INTRODUCTION
Despite decades of research, the biological pathways that initiate rheumatoid arthritis (RA) are unknown (1, 2). Without knowing the specific pathways that lead to RA, it is very difficult to develop novel therapies. Because genetic mutations are inherited prior to disease onset, human genetics provides prima facie evidence that a pathway is important in pathogenesis. Moreover, because human genetic strategies can be applied genome-wide, they offer an unbiased search of the human genome for insight into RA pathogenesis.
Since the 1970’s, more than 20 RA risk loci have been identified (Table 1). The first locus associated with RA risk was the major histocompatibility (MHC) locus, identified by mixed lymphocyte cultures between RA patients and controls (3, 4). Subsequently, Peter Gregersen and colleagues advanced the hypothesis that the multiple RA risk alleles within the HLA-DRB1 gene share a conserved amino acid sequence (5). This is now widely known as the “shared epitope” hypothesis, and the risk alleles are known as shared epitope (SE) alleles (6). With the sequence of the human genome (7) and improved understanding of human genetic diversity (8) came many additional genetic discoveries (9). Between 2003 and 2005, common alleles within the PADI4, PTPN22 and CTLA4 genes were found to be reproducibly associated with risk of RA (10–12). More recently, genome-wide association studies (GWAS) – which in the contemporary form test hundreds of thousands of single nucleotide polymorphisms (SNPs) across the genome – have been systematically performed in large case-control collections. These GWAS have identified more than 20 common alleles (where an allele is one of the two base pairs of a SNP) that confer a 10–20% increase in disease risk per copy of the risk allele (13–23). Collectively, these risk alleles explain approximately 15–20% of the overall disease burden (15, 16).
Table 1.
Known RA SNP Associations
| SNP | Locus | Candidate Gene | OR | Allele Frequency | References |
|---|---|---|---|---|---|
| rs3890745 | 1p36.2 | TNFSF14 | 0.920 | 0.320 | Raychaudhuri 2008 |
| rs2240340a | 1p36.13 | PADI4 | 1.40 | 0.373 | Suzuki 2003 |
| rs2476601 | 1p13.2 | PTPN22 | 1.750 | 0.100 | Begovich 2004 |
| rs11586238 | 1p13.1 | CD2, IGSF2, CD58 | 1.120 | 0.227 | Raychaudhuri 2009 |
| rs7528684a | 1q23.1 | FCLR3 | 1.2 | 0.35 | Kochi 2005 |
| rs12746613 | 1q23.2 | FCGR2A | 1.100 | 0.124 | Raychaudhuri 2009 |
| rs3766379a | 1q23.3 | CD244 | 1.31 | 0.53 | Suzuki 2008 |
| rs10919563 | 1q31.3 | PTPRC | 0.900 | 0.132 | Raychaudhuri 2009 |
| rs13031237 | 2p16.1 | REL | 1.207 | 0.340 | Gregersen 2009 |
| rs934734 | 2p14 | SPRED2 | 1.13 | 0.51 | Stahl 2010 |
| rs10865035 | 2q11.2 | AFF3 | 1.140 | 0.460 | Barton 2009 |
| rs7574865 | 2q32.3 | STAT4 | 1.320 | 0.180 | Remmers 2007 |
| rs1980422 | 2q33.2 | CD28 | 1.100 | 0.238 | Raychaudhuri 2009 |
| rs3087243 | 2q33.2 | CTLA4 | 1.136 | 0.560 | Plenge 2005 |
| rs13315591 | 3p14 | PXK | 1.13 | 0.08 | Stahl 2010 |
| rs874040 | 4p15 | RBPJ | 1.18 | 0.30 | Stahl 2010 |
| rs6822844 | 4q27 | IL2/IL21 | 1.389 | 0.710 | Zhernakova 2007 |
| rs6859219 | 5q11 | ANKRD55 | 0.85 | 0.22 | Stahl 2010 |
| rs26232 | 5q21 | C5orf13 | 0.93 | 0.32 | Stahl 2010 |
| rs2395175 (and others) | 6p21.32 | MHC | 1.000 | 0.021 | Gregersen 1987 |
