Abstract
We describe the direct detection of DNA methylation, without bisulfite conversion, through single-molecule real-time (SMRT) sequencing. In SMRT sequencing, DNA polymerases catalyze the incorporation of fluorescently labeled nucleotides into complementary nucleic acid strands. The arrival times and durations of the resulting fluorescence pulses yield information about polymerase kinetics and allow direct detection of modified nucleotides in the DNA template, including N6-methyladenosine, 5-methylcytosine, and 5-hydroxymethylcytosine. Measurement of polymerase kinetics is an intrinsic part of SMRT sequencing and does not adversely affect determination of the primary DNA sequence. The various modifications affect polymerase kinetics differently, allowing discrimination between them. We utilize these kinetic signatures to identify adenosine methylation in genomic samples and show that, in combination with circular consensus sequencing, they can enable single-molecule identification of epigenetic modifications with base-pair resolution. This method is amenable to long read lengths and will likely enable mapping of methylation patterns within even highly repetitive genomic regions.
Introduction
DNA methylation, in its various forms, has been implicated in the regulation of a variety of biological processes across virtually every branch of the taxonomic tree. For example, in certain bacteria, N6-methyladenosine (mA) appears primarily within GATC sequence contexts and helps regulate replication, the mismatch repair pathway, and the expression of certain genes1. In plants, 5-methylcytosine (mC) appears in multiple sequence contexts, each controlled by separate genetic mechanisms2, 3. 5-methylcytosine within vertebrates usually occurs at CG dinucleotides, which often cluster in regions called CpG islands that are at or near transcription start sites4-6. Methylation within these islands regulates gene expression within cells7 and can also confer epigenetic heritability in offspring8, 9. Changes in mC patterns play a crucial role in development10, 11 and have been associated with cancer12, 13 and other diseases14. Abundant cytosine methylation in non-CG contexts was recently found in human embryonic stem cells but not in differentiated cells, suggesting that it is a distinct type of methylation involved in the maintenance of the pluripotent state15. Finally, 5-hydroxymethylcytosine (hmC) is a newly identified epigenetic mark whose biological function is not yet understood, found thus far in mouse Purkinje neurons16 and embryonic stem cells17.
Because of the key role it plays in human health and disease, cytosine methylation is the most widely studied of the DNA modifications described above, and there is much interest in mapping genome-wide mC patterns across different cell types and in response to various environmental influences18. Currently, the most common technique for studying cytosine methylation involves bisulfite treatment (which transforms epigenetic information to genetic information by converting cytosine, but not methylcytosine, to uracil) followed by massively parallel DNA sequencing18. Using this approach, researchers have recently constructed single-base resolution methylation maps for the Arabidopsis thaliana genome2, 3, for a subset of the mouse genome19, and for both fibroblasts and embryonic stem cells throughout the majority of the human genome15. Despite these advances, enabled by bisulfite sequencing, there remain several drawbacks to the technique. For example, the sample preparation steps associated with bisulfite sequencing can be costly and time-consuming, and the harsh reaction conditions necessary for complete conversion can degrade DNA. In addition, the reduction of complexity in converted genomes constrains primer design for subsequent PCR amplification20 and also complicates alignment to a reference genome6. Finally, discrimination between C, mC, and hmC cannot be accomplished with bisulfite sequencing16, 17, 21, 22.
Direct detection of methylation is possible for nucleotides in solution using techniques such as thin layer chromatography16, 17, high performance liquid chromatography16, 23, mass spectrometry16, 17, 23, and nanopore amperometry24. No high-throughput method, however, has been demonstrated that allows the determination of primary sequence at the same time as methylation status. In this paper we present a method to directly detect DNA methylation during single-molecule, real-time (SMRT) DNA sequencing, an emerging technique for studying nucleic acid sequence and structure25. In this technique, single DNA polymerase molecules are observed in real time while catalyzing the incorporation of fluorescently labeled nucleotides complementary to a template nucleic acid strand. These reactions are measured simultaneously within thousands of arrayed zero-mode waveguides (ZMWs)26, nanophotonic structures that reduce background fluorescence, thereby enabling use of the high concentrations of labeled nucleotides necessary to support fast and processive DNA sequencing-by-synthesis. Incorporation of a nucleotide is detected as a pulse of fluorescence whose color identifies that nucleotide. The pulse ends when the fluorophore, linked to the nucleotide's terminal phosphate, is cleaved by the polymerase before translocation to the next base in the DNA template. Typical polymerase synthesis rates in SMRT sequencing are currently 1-3 bases per second25.
