Abstract
The pathogenic bacteria Pseudomonas aeruginosa and Bordetella pertussis contain in their outer membranes the rare sugar 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid. Five enzymes are required for the biosynthesis of this sugar starting from UDP-N-acetylglucosamine. One of these, referred to as WlbB, is an N-acetyltransferase that converts UDP-2-acetamido-3-amino-2,3-dideoxy-D-glucuronic acid (UDP-GlcNAc3NA) to UDP-2,3-diacetamido-2,3-dideoxy-D-glucuronic acid (UDP-GlcNAc3NAcA). Here we report the three-dimensional structure of WlbB from Bordetella petrii. For this analysis, two ternary structures were determined to 1.43-Å resolution: one in which the protein was complexed with acetyl-CoA and UDP and the second in which the protein contained bound CoA and UDP-GlcNAc3NA. WlbB adopts a trimeric quaternary structure and belongs to the LβH superfamily of N-acyltransferases. Each subunit contains 27 β-strands, 23 of which form the canonical left-handed β-helix. There are only two hydrogen bonds that occur between the protein and the GlcNAc3NA moiety, one between Oδ1 of Asn 84 and the sugar C-3' amino group and the second between the backbone amide group of Arg 94 and the sugar C-5' carboxylate. The sugar C-3' amino group is ideally positioned in the active site to attack the si face of acetyl-CoA. Given that there are no protein side chains that can function as general bases within the GlcNAc3NA binding pocket, a reaction mechanism is proposed for WlbB whereby the sulfur of CoA ultimately functions as the proton acceptor required for catalysis.
N-acetyltransferases catalyze the transfer of acetyl groups from acetyl-CoA to primary amine acceptors. They are distributed throughout nature where they play key roles in such biological processes as the sleep-wake cycles of vertebrates, the reversible modifications of histone proteins, and the inactivations of aminoglycoside antibiotics (1, 2). Substrates for these enzymes range from simple small molecules to large proteins. Thus far two major classes of N-acyltransferases have been identified and are referred to as the GNAT and LβH superfamilies. Members in GNAT superfamily adopt an overall fold built around a central mixed β-sheet, which is typically composed of six β-strands (1, 2). As the name implies, proteins that belong to the LβH superfamily have a structure dominated by a stunning parallel β-helix with repeating isoleucine-rich hexapeptide motifs and rare left-handed crossover connections (3).
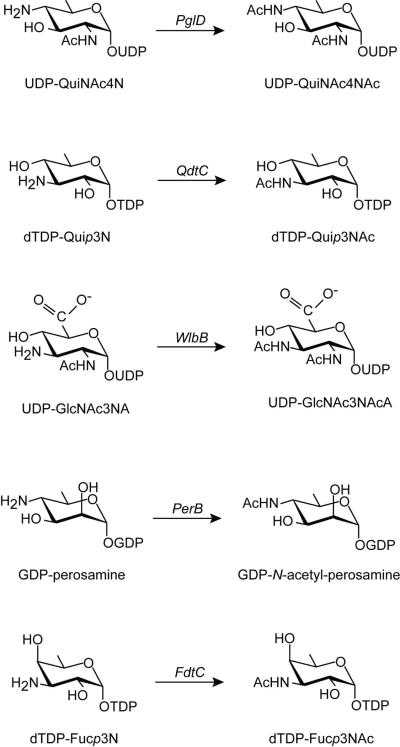
Within the last two years the molecular architectures of two intriguing N-acetyltransferases that function on nucleotide-linked sugars have been reported. The first of these was that of PglD, from Campylobacter jejuni, which specifically catalyzes the N-acetylation of UDP-2-acetamido-4-amino-2,4,6-trideoxy-D-glucose to yield UDP-2,4-diacetamido-2,4,6-D-trideoxyglucose or UDP-QuiNAc4NAc as indicated in Scheme 1 (4–6). C. jejuni, itself, is a highly unusual Gram-negative bacterium that contains N-glycosylated proteins, and QuiNAc4NAc serves as the bridge between the asparagine residue of a given glycosylated protein and the attached N-glycan moiety (4, 7). PglD functions as a trimer with each subunit folding into two separate domains, an N-terminal region containing a three-stranded parallel β-sheet flanked by two α-helices, and a C-terminal domain dominated by a left-handed β-helix motif (5). From detailed structural and kinetic studies it has been proposed that His 125 functions as an active site base to remove a proton from the amino group of the UDP-linked sugar as it attacks the carbonyl carbon of acetyl-CoA (5, 6). The actual role of His 125 is still open to debate, however, given the recent biochemical data suggesting that while it is important for the catalysis, it is not absolutely essential (8).
Scheme 1.
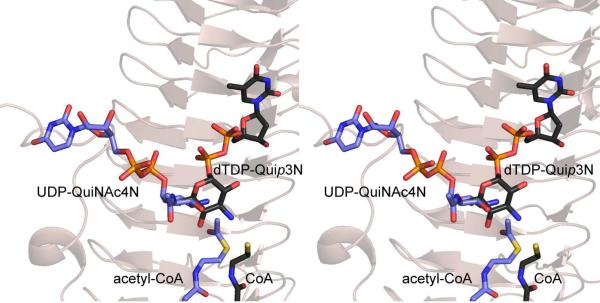
The second N-acetyltransferase structure recently reported was that of QdtC from Thermoanaerobacterium thermosaccharolyticum E207-71 (9). As indicated in Scheme 1, QdtC catalyzes the last step in the biosynthesis of dTDP-3-acetamido-3,6-dideoxy-D-glucose or Quip3NAc, an unusual dideoxysugar found in the O-antigens of some Gram-negative bacteria (10) and in the S-layer glycoprotein glycans of some Gram-positive bacteria (11). QdtC, like PglD, functions as a trimer. Each subunit of QdtC is dominated by 32 β-strands that form the canonical LβH motif. QdtC lacks, however, the N-terminal domain observed in PglD. Also, as opposed to PglD, there are no potential catalytic bases located within the active site cleft of QdtC. Indeed PglD and QdtC accommodate their nucleotide-linked sugars in completely different orientations as shown in Figure 1. Given the lack of a potential catalytic base in QdtC and in light of subsequent site-directed mutagenesis experiments and enzymatic activity assays, a novel catalytic mechanism for it was proposed (9). Specifically the sulfur of acetyl-CoA ultimately serves as the catalytic base by accepting a proton from the sugar amino group as it attacks the carbonyl carbon of acetyl-CoA.
