Abstract
Two novel landomycin compounds, landomycins I and J, were generated with a new mutant strain of Streptomyces cyanogenus in which the glycosyltransferase that is encoded by lanGT3 was over-expressed. This mutant also produced the known landomycins A, B, and D. All these compounds consist ofthe same polyketide-derived aglycon but differ in their sugar moieties, which are chains of different lengths. The major new metabolite, landomycin J, was found to consist of landomycinone with a tetrasaccharide chain attached. Combined with previous results ofthe production oflandomycin E (which contains three sugars) by the LanGT3– mutant strain (obtained by targeted gene deletion of lanGT3), it was verified that LanGT3 is a d-olivosyltransferase responsible for the transfer of the fourth sugar required for landomycin A biosynthesis. The experiments also showed that gene over-expression is a powerful method for unbalancing biosynthetic pathways in order to generate new metabolites. The cytotoxicity ofthe new landomycins—compared to known ones—was assessed by using three different tumor cell lines, and their structure–activity relationship (SAR) with respect to the length ofthe deoxysugar side chain was deduced from the results.
Keywords: biosynthesis, combinatorial biosynthesis, glycosylation, landomycins, polyketides
Introduction
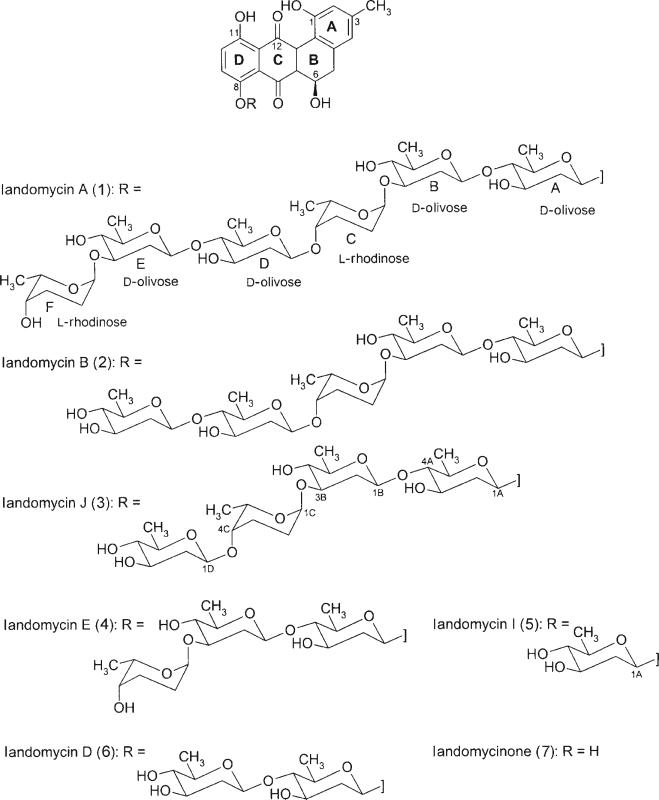
The landomycins (Las) are a growing subgroup of the large family of angucycline antibiotics that now includes far more then 100 members.[1–4] These have diverse biological effects, such as antitumor, antibacterial, and enzyme inhibitory activity.[5,6] Landomycin A (LaA, 1; Scheme 1) in particular has been studied as a potential antitumor agent. It possesses an interesting spectrum of bioactivity against 60 cancer cell lines.[4–6] Over the past decade, two gene clusters that encode for land-omycin biosynthesis have been cloned and sequenced: one encodes the hexasaccharide LaA (1), which is generated by Streptomyces cyanogenus S136 (lan), and the other encodes the trisaccharide, landomycin E (4; Scheme 1), which is produced by S. globisporus 1912 (lnd).[7,8] The clusters are very similar, both at the level of gene organization and nucleic acid sequence.[9]
Scheme 1.
The chemical structures of landomycins A, B, J, E, I, and D (1–6), and landomycinone (7).
Much of our recent studies have focused on the post-PKS (“tailoring”) steps of landomycin biosynthesis,[10,11] especially on oxygenation and glycosylation sequences during the biosynthesis of landomycins, which contribute considerably to the biological activity of many polyketides.[12] For example, the gene lndM2 was found to encode a bifunctional oxygenase–reductase that is responsible for the introduction of the 6-O atom in LaE,[11] and lndZ4/Z5 were found to be responsible for the addition of the 11-OH group in LaE.[10,13] Most recently, inactivation of the lndE gene in S. globisporus led to the accumulation of prejadomycin (2,3-dehydro-UWM6), which proved that oxygenase LndE is responsible for the generation of the p-quinone moiety through oxidation of the 12-position.[14] Thus, all oxygenases that incorporate oxygen atoms into the landomycin aglycon have been identified.