Fluorescence pulses in SMRT sequencing are characterized not only by their emission spectra, but also by their duration and by the interval between successive pulses25. These metrics, defined here as pulse width (PW) and interpulse duration (IPD), add valuable information about DNA polymerase kinetics. PW is a function of all kinetic steps after nucleotide binding and up to fluorophor release, while IPD is determined by the kinetics of nucleotide binding and polymerase translocation27. We have previously demonstrated that SMRT sequencing polymerase synthesis rates are sensitive to DNA primary and secondary structure25. Therefore, we hypothesized that methylated bases in a DNA template might be detected directly on the principle that their presence affects polymerase kinetics during SMRT sequencing (Fig. 1). Curiously, to our knowledge the kinetics of nucleotide incorporation against methylated templates have not been studied previously, even in bulk, despite evidence that other types of modified nucleotides do, in fact, alter DNA polymerase kinetics28.
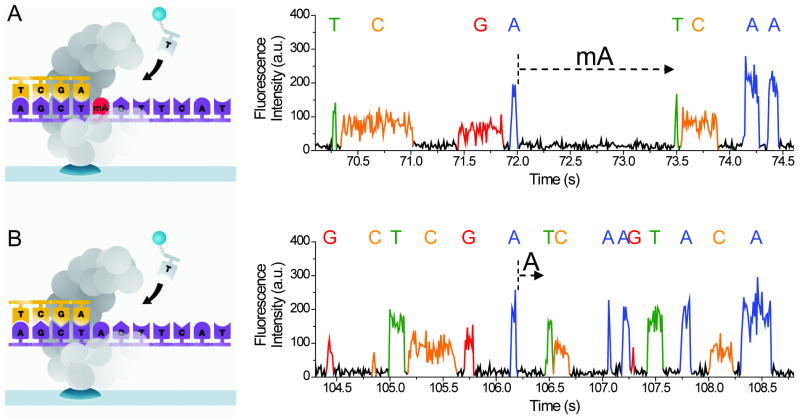
Figure 1.
Principle and corresponding example of detecting DNA methylation during SMRT sequencing. (a) Schematics of polymerase synthesis of DNA strands containing a methylated (top) or unmethylated (bottom) adenosine. (b) Typical SMRT sequencing fluorescence traces from these templates. Letters above the fluorescence trace pulses indicate the identity of the nucleotide incorporated into the growing complementary strand. The dashed arrows indicate the IPD before incorporation of the cognate T, and, for this typical example, the IPD is ∼5× larger for mA in the template compared to A.
Results
Effects of methylation on polymerase kinetics
In order to test this hypothesis, we designed several synthetic DNA templates that were identical except for their methylation status at specific sites. One template served as a control and contained no methylation, while the other templates contained several mA, mC, or hmC bases. Because the methylated bases could, in principle, affect the kinetics of DNA synthesis over a range of several nearby bases, we separated them by no less than 11 bases29. In all cases, mA was located within a GATC context, while mC and hmC were located within CG contexts. We sequenced these templates and then compared the average IPD in each of the methylated templates to the average IPD in the control template by computing their ratio at every template position. For all three methylation types, there is a clear excursion in polymerase synthesis kinetics in the vicinity of the methylated bases (Fig. 2). The methylated base is in contact with the polymerase for several bases prior and subsequent to occupying the active site29. Consistent with this model, the kinetic impact of methylation is not restricted to the nucleotide incorporation opposite the modified base. In addition, the IPD ratio patterns differ between the two methylated positions. As the only source of difference between these two loci in all three template types is the local sequence context (see Supplementary Note for template sequences), we conclude that, in general, the kinetic signatures of methylation will be sequence context dependent.
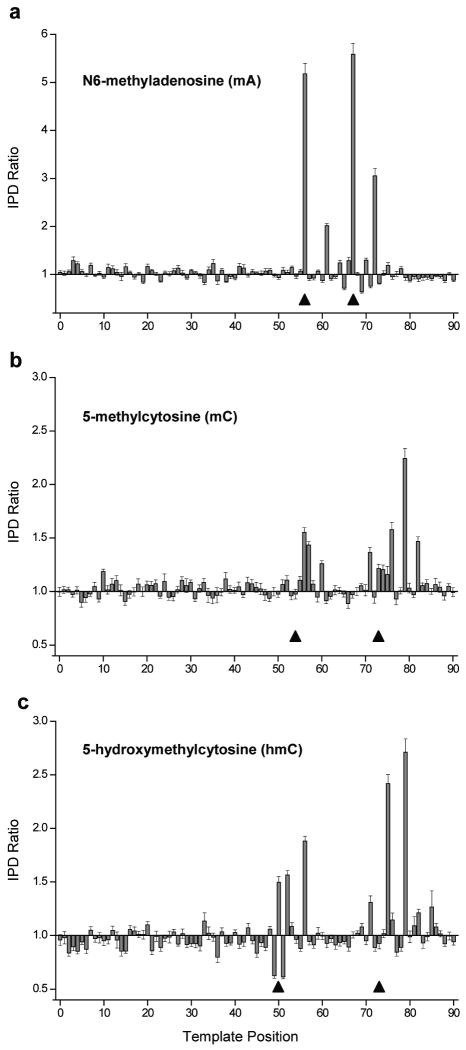
Figure 2.