Figure 1.
Differences in the orientations of the nucleotide-linked sugar substrates when bound to PglD or QdtC. The ribbon representation in the background corresponds to that for QdtC. The substrates for PglD and QdtC are highlighted in blue and black bonds, respectively. All figures were prepared with the software package PyMOL (31)
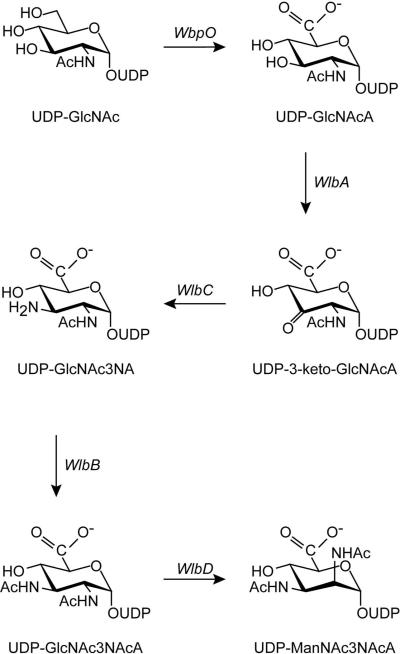
Herein we report the molecular architecture of WlbB, an N-acetyltransferase from Bordetella petrii. WlbB catalyzes the penultimate step in the biosynthesis of UDP-2,3-diacetamido-2,3-dideoxy-D-mannuronic acid or UDP-ManNAc3NAcA (Scheme 2). Specifically, as shown in Schemes 1 and 2, WlbB catalyzes the N-acetylation of UDP-2-acetamido-3-amino-2,3-dideoxy-D-glucuronic acid or UDP-GlcNAc3NA to yield UDP-GlcNAc3NAcA (12). Although a reasonably rare di-N-acetylated sugar, ManNAc3NAcA is found not only in B. petrii, but also in the B-band O-antigen of the opportunistic bacterium Pseudomonas aeruginosa, a major source of nosocomial infections and in the A-band trisaccharide of the bacterium Bordetella pertussis, the causative agent of whopping cough (13). Interestingly, mutant strains of B. pertussis that lack the A-band trisaccharide have been shown to be defective in colonization of the respiratory tracts of BALB/c mice (14). In addition, the O-antigen in P. aeruginosa is thought to protect the bacterium from phagocytosis (15) and from serum-mediated killing (16).
Scheme 2.
For the investigation described here, WlbB was crystallized in the presence of either acetyl-CoA and UDP or CoA and its substrate UDP-GlcNAc3NA. The structures were solved and refined to a nominal resolution of 1.43 Å. The binding mode for the UDP-GlcNAc3NA ligand is similar to that observed for QdtC. WlbB is considerably smaller than QdtC, however, and thus its LβH motif is shorter by nine β-strands. The pyranosyl moiety of UDP-GlcNAc3NA is accommodated within the active site region of WlbB via only two specific interactions: one between Oδ1 of Asn 84 and the sugar C-3' amino group and the second between the backbone amide group of Arg 94 and the sugar C-5' carboxylate. Importantly, the sugar C-3' amino nitrogen is in the proper position to attack the si face of acetyl-CoA. Details concerning the overall active site architecture of WlbB are presented and implications for its catalytic mechanism are discussed.
Materials and Methods
Cloning, Expression, and Purification
Genomic DNA from B. petrii (ATCC BAA-461) was isolated by standard procedures. The wlbB gene was PCR-amplified from genomic DNA such that the forward primer 5'-AAACATATGGCCACTATCCATCCTACCGCCATCG and the reverse primer 5'-AAAACTCGAGTCAGGCCAGTCGGCATACGCCGTCAG added NdeI and XhoI cloning sites, respectively. The purified PCR product was A-tailed and ligated into a pGEM-T (Promega) vector for screening and DNA sequencing. A WlbB-pGEM-T vector construct of the correct sequence was then appropriately digested and ligated into a pET28b(+) (Novagen) plasmid that had been previously modified for the production of a protein with a TEV protease cleavable N-terminal hexahistidine tag (17).
The WlbB-pET28jt plasmid was used to transform Rosetta(DE3) Escherichia coli cells (Novagen). The culture in LB media was grown at 37°C with shaking until the absorbance at 600 nm reached 0.7. The cultures were subsequently cooled in an ice bath, induced with 1.0 mM IPTG, and the cells were allowed to express protein at 16°C for 24 hours after induction. WlbB was purified by a modification to the standard procedures for Ni-NTA chromatography. Specifically, all buffers contained 500 mM NaCl, the lysis buffer contained 10% glycerol, and all steps, other than the initial sonication of the cell pellet at 4°C, were performed at room temperature. Following purification via Ni-NTA chromatography, TEV protease was added to the pooled protein (at a 1:50 molar ratio of TEV protease:WlbB), and this mixture was dialyzed against 50 mM sodium phosphate, 500 mM NaCl, 40 mM imidazole, and 1 mM DTT at pH 8.0 for 24 hours. The cleaved and intact versions of WlbB, as well as the TEV protease itself, were separated by passage through a column of Ni-NTA resin. The cleaved protein was dialyzed against 10 mM Tris and 400 mM NaCl at pH 8.0, and then concentrated to 12.5 mg/ml using an extinction coefficient of 0.83 (mg/ml)−1 cm−1 at 280 nm.