Of similar interest was the study of the glycosyltransferases involved in landomycin biosynthesis. Structure–activity relationship (SAR) studies suggested that the high cytotoxic activity of LaA was dependent on its hexasaccharide chain, although its mode of action has not been clearly established yet.[10] Other landomycins with shorter sugar side chains showed lower cytotoxic activities.[10] Four glycosyltransferases (LanGT1, LanGT2, LanGT3, and LanGT4) are involved in the biosynthesis of LaA's hexasaccharide chain. LanGT1 and LanGT4 were identified as olivosyl- and rhodinosyltransferases,[15] respectively, and were found to act twice during hexasaccharide side-chain biosynthesis. While LandGT1 is responsible for the transfer of the second and fifth sugar moiety, LanGT4 transfers both rhodinoses into the third and sixth positions.[16] Most recently, LanGT2 was identified to catalyze the first glycosylation step (d-olivosyl),[17] and a targeted gene inactivation experiment on lanGT3 showed that the corresponding glycosyltransferase LanGT3 is an olivosyltransferase.[18] LanGT3 is most likely responsible for the attachment of the fourth sugar, which is the key step that distinguishes the biosyntheses of LaA and LaE. Note also that an analogue of lanGT3 is missing in the S. globisporus 1912 gene cluster (lnd). All previous studies combined led to the generation of various landomycins with altered oxygen patterns or oligosaccharide chains. These ranged from analogues with only one sugar (LaH, which also lacked the 11-OH group[10]) to the complete hexasaccharide chain compounds found in LaA (1). None of the experiments so far have yielded a landomycin with a tetrasaccharide chain. Herein, we report on the over-expression of lanGT3 in S. cyanogenus which was aimed at the production of a tetrasaccharide landomycin. The experiments indeed yielded a mutant strain that accumulated the new tetrasaccharide, named landomycin J (3), as its major new metabolite, and landomycin I (5) as a minor product. The latter compound is a monosaccharide with a complete oxygenation pattern that has so far not been observed in landomycin analogues. The results also offer further support that LanGT3 is responsible for the attachment of the fourth sugar of the growing side chain of LaA. These findings therefore complete the elucidation of the enigmatic biosynthetic pattern of “six sugars, four glycosyltransferase-encoding genes” which occur in LaA biosynthesis. The experiments show that gene over-expression is a powerful method for unbalancing biosynthetic pathways in order to generate new metabolites. Besides the two new compounds LaI (5) and LaJ (3), the mutant strain also produced the three known compounds LaA (1), LaB (2), and LaD (6).
More importantly, the generation of five landomycins that differ only with respect to the length of their saccharide chain, allowed us to study the SAR with respect to the length of the deoxysugar chain. The results of the cytotoxicity studies clearly showed that the activities of the landomycins do not increase linearly with the length of their deoxysugar chain.
