of the amount of protein, and the assumption that all proteins form open channels or the knowledge of the fraction of open channels.
For in-vitro liposome assays, several additional factors such as the orientation of the protein in the membrane, the size and buffering capacity of the liposomes, and the fluxes of other monovalent ions (Na+ and K+) can affect the measured conductance. One of the most reliable liposome assays employed an intravesicular pH-sensitive dye to measure proton fluxes, and obtained a single-channel current of 2.7 aA at pH 5.7 and 18°C (conductance = 29 aS) for full-length M2 in DMPC/DMPS vesicles (9). These values agree well with the oocyte result (4). Several other liposome assays gave more divergent values due to different experimental designs and protein constructs (10–12). However, the general consensus is that the single-channel conductance of M2 ranges from tens of aS to a few hundred aS, indicating low transport rates of ten to several hundred protons per second at mildly acidic pH. It is thought that the low currents are a result of the low physiological proton concentration (9), although the idea that M2 may have some characteristics of transporters has also been proposed (13).
There is general consensus that AM2 is selective for protons over Na+ and K+ by a factor of 1.5–3.0 × 106 (9, 14, 15), and that the increased proton current at low exterior pH (pHout) is due to true activation of the channel rather than mere mass action, since increased pHout stops the outward current of acidified cells (15, 16). The site of activation is His37, the only residue in the TM domain that can be protonated within the relevant pH range. Mutation of His37 to either Gly or Glu results in channels that are no longer modulated by pHout (17). Solid-state NMR (SSNMR) studies of His37 in the TM peptide M2(22–46) suggest that it is the third protonation at pH 6.3 that opens the channel (18).
His37 selectivity filter and Trp41 gate in the TM domain
His37 and Trp41 are the two most conserved residues in the M2 protein of influenza A strains (19) and are essential for channel function. His37 protonation is responsible for the activation and selectivity of the channel. The wild-type (WT) A/Udorn/72 M2 protein has 50-fold conductance difference between pHout of 4.5 and 8.2. In contrast, many His37 mutants, while still forming channels, are toxic to oocytes and have pH-independent currents, indicating non-specific conductance of other cations (17). These mutants can be neither activated effectively by low pHout (H37G, H37S and H37T) (20) nor inactivated by high pHout (H37G, H37E and H37A) (17). The mutant channels are not proton selective and are only partially and reversibly blocked by amantadine (20), in contrast to the WT protein (16). Although the H37 mutants are still tetrameric in oocytes (20), analytical ultracentrifugation (AUC) experiments indicate that H37A and H37F mutants are 3.1 and 1.8 kcal/mol less stable than the WT peptide in DPC micelles (21).
When a high concentration of imidazole buffer is added to the bathing solution, proton-selective conductance and gating are partially restored to the His37 mutants (20). This chemical rescue is specific to imidazole and does not require interaction with Trp41. Thus, His37 was proposed to be an intermediate proton acceptor and donor, consistent with SSNMR titration data of the TM peptide (18).
The M2 proton channel responds exclusively to external pH: low pHout activates the channel and high pHout closes the channel, regardless of the interior pH. Mutagenesis data indicates that Trp41 acts as the gate in concert with His37 (22). At high pHout and low pHin, the WT protein does not exhibit outward current while W41A/C/F mutants do, indicating loss of gating by the mutants (22). Further, intracellular Cu2+ inhibited the open W41A channel but not the WT channel, indicating that Trp41 shields His37 from coordination by Cu2+ (22, 23). These data suggest that the indole rings adopt distinct conformations at different pHout to obstruct or open the pore. Resonance Raman data indicate that the Trp41 conformational change must be small to keep its environmental hydrophobicity unchanged (24). Concerted conformational changes seem necessary for His37 to act as the selectivity filter and Trp41 as the gate. Compared to His37, the two Trp41 mutants that abolish gating (W41A and W41F) (22) actually have enhanced tetramer stability in DPC micelles (21), indicating that this residue has evolved for function rather than stability.
Mechanism of proton transport
Molecular dynamics (MD) simulations have been used extensively to study the M2 proton transport mechanism. The basic premise is that protons hop along a water wire via the Grotthuss mechanism, but the water wire is broken at the narrow His37 constriction (and possibly other locations along the channel). Thus, the mechanism of proton passage through the His37 tetrad is the central question. Two main models have been proposed: the gating model and the shuttle model. In the gating model, protonation of the His37 tetrad at low pH causes pore opening by electrostatic repulsion, so that a continuous water wire is formed (25). Conformational changes of the His37 and possibly Trp41 are necessary in this model. MD simulations suggest that a 60° change of the His37 χ2 angle may be sufficient to open the pore (26), and the excess proton may form a Zundel complex with two water molecules on the extra- and intracellular side of the histidine rings (27, 28). Early MD simulations postulated various charged states of His37 for channel activation (29). However, with SSNMR data indicating the third protonation of the His37 tetrad to be responsible for channel opening (18), recent simulations confirmed that the +3 state has the lowest proton permeation free energy (30). The channel water molecules that transport the protons were calculated to have three-fold lower diffusion coefficients than bulk water (27, 31).
The second proton transport model is the shuttle model, where the imidazole nitrogen facing the extracellular side binds one proton to form a biprotonated intermediate, then the other nitrogen releases the proton on the intracellular side and returns the imidazole to the neutral form (32). The initial state is then recovered by tautomerization or ring flip of the imidazole. This model explains the low proton transport rate and low-pH saturation of the channel, since both would be associated with the limiting rate of His37 sidechain torsional motion or tautomerization.
Simulations of the protein conformation at different pH also shed light on the mechanism of proton transport. One proposal is that in addition to His37, Val27 may act as a secondary gate that breaks the water wire, especially in the presence of amantadine (33). The two gates may not be static but instead may move synchronously in response to His37 protonation: at high pH the Val27 region opens while the His/Trp region narrows, and the opposite is found at low pH (13). Thus, instead of equating high pH with the closed state and vice versa, these simulations suggest a more complex view of an Openout-Closedin conformation at high pH and Closedout-Openin conformation at low pH. This model reconciles the different helix orientations and packing in different high-resolution structures (34–37).
Inhibition of the AM2 channel
Three classes of M2 inhibitors have been identified so far: the adamantane analogs (38, 39), the spirene-containing compounds (40), and Cu2+ ions. The FDA approved amantadine achieves its antiviral effect by inhibiting the M2 proton channel activity (3): 100 µM amantadine nearly irreversibly blocks > 90% of the channel activity of A/Udorn/72 (H3N2) M2 in several minutes at both acidic and neutral pH (16, 41, 42). For some other influenza strains the amantadine block is moderately stronger (50% - 2 fold) at neutral pH than at low pH (16). The Hill coefficient of the amantadine block is 1, consistent with the scenario that a single drug inhibits each functional channel (16).