| rs548234 | 6q21 | PRDM1 | 1.100 | 0.322 | Raychaudhuri 2009 |
| rs10499194 | 6q23.3 | TNFAIP3 (a) | 1.220 | 0.220 | Plenge 2007a |
| rs6920220 | 6q23.3 | TNFAIP3 (b) | 1.333 | 0.610 | Thomson 2007 |
| rs5029937 | 6q23.3 | TNFAIP3 (c) | 1.34 | 0.04 | Orozco 2009 |
| rs394581 | 6q25.3 | TAGAP | 0.930 | 0.286 | Raychaudhuri 2009 |
| rs3093023 | 6q27 | CCR6 | 1.11 | 0.43 | Stahl 2010 |
| rs10488631 | 7q32 | IRF5 | 1.25 | 0.10 | Stahl 2010 |
| rs2736340 | 8p23.1 | BLK | 1.122 | 0.243 | Gregersen 2009 |
| rs2812378 | 9p13.3 | CCL21 | 1.100 | 0.355 | Raychaudhuri 2008 |
| rs951005 | 9p13.3 | CCL21 | 0.87 | 0.15 | Stahl 2010 |
| rs3761847 | 9q33.1 | TRAF1 | 1.100 | 0.440 | Plenge 2007b; Kurreeman 2007 |
| rs2104286 | 10p15.1 | IL2RA | 0.92 | 0.28 | Thomson 2007; Kurreeman 2009 |
| rs706778 | 10p15.1 | IL2RA | 1.11 | 0.40 | Stahl 2010 |
| rs4750316 | 10p15.1 | PRKCQ | 0.910 | 0.183 | Raychaudhuri 2008; Barton 2008 |
| rs540386 | 11p12 | RAG1, TRAF6 | 0.920 | 0.144 | Raychaudhuri 2009 |
| rs1678542 | 12q13.3 | KIF5A | 0.890 | 0.351 | Barton 2008; Raychaudhuri 2008 |
| rs4810485 | 20q13.12 | CD40 | 0.910 | 0.231 | Raychaudhuri 2008 |
| rs3218253 | 22q12.3 | IL2RB | 1.110 | 0.730 | Barton 2008 |
The purpose of this review is to place these genetic discoveries in the context of current and future therapeutic strategies for patients with RA. More specifically, our review will focus on (1) a brief overview of genetic studies, (2) human genetics as an approach to identify the Achilles’ heel of disease pathways, (3) humans as the model organism for functional studies of human mutations, (4) pharmacogenetic studies to gain insight into the mechanism of action of drugs, and (5) next-generation patient registries to enable large-scale genotype-phenotype studies.
Brief overview of human genetics: from SNP to causal allele
There are approximately 10 million common SNPs in the human genome (8). A fundamental challenge in human genetics is to systematically test each of these 10 million common SNPs for its role in disease. Advances in genomic technology have made this feasible (24). Contemporary GWAS test several hundred thousand SNPs across the entire human genome, most of which are common (minor allele frequency >5%) in the general, healthy population. To test the remaining >9 million common SNPs, the GWAS approach relies on the correlation structure of nearby SNPs (24). That is, 9 out of 10 SNPs are highly correlated, and testing 1 SNP serves to tag the remaining 9 nearby SNPs. This concept is known as linkage disequilibrium (LD).
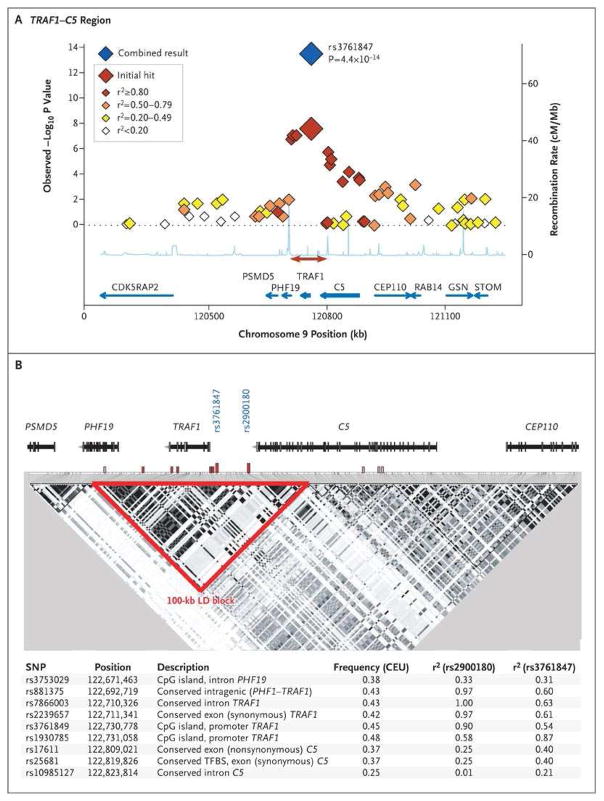
But the properties of LD that make it powerful for gene mapping also underscore the challenges that remain once a SNP is associated with disease risk: it is unknown if the SNP genotyped (and associated with risk in the genetic study) is the actual causal allele, or whether the genotyped/associated SNP is simply in LD with the causal allele. (Here we define causal allele as the single genetic mutation that is responsible for disrupting gene function and giving rise to the phenotype of interest.) Given the sheer number of common alleles, the genotyped/associated SNP is most likely just a proxy for the actual causal allele. An example of the correlation structure of an RA risk locus is shown in Figure 1 (14).