Production of UDP-2-acetamido-3-amino-2,3-dideoxy-D-glucuronic acid (UDP-GlcNAc3NA)
UDP-GlcNAc3NA was synthesized from UDP-GlcNAc. To aid in the purification process, the synthesis was divided into two steps. The first involved the oxidation of UDP-GlcNAc to UDP-GlcNAcA (Scheme 2). A typical 10 mL reaction mixture contained the following: 50 mM HEPPS (pH 8.5), 100 mM (NH4)2SO4, 100 mg UDP-GlcNAc (0.154 mmol), 310 mg NAD+ (0.462 mmol), and 10 mg WpbO (from B. petrii). The reaction was allowed to proceed at 37°C for 4 hours. The enzyme was removed via filtration with a 10-kDa cutoff centriprep concentrator, and the enzyme-free reaction products were diluted by 1:4 with water. Purification was achieved by chromatography using a 6 mL Resource-Q column and a 120 mL gradient from 0 – 500 mM ammonium bicarbonate at pH 8.5. The desired product was identified by mass spectrometry (m/z = 620.1). Fractions containing UDP-GlcNAcA were pooled and lyophilized until all traces of buffer were removed. The second step involved the oxidation and subsequent amination of UDP-GlcNAcA to yield UDP-GlcNAc3NA. A typical 400 mL reaction contained 0.8 mM UDP-GlcNAcA, 50 mM glutamate, 50 mM HEPPS (pH 8), 7 μM WlbA (from Thermus thermophilus) and 6 μM WlbC (from B. petrii). The reaction was allowed to proceed for 12 hours at 30°C. The enzymes were removed via filtration with a 10-kDa cutoff centriprep concentrator, and the enzyme-free reaction products were diluted by 1:3 with water. Purification was achieved by chromatography using a 6 mL Resource-Q column and a 150 mL gradient from 0 – 450 mM ammonium bicarbonate at pH 8.5. The desired product was identified by mass spectrometry (m/z = 619.0). Fractions containing the oxidized sugar product were pooled and lyophilized until all traces of buffer were removed.
The enzymatic activity of B. petrii WlbB was verified by its incubation (10 μM) with 0.5 mM UDP-GlcNAc3NA and 0.5 mM acetyl CoA at 30°C for 2 hours. Removal of the enzyme and purification of the resulting reaction products on a 1 mL resource Q column (20 mL gradient, 0 – 600 mM ammonium bicarbonate, pH 8.5) showed the disappearance of the peaks corresponding to UDP-GlcNAc3NA and acetyl CoA and the formation of peaks corresponding to CoA and a new uridine-containing product. Mass spectrometry of the uridine-containing product gave a m/z ratio of 661.1, which corresponds to UDP-GlcNAc3NAcA.
Structural Analysis of WlbB
Crystallization conditions were initially surveyed via the hanging drop method of vapor diffusion and using a sparse matrix screen developed in the laboratory. Trials were conducted with both the apoprotein or protein incubated with 10 mM acetyl-CoA. Diffraction quality crystals were subsequently grown via hanging drop by mixing in a 1:1 ratio the protein incubated with acetyl-CoA and a precipitant solution containing 15 – 20% poly(ethylene glycol) 8000 (100 mM MOPS, pH 7.5). The crystals belonged to the space group C2221 with unit cell dimensions of a = 85.8, b = 158.4, and c = 118.4 Å, and they contained two trimers per asymmetric unit.
An initial X-ray data set was measured from this orthorhombic crystal form at 4°C using a Bruker AXS HiSTAR area detector. The X-ray source was CuKα radiation from a Rigaku RU200 generator equipped with Supper mirrors and operated at 50 kV and 90 mA. These X-ray data were processed with SAINT version V7.06A (Bruker AXS Inc.) and internally scaled with SADABS version 2005/1 (Bruker AXS Inc.).
The structure was solved via single isomorphous replacement using a crystal soaked in 1 mM methylmercury(I) chloride for 24 hours. Six mercury binding sites were identified with the program SOLVE (18), giving an overall figure-of-merit of 0.23 to 3.0 Å resolution. Solvent flattening and six-fold averaging with RESOLVE (19, 20) generated an interpretable electron density map, which allowed for a preliminary model to be constructed using the software package COOT (21).
A second crystal form was obtained when 10 mM UDP was included with the acetyl-CoA in the crystallization experiments. These crystals were grown at pH 8.0 in the presence of 100 mM HEPPS, 16 – 20% poly(ethylene glycol) 3400 and 210 mM tetramethylammonium chloride. They belonged to the space group P21 with unit cell dimensions a = 68.5, b = 107.9, and c = 91.3 Å, β = 102.6° and two trimers per asymmetric unit. High-resolution X-ray data from these crystals were collected at Beamline 19-BM, Structural Biology Center, Argonne National Laboratory, and the data were processed and scaled with HKL-3000 (22). Prior to X-ray data collection the crystals were transferred to a stabilization solution containing 22% poly(ethylene glycol) 3400, 300 mM NaCl, 10 mM UDP, 210 mM tetramethylammonium chloride, 25 mM acetyl-CoA, 100 mM HEPPS (pH 8.0), and 18% ethylene glycol. For preparation of the ternary complex of WlbB with CoA and UDP-GlcNAc3NA, the crystals were soaked for 6 hours in a solution containing 17% poly(ethylene glycol) 3400, 300 mM NaCl, 100 mM HEPPS (pH 8.0), 210 mM tetramethylammonium chloride, 25 mM acetyl-CoA, and 25 mM UDP-GlcNAc3NA prior to transfer to the cryoprotectant. Although acetyl-CoA was included in the soaking solution, only CoA was observed in the electron density map. Most likely the UDP-sugar reacted with acetyl-CoA in the enzyme's active site, and the UDP-sugar product dissociated leaving bound CoA. A second UDP-sugar substrate entered the active site resulting in the trapping of an “abortive complex.” Relevant X-ray data collection statistics are presented in Table 1.