Naturally occurring mutations in the TM domain that confer amantadine resistance include L26, V27, A30, S31, and G34, all N-terminal to G34 (8, 38, 43, 44) (Fig. 1). Extensive whole-cell conductance studies have been carried out on these mutants. S31N, the most common resistant mutant (8), has an IC50 of 199.9 µM, more than ten times higher than that of the WT (IC50 = 16 µM) (41). The resistance is not due to altered channel activity, as S31N has very similar specific activity and acid activation as the WT (4, 42). L26F has ~35% higher channel activity than S31N but is more sensitive to amantadine (42). G34E has a 3-fold higher specific activity than S31N and is about 5-fold less sensitive to amantadine than S31N (4). V27 mutants have mixed phenotypes: V27A and V27S are amantadine resistant but V27T is amantadine sensitive, and all V27 mutants have significantly increased (~ 3 fold) channel activities than WT (4). In contrast, the A30T mutant has a greatly reduced but still detectable activity, while the A30P mutant is inactive (4). Thus, residue 30 appears to be important for maintaining a functional structure of the protein. Among the resistant mutants, S31N was found by thiol-disulfide exchange experiments to be stable in both bilayers and micelles (42), suggesting a reason for its dominance in current flu strains that is independent of the selective pressure of amantadine. The stability and similar channel activity of S31N to the WT protein argue against the model that the S31N variant is resistant to amantadine by destabilizing the closed state and favoring the open state of the channel (45). The latter hypothesis was made based on the larger NMR linewidths of S31N than WT proteins in the presence of rimantadine. However, since S31N does not bind rimantadine, the more relevant comparison should be between S31N and the apo WT protein, which is known to exhibit broad linewidths as well.
In the C-terminal region of the TM domain, L38F, W41A, and D44A mutants incorporated into influenza viruses showed similar growth as the WT protein in infected cells, and the growth is sensitive to amantadine (41) (despite the fact that the W41A mutant does not gate properly (22)). Electric recordings in oocytes indicate that the IC50 of D44A is 15.8 µM, indistinguishable from the WT protein. Thus, no amantadine-resistant mutations have been found C-terminal to G34, implicating the segment between L26 and G34 to be the amantadine-binding region (41). Further, when residues 6–18 of the amantadine-insensitive BM2 were replaced by AM2 residues 24–36, the chimeric protein showed amantadine-sensitive currents in oocytes (41), indicating that the C-terminus is not the pharmacologically relevant region associated with amantadine's antiviral effect. In contrast, a recent liposome proton flux assay of M2(18–60) found the D44A mutant to be resistant to rimantadine while the S31A mutant to be sensitive, supporting a C-terminal binding model (34, 45). This liposome assay data is in serious contradiction both with the chimeric protein result and with the finding from the “gold standard” assay that replication of live virus bearing the D44A mutation is inhibited by rimantadine (41).
The second class of M2 inhibitors are spirene-containing compounds, initially discovered from a growth impairment assay where the M2 toxicity to yeast cells due to its proton channel activity is reversed by inhibitors (40). The first compound, BL-1743 (2-[3-azaspiro(5,5)undecanol]-2-imidazoline), has similar inhibition rate, Hill coefficient, and spectra of resistance as amantadine (46). However, BL-1743 inhibition is reversible (~12 minutes) while amantadine block is irreversible on the experimental time scale (~30 minutes). BL-1743 also differs from amantadine in its interaction with I35: the I35T mutant is resistant to BL-1743 but is reversibly inhibited by amantadine (46). The cause of this difference is suggested by a recent SSNMR study of the analogous spirene-piperidine compound, 3-azaspiro (5,5) undecane hydrochloride (IC50 = 0.92 µM) (47). This compound reduces the dynamic disorder of the G34-I35 backbone of AM2, suggesting that spirene-amine inhibitors bind more extensively and strongly with the TM helices than the adamantane inhibitors.
B. High-resolution structure of AM2 TM domain
Orientation of the TM helices in lipid bilayers
The TM peptide of AM2 has been a prime target for high-resolution structure investigation by a number of biophysical techniques. Both global structural information such as helix orientation and site-specific information of key residues have been obtained. The orientation of the TM helices in lipid bilayers has been extensively studied by SSNMR, EPR and IR techniques. The majority of SSNMR studies measured orientation-dependent 15N tensors on glass-plate aligned M2(22–46) (37, 48–51) and to a small extent on aligned full-length M2 (52, 53). From 1D 15N chemical shift anisotropies (CSA), Cross and coworkers obtained a helix tilt angle of 33°–37° for M2(22–46) in DMPC bilayers (49, 50) and 33° in DOPC bilayers. 2D NMR spectra correlating 15N-1H dipolar couplings and 15N CSAs gave more precise orientational constraints, including both the tilt angle (τ) and the rotation angles (ρ) of individual residues. From these 2D spectra, the tilt angle of apo M2(22–46) in DMPC bilayers was refined to 38° (51), while the rotation angles around the helix axis are consistent with the pore- or lipid-facing positions of residues inferred from functional data (PDB: 1NYJ).
Prior to 2001, the oriented NMR studies were conducted on peptides that were first mixed with lipids in organic solvents before hydration. These samples were not pH controlled and may be acidic due to the TFA salt associated with the peptide. After 2001, the oriented membranes were prepared by an aqueous protocol where M2 was reconstituted into lipid bilayers by detergent dialysis at controlled pH before alignment on glass plates. The amantadine-bound M2(22–46) and the full-length M2 were studied in this way. In DMPC/DMPG lipid bilayers, the 2D spectra of uniformly 15N-labeled full-length M2 at pH 8.0 showed a TM helix with a 25° tilt, which is smaller than the tilt of the TM peptide alone, and an 80° tilted helix, which is attributed to an amphipathic helix C-terminal to the TM domain (52). Hydrogen/deuterium exchange of full-length M2 showed that the TM helix underwent fast H/D exchange, consistent with its participation in forming an aqueous pore, while the 80°-tilted helix resisted H/D exchange, suggesting that it is shielded from water. Full resonance assignment is necessary to elucidate the orientation and depth of this C-terminal amphipathic helix.
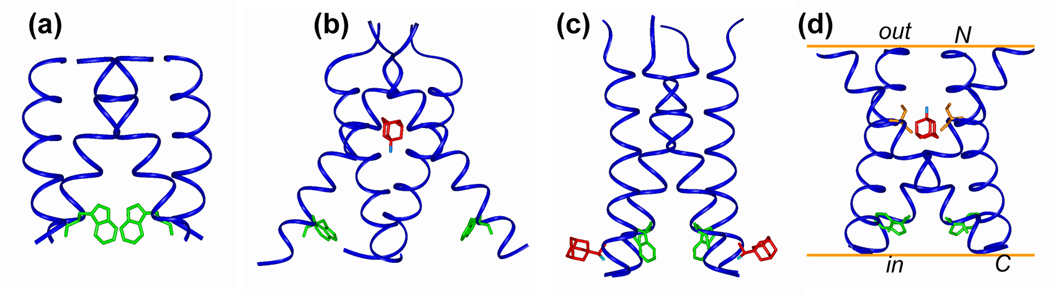
Upon amantadine binding, M2(22–46) exhibits a pronounced kink at G34 in DMPC bilayers, with a tilt angle of 31° for the N-terminal segment and 20° for the C-terminal segment (PDB: 2H95) (Fig. 5a) (48). While it is tempting to conclude that amantadine caused this kink, the bound peptide spectra were obtained from aqueous prepared samples at pH 8.8, under which condition there is no comparable apo peptide data. It is thus possible that the kink may already be present in the apo state, especially given the known propensity of Gly to perturb α-helices. Indeed, other high-resolution structures of AM2 suggest a G34 kink already in the apo state. For example, MAS-NMR data of DLPC-bound M2 suggest a kink of 5° for the bound peptide and 8° for the apo peptide (36), while the crystal structure of apo M2 at neutral pH shows a distribution of helix kink angles (35).
Fig. 5.