Results and Discussion
Over-expression of lanGT3 in S. cyanogenus—generation and structure elucidation of the new landomycins I and J
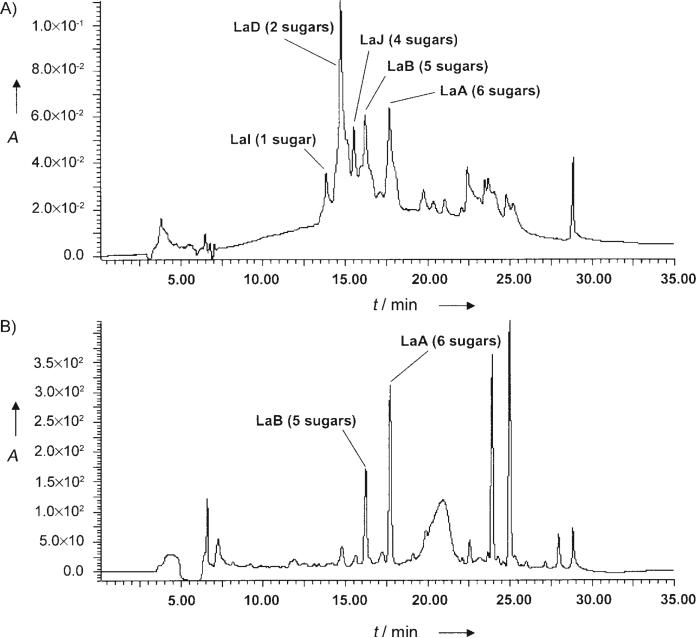
The 2.2 kb BamHI–SacI fragment from cosmid H2-26,[7] which contains the lanGT3 gene, was ligated into the same sites of pUC19 to yield plasmid pUClanGT3.[18] A plasmid for the over-expression of the lanGT3 mutant, named pKCGT3, was generated by ligating a 2.2 kb BamHI–EcoRI fragment from pUClanGT3 into the expression vector pKC1218ermE.[10] For the over-expression experiment plasmid pKCGT3, which consisted of lanGT3 ligated behind the ermE promoter, was introduced into the mutant strain by intergeneric conjugation. A culture of the resulting mutant strain, S. cyanogenus (lanGT3), produced five main metabolites (Figure 1), among which three were known compounds. These compounds were identified as LaA (1), LaB (2), and LaD (6) by using TLC, HPLC, and APCI-MS.[3,4] The structures of the novel landomycins I (5) and J (3) were determined by using mass spectrometry and NMR as well as UV spectroscopy. From the two new compounds, LaJ, which has four sugars attached to the aglycon moiety, was the major one. The chemical structures of all of these compounds are shown in Scheme 1.
Figure 1.
A) HPLC analysis of the crude extract of the S. cyanogenus (lanGT3) mutant strain, in which the glycosyltransferase LanGT3 was over-expressed, in comparison with B) the crude extract of the wild-type strain S. cyanogenus; the detection wavelength was 254 nm. Solvent A: 0.1% formic acid in H2O, solvent B: acetonitrile; flow rate: 0.5 mL min-1; 0–16 min, 75% A and 25% B to 30% A and 70% B (linear gradient), 16–18 min 30% A and 70% B to 0% A and 100% B (linear gradient), 18–22 min 0% A and 100%, 22–25 min 0% A and 100% to 75% A and 25% B (linear gradient), 25–35 min 75% A and 25% B.
Both of these two novel compounds displayed the same kind of color (dark red) as the typical landomycins and very similar UV spectra to those of the other three known landomycins, with three characteristic peaks at around λ = 213, 261, and 450 nm. APCI-MS analysis of LaI and LaJ showed a negative mode molecular ion peak at m/z 467 and 841, respectively. This suggests that the molecular weights for these two compounds are 468 and 842 g mol4-1, which correspond to one sugar (olivose) less than LaD (MW 598) and one sugar (olivose) more than LaE (MW 712), respectively. Subsequent high-resolution ESI-TOF-MS confirmed their molecular formulae to be C25H24O9 for LaI and C43H54O17 for LaJ. Moreover, the NMR spectra of these two new compounds (LaI and LaJ) were consistent in their aglycon moieties with those of the known landomycins, although they differ in their sugar moieties. The NMR spectra of LaJ (1H NMR, Table 1 and 13C NMR, Table 2) showed the presence of four O-glycosidic hexopyranoses, for example, four anomeric C signals (δ = 96.07–102.46) and four anomeric protons (δ = 4.59–5.24). However, there was only one corresponding signal for LaI, one anomeric C at δ = 97.88 and one proton at δ = 5.20. Similarly, the 1H NMR spectrum of LaJ showed four strong doublets (δ = 1.11–1.31) in the higher field, which corresponded to the methyl groups (C-6) of the four O-glycosidic hexopyranoses, while there was only one clear doublet in this area at δ = 1.17 for LaI. From the 1H,1H-COSY experiment, the assignments of all protons for every sugar moiety could be identified. These indicated three d-olivoses and one l-rhodinose in LaJ, and only one d-olivose moiety in LaI.
Table 1.