SMRT sequencing-mediated detection of methylated DNA bases. All three panels show the ratio of the average IPD in the methylated template to the average IPD in the control template, plotted versus DNA template position. In the region shown, the two templates are identical except at the two positions marked by triangles. Polymerase synthesis runs in the direction of increasing position number. While in all cases the two templates have a circular topology and are 199 bases in length, only 90 base segments surrounding the methylated regions are shown for clarity. Error bars indicate the s.e.m. IPD ratio at each template position (average n = 346 measurements for each position in (a), average n = 504 for each position in (b), and average n = 393 for each position in (c), computed using bootstrapping techniques (Online Methods).
There are, however, several features in common between the two instances of each methylation type, suggesting that there may be universal interactions between the methylated nucleobase and specific sites of the polymerase. For mA (Fig. 2a), the ratio of these IPDs is largest (ranging between 5-6) opposite the methylated positions themselves. The N6 position is involved in hydrogen bonding during complementary base pairing, and therefore it is possible that the methyl group of mA directly modifies nucleotide binding kinetics. Another characteristic common between the two mA positions is an excursion in the IPD ratio 5 bases after incorporation opposite the methylated base.
The templates containing mC (Fig. 2b) display considerable IPD increases 2, 3, and 6 bases after both methylated positions. The templates containing hmC (Fig. 2c) exhibit common IPD signals at 2 and 6, but not 3, bases following the methylated positions. Plots of PW ratios display excursions that are more pronounced for hmC than for mC (Supplementary Fig. 1). In fact, because each modification displays a unique IPD and PW signature within a given context (see position 73 in Fig. 2b-c and Supplementary Fig. 1), this approach opens up the intriguing possibility of directly distinguishing between C, mC, and hmC during real-time sequencing. To this end, we used principal component analysis to find the combination of weights for the IPD and PW signals at the various template positions near the putative modification that optimizes the resolution (Supplementary Table 1). The separation between projections of the kinetic signatures for each template onto the first two principal components (Fig. 3) demonstrates the discrimination amongst these three cytosine nucleobase types by utilizing information from multiple kinetic parameters at multiple template positions.
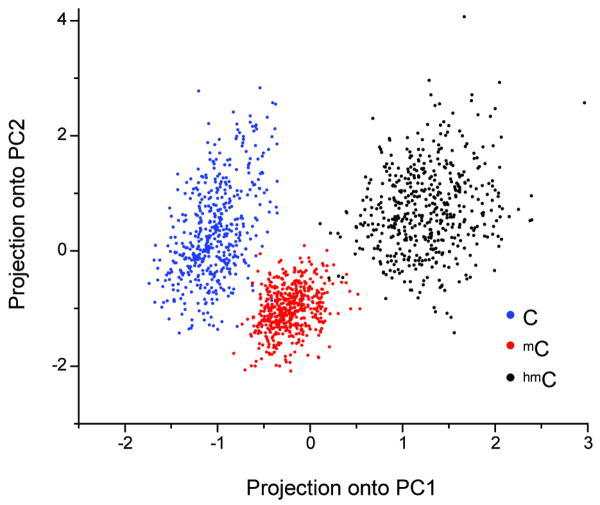
Figure 3.
Principal component analysis of C, mC, and hmC IPD and PW signatures. Each principal component is a linear combination of the mean IPD and PW at positions 71-79, which surround the variably modified template position 73. The weightings of IPD and PW at each position are shown in the Supplementary Table 1. Data points on the plot were computed by projecting a random 20% subsample of the IPD and PW values onto the first two principal components (PC1 and PC2) and then converting to a z-score (Online Methods).
