Abstract
Background and objectives: Diabetic nephropathy (DN) is a multifactorial complication characterized by persistent proteinuria in susceptible individuals with type 1 and type 2 diabetes. Disease burden in people of Mexican-American descent is particularly high, but there are only a few studies that characterize genes for DN in this ethnic group. Two genes, carnosine dipeptidase 1 (CNDP1) and engulfment and cell motility 1 (ELMO1) previously showed association with DN in other ethnic groups. CNDP1 and ELMO1 were examined along with eight other genes that are less well characterized for DN in a new study of Mexican-Americans.
Design, setting, participants, & measurements: The target sample was patients of Mexican-American ancestry collected from three centers: 455 patients with DN and 437 controls with long-term diabetes but no incident nephropathy. Forty-two, 227, and 401 single nucleotide polymorphisms (SNPs) in CNDP1, ELMO1, and the other eight genes, respectively, were examined.
Results: No region in CNDP1 or ELMO1 showed significant P values. Of the other eight candidate genes, an association of DN with a SNP pair, rs2146098 and rs6659783, was found in hemicentin 1 (HMCN1) (unadjusted P = 6.1 × 10−5). Association with a rare haplotype in this region was subsequently identified.
Conclusions: The associations in CNDP1 or ELMO1 were not replicable; however, an association of DN with HMCN1 was found. Additional work at this and other loci will enable refinement of the genetic hypotheses regarding DN in the Mexican-American population to find therapies for this debilitating disease.
Diabetic nephropathy (DN) is the main cause of ESRD in the United States (1). The disease burden in people of Mexican-American descent is particularly high (1), but there are only a limited number of studies that have characterized genes for DN in this ethnic group. Recently, two genes, carnosine dipeptidase 1 (CNDP1) and engulfment and cell motility 1 (ELMO1), were reported to be associated with DN (2–5). Janssen et al. (4) reported an association between DN and a microsatellite marker, D18S880, in CNDP1 among type 1 and type 2 diabetic patients from four different countries, and Freedman et al. (2) reported its replication among type 2 diabetic Caucasian patients. Shimazaki et al. (5) reported an association in the Japanese population between DN and ELMO1, which includes rs741301 as the most significant single nucleotide polymorphism (SNP).
Here, we study ten candidate genes for their association with DN in the Mexican-American population. We attempt to replicate the previous associations of CNDP1 and ELMO1 with a sample size that is similar or greater than previously used (2–5). In addition, we study the following eight genes, which are good biologic candidates but have not been studied extensively: hemicentin 1 (HMCN1), complement factor H (CFH), α-2Heremans-Schmid-glycoprotein (AHSG), caspase 3 (CASP3), heat shock 70-kD protein 1A (HSPA1A), heat shock 27-kD protein 1 (HSPB1), caspase 12 (CASP12), and heme oxygenase (decycling) 1 (HMOX1).
HMCN1 was shown to be associated with change in calculated GFR (6), but its role in DN has never been examined. CFH is long known to play a role in atypical hemolytic uremia and membranoproliferative GN, but its involvement in DN has not been evaluated. AHSG is reported to be associated with type 2 diabetes and dyslipidemia, it inhibits insulin-induced tyrosine phosphorylation of insulin receptor substrate-1 (7), and it has been identified as a marker of acute kidney injury (8). Its serum concentration is increased in nondialyzed patients with DN (9) and is low in patients with ESRD (10). High serum levels are associated with insulin resistance (11). HSPB1, also known as HSP27, is involved in the regulation of cell adhesion and invasion (12), regulates actin cytoskeleton turnover, and has anti-apoptotic and antioxidant properties in a wide variety of cells and tissues (13). A mutation in HSPB1 causing a variant of Charcot-Marie-Tooth disease is associated with the development of focal and segmental glomerulosclerosis (14). HMOX1, also known as HO-1, provides antioxidant adaptive functions in response to renal injury (15) and is associated with the degree of renal failure in DN (16). CASP3 and CASP12 mediate apoptotic cell death and were chosen as candidate genes because of their relevance to DN (17,18). Finally, HSPA1A was chosen because of its cellular protectant role in the unfolded protein response (19).