Figure 1.
Case–Control Association Results and Linkage Disequilibrium (LD) Structure in the TRAF1–C5 Locus.
Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, et al. TRAF1-C5 as a Risk Locus for Rheumatoid Arthritis -- A Genomewide Study. N Engl J Med 2007; 357(12):1199–209.
It should also be noted that there is often more than one gene in the region of LD that harbors the genotyped/associated SNP, which makes it difficult to pinpoint definitively which gene is the causal gene. On the other hand, there may be no nearby gene in the region of LD. Here, we define causal gene as the single gene that is altered by a mutation to give rise to the phenotype of interest (e.g., risk of RA). For convenience, the best biological gene, based on its known function, is often nominated as the “causal gene”. Figure 1 illustrates that there are three genes in a region of LD at the locus on chromosome 9, two of which are very strong biological candidate genes: TRAF1 (encoding tumor necrosis factor receptor–associated factor 1) and C5 (encoding complement component 5). Thus, we refer to this as the TRAF1-C5 RA risk locus.
For most of the >20 RA risk alleles shown in Table 1, the causal mutation and the causal gene have yet to be identified. Outside of the MHC, the one exception is PTPN22, where the associated mutation alters protein structure and function (25, 26). Although it may be reasonable to nominate the most likely biological candidate gene to be the causal gene, direct evidence is not yet available.
There are at least two reasons why it is important to identify (or “fine-map”) the causal mutation. First, knowing the causal mutation will help guide functional studies. For drug discovery, it is crucial to understand if the risk allele is a gain-of-function or loss-of-function allele. Second, knowing the causal allele provides more accurate estimates of risk, that could facilitate disease prediction. If the associated SNP is highly but not perfectly correlated with the causal allele, then risk estimates will be deflated.
A limitation of contemporary GWAS is that they only test common SNPs. For every common allele (defined as having an allele frequency of >5% in the general population), there is at least one – and likely many more – rare alleles (frequency <5%) (8). In addition, there are other forms of genetic variation besides SNPs, including copy number variants (where a gene may be duplicated or deleted). Next-generation sequencing and genotyping technologies will be required to identify and test rare variants and structural variants.
Drug discovery: risk alleles identify the Achilles’ heel of biological pathways
There are several effective drugs used to treat patients with active RA. Nonetheless, there are still needs that are unmet in the current pharmacological armamentarium. First, no single drug is effective in all patients – and most drugs induce disease remission in only one-third of patients. As a consequence, identifying the right medication for each patient continues to be a painstaking exercise in trial and error. Because irreversible erosive damage occurs early in the disease course, it is important to start RA patients on the right drug at disease onset to minimize long-term disability. Second, even for those drugs that are effective, many have unacceptable side effects (e.g., increased susceptibility to potentially life-threatening infections). Third, many of the drugs are extremely expensive, which places a large burden on families and the health care system. And finally, there is no drug that prevents or cures RA. Such a drug would radically change clinical practice.
How might human genetics help fill these unmet needs?
An important concept is human mutations that confer a modest increase in the risk of disease pinpoint critical components of disease pathways. To understand why, first consider two extreme examples of human mutations and their effect on biological pathways. If a gene is absolutely essential to a pathway, then a human mutation that alters gene function will likely be highly deleterious to the person carrying the mutation. Consequently, such a mutation is subject to negative evolutionary selection. In contrast, if a gene is not important at all to a pathway, then a human mutation will not exert any meaningful phenotype. Now consider the desired target of a drug. If a drug targets a pathway that is essential to survival, then the drug will likely have untoward side effects. And if a drug targets a pathway that is inconsequential, it will have no biological effect. Finally, consider common genetic variants that predispose to disease. They must affect the Achilles’ heel of a critical disease pathway since they predispose to disease, but at the same time these mutations have survived negative selection, and therefore have modest effects. These mutations are not so severe that they are highly deleterious to the individual who carries them, but they are also within a gene that is important to a disease-relevant biological pathway.
Effectively, human evolution has done the natural experiment, by introducing 10 million common mutations throughout the genome in an unbiased manner. It is up to geneticists to determine which of those 10 million common mutations are relevant to disease.
There is precedent for the concept that common alleles with a modest effect on disease risk identify effective drug targets. In each example below, the drug was developed before the genetic association was established. Still, these examples suggest that other human genetic mutations might identify the Achilles’ heel of a biological pathway, and that in turn these genetic mutations identify effective drug targets.