High-resolution structures of the TM domain of AM2. (a) Amantadine-bound orientational structure of M2(22–46) in DMPC bilayers at pH 8.8 from oriented-membrane SSNMR (PDB: 2H95) (48). The drug binding site was not directly studied but was implicitly assumed to be in the pore at the N-terminus side. (b) Amantadine-bound crystal structure of M2(22–46) in OG at pH 5.3 (PDB: 3C9J) (35). (c) Rimantadine-bound solution NMR structure of M2(18–60) in DHPC micelles at pH 7.5 (PDB: 2RLF) (34). Only the TM part is shown. (d) Amantadine-bound structure of M2(22–46) in DLPC bilayers at pH 7.5 from MAS SSNMR (PDB: 2KAD) (36). The amantadine position is hypothesized based on chemical shift perturbation of S31. The bilayer planes are drawn for reference. In each structure, two of the Trp41 indole rings (green) are shown, and the drug molecules are shown in red. IN (c), only two rimantadine molecules are shown for clarity.
Direct comparison of the orientation of amantadine-free and amantadine-bound M2(22–46) prepared under the same conditions was reported recently (54). Using unoriented DLPC vesicles and dialysis reconstituted M2(22–46), Hong and coworkers determined the helix orientations by exploiting the uniaxial diffusion of the helical bundles around the bilayer normal. The N-terminal half of the TM segment was found to have a tilt angle of 35° in the apo state and a slightly larger tilt of 38° in the amantadine-bound state (55) (Fig. 5d), indicating that the drug slightly enlarges the pore vestibule.
The TM helix orientation is also sensitive to membrane thickness. EPR spectra of N-terminus nitroxide labeled M2(22–46) reconstituted into DLPC, DMPC, DOPC, and POPC bilayers showed the least dipolar broadening in the thinnest DLPC membrane and the strongest broadening in the thickest POPC membrane (56). The broadening is attributed to shorter interhelical distances as the peptide decreases its tilt angle in the thicker membrane. The orientational change was detected directly in SSNMR spectra of unoriented M2(22–46). The TM helices showed a tilt angle of 35° in DLPC bilayers but a much smaller angle of 26° in the thicker POPC bilayers (54). These results are consistent with AM2 adjusting its TM helix orientation to minimize the hydrophobic mismatch with the lipid bilayer, as also observed for other membrane peptides (57, 58). These results revise an earlier conclusion that the M2 helix orientation is intrinsic to the peptide and unaffected by the membrane (50), made based on the fortuitously similar orientations of the peptide in DMPC and DOPC bilayers because of the similar thickness of these two bilayers.
The SSNMR and EPR orientational studies do not detect evidence of rotational angle change of M2 by pH: both the organic-solvent prepared samples, which likely have acidic pH, and the aqueous samples at high pH, gave the same rotation angles. These results are consistent with the functional requirement for a hydrophilic pore. Distances between the Trp41 indole 5-19F of neighboring helices were also found to be the same (11.8 Å) at pH 7.5 and pH 4.5 (Fig. 2), consistent with the lack of ρ change (59). Thus, it is surprising that a recent IR study of M2(22–46) in DMPC bilayers reported a one-residue (100°) rotation of the helices between low and high pH (60). The ρ angle of the low pH samples was found to agree well with early SSNMR data (37, 51) while that of the high pH samples differ by ~100°. When the ρ angles were combined with the +3 protonated His37 tetrad, MD simulations found that the rotated helices reduce His37 exposure to water at high pH, thus rationalizing channel closure (61). The ρ angles were extracted from diagonal linewidths of 2D IR spectra, which reflect the electrostatic environment of the 13C=18O groups, the main contributor of which is thought to be the water contact. However, the 2D lineshape analysis requires the distinction of homogeneous and inhomogeneous linewidths, consideration of population relaxation times, and consideration of water-independent electrostatic effects such as hydrogen bond network of the peptide. Thus the origin of the diagonal linewidth change is complex. In addition, linear FT-IR dichroism data for the high pH samples reported in this work do not show a clear sinusoidal dependence with the residue number, casting further doubt on the high pH ρ angle result (60).
Fig. 2.
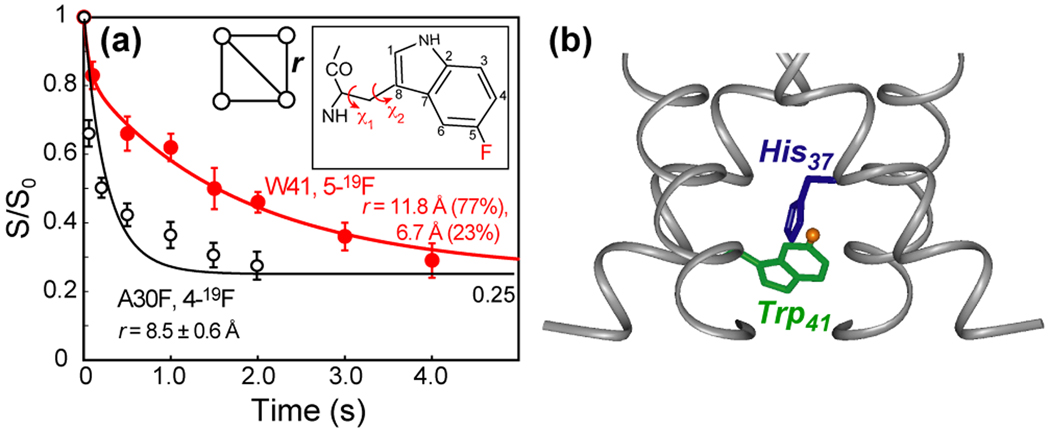
(a) Interhelical 19F-19F spin diffusion data of 5-19F-Trp41 WT M2 (red) and 4-19F-A30F M2 (black) in DMPC bilayers (59, 67). The best-fit distances confirm the tetrameric state of the peptide, and constrain the Trp41 sidechain (χ1, χ2) torsion angles (inset). (b) Trp41 t90 and His37 t-160 rotamers proposed from the distance data.
Tetramer stability and packing
In mammalian cells and oocytes the functional state of M2 is tetrameric, as shown by sucrose gradient, chemical cross-linking (62) and conductance experiments (5). In detergents and lipid bilayers, the TM peptide retains the tetrameric state under a wide range of conditions, and the monomer-tetramer equilibrium is readily shifted by membrane thickness, pH, amantadine binding, cholesterol, and amino acid sequence. The environmental effects on the free energy of tetramer association have been the focus of study using AUC in micelles (21, 63) and thiol disulfide exchange in lipid vesicles (64). It was shown that M2(19–46) tetramers are increasingly stabilized in the direction of detergents ≪ DLPC < DMPC < POPC. Among lipid bilayers the hydrophobic-thickness matched bilayers produce the most stable tetramers (64), but even the thinnest bilayer (DLPC) induces 100-fold tighter tetramer association than detergents. Both amantadine and cholesterol shift the equilibrium to tetramers (64, 65). Compared to external environmental factors, the sequence requirement for tetramer formation is much more lenient: Ala and Phe mutations of a number of pore residues retain stable M2(22–46) tetramers in DPC micelles, with the exception of the functionally essential His37 (21). However, more stable tetramers are often less functional tetramers, as manifested by their reduced ability to bind amantadine (63). For example, the most amantadine-resistant S31N mutant forms 0.4 kcal/mol more stable tetramers than the WT protein (42). This inverse relationship between stability and function suggests that the multiple M2 functions –proton conduction, gating and inhibitor binding – may require conformational flexibility, which cannot be provided by highly stable mutants locked in the wrong conformational minima.