Our study aimed to replicate the previous association of the two genes with DN and/or discover new associations at the other eight genes of biologic importance by contrasting the genotype frequencies of SNPs in these ten genes between cases and controls after allowing for relevant covariates.
Materials and Methods
The Family Investigation of Nephropathy and Diabetes (FIND) study used two study designs: genome-wide linkage analysis and mapping by admixture linkage disequilibrium (MALD) (20). The FIND MALD studies used a case-control design and enrolled individuals with African-American or Mexican-American ancestry to ascertain DN association on the basis of the ancestral inheritance of risk loci. This study was performed as an ancillary study to the Mexican-American MALD study in FIND. We used the same phenotypic definitions as previously reported in other FIND papers (20,21), with the exception that for this study all participants were required to have both parents and all four grandparents of Mexican-American ancestry. In addition, whereas in the FIND cohort, by definition, controls in this family study had siblings with DN, in the Mexican-American MALD study and in the subjects reported herein, all controls had long-term diabetes but no known first-degree relative with DN or other kidney disease.
Assessment of Diabetes
The definition of diabetes utilized in this study was identical to the definition used by the FIND consortium (20). Most of the diabetic subjects were, at the time of recruitment, either currently or previously treated with insulin, oral hypoglycemic drugs, or both. For diabetic subjects not treated with drugs, eligibility was determined by plasma hemoglobin Alc levels or, if possible, fasting plasma glucose concentrations at the time of study entry. A hemoglobin A1c level >6.0%, which is suggestive of diabetes, was followed by plasma glucose testing to confirm diabetes mellitus to be classified as diabetic for FIND. The date of diabetes diagnosis was obtained from the medical history, with confirmatory medical record review if possible. Persons with any type of diabetes were eligible for the study, but almost all had type 2 diabetes.
Assessment of Cases with DN
DN was defined by any one of three criteria. The greatest number of subjects with DN who entered the study had ESRD. For subjects with ESRD, eligibility for probands was based on (1) onset of diabetes ≥5 years before renal replacement therapy with diabetic retinopathy, (2) onset of diabetes ≥5 years before renal replacement therapy with historic 24-hour urine protein ≥3 g (protein-to-creatinine ratio ≥3.0 g/g), or (3) diabetic retinopathy with historic 24-hour urine protein >3 g (protein-to-creatinine ratio >3.0 g/g). The second largest group of subjects was those with chronic kidney disease from DN. For this group, eligibility was based on diabetic retinopathy with historic 24-hour urine protein ≥1 g (protein-to-creatinine ratio ≥1.0 g/g), or 24-hour urine protein excretion ≥3 g (protein-to-creatinine ratio ≥3.0 g/g) after diabetes duration ≥10 years. A few subjects were entered with biopsy confirmation of DN in association with proteinuria. DN was confirmed by biopsy if the following histologic criteria were present: (1) nodular and/or diffuse increase in mesangial matrix accumulation; (2) thickened glomerular basement membranes and/or arteriolar hyalinization; and (3) absence of mesangial Ig or paraprotein deposits by immunofluorescence microscopy, absence of amyloid deposits by Congo Red staining or electron microscopy, and absence of electron-dense deposits along the glomerular capillary wall.
Control Inclusion Criteria
Unrelated control samples were recruited with diabetes duration ≥10 years but without DN, as defined by normoalbuminuria, and no known prior history of microalbuminuria or overt nephropathy. Cases and controls were recruited through three centers: Los Angeles Biomedical Research Institute, University of California–Irvine (UCIrvine), and the University of Texas Health Science Center at San Antonio (UT). We sampled 455 diabetic patients with nephropathy (cases) and 437 patients without nephropathy (controls) (n = 892) (Tables 1 and 2).