Table 1.
X-ray Data Collection Statistics.
| Enzyme Complexed with Acetyl-CoA and UDP | Enzyme Complexed with CoA and UDP-GlcNAc3NA | |
|---|---|---|
| resolution limits | 50.0–1.43(1.48–1.43)b | 50.0–1.43(1.48–1.43)b |
| number of independent reflections | 226783(20999) | 213775(17996) |
| completeness (%) | 95.6(88.7) | 90.0(76.1) |
| redundancy | 3.9(2.7) | 2.9(2.1) |
| avg I/avg σ(I) | 38.2(8.9) | 28.3(6.6) |
| Rsym (%)a | 7.0(14.1) | 7.7(12.5) |
.
Statistics for the highest resolution bin.
The original structure solved from crystals belonging to the space group C2221 served as the search model for the subsequent three-dimensional analyses of the WlbB/acetyl-CoA/UDP and WlbB/CoA/UDP-GlcNAc3NA ternary complexes via molecular replacement with the software package PHASER (23). Each model was refined with the software package Refmac using maximum likelihood (24). Relevant refinement statistics are presented in Table 2.
Table 2.
Model Refinement Statistics.
| Enzyme Complexed with Acetyl-CoA and UDP | Enzyme Complexed with CoA and UDP-GlcNAc3NA | |
|---|---|---|
| resolution limits (Å) | 30.0–1.43 | 30.0–1.43 |
| aR-factor (overall)%/no. independent reflections | 19.2/226763 | 17.6/213775 |
| R-factor (working)%/no. independent reflections | 19.1/215388 | 17.4/203089 |
| R-factor (free)%/no. independent reflections | 21.7/11375 | 20.6/10686 |
| number of protein atoms | 8749b | 8796c |
| number of heteroatoms | 1945d | 2310e |
| average B values (Å2) | ||
| protein atoms | 12.2 | 10.7 |
| ligands | 17.0 | 9.7 |
| solvent | 26.0 | 25.9 |
| weighted RMS deviations from ideality | ||
| bond lengths (Å) | 0.012 | 0.012 |
| bond angles (°) | 2.18 | 2.12 |
| general planes (Å) | 0.009 | 0.007 |
R-factor = (Σ | Fo - Fc | / Σ | Fo |) × 100 where Fo is the observed structure-factor amplitude and Fc is the calculated structure-factor amplitude.
These include multiple conformations for Lys 57 and Arg 166 in Subunit A, Glu 36, Lys 57, Glu 159, and Glu 170 in Subunit B, Arg 21, Ile 121, and Glu 159 in Subunit C, Ile 16 in Subunit D, and Lys 57 and Glu 159 in Subunit E.
These include multiple conformations for Arg 49 and Arg 166 in Subunit A, Glu 33, Glu 36, Arg 55, Glu 93, and Glu 159 in Subunit B,Ile 22, Ile 121, Arg 149 and Glu 159 in Subunit C, Ile 16 and Ile 22 in Subunit D, Glu 33, Glu 93, and Glu 159 in Subunit E, and Glu 159 in Subunit F.
Heteroatoms include 6 acetyl CoA, 3 UDP, 3 UMP, 1144 waters, 2 phosphate ions, 4 ethylene glycol, 2 sodium ions, and one 2-(2-(2-(2-(2-(2-(2-ethoxy-ethoxy)-ethoxy)-ethoxy)-ethoxy)-ethoxy)-ethoxy)-ethanol.
Heteroatoms include 6 CoA, 6 UDP-GlcNAc3NA, 1698 waters, 2 phosphate ions, 12 ethylene glycol, 2 sodium ions, and one 2-(2-(2-(2-(2-(2-(2-ethoxy-ethoxy)-ethoxy)-ethoxy)-ethoxy)-ethoxy)-ethoxy)-ethanol.
Measurement of Enzymatic Activity
N-acetyltransferase activity was monitored spectrophotometrically by following the increase in absorbance at 412 nm due to the reaction of the sulfhydryl group of the CoASH product with 5,5'-dithiobis(2-nitrobenzoic acid) resulting in a disulfide interchange. This interchange leads to the formation of 5-thio-2-nitrobenzoic acid, which has a characteristic absorbance at 412 nm and an extinction coefficient of 14150 M−1 cm−1. The use of this compound for quantification of CoASH was first reported by Tomkins et al., (25), and our assay method was similar to that described in (26). Reactions were monitored continuously with a Beckman DU 640B spectrophotometer, and enzyme activities were calculated from the initial rates. Assay reactions were 100 μL in volume and contained, in addition to enzyme and substrates, 100 mM HEPES (pH 7.5) and 5 mM 5,5'-dithiobis(2-nitrobenzoic acid). The reactions were initiated by the addition of enzyme and monitored at 25°C. Kinetic parameters were determined by having one substrate at a saturating concentration (>10 Km) while varying the concentration of the second. Specifically, Km for acetyl-CoA was determined by varying its concentration from 0.01 to 5.0 mM using 5.0 mM UDP-GlcNAc3NA, and the Km for UDP-GlcNAc3NA was determined using 5.0 mM acetyl-CoA while varying the UDP-GlcNAc3NA concentration from 0.01 to 5.0 mM. Individual substrate kinetic data were fitted to equation 1 using Sigma Plot 8.
| (1) |
Calculated Km values were 83 ± 5 μM for UDP-GlcNAc3NA and 175 ± 10 μM for acetyl-CoA.