Direct distance constraints for M2 tetramers in lipid bilayers were recently reported based on 19F spin diffusion SSNMR experiments that probe 19F-19F distances in the ~10 Å range (66, 67). So far, four interhelical 19F-19F distances have been reported for M2(22–46) in DMPC bilayers. On 4-19F labeled A30F M2, the 19F spin diffusion equilibrium value of 1/4 directly proved the tetrameric state, and the decay trajectory yielded a distance of 8.5 ± 0.6 Å between nearest neighbor helices (Fig. 2a). This distance rules out helix tilt angles smaller than 20° and constrains the Phe χ1 angle. The 19F NMR data of V27F suggest incomplete tetramer formation (partial dimers), which is consistent with the reduced stability of this mutant seen in AUC experiments (21). The L38F NMR data also indicated distance heterogeneities (59).
His37 and Trp41 sidechain conformation
A semi-quantitative 13C-15N distance between His37 Nπ and Trp41 Cγ was measured on DMPC-bound M2(22–46) at pH 7 and 38°C using SSNMR REDOR experiments (37). Significant dipolar dephasing was observed that corresponded to a minimal coupling of 63±12 Hz (37) or a maximum distance of 3.9 Å. The limiting nature of this result is because the measurements were done at physiological temperature where M2(22–46) undergoes uniaxial diffusion around the bilayer normal, which averages the dipolar coupling (50, 54). Scanning the possible His and Trp rotameric states indicate that this C–N distance must be between helices i and i+1, and suggested that His37 adopts the t-160 (χ1=180°, χ2=−160°) rotamer while Trp41 adopts the t-105 rotamer (PDB code: 1NYJ) (37). Subsequent reexamination of the conformational space of the two residues indicates that other possible solutions also exist: His37 can adopt the t-160 or the t60 rotamer, while Trp41 can adopt the t-105 or the t90 rotamer. Density functional theory calculations and MD simulations suggest that the most stable combination is (t60, t90) (26).
19F spin diffusion NMR experiments provided a direct distance constraint on the Trp41 sidechain conformation. The i - i+1 distance between 5-19F-Trp41 of M2(22–46) was measured to be mainly 11.8 Å at pH 7.5 (59) (Fig. 2a), and the value does not change significantly with amantadine binding and pH. When the helix orientation is fixed to 35° and the channel diameter is fixed to 10.2 Å between G34 Cα, the only Trp41 rotamer that is consistent with the 19F-19F distance is t90, which is consistent with the MD prediction (26). Combined with the measured His37-Trp41 C–N distance, the (t-160, t90) rotamer pair was proposed for the closed state of His37 and Trp41 (59). This conformation suggests that it may not be Trp41 alone but rather the cation-π interaction between His37 and Trp41 that is responsible for gating and the shielding of His37 from intracellular ions (Fig. 2b).
A third Trp41 rotamer of (χ1=−100°, χ2=+100) was proposed for the high pH state of M2 based on analysis of 19F NMR lineshapes under the combined effects of 19F CSA and 19F-19F interhelical dipolar couplings (68). However, the presence of intermediate time scale broadening of the spectra and the lack of information on the orientational angles between the 19F chemical shift tensor and the F-F dipolar tensors make this analysis a severely under-determined problem. The resulting rotamers are also inconsistent with any of the computational predictions (26).
His37 protonation state
The charged state of His37 gives crucial information on the activation of the M2 proton channel. The pKa of His37 in micelle-bound M2(22–46) was measured by 1H solution NMR to be 6.8 for the monomer and 6.4 for the tetramer in the absence of drug (65). Amantadine decreases the tetramer pKa to 5.8. UV resonance Raman experiments on M2(22–46) in POPE/POPS membranes found a pKa of 5.7 based on the intensity of the 1407 cm−1 band (24). This imidazole band has the same intensity in liposomes and in SDS micelles at pH 4.0, indicating that all four His residues are protonated at pH 4.0 (24).
By far the most definitive study of His37 protonation states came from 15N SSNMR experiments of M2(22–46) in DMPC/DMPG bilayers by Cross and coworkers (18), who monitored the imidazole Nδ1 and Nε2 15N isotropic chemical shifts as a function of pH. Two neutral tautomers were found at pH 8.6, with the unprotonated nitrogen exhibiting a large isotropic shift of ~230 ppm. Between pH 8.0 and pH 7.0, two +1 dimer species, His-HisH+, were concluded. Based on the relatively large 15N linewidths in this pH range, which suggest intermediate time scale motion, low-barrier hydrogen bonds between imidazole and imidazolium within each dimer were proposed. Below pH 6.5 and pH 5.0, the third and fourth His are protonated. Quantification of the spectral intensities yielded four pKa’s at 8.2, 8.2, 6.3 and < 5.0. Thus, the first two histidines titrate cooperatively at higher pH than the aqueous pKa of His (6.0). Since the +1 dimers already exist at neutral pH, they are proposed to form a histidine-locked state that occludes the pore (18). The binding of the third proton below pH 6.3 was proposed to break the two-fold symmetry of the +1 dimers and thus activates the channel.
Amantadine binding changes the titration curve of the His37 tetrad (69). The 230 ppm unprotonated 15N peak persisted to a much lower pH of 6.0 in the presence of the drug. Intensity analysis shows that the bound peptide has only one pKa near 5.4. The nature of this apparently highly cooperative protonation event is still unknown. In addition, the 15N peak associated with the His-HisH+ dimer is no longer observed in the amantadine-bound peptide, suggesting that the neutral tautomers directly form biprotonated histidines in the presence of amantadine. Further studies are necessary to elucidate the amantadine-induced changes of His37 protonation.
Extensive conformational dynamics of AM2
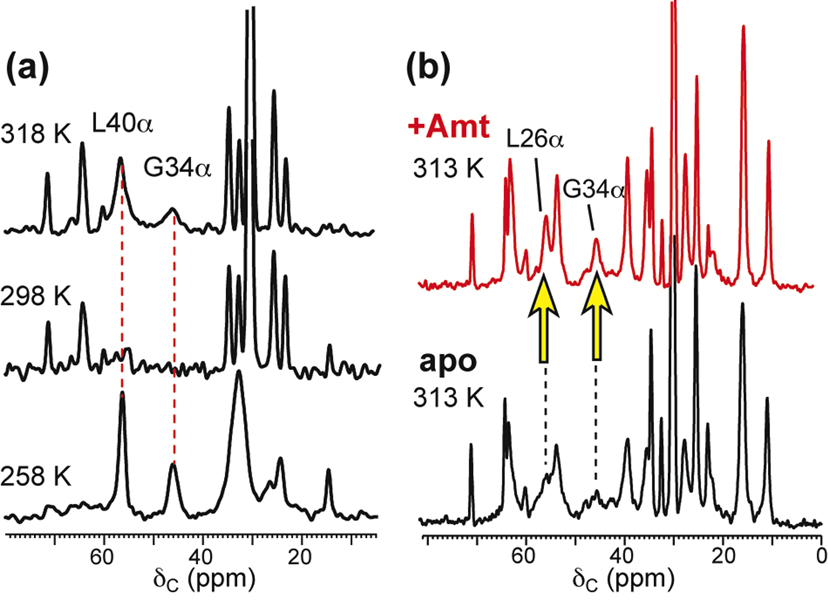
The M2 homo-tetramer exhibits extensive conformational dynamics in lipid bilayers and lipid-mimetic detergents, as well documented by SSNMR and solution NMR spectra. Membrane-bound apo M2(22–46) shows dynamically broadened lines in both unoriented vesicle spectra (36, 55) and oriented spectra (70) at ambient temperature in a range of model membranes: DLPC, DMPC/DMPG, and POPC. The fact that the line broadening is mainly dynamic in origin rather than due to static conformational distributions is proved by the temperature dependence of the linewidths. The broadest lines are observed between 298 K and 263 K (54), above and below which the lines narrow (Fig. 3a). Acidic pH broadens the lines compared to neutral pH for both membrane-bound M2(22–46) (70) and detergent-bound M2(18–60) (34). In all solvent environments and for both M2(22–46) and M2(18–60), amantadine noticeably narrows the linewidths (Fig. 3b). The line broadening or narrowing occur for all residues, thus it reflects the dynamic property of the whole protein rather than segmental motion.