Table 1.
Sample distribution
| LA Biomedical Research Institute | UC Irvine | UT | Total | |
|---|---|---|---|---|
| Diabetic patients with nephropathy (cases) | 361 | 83 | 11 | 455 |
| Diabetic patients without nephropathy (controls) | 320 | 36 | 81 | 437 |
| Total | 681 | 119 | 92 | 892 |
Table 2.
Clinical characteristic of cases and controls
| Characteristic | Diabetic Patients with Nephropathy (Cases) |
Diabetic Patients without Nephropathy (Controls) | |
|---|---|---|---|
| ESRD | Non-ESRD | ||
| N | 259 | 196 | 437 |
| Diabetes duration (years) | 20.2 ± 7.6 | 16.4 ± 5.9 | 15.7 ± 6.1 |
| Serum creatinine (mg/dl) | –a | 2.4 ± 1.3 | 0.8 ± 0.2 |
| Urine-albumin-to-creatinine ratio (g/g) | –a | 3.89 ± 2.74 | 0.01 ± 0.06 |
| ACEi use (%) | 34.9 | 39.7 | 39.2 |
| ARB use (%) | 10.2 | 12.4 | 4.4 |
| HbA1c (DCCT-HbA1c, %) | 6.7 ± 1.4 | 7.7 ± 1.8 | 8.2 ± 1.8 |
Data are mean ± SD or % phenotype group.
Data were not measured for this group.
Tagging SNPs and Genotyping
We chose tag SNPs to cover linkage disequilibrium (LD) bins crossing exons of the ten genes. Genotyping was performed by Perlegen Science, Inc., and the SNP data were filtered by criteria described in the supplementary information. Information on the tagging SNPs and genotyping results are given in Table 3. The LD plots of CNDP1 and ELMO1 obtained from HapMap phase 3 Mexican-Americans, along with our tag SNPs, are given in Figures 1 and 2. Multiple markers in our SNP set for CNDP1 are in high LD with the microsatellite marker D18S880 that showed association in the Janssen et al. study (4). The SNP rs741301 in ELMO1, for which association has been reported by Shimazaki et al. (5), is included in our SNP set for that gene. For quality control, 296 additional SNPs were chosen over the whole chromosome.
Table 3.
Details of Tag SNP at the candidate gene
| Gene | Chromosome | Size in kbp | Number of Tag SNPs | Number of SNPS Successfully Genotyped |
|---|---|---|---|---|
| CNDP1 | 18 | 70.6 | 12 | 9 |
| ELMO1 | 7 | 614.6 | 47 | 46 |
| HMCN1 | 1 | 476.4 | 27 | 21 |
| CFH | 1 | 115.5 | 23 | 18 |
| AHSG | 3 | 28.2 | 12 | 8 |
| CASP3 | 4 | 41.8 | 12 | 9 |
| HSPA1A | 6 | 22.4 | 11 | 8 |
| HSPB1 | 7 | 21.7 | 6 | 4 |
| CASP12 | 11 | 32.0 | 8 | 7 |
| HMOX1 | 22 | 33.1 | 10 | 10 |
| Total | 168 | 140 |
Figure 1.
Association test results for CNDP1 with (B) covariate configuration. Results for the two-marker tests are plotted on the midpoint of the two markers. Bonferroni corrected significance level of α = 0.05/42 is shown on the horizontal line.
Figure 2.
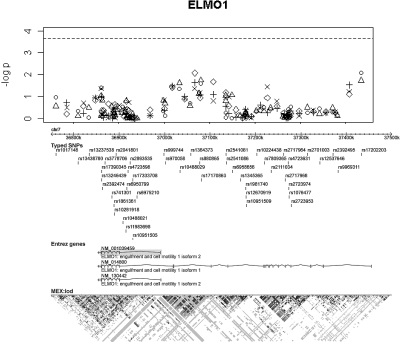
Association test results for ELMO1 with (B) covariate configuration. Results for the two-marker tests are plotted on the midpoint of the two markers. Bonferroni corrected significance level of α = 0.05/227 is shown on the horizontal line.