Results and Discussion
Structure of WlbB Complexed with Acetyl-CoA and UDP
The crystals utilized in this investigation belonged to the space group P21, contained two trimers per asymmetric unit, and diffracted to 1.43 Å resolution. These trimers were not related by a local twofold axis, but rather by a translation. The quality of the models for the six polypeptide chains in the asymmetric unit was excellent with 87%, 12.4% and 0.5% of the ϕ,ψ values lying within the core, allowed, and generously allowed regions of the Ramachandran plot (27). All six subunits demonstrated exceedingly similar architectures such that their α-carbons superimposed with root-mean-square deviations of between 0.08 Å and 0.19 Å. In light of this, the following discussion refers only to the first trimer in the X-ray coordinate file and specifically to the subunits designated “A” and “B.”
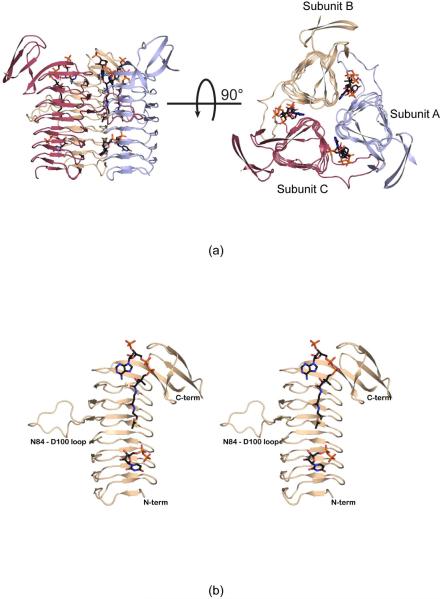
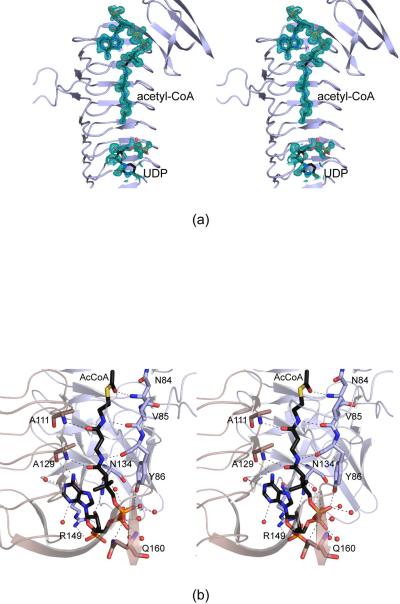
The WlbB trimer, as displayed in Figure 2a, has dimensions of ~70 Å × 66 Å × 67 Å, and each active site is wedged between two subunits. The surface area lost upon trimerization is ~2200 Å2. Each subunit contains all 190 amino acid residues of the crystallized protein, and Pro 147 adopts a cis-peptide conformation. A close-up view of one subunit is displayed in Figure 2b. There are 27 β-strands, 23 of which form the β-helix. The regularity of the β-helix is broken between β-strands 14 and 15 by a loop formed between Asn 84 and Asp 100. This loop reaches from one subunit into the next of the trimer and provides major binding interactions for both the acetyl-CoA and UDP-linked sugar ligands as described below. Following the last β-strand of the LβH motif, the polypeptide chain folds into a four-stranded anti-parallel β-barrel, which splays away from the main body of the trimer (Figure 2a).
Figure 2.
The structure of WlbB complexed with acetyl-CoA and UDP. A ribbon representation of the WlbB trimer is shown in (a) with the three subunits displayed in red, wheat, and light blue and the bound ligands shown as sticks. The overall three-dimensional fold of one subunit is presented in stereo in (b). The loop defined by Asn 84 to Asp 100 interrupts the regularity of the LβH motif.
For the initial structural analysis of WlbB, the protein was crystallized in the presence of acetyl-CoA and UDP. The electron densities corresponding to these bound ligands are shown in Figure 3a. As can be seen, the electron density for acetyl-CoA is extremely well ordered whereas that for UDP is somewhat less so. A close-up view of the binding pocket for acetyl-CoA is displayed in Figure 3b. The CoA adopts a curved conformation such that N6 of the adenine ring lies within 3.1 Å of one of the carbonyl oxygens of the pantothenate unit. The adenine ring is positioned within hydrogen bonding distance to the carbonyl oxygen of Ala 129 from subunit B and two waters whereas the ribose C-2 hydroxyl group is situated at 2.9 Å from Asn 134 in subunit A. The ribose adopts the C2'-endo pucker. A salt bridge occurs between the C-3 phosphoryl group on the ribose and the guanidinium group of Arg 149 of subunit A. The pyrophosphoryl group of the acetyl-CoA ligand interacts with seven waters, Oζ of Tyr 86 from subunit A, and Nε2 of Gln 160 from subunit B. Note that Tyr 86 lies in the loop that interrupts the regularity of the LβH motif. The pantothenate group adopts an extended structure much like a strand of β-sheet where it hydrogen bonds with the backbone carbonyl groups of Val 85 and Tyr 86 from subunit A and the backbone amide groups of Ala 111 and Ala 129 from subunit B. Finally the carbonyl oxygen of the cofactor's acetyl group interacts with the backbone amide of Asn 84 from subunit A, which initiates the extended loop (Figure 2b).
Figure 3.
Active site architecture for the WlbB/acetyl-CoA/UDP complex model. Shown in (a) is the initial electron density corresponding to the bound ligands (before they were included in the model). The map was contoured at ~1.5σ and calculated with coefficients of the form (2Fo - Fc), where Fo was the native structure factor amplitude and Fc was the calculated structure factor amplitude. Those amino acid residues lying within ~3.2 Å of acetyl-CoA are shown in (b). The active site is formed by residues contributed from two subunits in the trimer as indicated by the wheat and blue bonds and ribbon representations. Water molecules are depicted as red spheres, and potential hydrogen bonds are indicated by the dashed lines.