Fig. 3.
13C MAS spectra of DLPC-bound M2(22–46). (a) Variable temperature 13C MAS spectra indicate large-amplitude intermediate timescale motion at ambient temperature (54). (b) Amantadine narrows the linewidths of most residues at 313 K (55).
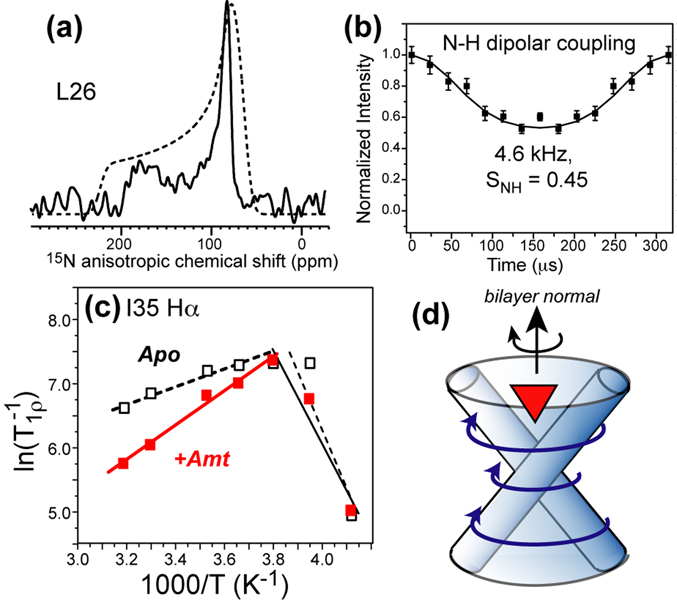
The origin of the broad linewidths and their narrowing by amantadine has been examined in detail by SSNMR (54, 71). The main motion accounting for the broad linewidth is the whole-body uniaxial diffusion of the tetramers around the bilayer normal, as shown by motionally averaged 2H, 15N and 13C NMR lineshapes (Fig. 4a, b) (54). The correlation time of this uniaxial diffusion has been estimated from relaxation studies to be ~3 µs (71) for DLPC-bound apo M2(22–46) at 313 K, which is very close to the time scale of the radio-frequency pulses and nuclear spin interactions (2–4 µs), thus causing intermediate time scale broadening. Amantadine binding reduces the correlation time by 2–3 fold at 313 K (71) (Fig. 4c), thus avoiding exchange broadening and narrowing the NMR lines. The larger diffusion rates strongly suggest that amantadine promotes the formation of more homogeneous helical bundles (Fig. 4d), which is consistent with the increased tetramer stability observed in thiol disulfide exchange experiments (64). The NMR relaxation data also indicate that excess amantadine in the bilayer increases the membrane viscosity. Since protein rotational diffusion is generally sensitive to membrane viscosity, cholesterol-rich virus-mimetic membranes immobilize this diffusion, and was found to suppress exchange broadening and give rise to high-resolution NMR spectra in a wide temperature range (72).
Fig. 4.
Uniaxial diffusion of M2(22–46) helical bundles around the bilayer normal. (a) 15N CSA of L26 is uniaxially averaged from the rigid-limit pattern (dashed line) (54). (b) L26 N–H dipolar coupling is reduced from the rigid-limit coupling. (c) Amantadine decreases 1H T1ρ relaxation rates at high temperature, indicating it speeds up protein motion (71). (d) Schematic of M2 uniaxial diffusion. Amantadine binding creates better packed tetramers, thus speeding up rotational diffusion.
Whether the whole-body uniaxial diffusion has functional importance is not known. If the viral envelope is in a liquid-ordered phase, then M2 is most likely immobilized. But if the virus envelope contains significant liquid-disordered domains, then the fact that M2 is poorly incorporated into raftlike microdomains (73) should promote uniaxial diffusion in the functional state. Most spectroscopic studies so far were carried out on M2 bound to non-cholesterol-containing membranes or bound to detergents, which favor the whole-body motion. Thus site-specific conformational dynamics have not been separated from the whole-body motion. In this light, the larger linewidths of M2 at low pH are at least partly due to global motional changes, and their interpretation in the context of pH activation needs to be considered carefully (34, 70).
Amantadine binding in high-resolution structures
Two atomic-resolution structures of TM-containing portions of AM2 were recently solved using X-ray crystallography and solution NMR, which shed light on the pH activation of the channel and led to two different models of M2 inhibition (74).
The crystal structures were solved on M2(22–46) in octyl-β-D-glucopyranoside (OG) at pH 7.3 in the absence of amantadine (PDB: 2BKD) (2.0 Å resolution) and at pH 5.3 in the presence of amantadine (PDB: 3C9J) (3.5 Å resolution) (35). In both structures, the N-terminal half of the TM domain has a tilt angle of ~35°, consistent with SSNMR data (51, 54). The orientation of the C-terminus half of the TM helix depends on the pH: at low pH the helices are uniformly straight and diverge toward the C-terminus, creating an open cavity, but at pH 7.3 two of the four helices bend and create a small pore at the His37/Trp41 region. The amantadine-bound structure, solved with a 1.3 : 1 protein : amantadine molar ratio, shows a single drug molecule in the pore, surrounded by residues that confer drug resistance (Fig. 5b). Thus, the amantadine binding site and stoichiometry agree with electrophysiology data (16), and the crystal structure supports a occlusion model for channel inhibition.
The solution NMR structure was solved on rimantadine-bound M2(18–60) in DHPC micelles at pH 7.5 (PDB: 2RLF) (34). The sample contains 0.75 mM protein, 40 mM rimantadine and 300 mM DHPC, thus the drug is in 53-fold excess to the protein or ~210 fold excess to the channel. The TM helix is tilted by ~15° from the bilayer normal and the C-terminal amphipathic helix is roughly perpendicular to the TM helix. Low pH broadens the linewidths and increases the millisecond motion of Trp41. Four protein-rimantadine NOEs were found to lipid-facing residues L40, I42, L43, and R45 near the C-terminus of the TM helix, with four rimantadine molecules per tetramer (Fig. 5c). Since this surface binding site is inconsistent with the large body of mutagenesis data that place drug-resistant mutations at pore-facing residues at the N-terminus, the authors proposed that rimantadine inhibits the channel by stabilizing its closed state and making the channel harder to open, whereas drug-resistant mutations destabilize the closed channel and make it easier to open. However, this allosteric inhibition model is inconsistent with the one-drug-per-channel stoichiometry of amantadine (16), and also contradicts virus replication and whole cell conductance data that showed D44A and R45A mutants to be amantadine sensitive (41). The drug – protein NOEs involve residues in the headgroup region of the micelle, exactly where the amphiphilic rimantadine is located based on paramagnetic relaxation NMR data (75, 76). At the high concentration of 40 mM, rimantadine constitutes 12 mol% of the micelle, making the surface binding site a likely secondary lipid binding site rather than the true inhibition site. Given the abundant reorientational dynamics of the peptide and amantadine discussed above (55, 71), the lack of NOEs between the N-terminal pore residues and rimantadine could arise from motion on unfavorable time scales that broadens the NMR signals and/or makes the NOE intensity vanish, or from micelle-altered helix orientation and packing that reject rimantadine in the pore. Thus, while this work shows the presence of rimantadine on the surface of the protein, they do not provide strong evidence against a pharmacologically relevant binding site in the pore.