In RA, a common allele within the CTLA4 gene is known to increase risk of RA by ~15% per copy of the risk allele (which corresponds to an odds ratio (OR) of ~1.15) (12, 23). Abatacept, which is a fusion protein composed of the extracellular domain of CTLA4 and an immunoglobulin domain, is very effective in treating RA patients with active disease (27). Following T-cell activation, CTLA4 expression is upregulated. The protein product traffics to the T-cell membrane, where it binds and inhibits co-stimulatory molecules CD80–CD86 (which are also known as B7 proteins) on the surface of antigen presenting cells (APCs) (28). Abatacept is thought to exert its effect through a high-affinity binding site for B7 molecules on the surface of APCs, thereby inhibiting the co-stimulatory signal to T-cells (29). Alternative splicing produces two CTLA4 protein isoforms: a full-length protein and a shorter, soluble isoform that is secreted from activated T-cells. Functional studies demonstrate that the CTLA4 risk allele results in lower mRNA levels of the soluble isoform of CTLA4 (30). Interestingly, common alleles in the co-stimulatory genes CD28 and CD40 have also been implicated in RA pathogenesis, indicating that these pathways are critical in RA pathogenesis (16, 31).
There are also examples in non-autoimmune diseases in which common alleles of modest effect on phenotype identify effective drug targets. A common allele in the HMGCoA reductase gene is associated with LDL cholesterol levels in the general population (32). Statins are highly effective cholesterol-lowering drugs that inhibit this enzyme, which is the rate-limiting enzyme of the mevalonate pathway of cholesterol synthesis. A common allele of PPARG is associated with risk of type 2 diabetes (T2D) (33).
Thiazolidinediones, a class of drugs used to treat T2D, act by binding to the PPARG gene product inside the cell nucleus. Another class of drugs to treat T2D, sulfonylureas, bind to an ATP-dependent potassium channel on the cell membrane of pancreatic beta cells. A common allele in the gene that codes for this receptor, KCNJ11, is associated with risk of T2D (34). Common alleles in genes associated with bone mineral density (RANKL and OPG) identify drugs (e.g., denosumab) that show promise in clinical trials (35–37).
Genotype to function: humans as the model organism for translational immunology
In order to turn genetic studies into effective medications, a major challenge is to take the expanding list of RA risk alleles and understand the effect on gene function. For example, it is important to understand whether the risk allele increases or decreases gene function (i.e., whether it is a gain-of-function or loss-of-function) and to understand the most relevant cell type. Armed with this insight, it should be possible to target novel therapies to clinically relevant biological pathways.
In order to go from genotype to function, there is a growing awareness of the importance of conducting experiments in human tissues of immunological relevance (38, 39). Essentially, a goal should be to use humans as the model organism for translational immunology. To do this, novel resources are becoming available.
Biorepositories have now been established to study the functional consequences of common genetic mutations in blood cells from healthy control subjects (40). Human immune cells are easily accessible through a simple blood draw. These immune cells are of direct relevance to pathogenesis of RA and other autoimmune diseases (1, 41). Human immune cells derived from healthy control subjects have been used successfully to gain insight into function of common mutations at several autoimmune genes: the missense mutation at PTPN22 has been shown to alter secretion of IL2 from T-cells stimulated via the T-cell receptor (25); a common multiple sclerosis risk mutation at CD58 can explain ~40% of the variance of CD58 cell surface expression on peripheral blood mononuclear cells (PBMCs) (42); and a common type 1 diabetes mutation in IL2RA alters IL2RA cell surface expression on CD4+ memory T-cells (43).
Another novel approach is to generate induced pluripotent stem (iPS) cells from patients who carry specific genetic mutations. First described in 2006 (44), several studies have shown that iPS cells can be derived from patients with human mutations (45). Protocols have been developed to differentiate human embryonic stem cells into B-cells, T-cells, natural killer (NK) cells, and other immune lineages (46–53). Whether iPS cells derived from patients with RA will be useful for functional studies of human genetic mutations is a hypothesis that needs to be rigorously tested.
Pharmacogenetics: insight into mechanisms of treatment failure
While the general mechanism of action of many RA drugs is known – for example, drugs that target the inflammatory cytokine tumor necrosis factor-alpha (TNF) – it is still not known why some patients respond and other patients do not. Given that most RA drugs induce remission in only one-third of patients, it would be very useful to identify genetic predictors of treatment response. Moreover, the exact mechanisms of action of other drugs, including the most commonly prescribed RA drug, methotrexate (MTX), are not entirely clear. Once these mechanisms are illuminated, it should be possible to develop more effective drugs.