1H NMR signals of landomycins I (5)[a] and J (3).[b] The couplings between some protons could be clearly observed by gCOSY,[g] and other couplings could be observed by NOESY.[h]
| Proton | LaI (5) | Multiplicity (J in Hz) | LaJ (3) | Multiplicity (J in Hz) |
|---|---|---|---|---|
| 1-OH | 9.63 | brs | 8.67 | brs |
| 2-H | 6.56 | brs | 6.68 | brs |
| 3-CH3 | 2.25 | s | 2.29 | s |
| 4-H | 6.63 | brs | 6.73 | brs |
| 5-Hα | 2.71 | dd (15, 4.5) | 2.88 | dd (15, 4.5) |
| 5-Hβ | 2.87 | dd (15, 5) | 3.04 | dd (15, 5) |
| 6-H | 4.96 | dd (5, 4.5) | 5.14 | dd (5, 4.5) |
| 9-H | 7.49 | d (9.6) | 7.60 | d (9.2) |
| 10-H | 7.30 | d (9.6) | 7.27 | d (9.2) |
| 11-OH | 11.69 | brs | 12.10 | brs |
| 1A-H | 5.20 | dd, (9.6, 1.6) | 5.24 | dd, (9.6, 2) |
| 2A-Ha | 1.66 | ddd (12, 12, 9.6) | 1.78 | ddd (12, 12, 9.6) |
| 2A-He | 2.30 | ddd (12, 4.8, 1.6) | 2.60 | ddd (12, 5, 2) |
| 3A-H | 3.45 | ddd (12, 8.5, 4.8) | 3.68[c] | ddd (12, 8.8, 5) |
| 4A-H | 2.83 | dd (8.5, 8.5) | 3.12[e] | dd (8.8, 8.8) |
| 5A-H | 3.30 | dq (8.5, 6) | 3.47[d] | dq (8.8, 6) |
| 5A-CH3 | 1.17 | d (6) | 1.27 | d (6) |
| 1B-H | 4.72 | dd (10, 2) | ||
| 2B-Ha | 1.48 | ddd (12, 12, 10) | ||
| 2B-He | 2.15 | ddd (12, 5, 2) | ||
| 3B-H | 3.44 | ddd (12, 8.8, 5) | ||
| 4B-H | 3.08[e] | dd (8.8, 8) | ||
| 5B-H | 3.48[d] | dq (8, 6) | ||
| 5B-CH3 | 1.31 | d (6) | ||
| 1C-H | 4.94 | brs | ||
| 2C-Ha | 1.41 | m | ||
| 2C-He | 1.98[f] | m | ||
| 3C-Ha | 1.95[f] | m | ||
| 3C-He | 2.01 | m | ||
| 4C-H | 3.54 | brs | ||
| 5C-H | 4.19 | dq (6.4, 2) | ||
| 5C-CH3 | 1.11 | d (6.4) | ||
| 1D-H | 4.59 | dd (10, 2) | ||
| 2D-Ha | 1.51 | ddd (12, 12, 10) | ||
| 2D-He | 2.37 | ddd (12, 5, 2) | ||
| 3D-H | 3.64[c] | ddd (12, 8.8, 5) | ||
| 4D-H | 2.99 | dd (8.8, 8) | ||
| 5D-H | 3.21 | dq (8, 6) | ||
| 5D-CH3 | 1.22 | d (6) |
Landomycin I: [D6]DMSO, 400 MHz
landomycin J: [D6]acetone, 400 MHz.
These signals overlapped with each other.
These signals overlapped with each other.
These signals overlapped with each other.
These signals overlapped with each other.
(1A-H, 2A-Ha); (1B-H, 2B-Ha); (1C-H, 2C-He); (1 D-H, 2D-Ha); (5Hα, 6-H); (5Hβ, 6-H); (3A-H, 4A-H); (3A-H, 2A-He); (3B-H, 2B-He); (1B-H, 2B-Ha); (3C-He, 4C-H); (1D-H, 2D-Ha); (3D-H, 2D-He); (5A-H, 5A-CH3); (5B-H, 5B-CH3); (5C-H, 5C-CH3); (5D-H, 5D-CH3).
(4-H, 5-Hα); (3A-H, 1B-H); (1C-H, 3B-H).