Structure of WlbB Complexed with CoA and UDP-GlcNAc3NA
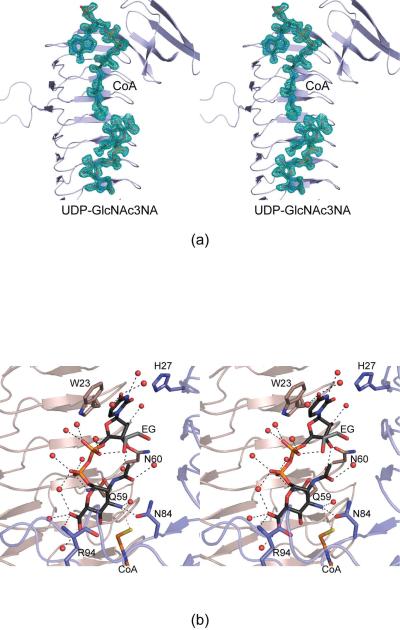
The binding of a nucleotide-linked sugar to WlbB resulted in very little overall structural perturbations. Indeed, the α-carbons for the structures, with or without the UDP-linked sugar, superimpose with a root-mean-square deviation of ~0.8 Å, and the side chain conformations within the active site regions of these two complexes are virtually identical. The only notable difference is that in the WlbB/acetyl-CoA/UDP complex, water molecules occupy the position assumed by the sugar in the WlbB/CoA/UDP-sugar complex.
For the structural analysis of this ternary complex, crystals were prepared in the presence of acetyl-CoA and UDP-GlcNAc3NA, but only CoA and UDP-GlcNAc3NA were visible in the electron density map (Figure 4a). This also occurred in our previous structural analysis of QdtC where crystals were grown in the presence of acetyl-CoA and a nucleotide-linked sugar, but only CoA was observed (9). As a consequence, the structure of the WlbB ternary complex reported in this section represents that of an abortive complex. A close-up view of the active site surrounding the UDP-linked sugar is presented in Figure 4b. The uridine ring lies within 3.2 Å of three water molecules. The ribose, which adopt the C3'-endo pucker, hydrogen bonds to two waters and the carboxamide side chain of Asn 60 in subunit B. The carboxamide side chain of Gln 59 in subunit B, as well as numerous solvent molecules, surround the pyrophosphoryl group of the UDP-linked sugar. The pyranose of UDP-GlcNAc3NA assumes the 4C1 chair conformation. Only two specific interactions occur between the protein (subunit A) and the GlcNAc3NA moiety: the backbone amide group of Arg 94 interacts with the sugar carboxylate group at C-5' and Oδ1 of Asn 84 lies within 2.8 Å of the sugar C-3' amino group that is ultimately acetylated during the reaction. As indicated by the red dashed line in Figure 4b, the sugar C-3' amino group is positioned within 3.2 Å of the CoA sulfur atom. The lack of specific interactions between the pyranosyl moiety and the protein suggests that WlbB may accept alternative substrates. Experiments to test this are presently underway.
Figure 4.
Active site architecture for the WlbB/CoA/UDP-GlcNAc3NA complex model. Shown in (a) is the initial electron density corresponding to the bound ligands (before they were included in the model). The map was contoured at ~1.5σ and calculated with coefficients of the form (2Fo - Fc), where Fo was the native structure factor amplitude and Fc was the calculated structure factor amplitude. Those amino acid residues lying within ~3.2 Å of UDP-GlcNAc3NA are shown in (b). Water molecules are depicted as red spheres, and potential hydrogen bonds are indicated by the dashed lines. Note that the water molecule located within hydrogen bonding distance of the sugar C-3' amino group is displaced when acetyl-CoA is bound in the active site.
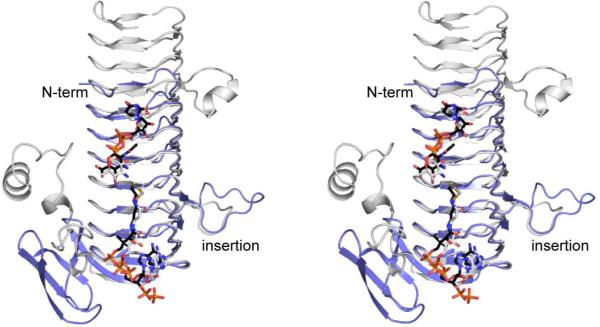
Comparison of WlbB to QdtC
As discussed previously, QdtC is an N-acetyltransferase that catalyzes the acetylation of the C-3' amino group of dTDP-Quip3N (Scheme 1). The overall amino acid sequence identity between QdtC and WlbB is 36%. A superposition of the two structures is presented in Figure 5. Whereas WlbB is considerably smaller than QdtC (190 versus 265 amino acid residues), both display nearly identical binding orientations of the nucleotide-linked sugars. The two proteins initially align at Ala 2 in WlbB and Ala 77 in QdtC. A five-residue insertion occurs after Pro 88 in WlbB such that the two proteins begin to align again at Glu 97 in WlbB and Glu 167 in QdtC. Their close structural correspondence continues until Met 154 in WlbB and Ile 224 in QdtC. Indeed, the α-carbons for 123 target pairs between WlbB and QdtC superimpose with a root-mean-square deviation of 0.71 Å. The specific interactions between WlbB or QdtC and their respective nucleotide-linked sugars are quite different, however. In QdtC, three amino acid side chains are located within hydrogen bonding distance to the sugar's amino and hydroxyl groups, namely Glu 141, Asn 159, and Asp 160 (9). These residues were tested for their roles in catalysis by preparing the following site-directed mutant proteins: E141Q, E141A, N159D, N159A, D160N, and D160A (9). All of these proteins retained catalytic activity, albeit the activity was seriously reduced for the D160A but not the D160N mutant enzyme. In WlbB, these residues correspond to Asp 66, Asn 84, and Val 85. The only conserved interaction between the nucleotide-linked sugars and these two proteins is the hydrogen bond between the side chain of Asn 84 in WlbB (Asn 159 in QdtC) and the C-3' amino group. As in the case of QdtC, there are no potential catalytic bases within the active site of WlbB to deprotonate the C-3' amino group for its attack at the carbonyl carbon of acetyl CoA.