Methylation detection by circular consensus sequencing
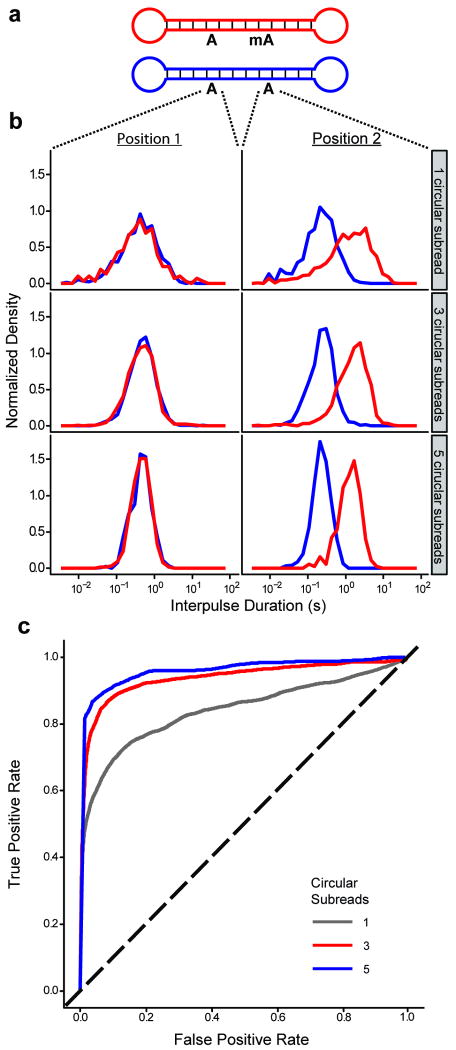
While the previous experiments establish the principle of methylation detection in populations of identical molecules, in practice individual positions in a genomic sample might be methylated in only a fraction of the molecules present. When a genome position is not always methylated, the aggregate kinetic data over an ensemble will be a linear superposition of the kinetic signatures for the methylated and unmethylated cases. Partial methylation can be quantitated by fitting the data to a two-component model, but for the highly overlapping IPD distributions that result from a single, rate-limiting step25, it can be preferable to sequence a smaller number of molecules, but multiple times each. To enable reading of individual molecules multiple times, we exploited the circular topology of our DNA templates, achieved by the ligation of hairpin adaptors to both ends of a double-stranded DNA insert (Fig. 4a). A strand-displacing DNA polymerase can carry out multiple laps of DNA synthesis around such a DNA template and enable repeated, or circular consensus, sequencing of the same DNA molecule. Repeated measurements (which we call circular subreads) of IPD at a particular DNA template position yield a mean IPD at that position that follows a gamma distribution, which is narrower than the underlying exponential distribution. As more circular subreads are collected, the distributions of mean IPD for methylated and unmethylated bases become better separated (Fig. 4b). This substantially improves the discrimination between them, as shown by receiver operating characteristics (ROC) curves for calling A versus mA (Fig. 4c) within a particular context. The normalized area under the ROC curve is 0.80 after the first circular subread but increases to 0.92 and 0.96 after three and five circular subreads, respectively. In fact, after five subreads, >85% of mA bases can be detected at this template position with a false positive rate of only ∼5%, and additional subreads enabled by longer read lengths would yield even better discrimination. The discrimination can be better still if all of the template positions affected by the methylated site are taken into consideration (as seen with mC and hmC in Fig. 3). Interestingly, there is a similarity between using multiple observations of the same template position through circular consensus and using multiple affected positions from the same subread. Both lead to transitions from highly overlapping exponential distributions to better separated modal distributions. The combination of circular consensus sequencing and methods (such as principal component analysis) to combine all available information will greatly aid in the extension of this technique to quantitation of variably methylated genomic sites with base-pair resolution.
Figure 4.
IPD distributions for A and mA in synthetic DNA templates. (a) Schematic of the DNA templates with a total length of 199 bases. (b) IPD distributions at the indicated positions for both templates. For each row, the histograms depict the distributions of mean IPD (averaged over the indicated number of circular subreads). (c) Receiver operating characteristic (ROC) curves, based on the IPD distributions from the differentially methylated position in (b) and parameterized by IPD threshold, for assigning a methylation status to an adenosine nucleotide after one (gray), three (red), or five (blue) circular consensus sequencing subreads. The black dashed line depicts the ROC curve for randomly guessing the methylation status. Note that because the templates have a length of 199 bases, five full circular subreads correspond to read lengths of nearly 1000 bases.