The field of pharmacogenetics is at a much earlier state than the field of genetic predictors of disease risk (54). In short, there is no reproducible genetic variant that is associated with response to any drugs used to treat RA. Several studies have identified alleles in the enzyme 5,10- methylenetetrahydrofolate reductase (MTHFR) that reduce enzyme activities (C677T and A1298C) and that have suggestive associations with efficacy or toxicity of MTX. However, these studies have been small and have not achieved overwhelming evidence of statistical significance (55–61). Several studies have identified alleles associated with response to anti-TNF therapy (reviewed in (54)). As with the MTX studies, these studies have identified only suggestive, but no confirmed associations.
Although it might be possible to use human genetics to help target the right therapy to the right patient – so-called personalized medicine – the predictive value of common alleles of modest effect sizes are likely to have little discriminating value (62). As an example, a composite genetic risk score (GRS) based on all known RA susceptibility alleles is able to risk-stratify patients across a 10-fold range. However, a GRS has little clinical application in patients without symptoms (as there are no preventative measures). Once alleles that influence treatment response are identified, it will be important to determine whether or not they add to predictive models of treatment response.
Next-generation patient registries: enabling large-scale genotype-phenotype studies
Currently, the main bottleneck in pharmacogenetic and other genotype-phenotype studies is access to large sample collections that have both DNA and detailed clinical data. The largest pharmacogenetic studies to date, conducted by the Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate (BRAGGSS) based in the United Kingdom, have analyzed just over 1,000 RA patients (63–65). Most genetic studies of patient outcomes (e.g., cardiovascular events, infections, malignancy) are conducted on fewer patients. In contrast, the largest genetic association studies of RA susceptibility have been conducted in >20,000 combined case-control samples (16).
Traditional patient registries and clinical trials – the workhorse for sample collection over the past decades – are unlikely to achieve the size required to obtain thousands of patient samples with DNA and detailed clinical data. Novel approaches – next-generation registries – will be required to break this bottleneck. In theory, it should be possible to collect data as part of routine patient care. Medical informatic tools are available to collect basic clinical information (e.g., components of the DAS28, medications, adverse events) every time a physician sees a patient (66, 67). Increased utilization of electronic medical records (EMRs) (68) and novel approaches to mine clinical data from EMRs (69) represents one exciting approach to expand sample collections.
CONCLUSIONS
The greatest impact of human genetics will likely be insight into the Achilles’ heel of biological pathways that lead to RA and a better understanding of mechanism of action of drugs to treat RA. For drug discovery, it will be important to understand the functional relevance of the risk alleles identified to date (and the many alleles that remain to be discovered). This will require using humans as a model organism for translational immunology. For pharmacogenetic studies, novel resources are required to build large patient registries with detailed clinical data linked with biospecimens for genomic studies.
Panel A shows results for SNPs genotyped across 1 Mb as part of the original genome-wide association scan in samples from 1522 case subjects with anti–CCP-positive rheumatoid arthritis and 1850 control subjects. Each diamond indicates a genotyped SNP; the color of each diamond is based on the correlation coefficient (r2) with the CEU HapMap with the most significant SNP in our study (rs3761847). The blue diamond indicates the P value for all samples in our study (the original scan plus replication samples), as determined by the Cochran–Mantel–Haenszel method in both NARAC and EIRA samples. The recombination rate (in centimorgans per megabase) with the CEU HapMap is shown in light blue along the x axis; the red arrow indicates the block of linkage disequilibrium shown in Panel B. The blue arrows indicate gene location. Panel B shows the linkage-disequilibrium (LD) structure across 200 kb of the TRAF1–C5 locus, based on pairwise r2 with the CEU HapMap. The intron–exon structure of each gene is at the top of the figure. Putative functional SNPs in linkage disequilibrium with either rs3761847 or rs2900180 are indicated by hatched bars, in which red indicates r2>0.80 and pink indicates r2=0.20 to 0.80; the specific SNPs, frequency, pairwise r2 with the CEU HapMap, and the putative annotated function are listed at the bottom of the figure. CpG denotes cytidine and guanosine joined by a phosphodiester bond.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423(6937):356–61. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 2.Klareskog L, Catrina AI, Paget S. Rheumatoid arthritis. Lancet. 2009;373(9664):659–72. doi: 10.1016/S0140-6736(09)60008-8. [DOI] [PubMed] [Google Scholar]