A QQ plot of the χ2 statistics was examined (Supplementary Information). There was no significant inflation of the statistic (mean: 1.038 and not significantly different from 1); therefore, our association test results using the logistic regression model will not be invalidated by spurious association as a result of unexplained population stratification or differential bias in genotyping.
Adjusting for Population Stratification
Population stratification among the samples was adjusted for by using principal components (22). Principal components were computed from 311 SNPs, chosen by the method described in the supplementary information. We initially included the first four principal components (PC1 through PC4) in the logistic regression model as covariates. When applied to HapMap data, they discriminated well the Caucasian (CEU), Japanese/Chinese (JPT/CHB), and West African samples (YRI) (Supplementary Information).
Covariates
To assess the effects of the three recruitment centers, two dummy covariates were formed: an indicator variable for UCIrvine samples and an indicator variable for UT samples. The significances of these covariates, which represent UCIrvine samples versus LA Biomedical Research Institute samples (UCIrvine) and UT samples versus LA Biomedical Research Institute samples (UT), respectively, were tested together with other covariates, as detailed below.
The covariates, other than recruitment center (Site), were the patient gender (Gender), age (Age), duration of diabetes (DM_Duration), use of angiotensin converting enzyme inhibitors (ACEi), and use of angiotensin II receptor blockers (ARB). We included ACEi and ARB use because by lowering albuminuria, they affect our ability to diagnose DN, since the finding of albuminuria is a key component of the diagnostic designation.
The covariates were tested under a null logistic model that does not include candidate SNPs (Table S-1 in Supplemental Information). The following covariates were significant at α = 0.1 and so incorporated as covariates in the genetic association analysis: gender (Gender); the two sites (UCIrvine and UT); diabetes duration (DM_Duration); age (Age); the first, second, and fourth principal components (PC1, PC2 and PC4); and ARB use (ARB). The third principal component (PC3) and ACEi use (ACEi) were not significant. Because medication use (ARB) records were unavailable on a portion of the samples (11.0%) despite careful medical record review, we performed the analyses with two different covariate sets, one on a smaller sample that included the covariate for ARB use and the other on the whole sample, excluding it. The two sets of covariates are thus:
(A) Gender, Age, DM_Duration, UCIrvine, UT, PC1, PC2, PC4
(B) (A) + ARB
Association Tests in a Logistic Regression Model
Two single-marker tests and three two-marker tests (23) were performed in a logistic regression model (Supplementary Information). Consecutive SNP markers were paired, and the two-marker tests were performed with these (overlapping) pairs. The likelihood ratio test (LRT) P value of the genotype term is reported for each SNP and each SNP pair. This strategy is appropriate for detecting nonadditive effects [e.g., dominant, recessive, and heterozygote (dis)advantage effects] as well as additive effects and takes the multidimensional LD structure into account, similar to a haplotype-based analysis (23).
Because CNDP1 and ELMO1 are candidates that have previously shown evidence of association, we used a sequential procedure to adjust for the number of tests performed. Thus, we first conducted our association testing on CNDP1, followed by ELMO1, and then followed this up by testing the eight other candidate genes. In each study we set a significance level adjusted for multiple tests according to the number of tests performed in that study.
Haplotype-Based Analysis
The same logistic regression model was constructed with a haplotype G-A count variable instead of a genotype variable after estimating haplotype frequencies by the expectation-maximization algorithm. Then we performed a LRT with this model to test the effect of the haplotype. Note that, because the haplotype G-A was observed as only one copy or no copies on each member of the sample, there is no difference between the additive and dominant model.