Adenosine methylation in E. coli
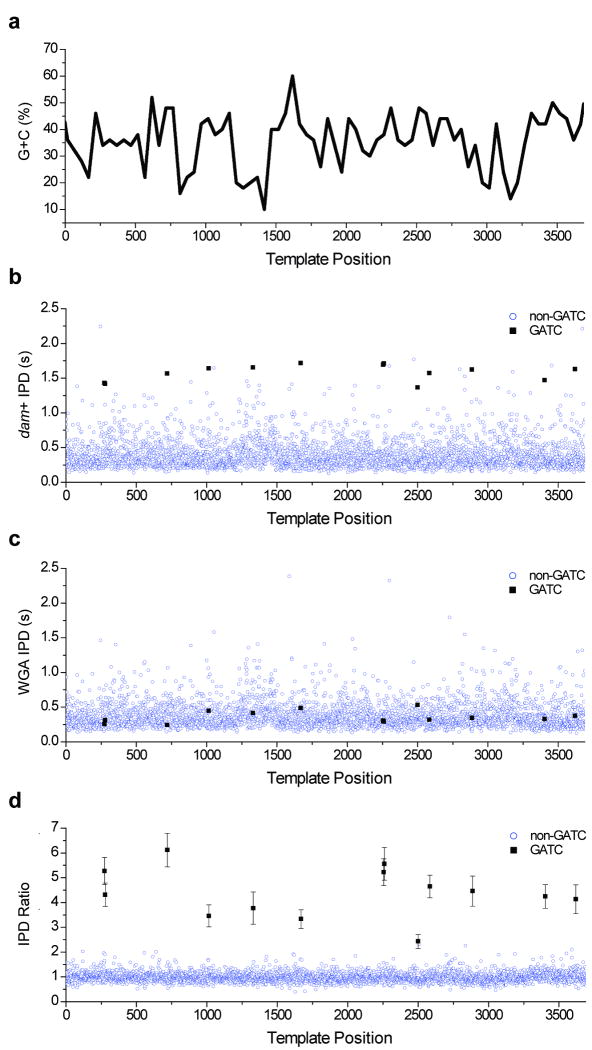
To demonstrate the application of direct methylation detection to genomic DNA, we mapped the dependence of IPD on sequence context for a C. elegans fosmid, isolated from a DNA adenosine methyltransferase positive (dam+) E. coli strain. To provide an unmethylated control template, a portion of the sample was subjected to whole genome amplification (WGA), which is expected to erase any methylation signatures (Supplementary Fig. 2). The sequencing kinetics of a 3.7-kb section of this fosmid were examined in detail (Fig. 5, Supplementary Fig. 3, Supplementary Data). Over a range of sequence contexts with varying GC content (Fig. 5a), the dam+ samples have average IPDs at GATC positions that are generally greater than those at non-GATC positions (Fig. 5b). In contrast, average IPDs were similar at all template positions within the WGA samples (Fig. 5c). The resulting ratio of average IPDs between the two samples (Fig. 5d) demonstrates that polymerase kinetics are altered substantially by adenosine methylation in a wide variety of surrounding sequence contexts (Table 1). The IPD ratio increase is similar over different GC-content levels for the range represented in this sample (Supplementary Fig. 4). The increase in average IPD caused by adenosine methylation in E. coli was consistent with the range of IPD ratios measured in the synthetic mA templates. Average IPDs over all possible 4-mer sequence contexts in the entire fosmid sample (48 kb including the vector, see Supplementary Fig. 5) show notable context dependence, evident as a non-random profile in the dam+ and WGA heat maps, highlighting the sensitivity of the method for studying DNA polymerase kinetics. The high degree of similarity between the two maps demonstrates the robustness of SMRT sequencing IPD measurements. The notable exception to their similarity is the sequence context GATC, which has a mean IPD ∼4× larger in the dam+ samples than in the WGA samples. Extension to much larger genomic samples will be straightforward using future commercial versions of SMRT sequencing instrumentation that have ∼102× greater throughput than the prototype instrument25, 30 used in these experiments.
Figure 5.
Comparison of SMRT sequencing kinetics for DNA samples propagated within dam+ E. coli and for the same samples after whole-genome amplification (WGA). The sample comprises a 3.7-kb subregion of a C. elegans fosmid cloned into an E. coli vector. (a) 50-bp window GC-content of the sample, plotted versus template position. (b) Average IPD at each template position within the dam+ sample. (c) Average IPD at each template position within the WGA sample. (d) Ratio of the average IPDs (dam+ in (b) divided by WGA in (c)), plotted versus template position. Positions with a GATC context, where methylation of adenine at the sequence motif GATC is expected, are denoted by black squares, and all other positions are denoted by open blue circles. Error bars at the GATC positions denote the s.e.m. IPD ratio at those positions (average n = 106 measurements at each position). For comparison, the mean ± s.d. of all IPD ratios at non-GATC positions (open blue circles) is 1.00 ± 0.24 (n is ∼389,000 measurements). Average sequencing coverage across this fosmid region was 121-fold for the dam+ sample and 91-fold for the WGA sample.
Table 1.