It is unlikely for the different protein lengths to be the cause for the drug binding site difference, since the TM peptide is capable of binding amantadine, folding into appropriate tetramers, and having at least 50% of the proton conductance of full-length M2 (77).
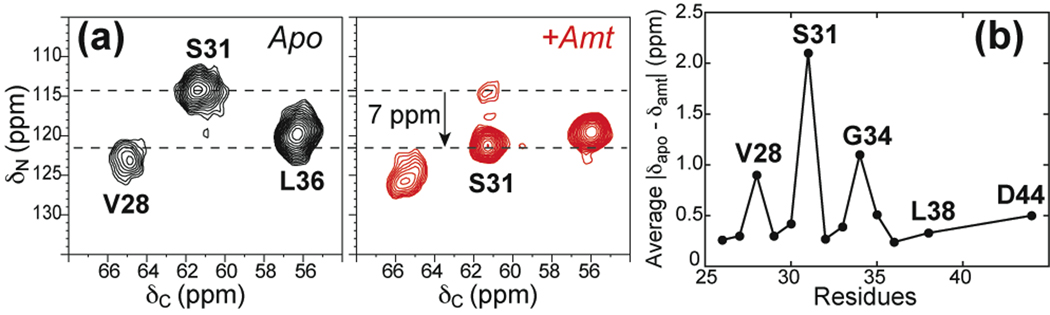
The above two high-resolution structures were solved in detergents and micelles. Recently, an amantadine-bound structure of M2(22–46) in DLPC bilayers was reported based on MAS SSNMR data (36, 55). The experiments were carried out at a peptide : amantadine : DLPC molar ratio of 1 : 8 : 15. The main conformational constraints came from 13C and 15N isotropic chemical shifts (CS) measured at 243 K where the protein dynamics is frozen. Comparison of the apo and bound peptide chemical shifts indicated that the M2(22–46) conformation is most perturbed by amantadine at S31, with a large 15N CS change of 7 ppm, strongly suggesting drug binding near S31 (Fig. 6a). The CS perturbation is also high for G34 and V28, while the C-terminal L38 and D44 are minimally affected (Fig. 6b). The chemical-shift predicted (φ, ψ) angles for the apo and bound monomers also show that the apo peptide has a slightly larger kink than the amantadine-bound peptide. The monomer structure was combined with the helix orientation and inter-helical 19F-19F distances to lead to a tetramer structure for M2(22–46) at pH 7.5 (PDB: 2KAD). The tetramer has larger N-terminus vestibule than the other structures, and differs in the Trp41 conformation (36) (Fig. 5d). SSNMR studies also found that amantadine slightly perturbs the sidechain conformations of V27 and V28 (78).
Fig. 6.
SSNMR evidence for amantadine binding near S31. (a) 2D 15N-13C correlation spectra of DLPC-bound M2(22–46) in the absence (black) and presence (red) of amantadine (36). (b) Chemical shift perturbation by amantadine in bilayer-bound M2(22–46).
C. Future directions
While a large amount of biochemical and structural information have been obtained for the M2 proton channel, many open questions remain about this fascinating multi-functional protein. So far the most complete three-dimensional structures were solved in detergents and micelles, which are only partial mimics of lipid bilayers. Given the plasticity of the protein, high-resolution structural studies in lipid bilayers that mimic the virus envelope will be very valuable. Most structural studies so far used partial constructs of the full-length protein, thus future studies of the full-length protein will clearly be desirable, especially to understand the role of the C-terminal amphipathic helix, which was found to be able to affect ion channel function by interacting with the TM helix (79). Elucidating how M2 changes its conformation and dynamics between high pH and low pH is central to understanding its activation, and will require atomic-resolution information of His37 and Trp41 conformation as a function of pH. Clearly, protein-drug distances in lipid bilayers will be essential for resolving the controversy of M2 inhibition. Finally, the ultimate public health benefit of this research is to develop new antiviral drugs to target the dominant mutants of influenza A viruses to prevent future influenza pandemics. Achieving this end will require a combination of pharmacological, biochemical, and structural studies.
References
- 1.Lamb RA, Holsinger KJ, Pinto LH. The Influenza A virus M2 ion channel protein and its role in the influenza virus life cycle. In: Wemmer E, editor. Cellular Receptors of Animal Viruses. Plainview, NY: Cold Spring Harbor Lab Press; 1994. pp. 303–321. [Google Scholar]
- 2.Sugrue RJ, Bahadur G, Zambon MC, Hall-Smith M, Douglas AR, et al. Specific structural alteration of the influenza haemagglutinin by amantadine. EMBO J. 1990;9:3469–3476. doi: 10.1002/j.1460-2075.1990.tb07555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 4.Holsinger LJ, Nichani D, Pinto LH, Lamb RA. Influenza A virus M2 ion channel protein: a structure-function analysis. J. Virol. 1994;68:1551–1563. doi: 10.1128/jvi.68.3.1551-1563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakaguchi T, Tu Q, Pinto LH, Lamb RA. The active oligomeric state of the minimalistic influenza virus M2 ion channel is a tetramer. Proc. Natl. Acad. Sci. USA. 1997;94:5000–5005. doi: 10.1073/pnas.94.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinto LH, Lamb RA. The M2 Proton Channels of Influenza A and B Viruses. J. Biol. Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 7.Pinto LH, Lamb RA. Controlling influenza virus replication by inhibiting its proton channel. Mol. Biosyst. 2007;3:18–23. doi: 10.1039/b611613m. [DOI] [PubMed] [Google Scholar]
- 8.Bright RA, Medina MJ, Xu X, Perez-Oronoz G, Wallis TR, et al. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- 9.Lin TI, Schroeder C. Definitive assignment of proton selectivity and attoampere unitary current to the M2 ion channel protein of influenza A virus. J. Virol. 2001;75:3647–3656. doi: 10.1128/JVI.75.8.3647-3656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moffat JC, Vijayvergiya V, Gao PF, Cross TA, Woodbury DJ, et al. Proton transport through influenza A virus M2 protein reconstituted in vesicles. Biophys. J. 2008;94:434–445. doi: 10.1529/biophysj.107.109082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijayvergiya V, Wilson R, Chorak A, Gao PF, Cross TA, et al. Proton conductance of influenza virus M2 protein in planar lipid bilayers. Biophys. J. 2004;87:1697–1704. doi: 10.1529/biophysj.104.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duff KC, Ashley RH. The transmembrane domain of influenza A M2 protein forms amantadine-sensitive proton channels in planar lipid bilayers. Virology. 1992;190:485–489. doi: 10.1016/0042-6822(92)91239-q. [DOI] [PubMed] [Google Scholar]
- 13.Khurana E, Dal Peraro M, DeVane R, Vemparala S, DeGrado WF, et al. Molecular dynamics calculations suggest a conduction mechanism for the M2 proton channel from influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 2009;106:1069–1074. doi: 10.1073/pnas.0811720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chizhmakov IV, Geraghty FM, Ogden DC, Hayhurst A, Antoniou M, et al. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J. Physiol. 1996;494:329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mould JA, Drury JE, Frings SM, Kaupp UB, Pekosz A, et al. Permeation and activation of the M2 ion channel of influenza A virus. J. Biol. Chem. 2000;275:31038–31050. doi: 10.1074/jbc.M003663200. [DOI] [PubMed] [Google Scholar]