Figure 5.
Superposition of WlbB and QdtC. Ribbon representations for the WlbB and QdtC subunits are highlighted in light blue and white, respectively. The CoA and nucleotide-linked sugars, when bound to WlbB and QdtC, are displayed in black and white bonds, respectively.
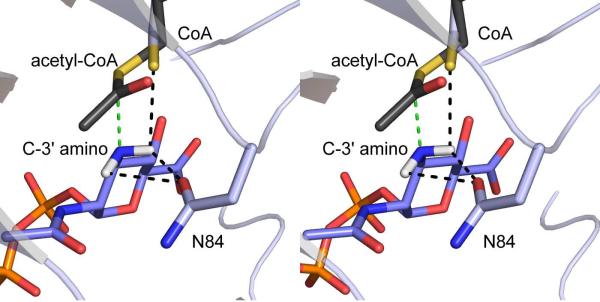
Implications for the Catalytic Mechanism of WlbB
The two complex structures of WlbB described here show the binding conformations for acetyl-CoA and CoA, and thus represent the “pre” and “post” reaction states of the cofactor. A superposition of these two “states” is presented in Figure 6. Note that the sulfur atom of the cofactor moves in the active site by ~2 Å upon departure of the acetyl group. In Figure 6, these two conformations of the cofactor are displayed as well as the binding position of the UDP-GlcNAc3NA substrate. Assuming that the amino group of the UDP-GlcNAc3NA substrate enters the WlbB active site in an unprotonated state, the hypothetical positions for the two hydrogens attached to nitrogen are also included in Figure 6. A potential catalytic mechanism can thus be envisioned as follows. The C-3' amino nitrogen of the substrate attacks the si face of the acetyl moiety of acetyl-CoA to produce a tetrahedral oxyanion intermediate. This intermediate is most likely stabilized by interaction of the oxyanion with the backbone amide group of Asn 84 (Figure 3b). In the model shown in Figure 6, the amino nitrogen is located within 2.2 Å of the carbonyl carbon of acetyl CoA and is oriented such that its lone pair of electrons is in the proper location for nucleophilic attack. The two hydrogens attached to the amino nitrogen lie within hydrogen bonding distance of Asn 84, which most likely plays a key role in proper substrate positioning. As the oxyanion intermediate collapses, the bond between the carbon and the sulfur of acetyl-CoA breaks, the sulfur shifts its position in the active site as indicated in Figure 6, and accepts a proton from the C-3' amino group.
Figure 6.
Close-up view of the WlbB active site. For this figure, the two structures described in this report were superimposed, and hydrogens were added to the C-3' amino group of the sugar substrate. The green dashed line represents the hypothetical approach of the lone pair of electrons on the amino nitrogen towards the si face of acetyl-CoA. The black dashed lines do not specifically imply hydrogen bonds, but merely indicate distances of 2.2 Å to 2.9 Å between the hydrogens on the C-3' nitrogen and either Oδ1 of Asn 84 or the sulfur of CoA.
We previously suggested a similar reaction mechanism for QdtC on the basis of X-ray crystallographic analyses, site-directed mutagenesis, and kinetic experiments (9). At the time we assumed that the catalytic mechanism of QdtC might be an “outlier” among sugar-modifying N-acetyltransferases in light of the structure of PglD, which contains a putative catalytic base. However, from this study we now know that QdtC and WlbB accommodate their sugar substrates in nearly identical fashions and in marked contrast to that observed for PglD (Figure 1). In addition, both QdtC and WlbB contain asparagine residues that hydrogen bond to the substrate amino groups. Interestingly, these enzymes operate on amino groups attached to the C-3' carbons of the sugar substrates (Scheme 1). PglD, on the other hand, acetylates an amino group attached to the C-4' carbon. On the basis of these observations, we suggest that there are two subfamilies of the LβH N-acetyltransferases that operate on nucleotide-linked sugars: those that belong to the “PglD” group, which function on amino groups attached to C-4' carbons and those that belong to the “QdtC” family, which operate on substrates containing C-3' amino groups.
As shown in Scheme 1, GDP-perosamine N-acetyltransferase (PerB) catalyzes the acetylation of GDP-perosamine to yield GDP-N-acetyl-perosamine, a 6-deoxyhexose found on the lipopolysaccharide of E. coli O157:H7, the causative agent of enterohemorrhagic diarrhea first identified in contaminated hamburgers and subsequently in other foods including spinach (28, 29). We suggest that its structure will belong to the “PglD” subfamily. Another N-acetyltransferase, FdtC from Aneurinibacillus thermoaerophilus L420-91T catalyzes the acetylation of dTDP-3-amino-3,6-dideoxy-galactose or dTDP-Fucp3N to yield dTDP-Fucp3NAc (11). This unusual dideoxysugar is found in the S-layer of many Gram-positive and Gram-negative bacteria (30) and in the O-antigens of some Gram-negative bacteria (10). Given that FdtC functions on a C-3' amino group, we would predict that it belongs to the “QdtC” subfamily. X-ray crystallographic analyses of these two enzymes are presently underway.