Sequence context and IPD ratios for each fosmid GATC motif For each GATC motif in the 3.7 kb fosmid subregion (Fig. 5), the local sequence context, IPD ratio (average IPD from dam+ sample divided by the average IPD from the WGA sample), and p-value are shown. The p-value was derived by performing a two-sample Kolmogorov-Smirnov goodness-of-fit test, which compares the IPD data at each position within the dam+ and WGA samples and tests the likelihood that they are drawn from the same underlying distribution (the null hypothesis). Lower p-values indicate greater confidence that the null hypothesis should be rejected. The central GATC motif within each local context is underlined.
| # | Position | Sequence | IPD Ratio | P-value |
|---|---|---|---|---|
| 1 | 273 | TGCCATGATCTAGATC | 5.28 | 2.84 × 10-18 |
| 2 | 279 | GATCTAGATCATCGTG | 4.32 | 4.41 × 10-16 |
| 3 | 720 | TTCTATGATCAGGGAG | 6.12 | 7.00 × 10-21 |
| 4 | 1015 | GCGTGGGATCTGTATG | 3.46 | 6.25 × 10-13 |
| 5 | 1329 | TATCACGATCTCATTA | 3.78 | 2.35 × 10-12 |
| 6 | 1668 | TAGTTGGATCAAGAGA | 3.33 | 2.61 × 10-13 |
| 7 | 2256 | CTTTTGGATCAGATCC | 5.22 | 1.70 × 10-19 |
| 8 | 2261 | GGATCAGATCCAATTA | 5.56 | 1.43 × 10-22 |
| 9 | 2499 | CAGATGGATCAATCAA | 2.43 | 4.44 × 10-7 |
| 10 | 2583 | ATTTTTGATCTAGTTT | 4.65 | 2.00 × 10-21 |
| 11 | 2887 | ATTCGCGATCTCCACA | 4.46 | 1.57 × 10-14 |
| 12 | 3402 | CCTCAAGATCATCATC | 4.24 | 2.04 × 10-15 |
| 13 | 3619 | GCCAGCGATCATATTT | 4.13 | 2.05 × 10-10 |
Discussion
In the experiments described in this paper, both the methylated and control DNA were sequenced for each experiment, but in resequencing applications unmethylated kinetic reference data could be collected just once and tabulated for all subsequent studies of the same species. In the future, de novo detection of methylation may also be possible through tabulation of expected kinetics over a suitable number of contexts or by taking advantage of heuristics that embody the observed trends in SMRT sequencing kinetics.
In SMRT DNA sequencing, measurement of polymerase kinetics occurs directly alongside primary sequence determination and does not require any additional sample preparation steps. We have shown that several forms of methylation in the DNA template, namely mA, mC, and hmC, all alter incorporation kinetics. Based on the same principle, we expect that other epigenetic modifications, as well as various forms of DNA damage, may also be detected by this method. Unique kinetic signatures displayed by each modification will permit discrimination between them within the same DNA sample. By enabling repeated interrogation of individual molecules, circular consensus sequencing allows base-pair resolution and single-molecule sensitivity for detection of mA. For mC and hmC, enhancements of kinetic sensitivity will likely be required. Such improvements could come from optimized solution conditions, polymerase mutations, and algorithmic approaches that take advantage of the kinetic signatures' spread over multiple template positions, while deconvolution techniques will help resolve neighboring mC bases (Supplementary Fig. 6). The long read lengths of SMRT sequencing will likely permit methylation profiling in highly-repetitive genomic regions, where a substantial fraction of mC residues resides6. Combined with single-molecule sensitivity, these long reads will also allow phasing of methylation status between different genomic positions. As we continue to refine this technique, de novo methylation profiling may become possible.
Online Methods
Zero-mode waveguide (ZMW) fabrication
ZMW nanostructures were fabricated and functionalized as previously described24, 31, 32. Sequencing experiments were performed using arrays of 3,000 ZMWs monitored simultaneously24, 31, 32.
Preparation of DNA templates
Sets of ∼35-base ssDNA oligonucleotides (Figs. 1-4 and Supplementary Fig. 1) were purchased (Trilink Biotechnologies, San Diego, CA). Presence of base modifications within these single-stranded oligonucleotides was verified by mass spectrometry. After hybridization and ligation, each end of the resulting dsDNA oligonucleotides was ligated to a hairpin oligonucleotide. Samples were treated with exonucleases to remove any molecules that were not covalently closed. Sequences for the resulting DNA templates, which were 199 bases in length and consisted of a central 84-bp double-stranded region with single-stranded loops at each end, are shown (Supplementary Note).
For sequencing of the full fosmid (Supplementary Figs. 2 and 5), a fosmid clone (clone id: WRM0639cE06) containing an ∼40 kb C. elegans genomic insert was obtained from Geneservice (Cambridge, UK, http://www.geneservice.co.uk/products/clones/Celegans_Fos.jsp) in dam+ E. coli strain EPI300, and cultured and amplified using the inducible origin (CopyControl system, Epicentre, Madison, WI). Fosmid DNA was purified using standard methods. DNA templates were then created directly from fosmid DNA or from whole genome amplified (WGA) fosmid DNA. For WGA libraries, 25 ng of fosmid DNA was amplified using the manufacturer recommended conditions in the GenomiPhi HY DNA Amplification Kit (GE Healthcare, U.K.).