Table 2.
| Carbon no. | LaI, 5 | LaJ, 3 | Carbon No. | LaI, 5 | LaJ, 3 |
|---|---|---|---|---|---|
| C-1 | 154.88 | 156.34 | C-4A | 76.52 | 78.57 |
| C-2 | 115.84 | 120.52 | C-5A | 69.99 | 71.58 |
| C-3 | 141.14 | 142.90 | C-6A | 18.03 | 18.34 |
| C-4 | 124.70 | 125.99 | C-1B | 101.80 | |
| C-4A | 138.28 | 139.30 | C-2B | 37.44 | |
| C-5 | 36.61 | 39.13 | C-3B | 73.17 | |
| C-6 | 57.33 | 60.16 | C-4B | 77.89 | |
| C-6A | 140.29 | 140.53 | C-5B | 72.28 | |
| C-7 | 181.02 | 182.39 | C-6B | 18.13 | |
| C-7A | 113.20 | 114.57 | C-1C | 96.07 | |
| C-8 | 149.50 | 151.17 | C-2C | 25.56 | |
| C-9 | 120.98 | 123.10 | C-3C | 25.24 | |
| C-10 | 128.24 | 131.40 | C-4C | 76.63 | |
| C-11 | 155.34 | 158.27 | C-5C | 67.54 | |
| C-11A | 119.12 | 117.77 | C-6C | 17.46 | |
| C-12 | 187.88 | 191.13 | C-1D | 102.46 | |
| C-12A | 141.74 | 145.15 | C-2D | 40.57 | |
| C-12B | 115.43 | 116.56 | C-3D | 72.73 | |
| C-3CH3 | 21.10 | 21.40 | C-4D | 88.73 | |
| C-1A | 97.88 | 99.96 | C-5D | 70.03 | |
| C-2A | 29.61 | 37.66 | C-6D | 18.51 | |
| C-3A | 72.01 | 75.97 |
Landomycin I: [D6]DMSO, 100 MHz
landomycin J: [D6]acetone, 100 MHz.
The production of the three known landomycins is quite reasonable since the entire lan gene cluster exists in the new mutant strain; the only difference to the wild-type strain was the level of LanGT3 expression. The generation of the new major metabolite LaJ, the first tetrasaccharidal landomycin, not only supports the previously suggested assignment of the role of LanGT3 for the biosynthesis of the landomycin saccharide chain (see also below),[18] but also shows that gene over-expression is a suitable method for altering biosynthetic pathways and generating new metabolites. Note that we previously reported the accumulation of novel urdamycin analogues with altered saccharide patterns after over-expressing glycosyltransferase UrdGT1c in the urdamycin producer, S. fradiae Tü2717.[19] The minor compound LaI, with only one olivose attached to the aglycon moiety, was the first ever monosaccharidal landomycin produced by a S. cyanogenus strain, after its 11-deoxy analogue LaH (11-deoxy-LaI) was previously generated in a S. globisporus mutant.[10] While the accumulation of the tetrasaccharide 3 was anticipated and appeared to be an obvious result of the over-expression of LanGT3, the moderate accumulation of the monosaccharide 5 in S. cyanogenus (lanGT3) seemed surprising. However, this can be explained through depletion of the other olivosyltransferases of their NDP-activated sugar donor substrate, which in turn disturbs the normal flux of oligosaccharide chain formation. Particularly LanGT1 seems to be affected, which is responsible for the addition of the second d-olivose moiety.
Function of LanGT3
A previous mutation of the lanGT3 gene in S. cyanogenus yielded a mutant that accumulated the known compound LaE, which consists of a trisaccharide side chain attached to the polyketide derived aglycon moiety. The expression of lanGT3 in this mutant restored landomycin A production.[18] Herein, through the over-expression of lanGT3 in S. cyanogenus, a major novel tetrasaccharidal metabolite LaJ was produced. This provides further evidence that LanGT3 is an olivosyltransferase that catalyzes the fourth sugar attachment during biosynthesis of landomycins A, B, and J. The combination of previous studies[10,15,17,18] with the present one allows the unambiguous assignment of the function of all four glycosyltransferases involved in the step-by-step formation of the hexasaccharide chain of LaA: LanGT2 catalyzes the addition of the first d-olivose; LanGT1 is responsible for the attachment of the second and fifth sugar (each a d-olivose); LanGT4 attaches the third and sixth sugar (each an l-rhodi-nose); and LanGT3 catalyzes the addition of the fourth sugar (another d-olivose).
Cytotoxicity assays of the land-omycins and SAR studies
Cytotoxicity studies were performed with NCI-H460 human lung cancer and MCF7 human breast cancer cell lines, as well as the murine Lewis lung cancer cell line LL/2 by using the reliable sulforhodamine B assay.[20–22] The results allow conclusions to be made with regard to the SAR of the deoxysugar chain length within the landomycin family. The results of these assays are listed in Table 3.
Table 3.