Results
The association test results for CNDP1 and ELMO1 adjusting for ARB use are shown in Figures 1 and 2. For the CNDP1 gene, we considered nine SNPs and eight SNP pairs. Two and three tests were performed for each SNP and each SNP pair, respectively; therefore, 42 (9 × 2 + 8 × 3 = 42) tests were performed for each covariate configuration (A) and (B). In the same way, 227 (42 × 2 + 41 × 3 = 227) tests were performed for the ELMO1 gene. Hence, Bonferroni corrected significance levels corresponding to α = 0.05 for a single test are α = 0.05/42 and α = 0.05/227 for CNDP1 and ELMO1, respectively. These are shown as horizontal lines in Figures 1 and 2.
These figures show that our study did not replicate the associations of CNDP1 and ELMO1 with DN, including at the SNP marker rs741301 in ELMO1 (uncorrected P values are 0.804 and 0.274 for the allelic and genotypic test, respectively). Therefore, common variants in these genes do not play a strong role in DN in Mexican-Americans.
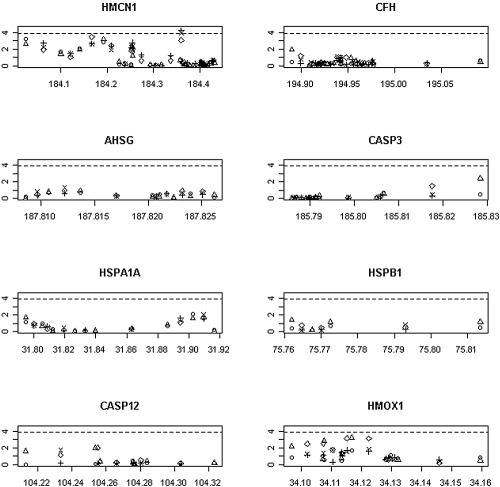
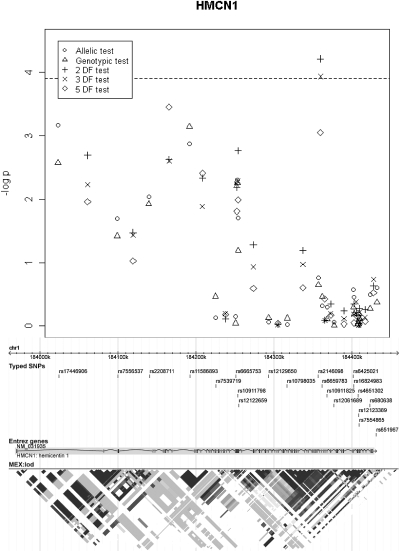
Four hundred one (85 × 2 + 77 × 3 = 401) tests were performed to test the eight other candidate genes, and the results are shown in Figure 3. Among them, two tests performed with one SNP pair in the HMCN1 gene (rs2146098, rs6659783) provided significant results at the α = 0.05/401 level after adjusting for all of the significant covariates including ARB use (Figure 4) (P values of the 2 degrees of freedom (DF) test and the 3-DF test were 6.1 × 10−5 and 1.2 × 10−4, respectively). These SNP pairs also had strong signals in the tests without adjusting for ARB use (e.g., P value of the 3-DF-test was 6.8 × 10−4), but those tests failed to reach the same significance levels.
Figure 3.
Results for the eight other candidate genes. Significance levels of α = 0.05/401 are shown as horizontal lines.
Figure 4.
Association test results for HMCN1 with (B) covariate configuration.