For sequencing of the subsection of the fosmid (Fig. 5 and Supplementary Figs. 3 and 4), an ∼3.7 kb segment (corresponding to positions 12797-16484 within the fosmid) containing 13 instances of the GATC sequence context was PCR amplified from the fosmid using Phusion High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA) and the following primers: Forward 5′-AGTCCTGATGCTTTCACCAAAT-3′; Reverse 5′-ATTTAGATTGCCAAAGCCGTAA-3′. PCR products were cloned into the pCR-Blunt vector using the Zero Blunt PCR Cloning Kit (Invitrogen, Carlsbad, CA) and propagated in the dam+ E. coli strain TOP10 (Invitrogen, Carlsbad, CA). Approximately 25 ng of the DNA was amplified using the REPLI-g Mini Kit (QIAGEN, Valencia, CA) for the generation of the unmethylated control sample.
Fosmid DNA, or an equivalent quantity of WGA fosmid DNA, was sheared to a mean size of 500 bp (Fig. 5 and Supplementary Figs. 3 and 4) or 200 bp (Supplementary Fig. 5) using an ultrasonicator (Covaris Inc, Woburn, MA). Sheared DNA was then end-repaired with a cocktail of T4 DNA polymerase and T4 polynucleotide kinase, purified, and subjected to 3′ A-tailing with Klenow(exo-). The A-tailed fragments were ligated to hairpin oligonucleotides that contained a single 3′ T overhang and 5′ phosphate. Samples were treated with a mixture of exonucleases to remove any molecules that were not covalently closed. The resulting DNA templates were purified using SPRI magnetic beads (AMPure, Agencourt Bioscience, Beverly, MA) and annealed to a two-fold molar excess of a sequencing primer (5′-GGAGGAGGAGGA -3′) that specifically bound to the single-stranded loop region of the hairpin adapters.
Preparation of DNA polymerase and phospholinked dNTP, and DNA sequencing assays
DNA polymerases were generated as described24, 33. Phospholinked dNTPs were generated as described24, 33, with the exception of replacing Alexa Fluor 660 with a modified Cy5.5 fluorophore. Additional modifications included permuting the nucleobases associated with each dye to the following configuration: A555-dT, A568-dG, A647-dA, Cy5.5-dC. The excitation laser lines used were the same as described24, 33. Protocols for DNA polymerase/template complex formation, complex immobilization on the ZMW array, and sequencing reactions were similar to those described previously.
Data collection & analysis
Data collection was performed on a highly parallel confocal fluorescence detection instrument, as previously described24, 30. Pulse calling, which utilized a threshold algorithm on the dye-weighted intensities of fluorescence emissions, and read alignments, achieved using a Smith-Waterman algorithm, have been described24. Reads were filtered after alignment to remove low quality sequences derived from doubly-loaded ZMWs. Interpulse duration (IPD) values were tabulated from consecutive pairs of correctly aligning template positions and were assigned to the second template position in the pair. Pulse width (PW) values were computed as the duration of the pulses associated with correctly aligning base calls. To avoid outlier effects, the smallest and largest five percent of IPDs and PWs at each position were excluded from all analyses.
Bar plot error bars (Fig. 2) represent an estimate of the standard error of the mean IPD ratio, computed by bootstrapping 10 randomly selected subsamples of 10% of the data. It can be seen that the error bars underestimate the error somewhat, as small excursions do occur at positions far away from any modification, where differences from the control are not expected. Molecular coverage varied between template, with an average coverage of 346, 504, and 393 for the 10% subsamples of the mA, mC, and hmC templates, respectively. No bootstrapping was performed for creation of the PW plots (Supplementary Fig. 1), for which all molecules were used.
Standard principal component analysis34 was carried out using the prcomp function from the Stats Package of the statistical computing program, R35. Input variables were scaled to have zero mean and unit variance, and the resulting first and second principal components were determined from the entire data set (see Supplementary Table 1). To generate the principal component scatter plot (Fig. 3), 500 subsets of 20% of the data for each template were first projected onto these first two principal components. These values were then converted into a z-score by subtracting the mean and dividing by the standard deviation of all 1500 data points for each principal component.
IPD distributions (Fig. 4) were determined by averaging multiple IPD measurements at the same template position within single molecules. All molecules from the mA experiment were used. The corresponding ROC curves were generated by sliding a threshold value across the full range of observed average IPDs. The true positive rate was computed for each threshold as the fraction of methylated observations with an average IPD larger than the threshold. Similarly, the false positive rate was determined by the fraction of non-methylated observations with an average IPD larger than the threshold.