Cytotoxicity assays[a] of landomycins with variable saccharide chain lengths.
| Landomycins (No. of sugars) | Cytotoxicity GI50 [μm] | ||
|---|---|---|---|
| NCI-H460 | MCF7 | LL/2 | |
| landomycinone 7 (0) | 5.9±1.3 | 2.9±1.6 | 2.9±0.7 |
| landomycin I 5 (1) | 13.9±8.5 | 3.7±0.8 | 3.5±0.9 |
| landomycin D 6 (2) | 11.5±4.6 | 4.2±1.0 | 2.1±0.2 |
| landomycin E 4 (3) | 5.4±1.5 | 13.0±5.8 | 0.7±0.1 |
| landomycin J 3 (4) | 10.2±2.7 | 4.5±2.2 | 1.4±0.6 |
| landomycin B 2 (5) | 6.6±3.4 | 2.1±0.2 | 1.5±0.3 |
| landomycin A 1 (6) | 2.1±1.0 | 1.8±0.8 | 0.7±0.6 |
This study completes the generation of different landomycin members that consist of the same polyketide derived aglycon moiety (landomycinone), but which differ in the number of sugars attached to them. This has, for the first time, allowed SAR studies with respect to the length of the oligosaccharide chain. For comparison, LaE (4, from S. globisporus 1912) and the aglycon 7 (obtained through hydrolysis of the landomycin mixture) were also included. The SAR study showed that all the landomycins possess activity against lung cancer (H460, LL/2) or breast cancer (MCF7) cell lines. With the cell lines tested here, the activity against murine LL/2 was generally the highest. Unexpectedly, the antitumor activities of the landomycins do not increase steadily with the growing length of the deoxysugar chain. The compounds with longer sugar chains (LaA and LaB—LaA was the overall most potent compound) in general displayed higher activities than the analogues with short and medium saccharide chain lengths (1–4 sugars). Exceptions here are the high activities of the trisaccharide LaE (GI50 0.7 mm) and the tetrasaccharide LaJ (GI50 1.4 μm) against the murine Lewis lung cancer cell line, and the high activity of LaE (GI50 5.4 μm) against the human lung cancer cell NCI-H460. On the other hand, LaE shows the relatively lowest activity against the breast cancer cell line MCF7. Surprisingly, the aglycon landomycinone (7) was quite active against all three cancer cell lines. This could indicate a change in the mechanism of action, and landomycinone might act in a different way (e.g., different type of binding to DNA, inhibition of a different signal transduction pathway) than its saccharidal analogues—the optimal activity of which appears to be more dependent on a long sugar chain. However, except for the cell-cycle analysis studies by Crow et al.,[6] the mode of action of the landomycins remains unknown.
Conclusions
In-depth study of glycosyltransfer (GT) in the landomycin biosynthetic pathway allows the harnessing of selected genes for the targeted generation of novel bioactive compounds through combinatorial biosynthesis, as shown here. With the over-expression of lanGT3 in S. cyanogenus 1912, two new landomycins (LaI and LaJ) in addition to three known ones (LaA, LaB, and LaD) were generated. The occurrence of the two new landomycins, LaI and LaJ, can be explained by sugar-donor substrate depletion of LanGT1, which slows down the second GT step catalyzed by this enzyme, and simultaneously accelerates the fourth GT step catalyzed by LanGT3. The addition of novel compounds to the landomycin family allowed a more detailed SAR study. The results revealed that the anti-cancer activity of landomycins did not increase simultaneously with the elongation of their oligosaccharide chain lengths, as suggested before.[10]
ExperimentalSection
Bacterial strains and culture conditions
E. coli DH5α (MBI Fermentas, Lithuania) was used as host for subcloning. E. coli ET12567 which carries plasmid pUB307 (dam–dcm–hsdS–CmRKmR) was used to perform intergeneric conjugations; this strain was a gift from C. P. Smith (University of Manchester, UK). E. coli strains were grown under standard conditions.[23] Apramycin (25 μgmL-1), chloramphenicol (25 μgmL-1), ampicillin (100 μgmL-1), kanamycin (50 μgmL-1), or nalidixic acid (50 μgmL-1) were used for antibiotic selection as required. The S. cyanogenus strain was grown in tryptone soya broth (TSB) medium for conjugation and DNA isolation, and in soypeptone-glucose (SG) medium for the landomycin A production as described.[10]
Generalgenetic manipulations
Routine methods were performed as described.[23,24] Intergeneric E. coli–Streptomyces conjugation was used to introduce plasmid DNA into S. globisporus strains. Mating was performed as described previously.[10] Restriction enzymes were purchased from NEB; plasmids were obtained from Invitro-gene or Boehringer Mannheim; Immobilon-Ny + transfer-membranes were obtained from Millipore. All products were used according to manufacturer's directions.