Because the two SNPs rs2146098 and rs6659783 are in high LD with each other (r2 = 0.96), their haplotypes could be determined almost unambiguously. Haplotype distributions in cases and controls are given in Table 4. In particular, the table shows that 13 G-A haplotypes are found in the cases, but only one G-A haplotype is present in the controls. A subsequent haplotype-based test showed evidence of association for the rare haplotype G-A (P values are 2.1 × 10−4 and 2.1 × 10−5 for covariate set A and B, respectively). We also assessed this finding using Fisher's exact test applied to a 2 × 2 table, which cannot include covariates. A less significant result was obtained by the exact test (P = 0.00175). This reduction in significance may well be due to exclusion of covariates because it was also observed in the LRT analysis without covariates (P = 0.00057). Lastly, we assessed the interaction between the haplotype and ARB use (including a category for missing use status) using a log-linear model. The three-way interaction term of haplotype, phenotype, and ARB use was not significant (P = 0.966), which indicates that the association of haplotype and phenotype is not significantly modified by ARB use or its status being missing.
Table 4.
The haplotype distribution on rs2146098-rs6659783 region in HMCN1
| Haplotype | Cases | Controls |
|---|---|---|
| G-T | 533 (58.6%) | 511 (58.5%) |
| G-A | 13 (1.4%) | 1 (0.1%) |
| A-A | 364 (40.0%) | 362 (41.4%) |
| Total | 910 | 874 |
The LD plot of HapMap Mexican-Americans in HMCN1, together with our tagged SNPs, is given in Figure 4. The 21 tagged SNPs covered the exonic regions of this gene quite well. Although we have only reported the best SNP and the markers within a rare haplotype that tagged disease risk, many other markers that were in moderate LD with these SNPs also showed evidence of association (Figure 4).
Our study did not discover an association with any other genes except HMCN1, although there is one region in HMOX1 that has moderate signals that did not reach the significance level (Figure 3).
Discussion
In a cohort of 892 Mexican-American probands with DN and long-term diabetic controls without nephropathy, we found a new association of DN with SNPs in the HMCN1 gene. A series of subsequent analyses on this region identified the strong association of the rare haplotype G-A in the SNPs rs2146098 and rs6659783. HMCN1, which is located in 1q25.3-q31.1, codes for extracellular protein in the Ig superfamily. It has been suspected to contain mutations associated with age-related macular degeneration (6,24). Here, we found a strong association between HMCN1 and DN. Further study controlling for DN and age-related macular degeneration would allow better understanding of the underlying pathway. Additional work at this and other loci will enable us to refine the genetic hypotheses regarding DN in the Mexican-American population.
We tried to replicate the previously reported association between DN and either gene CNDP1 or ELMO1 in individuals of European ancestry with Mexican-Americans but failed to do so. We further determined the power our study had to replicate each of the previous findings for CNDP1 and ELMO1 to be more than 91.5% and 57.3%, respectively (Supplementary Information). Mexican-Americans are an admixed population with substantial American-Indian ancestry. The latter, in turn, have Asian ancestral relatedness. Therefore, our negative results may be explained by genetic heterogeneity, population structure, type 1 error in the original papers, and/or type 2 error in our cohort analysis.
Interestingly, many of the SNPs in the ELMO1 gene reported by Shimazaki et al. (5), including the most significant SNP, rs741301, show severe deviation from Hardy–Weinberg proportions in their controls (e.g., P value for rs741301 is 7.7 × 10−5 for their largest control group). We noticed that the minor allele homozygote genotype frequencies in their three control groups (3.3∼6.3%) are quite different from the estimates for HapMap Japanese data (12.8% for merged Phase II and III data), whereas their cases and our case and control conform to Hardy–Weinberg proportions quite well [P = 0.79 (their largest case group), 0.39 and 0.77, respectively]. Although there could be other reasons for the difference between the two results, this discrepancy would strongly suggest that their finding was due to a systematic differential bias in genotyping or population stratification within their samples.
In conclusion, our study did not replicate an association between DN and either ELMO1 or CNDP1; however, it discovered a new association with HMCN1 and subsequently identified an association of a rare haplotype in this gene.
Disclosures
None.