- 3.Stastny P. Mixed lymphocyte cultures in rheumatoid arthritis. J Clin Invest. 1976;57(5):1148–57. doi: 10.1172/JCI108382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stastny P. Association of the B-cell alloantigen DRw4 with rheumatoid arthritis. N Engl J Med. 1978;298(16):869–71. doi: 10.1056/NEJM197804202981602. [DOI] [PubMed] [Google Scholar]
- 5.Gregersen PK, Moriuchi T, Karr RW, Obata F, Moriuchi J, Maccari J, et al. Polymorphism of HLA-DR beta chains in DR4, −7, and −9 haplotypes: implications for the mechanisms of allelic variation. Proc Natl Acad Sci U S A. 1986;83(23):9149–53. doi: 10.1073/pnas.83.23.9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30(11):1205–13. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 7.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Consortium IH, Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322(5903):881–8. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34(4):395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 11.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75(2):330–7. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plenge RM, Padyukov L, Remmers EF, Purcell S, Lee AT, Karlson EW, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77(6):1044–60. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, et al. TRAF1-C5 as a Risk Locus for Rheumatoid Arthritis -- A Genomewide Study. N Engl J Med. 2007;357(12):1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40(10):1216–23. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41(12):1313–8. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357(10):977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhernakova A, Alizadeh BZ, Bevova M, van Leeuwen MA, Coenen MJ, Franke B, et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 2007;81(6):1284–8. doi: 10.1086/522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barton A, Eyre S, Ke X, Hinks A, Bowes J, Flynn E, et al. Identification of AF4/FMR2 family, member 3 (AFF3) as a novel rheumatoid arthritis susceptibility locus and confirmation of two further pan-autoimmune susceptibility genes. Hum Mol Genet. 2009;18(13):2518–22. doi: 10.1093/hmg/ddp177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barton A, Thomson W, Ke X, Eyre S, Hinks A, Bowes J, et al. Re-evaluation of putative rheumatoid arthritis susceptibility genes in the post-genome wide association study era and hypothesis of a key pathway underlying susceptibility. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barton A, Thomson W, Ke X, Eyre S, Hinks A, Bowes J, et al. Rheumatoid arthritis susceptibility loci at chromosomes 10p15, 12q13 and 22q13. Nat Genet. 2008;40(10):1156–9. doi: 10.1038/ng.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson W, Barton A, Ke X, Eyre S, Hinks A, Bowes J, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39:1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregersen PK, Amos CI, Lee AT, Lu Y, Remmers EF, Kastner DL, et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41(7):820–3. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 25.Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37(12):1317–9. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 26.Vang T, Miletic AV, Bottini N, Mustelin T. Protein tyrosine phosphatase PTPN22 in human autoimmunity. Autoimmunity. 2007;40(6):453–61. doi: 10.1080/08916930701464897. [DOI] [PubMed] [Google Scholar]
- 27.Moreland L, Bate G, Kirkpatrick P. Abatacept. Nat Rev Drug Discov. 2006;5(3):185–6. doi: 10.1038/nrd1989. [DOI] [PubMed] [Google Scholar]
- 28.Gough SC, Walker LS, Sansom DM. CTLA4 gene polymorphism and autoimmunity. Immunol Rev. 2005;204:102–15. doi: 10.1111/j.0105-2896.2005.00249.x. [DOI] [PubMed] [Google Scholar]
- 29.Ruderman EM, Pope RM. The evolving clinical profile of abatacept (CTLA4-Ig): a novel co-stimulatory modulator for the treatment of rheumatoid arthritis. Arthritis Res Ther. 2005;7 (Suppl 2):S21–5. doi: 10.1186/ar1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423(6939):506–11. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 31.Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5(6):e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kathiresan S, Musunuru K, Orho-Melander M. Defining the spectrum of alleles that contribute to blood lipid concentrations in humans. Curr Opin Lipidol. 2008;19(2):122–7. doi: 10.1097/MOL.0b013e3282f70296. [DOI] [PubMed] [Google Scholar]