Metabolite analysis
Two liters of S. cyanogenus (lanGT3) were cultured by using liquid SG medium supplemented with apramycin (25 μg mL-1). A preculture was grown at 30°C and 200 rpm for 24 h. This was subsequently inoculated into the main culture (20 Erlenmeyer flasks, each containing 100 mL medium) which had the same composition and culture conditions and which was harvested after 40 h incubation. (The metabolites decomposed when cells were cultured longer than 48 h). The culture broth was extracted three times with ethyl acetate (each 100 mL), the combined organic extracts were dried in vacuo, and dissolved in methanol for TLC, HPLC, and LC-MS analysis. TLC analysis was carried out on SilG-25 and silica 60 F254 silica gel plates (Merck, Darmstadt, Germany) with UV fluorescence indicator. HPLC-MS was performed by using a Waters Alliance 2695 system with a Waters 2996 photodiode array detector and a Micromass ZQ 2000 mass spectrometer equipped with an APCI (atmospheric pressure chemical ionization) probe. The column used in the LC-MS system was a Waters Symmetry C18 (4.6 × 50 mm, 5 μm particles) with solvent A (0.1% formic acid in H2O) and solvent B (acetonitrile); flow rate, 0.5 mL min-1;0–12 min, 75% A and 25% B to 30% A and 70% B (linear gradient), 12–15 min, 30% A and 70% B to 100% B (linear gradient). The combined crude ethyl acetate extract of the main cultures was purified by repeated (twice) semipreparative HPLC on a Waters Delta 600 instrument with a Waters 996 photodiode array detector (solvent A: 0.1% trifluoroacetic acid in H2O; solvent B, acetonitrile); flow rate, 10.0 mL min-1; 0–4 min 100% A to 75% A and 25% B (linear gradient), 4–30 min 75% A and 25% B to 100% B (linear gradient), 30–33 min 100% B, 33–35 min 100% B to 100% A (linear gradient), 35–42 min 100% A. The column used was a Waters SymmetryPrep C18, 19 × 150 mm, 7 mm particles. The yields were: LaA, 5.5 mg L-1; LaB, 7 mg L-1; LaD, 10 mg L-1; LaI, 1 mg L-1; and LaJ, 3.3 mg L-1.
The structures of the two new landomycins (LaI and LaJ) were identified by using a combination of the NMR spectra, the UV- and MS data. NMR spectra were acquired on a Varian Inova 400 instrument at a magnetic field strength of B0 9.4 T. For detailed data see Tables 1 and 2, chemical shifts are reported in ppm relative to internal TMS as standard.
Acknowledgements
This work was partly supported by grants from the Kentucky Lung Cancer Research Foundation and the US National Institutes ofHealth CA91901 and CA102102 (to J.R.), as well as by a Deutsche Forschungsgemeinschaft (DFG Schwerpunkt Evolution metabolischer Diversität) grant to A.B. The NMR spectroscopy and mass spectrometry core facilities of the University of Kentucky are acknowledged for use of their instruments.