Acknowledgments
This work was supported by a General Clinic Research Center National Institute of Health grant (M01-RR00425), grants to S.G.A. (R01-DK069844 and U01-DK57249), U.S. Public Health Service Resource grant (RR03655) from the National Center for Research Resources, research grant (GM28356) from the National Institute of General Medical Sciences, and a Cancer Center Support grant (P30CAD43703) from the National Cancer Institute. Recruitment was enhanced by the participation of the Little Company of Mary Research Institute. DaVita Clinical Research supported the clinical trial effort. Genotyping was performed by Perlegen Science, Inc.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
Supplemental information for this article is available online at http://www.cjasn.org/.
References
- 1.U.S. Renal Data System: 2008 Annual Data Report: Atlas of End-Stage Renal Disease in the United States Bethesda MD, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2008 [Google Scholar]
- 2.Freedman BI, Hicks PJ, Sale MM, Pierson ED, Langefeld CD, Rich SS, Xu J, McDonough C, Janssen B, Yard BA, van der Woude FJ, Bowden DW: A leucine repeat in the carnosinase gene CNDP1 is associated with diabetic end-stage renal disease in European Americans. Nephrol Dial Transplant 22: 1131–1135, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Leak TS, Perlegas PS, Smith SG, Keene KL, Hicks PJ, Langefeld CD, Mychaleckyj JC, Rich SS, Kirk JK, Freedman BI, Bowden DW, Sale MM: Variants in intron 13 of the ELMO1 gene are associated with diabetic nephropathy in African Americans. Ann Hum Genet 73: 152–159, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janssen B, Hohenadel D, Brinkkoetter P, Peters V, Rind N, Fischer C, Rychlik I, Cerna M, Romzova M, de Heer E, Baelde H, Bakker SJ, Zirie M, Rondeau E, Mathieson P, Saleem MA, Meyer J, Koppel H, Sauerhoefer S, Bartram CR, Nawroth P, Hammes HP, Yard BA, Zschocke J, van der Woude FJ: Carnosine as a protective factor in diabetic nephropathy: Association with a leucine repeat of the carnosinase gene CNDP1. Diabetes 54: 2320–2327, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, Tsunoda T, Koya D, Babazono T, Tanaka Y, Matsuda M, Kawai K, Iiizumi T, Imanishi M, Shinosaki T, Yanagimoto T, Ikeda M, Omachi S, Kashiwagi A, Kaku K, Iwamoto Y, Kawamori R, Kikkawa R, Nakajima M, Nakamura Y, Maeda S: Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes 54: 1171–1178, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Thompson CL, Klein BE, Klein R, Xu Z, Capriotti J, Joshi T, Leontiev D, Lee KE, Elston RC, Iyengar SK: Complement factor H and hemicentin-1 in age-related macular degeneration and renal phenotypes. Hum Mol Genet 16: 2135–2148, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Andersen G, Burgdorf KS, Sparso T, Borch-Johnsen K, Jorgensen T, Hansen T, Pedersen O: AHSG tag single nucleotide polymorphisms associate with type 2 diabetes and dyslipidemia: Studies of metabolic traits in 7,683 white Danish subjects. Diabetes 57: 1427–1432, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, Pisitkun T, Aponte A, Yuen PST, Hoffert JD, Yasuda H, Hu X, Chawla L, Shen RF, Knepper MA, Star RA: Exosomal fetuin-A identified by proteomics: A novel urinary biomarker for detecting acute kidney injury. Kidney Int 70: 1847–1857, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehrotra R, Westenfeld R, Christenson P, Budoff M, Ipp E, Takasu J, Gupta A, Norris K, Ketteler M, Adler S: Serum fetuin-A in nondialyzed patients with diabetic nephropathy: Relationship with coronary artery calcification. Kidney Int 67: 1070–1077, 2005 [DOI] [PubMed] [Google Scholar]