- 16.Wang C, Takeuchi K, Pinto LH, Lamb RA. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J. Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Lamb RA, Pinto LH. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys. J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J, Fu R, Nishimura K, Zhang L, Zhou HX, et al. Histidines, heart of the hydrogen ion channel from influenza A virus: toward an understanding of conductance and proton selectivity. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito T, Gorman OT, Kawaoka Y, Bean WJ, Webster RG. Evolutionary analysis of the influenza A virus M gene with comparison of the M1 and M2 proteins. J. Virol. 1991;65:5491–5498. doi: 10.1128/jvi.65.10.5491-5498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkataraman P, Lamb RA, Pinto LH. Chemical rescue of histidine selectivity filter mutants of the M2 ion channel of influenza A virus. J. Biol. Chem. 2005;280:21463–21472. doi: 10.1074/jbc.M412406200. [DOI] [PubMed] [Google Scholar]
- 21.Howard KP, Lear JD, DeGrado WF. Sequence determinants of the energetics of folding of a transmembrane four-helix-bundle protein. Proc. Natl. Acad. Sci. USA. 2002;99:8568–8572. doi: 10.1073/pnas.132266099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y, Zaitseva F, Lamb RA, Pinto LH. The Gate of the Influenza Virus M2 Proton Channel Is Formed by a Single Tryptophan Residue. J. Biol. Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 23.Gandhi CS, Shuck K, Lear JD, Dieckmann GR, DeGrado WF, et al. Cu(II) inhibition of the proton translocation machinery of the influenza A virus M2 protein. J. Biol. Chem. 1999;274:5474–5482. doi: 10.1074/jbc.274.9.5474. [DOI] [PubMed] [Google Scholar]
- 24.Okada A, Miura T, Takeuchi H. Protonation of Histidine and Histidine-Tryptophan Interaction in the Activation of the M2 Ion Channel from Influenza A Virus. Biochemistry. 2001;40:6053–6060. doi: 10.1021/bi0028441. [DOI] [PubMed] [Google Scholar]
- 25.Sansom MS, Kerr ID, Smith GR, Son HS. The influenza A virus M2 channel: a molecular modeling and simulation study. Virology. 1997;233:163–173. doi: 10.1006/viro.1997.8578. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y, Voth GA. A Computational Study of the Closed and Open States of the Influenza A M2 Proton Channel. Biophys. J. 2005;89:2402–2411. doi: 10.1529/biophysj.105.066647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smondyrev AM, Voth GA. Molecular dynamics simulation of proton transport through the influenza A virus M2 channel. Biophys. J. 2002;83:1987–1996. doi: 10.1016/S0006-3495(02)73960-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Voth GA. Computational studies of proton transport through the M2 channel. FEBS Lett. 2003;552:23–27. doi: 10.1016/s0014-5793(03)00779-8. [DOI] [PubMed] [Google Scholar]
- 29.Zhong Q, Newns DM, Pattnaik P, Lear JD, Klein ML. Two possible conducting states of the influenza A virus M2 ion channel. FEBS Lett. 2000;473:195–198. doi: 10.1016/s0014-5793(00)01522-2. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Wu Y, Voth GA. Proton transport behavior through the influenza A M2 channel: insights from molecular simulation. Biophys. J. 2007;93:3470–3479. doi: 10.1529/biophysj.107.105742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forrest LR, Kukol A, Arkin IT, Tieleman DP, Sansom MSP. Exploring models of the influenza A M2 channel: MD simulations in a phospholipid bilayer. Biophys. J. 2000;78:55–69. doi: 10.1016/s0006-3495(00)76572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, et al. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc. Natl. Acad. Sci. USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi M, Cross TA, Zhou HX. A secondary gate as a mechanism for inhibition of the M2 proton channel by amantadine. J. Phys. Chem. B. 2008;112:7977–7979. doi: 10.1021/jp800171m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cady SD, Mishanina TV, Hong M. Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding. J. Mol. Biol. 2009;385:1127–1141. doi: 10.1016/j.jmb.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishimura K, Kim S, Zhang L, Cross TA. The closed state of a H+ channel helical bundle combining precise orientational and distance restraints from solid state NMR. Biochemistry. 2002;41:13170–13177. doi: 10.1021/bi0262799. [DOI] [PubMed] [Google Scholar]
- 38.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grambas S, Bennett MS, Hay AJ. Influence of amantadine resistance mutations on the pH regulatory function of the M2 protein of influenza A viruses. Virology. 1992;191:541–549. doi: 10.1016/0042-6822(92)90229-i. [DOI] [PubMed] [Google Scholar]
- 40.Kurtz S, Luo G, Hahnenberger KM, Brooks C, Gecha O, et al. Growth impairment resulting from expression of influenza virus M2 protein in Saccharomyces cerevisiae: identification of a novel inhibitor of influenza virus. Antimicrob. Agents Chemother. 1995;39:2204–2209. doi: 10.1128/aac.39.10.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jing X, Ma C, Ohigashi Y, Oliveira FA, Jardetzky TS, et al. Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc. Natl. Acad. Sci. USA. 2008;105:10967–10972. doi: 10.1073/pnas.0804958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stouffer AL, Ma C, Cristian L, Ohigashi Y, Lamb RA, et al. The interplay of functional tuning, drug resistance, and thermodynamic stability in the evolution of the M2 proton channel from the influenza A virus. Structure. 2008;16:1067–1076. doi: 10.1016/j.str.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidtke M, Zell R, Bauer K, Krumbholz A, Schrader C, et al. Amantadine resistance among porcine H1N1, H1N2, and H3N2 influenza A viruses isolated in Germany between 1981 and 2001. Intervirology. 2006;49:286–293. doi: 10.1159/000094244. [DOI] [PubMed] [Google Scholar]
- 44.Tang JW, Ngai KL, Wong JC, Lam WY, Chan PK. Emergence of adamantane-resistant influenza A(H3N2) viruses in Hong Kong between 1997 and 2006. J. Med. Virol. 2008;80:895–901. doi: 10.1002/jmv.21155. [DOI] [PubMed] [Google Scholar]
- 45.Pielak RM, Schnell JR, Chou JJ. Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7379–7384. doi: 10.1073/pnas.0902548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tu Q, Pinto LH, Luo G, Shaughnessy MA, Mullaney D, et al. Characterization of inhibition of M2 ion channel activity by BL-1743, an inhibitor of influenza A virus. J. Virol. 1996;70:4246–4252. doi: 10.1128/jvi.70.7.4246-4252.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Cady SD, Balannik V, Pinto LH, DeGrado WF, et al. Discovery of spiro-piperidine inhibitors and their modulation of the dynamics of the M2 proton channel from influenza A virus. J. Am. Chem. Soc. 2009;131:8066–8076. doi: 10.1021/ja900063s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu J, Asbury T, Achuthan S, Li C, Bertram R, et al. Backbone Stucture of the Amantadine-Block Trans-Membrane Domain M2 Proton Channel from Influenza A Virus. Biophys. J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kovacs FA, Cross TA. Transmembrane four-helix bundle of influenza A M2 protein channel: structural implications from helix tilt and orientation. Biophys. J. 1997;73:2511–2517. doi: 10.1016/S0006-3495(97)78279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kovacs FA, Denny JK, Song Z, Quine JR, Cross TA. Helix tilt of the M2 transmembrane peptide from influenza A virus: an intrinsic property. J. Mol. Biol. 2000;295:117–125. doi: 10.1006/jmbi.1999.3322. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Kim S, Kovacs F, Cross TA. Structure of the transmembrane region of the M2 protein H(+) channel. Protein Sci. 2001;10:2241–2250. doi: 10.1110/ps.17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian C, Tobler K, Lamb RA, Pinto LH, Cross TA. Expression and initial structural insights from solid-state NMR of the M2 proton channel from influenza A virus. Biochemistry. 2002;41:11294–11300. doi: 10.1021/bi025695q. [DOI] [PubMed] [Google Scholar]