- 33.Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26(1):76–80. doi: 10.1038/79216. taf/dynapage.taf?file=/ncb/genetics/v26/n1/full/ng0900_76.html taf/dynapage.taf?file=/ncb/genetics/v26/n1/abs/ng0900_76.html. [DOI] [PubMed]
- 34.Florez JC, Burtt N, de Bakker PI, Almgren P, Tuomi T, Holmkvist J, et al. Haplotype structure and genotype-phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes. 2004;53(5):1360–8. doi: 10.2337/diabetes.53.5.1360. [DOI] [PubMed] [Google Scholar]
- 35.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, et al. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358(22):2355–65. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 36.Smith MR, Egerdie B, Hernandez Toriz N, Feldman R, Tammela TL, Saad F, et al. Denosumab in men receiving androgen-deprivation therapy for prostate cancer. N Engl J Med. 2009;361(8):745–55. doi: 10.1056/NEJMoa0809003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cummings SR, San Martin J, McClung MR, Siris ES, Eastell R, Reid IR, et al. Denosumab for prevention of fractures in postmenopausal women with osteoporosis. N Engl J Med. 2009;361(8):756–65. doi: 10.1056/NEJMoa0809493. [DOI] [PubMed] [Google Scholar]
- 38.Davis MM. A prescription for human immunology. Immunity. 2008;29(6):835–8. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayday AC, Peakman M. The habitual, diverse and surmountable obstacles to human immunology research. Nat Immunol. 2008;9(6):575–80. doi: 10.1038/ni0608-575. [DOI] [PubMed] [Google Scholar]
- 40.Gregersen PK. Closing the gap between genotype and phenotype. Nat Genet. 2009;41(9):958–9. doi: 10.1038/ng0909-958. [DOI] [PubMed] [Google Scholar]
- 41.Gregersen PK, Behrens TW. Genetics of autoimmune diseases--disorders of immune homeostasis. Nat Rev Genet. 2006;7(12):917–28. doi: 10.1038/nrg1944. [DOI] [PubMed] [Google Scholar]
- 42.De Jager PL, Baecher-Allan C, Maier LM, Arthur AT, Ottoboni L, Barcellos L, et al. The role of the CD58 locus in multiple sclerosis. Proc Natl Acad Sci U S A. 2009;106(13):5264–9. doi: 10.1073/pnas.0813310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dendrou CA, Plagnol V, Fung E, Yang JH, Downes K, Cooper JD, et al. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat Genet. 2009;41(9):1011–5. doi: 10.1038/ng.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 45.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134(5):877–86. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vodyanik MA, Bork JA, Thomson JA, Slukvin II. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105(2):617–26. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- 47.Kaufman DS, Hanson ET, Lewis RL, Auerbach R, Thomson JA. Hematopoietic colony-forming cells derived from human embryonic stem cells. Proc Natl Acad Sci U S A. 2001;98(19):10716–21. doi: 10.1073/pnas.191362598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chadwick K, Wang L, Li L, Menendez P, Murdoch B, Rouleau A, et al. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood. 2003;102(3):906–15. doi: 10.1182/blood-2003-03-0832. [DOI] [PubMed] [Google Scholar]
- 49.Zambidis ET, Peault B, Park TS, Bunz F, Civin CI. Hematopoietic differentiation of human embryonic stem cells progresses through sequential hematoendothelial, primitive, and definitive stages resembling human yolk sac development. Blood. 2005;106(3):860–70. doi: 10.1182/blood-2004-11-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin CH, Woll PS, Ni Z, Zuniga-Pflucker JC, Kaufman DS. Differences in lymphocyte developmental potential between human embryonic stem cell and umbilical cord blood-derived hematopoietic progenitor cells. Blood. 2008;112(7):2730–7. doi: 10.1182/blood-2008-01-133801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woll PS, Martin CH, Miller JS, Kaufman DS. Human embryonic stem cell-derived NK cells acquire functional receptors and cytolytic activity. J Immunol. 2005;175(8):5095–103. doi: 10.4049/jimmunol.175.8.5095. [DOI] [PubMed] [Google Scholar]
- 52.Galic Z, Kitchen SG, Kacena A, Subramanian A, Burke B, Cortado R, et al. T lineage differentiation from human embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103(31):11742–7. doi: 10.1073/pnas.0604244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galic Z, Kitchen SG, Subramanian A, Bristol G, Marsden MD, Balamurugan A, et al. Generation of T lineage cells from human embryonic stem cells in a feeder free system. Stem Cells. 2009;27(1):100–7. doi: 10.1634/stemcells.2008-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plenge RM, Criswell LA. Genetic variants that predict response to anti-tumor necrosis factor therapy in rheumatoid arthritis: current challenges and future directions. Curr Opin Rheumatol. 2008;20(2):145–52. doi: 10.1097/BOR.0b013e3282f5135b. [DOI] [PubMed] [Google Scholar]