Acknowledgments
A portion of the research described in this paper was performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source (United States Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357). We gratefully acknowledge Dr. Norma E. C. Duke for assistance during the X-ray data collection at Argonne and Professor W. W. Cleland for helpful discussions.
This research was supported in part by an NIH grant (DK47814 to H. M. H.)
Abbreviations
- CoA
Coenzyme A
- HEPES
4-(2-hydroxyethyl)-1-piperazine-ethane sulfonic acid
- HEPPS
3-(4-(2-hydroxyethyl)-1-piperazine-propane sulfonic acid
- IPTG
isopropyl-β-D-thiogalactopyranoside
- LB
Luria-Bertani
- MOPS
3-(N-morpholino-propanesulfonic acid)
- NiNTA
nickel-nitrilotriacetic acid
- PCR
polymerase chain reaction
- RMS
root-mean-square
- TB
terrific broth
- UDP
uridine diphosphate
- UMP
uridine monophosphate
- Tris
tris-(hydroxymethyl)aminomethane
Footnotes
X-ray coordinates have been deposited in the Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, N. J., (accession nos. 3MQG and 3MQH).
References
- 1.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: a structural overview. Annu Rev Biophys Biomol Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vetting MW, LP S. d. C., Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Raetz CR, Roderick SL. A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 4.Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 5.Rangarajan ES, Ruane KM, Sulea T, Watson DC, Proteau A, Leclerc S, Cygler M, Matte A, Young NM. Structure and active site residues of PglD, an N-acetyltransferase from the bacillosamine synthetic pathway required for N-glycan synthesis in Campylobacter jejuni. Biochemistry. 2008;47:1827–1836. doi: 10.1021/bi702032r. [DOI] [PubMed] [Google Scholar]
- 6.Olivier NB, Imperiali B. Crystal structure and catalytic mechanism of PglD from Campylobacter jejuni. J Biol Chem. 2008;283:27937–27946. doi: 10.1074/jbc.M801207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerry P, Szymanski CM. Campylobacter sugars sticking out. Trends Microbiol. 2008;16:428–435. doi: 10.1016/j.tim.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Demendi M, Creuzenet C. Cj1123c (PglD), a multifaceted acetyltransferase from Campylobacter jejuni. Biochem Cell Biol. 2009;87:469–483. doi: 10.1139/o09-002. [DOI] [PubMed] [Google Scholar]
- 9.Thoden JB, Cook PD, Schaffer C, Messner P, Holden HM. Structural and functional studies of QdtC: an N-acetyltransferase required for the biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose. Biochemistry. 2009;48:2699–2709. doi: 10.1021/bi802313n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng L, Wang W, Tao J, Guo H, Krause G, Beutin L, Wang L. Identification of Escherichia coli O114 O-antigen gene cluster and development of an O114 serogroup-specific PCR assay. J Clin Microbiol. 2004;42:3799–3804. doi: 10.1128/JCM.42.8.3799-3804.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfoestl A, Hofinger A, Kosma P, Messner P. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-galactose in Aneurinibacillus thermoaerophilus L420-91T. J Biol Chem. 2003;278:26410–26417. doi: 10.1074/jbc.M300858200. [DOI] [PubMed] [Google Scholar]
- 12.Westman EL, Preston A, Field RA, Lam JS. Biosynthesis of a rare di-N-acetylated sugar in the lipopolysaccharides of both Pseudomonas aeruginosa and Bordetella pertussis occurs via an identical scheme despite different gene clusters. J Bacteriol. 2008;190:6060–6069. doi: 10.1128/JB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kochetkov NK, Knirel YA. Structure of lipopolysaccharides from Gram-negative bacteria. III. Structure of O-specific polysaccharides. Biochemistry (Moscow) 1994;59:1325–1383. [PubMed] [Google Scholar]
- 14.Harvill ET, Preston A, Cotter PA, Allen AG, Maskell DJ, Miller JF. Multiple roles for Bordetella lipopolysaccharide molecules during respiratory tract infection. Infect Immun. 2000;68:6720–6728. doi: 10.1128/iai.68.12.6720-6728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engels W, Endert J, Kamps MA, van Boven CP. Role of lipopolysaccharide in opsonization and phagocytosis of Pseudomonas aeruginosa. Infect Immun. 1985;49:182–189. doi: 10.1128/iai.49.1.182-189.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dasgupta T, de Kievit TR, Masoud H, Altman E, Richards JC, Sadovskaya I, Speert DP, Lam JS. Characterization of lipopolysaccharide-deficient mutants of Pseudomonas aeruginosa derived from serotypes O3, O5, and O6. Infect Immun. 1994;62:809–817. doi: 10.1128/iai.62.3.809-817.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thoden JB, Timson DJ, Reece RJ, Holden HM. Molecular structure of human galactokinase: implications for Type II galactosemia. J Biol Chem. 2005;280:9662–9670. doi: 10.1074/jbc.M412916200. [DOI] [PubMed] [Google Scholar]
- 18.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 8):965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 23.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Alpers DH, Appel SH, Tomkins GM. A spectrophotometric assay for thiogalactoside transacetylase. J Biol Chem. 1965;240:10–13. [PubMed] [Google Scholar]
- 26.Magalhaes ML, Blanchard JS. The kinetic mechanism of AAC3-IV aminoglycoside acetyltransferase from Escherichia coli. Biochemistry. 2005;44:16275–16283. doi: 10.1021/bi051777d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 28.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen ML. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 29.Albermann C, Beuttler H. Identification of the GDP-N-acetyl-D-perosamine producing enzymes from Escherichia coli O157:H7. FEBS Lett. 2008;582:479–484. doi: 10.1016/j.febslet.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Pfostl A, Zayni S, Hofinger A, Kosma P, Schaffer C, Messner P. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose. Biochem J. 2008;410:187–194. doi: 10.1042/BJ20071044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA. [Google Scholar]