- 10.Ketteler M, Bongartz P, Westenfeld R, Wildberger JE, Mahnken AH, Bohm R, Metzger T, Wanner C, Jahnen-Dechent W, Floege J: Association of low fetuin-A (AHSG) concentrations in serum with cardiovascular mortality in patients on dialysis: A cross-sectional study. Lancet 361: 827–833, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Stefan N, Hennige AM, Staiger H, Machann J, Schick F, Kröber SM, Machicao F, Fritsche A, Häring HU: Alpha2 Heremans-Schmid glycoprotein/fetuin-A is associated with insulin resistance and fat accumulation in the liver in humans. Diabetes Care 29: 853–857, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Lee JW, Kwak HJ, Lee JJ, Kim YN, Lee JW, Park MJ, Jung SE, Hong SI, Lee JH, Lee JS: HSP27 regulates cell adhesion and invasion via modulation of focal adhesion kinase and MMP-2 expression. Eur J Cell Biol 87: 377–387, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Garrido C: Size matters: of the small HSP27 and its large oligomers. Cell Death Differ 9: 483–485, 2002 [DOI] [PubMed] [Google Scholar]
- 14.Gherardi R, Belghiti-Deprez D, Hirbec G, Bouche P, Weil B, Lagrue G: Focal glomerulosclerosis associated with Charcot-Marie-Tooth disease. Nephron 40: 357–361, 1985 [DOI] [PubMed] [Google Scholar]
- 15.Agarwal A, Nick HS: Renal response to tissue injury lessons from heme oxygenase-1 gene ablation and expression. Am Soc Nephrol 11: 965–973, 2000 [DOI] [PubMed] [Google Scholar]
- 16.Calabrese V, Mancuso C, Sapienza M, Puleo E, Calafato S, Cornelius C, Finocchiaro M, Mangiameli A, Di Mauro M, Stella AM, Castellino P: Oxidative stress and cellular stress response in diabetic nephropathy. Cell Stress Chaperones 12: 299–306, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar D, Robertson S, Burns KD: Evidence of apoptosis in human diabetic kidney. Mol Cell Biochem 259: 67–70, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Isermann B, Vinnikov IA, Madhusudhan T, Herzog S, Kashif M, Blautzik J, Corat MAF, Zeier M, Blessing E, Oh J: Activated protein C protects against diabetic nephropathy by inhibiting endothelial and podocyte apoptosis. Nat Med 13: 1349–1358, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Kaufman RJ: Orchestrating the unfolded protein response in health and disease. Am Soc Clin Investig 110: 1389–1398, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knowler WC, Coresh J, Elston RC, Freedman BI, Iyengar SK, Kimmel PL, Olson JM, Plaetke R, Sedor JR, Seldin MF: The Family Investigation of Nephropathy and Diabetes (FIND): Design and methods. J Diabetes Complications 19: 1–9, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Iyengar SK, Abboud HE, Goddard KA, Saad MF, Adler SG, Arar NH, Bowden DW, Duggirala R, Elston RC, Hanson RL, Ipp E, Kao WH, Kimmel PL, Klag MJ, Knowler WC, Meoni LA, Nelson RG, Nicholas SB, Pahl MV, Parekh RS, Quade SR, Rich SS, Rotter JI, Scavini M, Schelling JR, Sedor JR, Sehgal AR, Shah VO, Smith MW, Taylor KD, Winkler CA, Zager PG, Freedman BI: Genome-wide scans for diabetic nephropathy and albuminuria in multiethnic populations: The Family Investigation of Nephropathy and Diabetes (FIND). Diabetes 56: 1577–1585, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D: Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Kim S, Morris NJ, Won S, Elston RC: Single-marker and two-marker association tests for unphased case-control genotype data, with a power comparison. Genet Epidemiol 34: 67–77, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz DW, Weleber RG, Lawrence G, Barral S, Majewski J, Acott TS, Klein ML: HEMICENTIN-1 (FIBULIN-6) and the 1q31 AMD locus in the context of complex disease: Review and perspective. Ophthalmic Genet 26: 101–105, 2005 [DOI] [PubMed] [Google Scholar]