- 54.Cady SD, Goodman C, Tatko CD, DeGrado WF, Hong M. Determining the orientation of uniaxially rotating membrane proteins using unoriented samples: a 2H, 13C, and 15N solid-state NMR investigation of the dynamics and orientation of a transmembrane helical bundle. J. Am. Chem. Soc. 2007;129:5719–5729. doi: 10.1021/ja070305e. [DOI] [PubMed] [Google Scholar]
- 55.Cady SD, Hong M. Amantadine-Induced Conformational and Dynamical Changes of the Influenza M2 Transmembrane Proton Channel. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1483–1488. doi: 10.1073/pnas.0711500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duong-Ly KC, Nanda V, DeGrado WF, Howard KP. The conformation of the pore region of the M2 proton channel depends on lipid bilayer environment. Protein Sci. 2005;14:856–861. doi: 10.1110/ps.041185805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Planque MR, Greathouse DV, Koeppe RE, Schafer H, Marsh D, et al. Influence of lipid/peptide hydrophobic mismatch on the thickness of diacylphosphatidylcholine bilayers. A 2H NMR and ESR study using designed transmembrane alpha-helical peptides and gramicidin A. Biochemistry. 1998;37:9333–9345. doi: 10.1021/bi980233r. [DOI] [PubMed] [Google Scholar]
- 58.Park SH, Opella SJ. Tilt angle of a trans-membrane helix is determined by hydrophobic mismatch. J. Mol. Biol. 2005;350:310–318. doi: 10.1016/j.jmb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 59.Luo W, Mani R, Hong M. Sidechain conformation and gating of the M2 transmembrane peptide proton channel of influenza A virus from solid-state NMR. J. Phys. Chem. 2007;111:10825–10832. doi: 10.1021/jp073823k. [DOI] [PubMed] [Google Scholar]
- 60.Manor J, Mukherjee P, Lin YS, Leonov H, Skinner JL, et al. Gating mechanism of the influenza A M2 channel revealed by 1D and 2D IR spectroscopies. Structure. 2009;17:247–254. doi: 10.1016/j.str.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leonov H, Arkin IT. pH-driven helix rotations in the influenza M2 H(+) channel: a potential gating mechanism. Eur Biophys J. 2009 doi: 10.1007/s00249-009-0434-0. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 62.Holsinger LJ, Lamb RA. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- 63.Stouffer AL, Nanda V, Lear JD, DeGrado WF. Sequence determinants of a transmembrane proton channel: an inverse relationship between stability and function. J. Mol. Biol. 2005;347:169–179. doi: 10.1016/j.jmb.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 64.Cristian L, Lear JD, DeGrado WF. Use of thiol-disulfide equilibria to measure the energetics of assembly of transmembrane helices in phospholipid bilayers. Proc. Natl. Acad. Sci. USA. 2003;100:14772–14777. doi: 10.1073/pnas.2536751100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salom D, Hill BR, Lear JD, DeGrado WF. pH-dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus. Biochemistry. 2000;39:14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buffy JJ, Waring AJ, Hong M. Determination of Peptide Oligomerization in Lipid Membranes with Magic-Angle Spinning Spin Diffusion NMR. J. Am. Chem. Soc. 2005;127:4477–4483. doi: 10.1021/ja043621r. [DOI] [PubMed] [Google Scholar]
- 67.Luo W, Hong M. Determination of the oligomeric number and intermolecular distances of membrane protein assemblies by anisotropic 1H-driven spin diffusion NMR spectroscopy. J. Am. Chem. Soc. 2006;128:7242–7251. doi: 10.1021/ja0603406. [DOI] [PubMed] [Google Scholar]
- 68.Witter R, Nozirov F, Sternberg U, Cross TA, Ulrich AS, et al. Solid-state 19F NMR spectroscopy reveals that Trp41 participates in the gating mechanism of the M2 proton channel of influenza A virus. J. Am. Chem. Soc. 2008;130:918–924. doi: 10.1021/ja0754305. [DOI] [PubMed] [Google Scholar]
- 69.Hu J, Riqiang F, Cross TA. The Chemical and Dynamical Influence of the Anti-Viral Drug Amantadine on the M2 Proton Channel Transmembrane Domain. Biophys. J. 2007;93:276–283. doi: 10.1529/biophysj.106.102103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li C, Qin H, Gao FP, Cross TA. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim. Biophys. Acta. 2007;1768:3162–3170. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cady SD, Hong M. Effects of amantadine binding on the dynamics of membrane-bound influenza A M2 transmembrane peptide studied by NMR relaxation. J. Biomol. NMR. 2009 doi: 10.1007/s10858-009-9352-9. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo W, Cady SD, Hong M. Immobilization of the Influenza A M2 Transmembrane Peptide in Virus-Envelope Mimetic Lipid Membranes: A Solid-State NMR Investigation. Biochemistry. 2009 doi: 10.1021/bi900716s. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leser GP, Lamb RA. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 74.Miller C. Ion channels: coughing up flu's proton channels. Nature. 2008;451:532–533. doi: 10.1038/451532a. [DOI] [PubMed] [Google Scholar]
- 75.Wang J, Schnell JR, Chou JJ. Amantadine partition and localization in phospholipid membranes: a solution NMR study. Biochem. Biophys. Res. Commun. 2004;324:212–217. doi: 10.1016/j.bbrc.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 76.Li C, Yi M, Hu J, Zhou HX, Cross TA. Solid-state NMR and MD simulations of the antiviral drug amantadine solubilized in DMPC bilayers. Biophys. J. 2008;94:1295–1302. doi: 10.1529/biophysj.107.112482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma C, Polishchuk AL, Ohigashi Y, Stouffer AL, Scho_n A, et al. Identification of the Functional Core of the Influenza A Virus A/M2 Proton-Selective Ion Channel. Proc. Natl. Acad. Sci. U.S.A. 2009 doi: 10.1073/pnas.0905726106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hong M, Mishanina TV, Cady SD. Accurate measurement of methyl 13C chemical shifts by solid-state NMR for the determination of protein sidechain conformation: the influenza M2 transmembrane peptide as an example. J. Am. Chem. Soc. 2009;131:7806–7816. doi: 10.1021/ja901550q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nguyen PA, Soto CS, Polishchuk A, Caputo GA, Tatko CD, et al. pH-induced conformational change of the influenza M2 protein C-terminal domain. Biochemistry. 2008;47:9934–9936. doi: 10.1021/bi801315m. [DOI] [PMC free article] [PubMed] [Google Scholar]