References
- 1.Rohr J, Thiericke R. Nat. Prod. Rep. 1992;9:103. doi: 10.1039/np9920900103. [DOI] [PubMed] [Google Scholar]
- 2.Krohn K, Rohr J. Top. Curr. Chem. 1997;188:127. [Google Scholar]
- 3.Weber S, Zolke C, Rohr J, Beale JM. J. Org. Chem. 1994;59:4211. [Google Scholar]
- 4.Henkel T, Rohr J, Beale JM, Schwenen L. J. Antibiot. 1990;43:492. doi: 10.7164/antibiotics.43.492. [DOI] [PubMed] [Google Scholar]
- 5.Depenbrock H, Bornschlegl S, Peter R, Rohr J, Schmid P, Schweighart P, Block T, Rastetter J, Hanauske AR. Ann. Hematol. 1996;73(Supl II):A80/316. [Google Scholar]
- 6.Crow RT, Rosenbaum B, Smith R, 3rd, Guo Y, Ramos KS, Sulikowski GA. Bioorg. Med. Chem. Lett. 1999;9:1663. doi: 10.1016/s0960-894x(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 7.Westrich L, Domann S, Faust B, Bedford D, Hopwood DA, Bechthold A. FEMS Microbiol. Lett. 1999;170:381. doi: 10.1111/j.1574-6968.1999.tb13398.x. [DOI] [PubMed] [Google Scholar]
- 8.Fedorenko V, Basiliya L, Pankevych K, Dubitska L, Ostash B, Luzhetskyy A, Gromyko O, Krügel H. Bull. Inst. Agr. Microbiol. (Ukr.) 2000;8:27. [Google Scholar]
- 9.Rebets Y, Ostash B, Luzhetskyy A, Hoffmeister D, Braña AF, Méndez C, Salas JA, Bechthold A, Fedorenko V. FEMS Microbiol. Lett. 2003;222:149. doi: 10.1016/S0378-1097(03)00258-1. [DOI] [PubMed] [Google Scholar]
- 10.Ostash B, Rix U, Remsing Rix LL, Liu T, Lombó F, Luzhetskyy A, Gromyko O, Wang C, Braña AF, Méndez C, Salas JA, Fedorenko V, Rohr J. Chem. Biol. 2004;11:547. doi: 10.1016/j.chembiol.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Zhu L, Ostash B, Rix U, Nur-e-Alam M, Mayers A, Luzhetskyy A, Méndez C, Salas JA, Bechthold A, Fedorenko V, Rohr J. J. Org. Chem. 2005;70:631. doi: 10.1021/jo0483623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rix U, Fischer C, Remsing LL, Rohr J. Nat. Prod. Rep. 2002;19:542. doi: 10.1039/b103920m. [DOI] [PubMed] [Google Scholar]
- 13.Luzhetskyy A, Zhu L, Gibson M, Fedoryshyn M, Dürr C, Hofmann C, Hoffmeister D, Ostash B, Mattingly C, Adams V, Fedorenko V, Rohr J, Bechthold A. ChemBioChem. 2005;6:675. doi: 10.1002/cbic.200400316. [DOI] [PubMed] [Google Scholar]
- 14.Baig I, Kharel M, Kobylyanskyy A, Zhu L, Rebets Y, Ostash B, Luzhetskyy A, Bechthold A, Fedorenko V, Rohr J. Angew. Chem. 2006;118:8006–8010. doi: 10.1002/anie.200603176. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2006;45:7842–7846. doi: 10.1002/anie.200603176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trefzer A, Fischer C, Stockert S, Westrich L, Künzel E, Girreser U, Rohr J, Bechthold A. Chem. Biol. 2001;8:1239. doi: 10.1016/s1074-5521(01)00091-6. [DOI] [PubMed] [Google Scholar]
- 16.Luzhetskyy A, Fedoryshyn M, Dürr C, Taguchi T, Novikov V, Bechthold A. Chem. Biol. 2005;12:725. doi: 10.1016/j.chembiol.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Luzhetskyy A, Taguchi T, Fedoryshyn M, Dürr C, Wohlert SE, Novikov V, Bechthold A. ChemBioChem. 2005;6:1406. doi: 10.1002/cbic.200500018. [DOI] [PubMed] [Google Scholar]
- 18.Luzhetskyy A, Liu T, Fedoryshyn M, Ostash B, Fedorenko V, Rohr J, Bechthold A. ChemBioChem. 2004;5:1567. doi: 10.1002/cbic.200400123. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmeister D, Ichinose K, Domann S, Faust B, Trefzer A, Dräger G, Kirschning A, Fischer C, Künzel E, Bearden DW, Rohr J, Bechthold A. Chem. Biol. 2000;7:821. doi: 10.1016/s1074-5521(00)00029-6. [DOI] [PubMed] [Google Scholar]
- 20.Rubinstein LV, Shoemaker RH, Paull KD, Simon RM, Tosini S, Skehan P, Scudiero DA, Monks A, Boyd MR. J. Natl. Cancer Inst. 1990;82:1113. doi: 10.1093/jnci/82.13.1113. [DOI] [PubMed] [Google Scholar]
- 21.Boyd MR. In: Anticancer Drug Development Guide: Preclinical Screening, Clinical Trials, and Approval. Teicher B, editor. Humana; Totowa: 1997. p. 23. [Google Scholar]
- 22.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. J. Natl. Cancer Inst. 1990;82:1107. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- 24.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Norwich: 2000. [Google Scholar]