- 55.van Ede AE, Laan RF, Blom HJ, Huizinga TW, Haagsma CJ, Giesendorf BA, et al. The C677T mutation in the methylenetetrahydrofolate reductase gene: a genetic risk factor for methotrexate-related elevation of liver enzymes in rheumatoid arthritis patients. Arthritis Rheum. 2001;44(11):2525–30. doi: 10.1002/1529-0131(200111)44:11<2525::aid-art432>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 56.Urano W, Taniguchi A, Yamanaka H, Tanaka E, Nakajima H, Matsuda Y, et al. Polymorphisms in the methylenetetrahydrofolate reductase gene were associated with both the efficacy and the toxicity of methotrexate used for the treatment of rheumatoid arthritis, as evidenced by single locus and haplotype analyses. Pharmacogenetics. 2002;12(3):183–90. doi: 10.1097/00008571-200204000-00002. [DOI] [PubMed] [Google Scholar]
- 57.Kumagai K, Hiyama K, Oyama T, Maeda H, Kohno N. Polymorphisms in the thymidylate synthase and methylenetetrahydrofolate reductase genes and sensitivity to the low-dose methotrexate therapy in patients with rheumatoid arthritis. Int J Mol Med. 2003;11(5):593–600. [PubMed] [Google Scholar]
- 58.Weisman MH, Furst DE, Park GS, Kremer JM, Smith KM, Wallace DJ, et al. Risk genotypes in folate-dependent enzymes and their association with methotrexate-related side effects in rheumatoid arthritis. Arthritis Rheum. 2006;54(2):607–12. doi: 10.1002/art.21573. [DOI] [PubMed] [Google Scholar]
- 59.Kurzawski M, Pawlik A, Safranow K, Herczynska M, Drozdzik M. 677C>T and 1298A>C MTHFR polymorphisms affect methotrexate treatment outcome in rheumatoid arthritis. Pharmacogenomics. 2007;8(11):1551–9. doi: 10.2217/14622416.8.11.1551. [DOI] [PubMed] [Google Scholar]
- 60.Kooloos WM, Huizinga TW, Guchelaar HJ, Wessels JA. Pharmacogenetics in Treatment of Rheumatoid Arthritis. Curr Pharm Des. 2009 doi: 10.2174/138161210790112764. [DOI] [PubMed] [Google Scholar]
- 61.Kooloos WM, Wessels JA, van der Kooij SM, Allaart CF, Huizinga TW, Guchelaar HJ. Optimalization of the clinical pharmacogenetic model to predict methotrexate treatment response: the influence of the number of haplotypes of MTHFR 1298A-677C alleles on probability to respond. Ann Rheum Dis. 2009;68(8):1371. doi: 10.1136/ard.2008.096891. [DOI] [PubMed] [Google Scholar]
- 62.Kraft P, Wacholder S, Cornelis MC, Hu FB, Hayes RB, Thomas G, et al. Beyond odds ratios--communicating disease risk based on genetic profiles. Nat Rev Genet. 2009;10(4):264–9. doi: 10.1038/nrg2516. [DOI] [PubMed] [Google Scholar]
- 63.Potter C, Hyrich KL, Tracey A, Lunt M, Plant D, Symmons DP, et al. Association of rheumatoid factor and anti-cyclic citrullinated peptide positivity, but not carriage of shared epitope or PTPN22 susceptibility variants, with anti-tumour necrosis factor response in rheumatoid arthritis. Ann Rheum Dis. 2009;68(1):69–74. doi: 10.1136/ard.2007.084715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Potter C, Hyrich KL, Tracey A, Lunt M, Plant D, Symmons DP, et al. Association of RF and anti-CCP positivity, but not carriage of shared epitope or PTPN22 susceptibility variants, with anti-TNF response in RA. Ann Rheum Dis. 2008 doi: 10.1136/ard.2007.084715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bowes JD, Potter C, Gibbons LJ, Hyrich K, Plant D, Morgan AW, et al. Investigation of genetic variants within candidate genes of the TNFRSF1B signalling pathway on the response to anti-TNF agents in a UK cohort of rheumatoid arthritis patients. Pharmacogenet Genomics. 2009;19(4):319–323. doi: 10.1097/FPC.0b013e328328d51f. [DOI] [PubMed] [Google Scholar]
- 66.Collier DS, Kay J, Estey G, Surrao D, Chueh HC, Grant RW. A rheumatology-specific informatics-based application with a disease activity calculator. Arthritis Rheum. 2009;61(4):488–94. doi: 10.1002/art.24345. [DOI] [PubMed] [Google Scholar]
- 67.Hetland ML. DANBIO: a nationwide registry of biological therapies in Denmark. Clin Exp Rheumatol. 2005;23(5 Suppl 39):S205–7. [PubMed] [Google Scholar]
- 68.Steinbrook R. Personally controlled online health data--the next big thing in medical care? N Engl J Med. 2008;358(16):1653–6. doi: 10.1056/NEJMp0801736. [DOI] [PubMed] [Google Scholar]
- 69.Murphy S, Churchill S, Bry L, Chueh H, Weiss S, Lazarus R, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res. 2009;19(9):1675–81. doi: 10.1101/gr.094615.109. [DOI] [PMC free article] [PubMed] [Google Scholar]