For the fosmid experiments (Fig. 5, Supplementary Figs. 3-5), GATC positions are defined as those positions at which a T is incorporated opposite a template A that is within a template GATC context. Non-GATC positions correspond to all other positions. IPD ratios at each position were normalized by the ratio of the average IPD over all dam+ reads to the ratio of the average IPD over all WGA reads. Average sequencing coverage for the 3,688-bp fosmid region analyzed in this figure was 121-fold for the dam+ sample and 91-fold for the WGA sample. This coverage was obtained using nine ZMW arrays (SMRT™ Cells) and a total of ∼1.5 hours of sequencing for each sample (dam+ and WGA).
Local sequence context (Supplementary Fig. 5) was determined using the standard Smith-Waterman alignment algorithm. The ‘local context’ of detected bases was defined as the two bases previously detected (shown on the left axis), the detected base itself and the next base detected afterwards (both shown on the bottom axis). For example, in the local context 5′-GATC-3′, the mean IPD reported describes the average duration between the detected A (complementary to a T in the template DNA) and the detected T (complementary to an A or mA in the template). On average, 1890 observations (remaining after removal of the smallest and largest five percent) were used to compute the mean IPD for each of the 256 possible 4-mer contexts, corresponding to 10-fold coverage of the entire fosmid.
Supplementary Material
Acknowledgments
We thank the entire staff at Pacific Biosciences, in particular J. Londry and D. Kolesnikov for sample preparation; E. Mollova, M. Berhe, and J. Yen for running sequencing experiments; J. Sorenson, J. Chin, A. Kislyuk, and D. Holden for help with data analysis; and E. Schadt and J. Eid for helpful discussions. Supported by National Human Genome Research Institute grant 1RC2HG005618-01.
Footnotes
Author Contributions: B.A.F., K.T., J.K., J.L., and S.W.T. designed the experiments. E.O. and T.A.C. prepared fosmid library constructs. B.A.F. conducted the sequencing experiments. D.W. and B.A.F. performed data analysis. B.A.F., J.K., S.W.T., E.O, D.W., and T.A.C. wrote the manuscript.
Competing Interests Statement: The authors declare competing financial interests: all of the authors are employees of Pacific Biosciences, a private company commercializing DNA sequencing technology.
References
- 1.Marinus MG, Casadesus J. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol Rev. 2009;33:488–503. doi: 10.1111/j.1574-6976.2008.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 5.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47:142–150. doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 7.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 8.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 9.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 10.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 11.Razin A, Shemer R. DNA methylation in early development. Hum Mol Genet. 1995;4 Spec No:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- 12.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 13.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 14.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 15.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009 doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kriaucionis S, Heintz N. The Nuclear DNA Base 5-Hydroxymethylcytosine Is Present in Purkinje Neurons and the Brain. Science. 2009 doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lister R, Ecker JR. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark SJ, Statham A, Stirzaker C, Molloy PL, Frommer M. DNA methylation: bisulphite modification and analysis. Nat Protoc. 2006;1:2353–2364. doi: 10.1038/nprot.2006.324. [DOI] [PubMed] [Google Scholar]
- 21.Hayatsu H, Shiragami M. Reaction of bisulfite with the 5-hydroxymethyl group in pyrimidines and in phage DNAs. Biochemistry. 1979;18:632–637. doi: 10.1021/bi00571a013. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, et al. The Behaviour of 5-Hydroxymethylcytosine in Bisulfite Sequencing. PLoS ONE. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tardy-Planechaud S, Fujimoto J, Lin SS, Sowers LC. Solid phase synthesis and restriction endonuclease cleavage of oligodeoxynucleotides containing 5-(hydroxymethyl)-cytosine. Nucleic Acids Res. 1997;25:553–559. doi: 10.1093/nar/25.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 25.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 26.Levene MJ, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 27.Wong I, Patel SS, Johnson KA. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 28.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 29.Berman AJ, et al. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundquist PM, et al. Parallel confocal detection of single molecules in real time. Optics Letters. 2008;33:1026–1028. doi: 10.1364/ol.33.001026. [DOI] [PubMed] [Google Scholar]
- 31.Foquet M, et al. Improved fabrication of zero-mode waveguides for single-molecule detection. Journal Of Applied Physics. 2008;103:034301. [Google Scholar]
- 32.Korlach J, et al. Selective aluminum passivation for targeted immobilization of single DNA polymerase molecules in zero-mode waveguide nanostructures. Proc Natl Acad Sci U S A. 2008;105:1176–1181. doi: 10.1073/pnas.0710982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korlach J, et al. Long, Processive Enzymatic DNA Synthesis Using 100% Dye-Labeled Terminal Phosphate-Linked Nucleotides. Nucleosides, Nucleotides & Nucleic Acids. 2008;27 doi: 10.1080/15257770802260741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jolliffe IT. Principal Component Analysis, Edn. 2002:2. [Google Scholar]
- 35.R Foundation for Statistical Computing; 2009. R: A Language and Environment for Statistical Computing. http://www.R-project.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.